Abstract
Novel drugs are needed to treat a variety of persistent diseases caused by intracellular bacterial pathogens. Virulence pathways enable many functions required for the survival of these pathogens, including invasion, nutrient acquisition, and immune evasion. Inhibition of virulence pathways is an established route for drug discovery; however, many challenges remain. Here, we propose the biggest problems that must be solved to advance the field meaningfully. While it is established that we do not yet understand the nature of chemicals capable of permeating into the bacterial cell, this problem is compounded when targeting intracellular bacteria because we are limited to only those chemicals that can permeate through both human and bacterial outer envelopes. Unfortunately, many chemicals that permeate through the outer layers of mammalian cells fail to penetrate the bacterial cytoplasm. Another challenge is the lack of publicly available information on virulence factors. It is virtually impossible to know which virulence factors are clinically relevant and have broad cross-species and cross-strain distribution. In other words, we have yet to identify the best drug targets. Yes, standard genomics databases have much of the information necessary for short-term studies, but the connections with patient outcomes are yet to be established. Without comprehensive data on matters such as these, it is difficult to devise broad-spectrum, effective anti-virulence agents. Furthermore, anti-virulence drug discovery is hindered by the current state of technologies available for experimental investigation. Antimicrobial drug discovery was greatly advanced by the establishment and standardization of broth microdilution assays to measure the effectiveness of antimicrobials. However, the currently available models used for anti-virulence drug discovery are too broad, as they must address varied phenotypes, and too expensive to be generally adopted by many research groups. Therefore, we believe drug discovery against intracellular bacterial pathogens can be advanced significantly by overcoming the above hurdles.
Keywords: intracellular bacteria, virulence, persistence, drug discovery, infection
1. Introduction
Communicable diseases continue to burden the globe [1]. The cost of infections in terms of lives and financial burden is huge [1,2,3]. Six of the bacteria identified by the World Health Organization (WHO) and the U.S. Centers for Disease Control and Prevention (CDC) as threats to human health are intracellular bacteria [4,5]. Intracellular bacteria often cause chronic or latent infections, with difficult diagnosis and treatment (Supplementary Materials, Section SI) [6]. Clinically relevant intracellular bacteria can be seen in Supplementary Table S1. Evasion of host immune response by intracellular bacteria [7] makes antibiotic therapy especially important, even in immunocompetent hosts.
Bacterial resistance is a major problem [5,8,9], and resistance is inevitable with increased use [10]. Infections caused by intracellular organisms often require longer treatment durations than non-persistent infections [11,12,13,14] with multiple antibiotics [11,12,14,15], leading to resistance [4,5]. Only a limited selection of antimicrobials (e.g., aminoglycosides, fluoroquinolones, macrolides, rifamycins, tetracyclines) are useful against intracellular pathogens (Supplementary Table S1). This limited armamentarium means that each time an antibiotic succumbs to resistance, our options for appropriate treatment reduce drastically.
Novel, nontraditional agents (such as anti-virulence agents) are a promising alternative. It has been repeatedly demonstrated (e.g., [16,17,18,19]) that intracellular lifestyles are facilitated by virulence factors, blocking which will prove to be avenues for therapy. See Supplementary Materials, Section SII for some detailed examples. Clearly, if we can identify virulence factors that enable intracellular infections, and target them successfully, we could theoretically build an array of frontline treatments for infections. The powerful advantage of these medications would be that they do not require bacterial cell division for an effect to take place: traditional antimicrobials (i.e., antibiotics) are only useful against bacteria undergoing continuous cell division, such as intracellular bacterial colonies (IBCs). They would not function against a subpopulation of non-dividing bacteria, such as persisters or quiescent cells. Anti-virulence drugs would function by eliminating the pathogen’s ability to survive intracellularly, regardless of whether they were rapidly dividing or not. Yet, is it so easy?
It is well recognized that antimicrobials are hard to find because this has been the major focus of the drug discovery and development community, but the challenges we face in drug discovery against intracellular bacteria have rarely been addressed.
2. Challenges in Drug Discovery against Intracellular Bacteria
Several hurdles stand in the way of advancements in antibacterial treatments: (1) we do not yet fully understand the type of chemicals most suitable to become antimicrobials (this also applies to anti-virulence agents), (2) standardized databases containing information on virulence drug targets to guide drug discovery are virtually absent, and (3) we currently lack standardized and inexpensive techniques to identify and assess virulence in intracellular bacteria. These challenges will need to be met and overcome to make anti-virulence drug discovery successful.
2.1. Chemical Space Is a Severe Limitation
Pathogenic outer layers are not the same as ours, and hence chemicals that permeate into their cells are different. This is why chemical libraries optimized to penetrate human cells do not serve the needs of antimicrobial drug discovery [20,21,22]. O’Shea and Moser demonstrated that current antimicrobials are extremely different from typical chemicals most likely to be found in pharmaceutical screening libraries [23], which are compliant with Lipinski’s rules-of-5 (Ro5) [24]. Very importantly, they also demonstrated that the chemicals that permeate Gram-positive bacteria were different from those that permeate Gram-negative bacteria. However, further studies have shown that many Ro5-compliant chemicals are permeators as well [25,26]. Even though recent work by multiple groups [27,28,29,30,31,32,33,34,35,36,37], including us [26], have made tremendous advances in understanding which chemicals are most likely to penetrate bacterial cells (or even which chemicals are most likely to be extruded by efflux pumps), we are still far away from accurately predicting the chemical space most likely to contain the next generation of antimicrobials [38]. This situation is further complicated by the fact that differences between bacterial species (outer membrane composition, efflux pump efficiencies, and so on) make it virtually certain that our current knowledge of bacterial outer membrane permeability is insufficient. Moreover, there is no guarantee that methods built on a particular bacterial strain will work on another strain of the same species—bacteria are notorious for being “exceptions to the rules”.
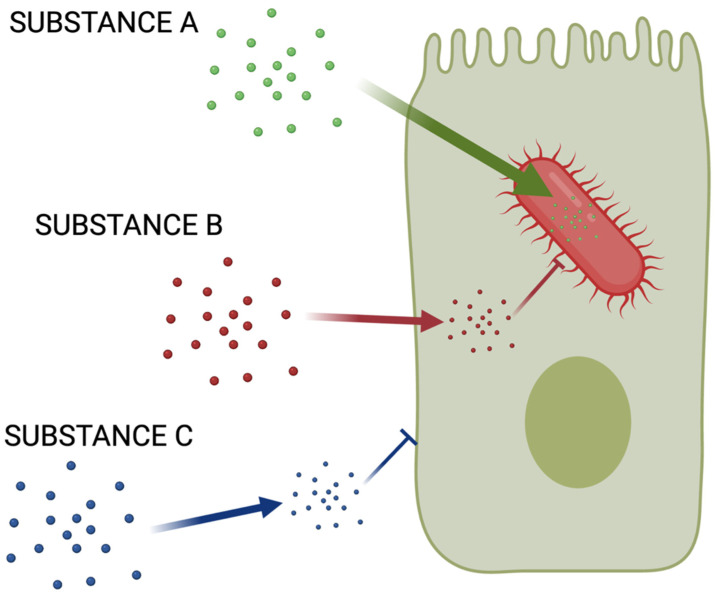
Now, consider that drugs that act against virulence factors of intracellular bacteria must permeate both, human and bacterial outer membranes. There is no escaping this fact (Figure 1). This means there are only two types of chemicals we can consider favorably—Ro5-compliant chemicals that easily penetrate the cytosol of mammalian cells or are facilitated to enter the cytosol through transporters. This is, perhaps, the biggest challenge we must face.
Figure 1.
The chemical space conundrum. It is already difficult to identify chemicals that permeate bacterial outer envelopes. While recent work [27,28,29,30,31,32,33,34,35,36,37] has identified plausible characteristics enabling penetration of a few Gram-negative cells, Gram-positive permeability rules are virtually undiscovered. Additionally, only a fraction of chemicals capable of permeating the human cell can penetrate the bacterial cell to reach the cytosol. Chemicals that target intracellular bacteria must penetrate both (Substance A). Substance A must be both, Ro5-compliant, as well as bacterial cell-penetrating, which are difficult to find. Substance B, on the other hand, is useless against intracellular pathogens because it penetrates human cells but fails to reach bacterial targets. Substance C would be useless because it would not penetrate the human cell, even if it was able to reach the bacterial cytoplasm.
2.2. Finding Good Drug Targets
There are 3 different aspects of drug target selection: (1) identifying a pathway that is critical to the outcome of interest (in this case, virulence and intracellular survival), (2) identifying key enzymes of a tractable nature (i.e., enzymes whose disruption with organic chemicals is possible), and unique in the case of antimicrobials, (3) determining whether these targets are useful when attempting to design broad-spectrum agents.
Identifying key virulence pathways. Amongst these, our knowledge of microbial virulence factors is perhaps the most promising. We can identify genes that are directly responsible for pathogenicity or survival, as well as their mechanisms of regulation [39,40,41]. Techniques such as proteomics have helped evaluate such matters quite thoroughly (see Supplementary Materials, Section SIII for an example of application in understanding bacterial virulence). Other methods, including transcriptomics are also likely to help elucidate the basic biology dictating bacterial behavior during intracellular infections. There is no doubt that compilation of such data is ongoing, which will add tremendously to our knowledge of microbial virulence factors and their role(s) in intracellular survival. Some examples of genomics databases include the Virulence Factor Database (VFDB) [42,43] and MvirDB [44]. There are even databases to connect virulence factors to protein-protein interaction networks [45], which provide key information on the regulation of virulence as a process. While none of these databases are specifically focused on identifying drug targets in intracellular bacteria, they contain relevant data of tremendous use. Additional work will certainly provide data to aid in anti-virulence drug discovery.
At the same time, it is important to acknowledge that very little is truly understood about genetic variability for different strains within the same species. Even in the best-studied pathogens such as E. coli and S. aureus, only a handful of strains have been genotyped and made available. This is not surprising. While humans evolve over several thousand years, the time scale is literally minutes for bacteria—thus, a very large number of strains exist for each species. Enough samples to represent the genetic variability among even a single bacterial species could potentially constitute a mind-boggling amount of data. It may, however, be useful to systematically catalog strains from clinical isolates, and correlate them with patient outcomes such as clinical failure, recurrence, or even death. This may limit the number of strains for which data must be obtained as priority could be given to clinically virulent strains. Studying patients over time could also help understand how pathogens evolve within hosts to facilitate intracellular survival.
Chemical tractability of targets. The next challenge is to segregate tractable drug targets from non-tractable. For the purposes of this article, we will only refer to tractability by synthetic, organic drugs because other medications such as biologics are unlikely to act directly on intracellular bacteria. Tractability by drug-like chemicals has been the subject of significant work across the past two decades [46,47,48,49,50,51,52,53,54,55]; it is also sometimes referred to as “druggability”. It is well-known that certain drug targets are much more likely to bind drug-like small molecules with high affinity [56]. The techniques are sophisticated enough to successfully identify tractable binding pockets with approximately 80%–90% accuracy, and hence can be easily used for target discovery in intracellular bacteria. Such information, when curated into a database, can provide critical clues regarding drug targets for discovery of drug targets in intracellular bacteria. However, to the best of our knowledge, only Sarkar and Brenk have used a chemical tractability method [57] to a pathogen (P. aeruginosa [58]).
Broad-spectrum or narrow-spectrum? Virulence pathways are, by definition, not critical for survival in broth—they are required only during the process of invasion and infection in a living host. These are essentially abilities developed by pathogens later in the evolutionary timeline after they encountered hosts. Thus, virulence-related pathways are not conserved across species, or even strains. For example, S. aureus strains have significantly varied genetic makeup, and the virulence pathways involved in infections are often different. The community-acquired methicillin-resistant S. aureus USA300 clone, which currently dominates skin and soft tissue infections in the U.S., does so in part because of newly acquired virulence factors lacking in the hospital-acquired S. aureus clones that previously dominated [59]. Therefore, it may be difficult to identify broad-spectrum anti-virulence agents. This can be countered by our knowledge of disease etiology. For example, since staphylococci are responsible for infective endocarditis in the U.S. and Europe, while streptococci are responsible for the same in other parts of the world, different narrow-spectrum agents could be used depending on local etiology [59].
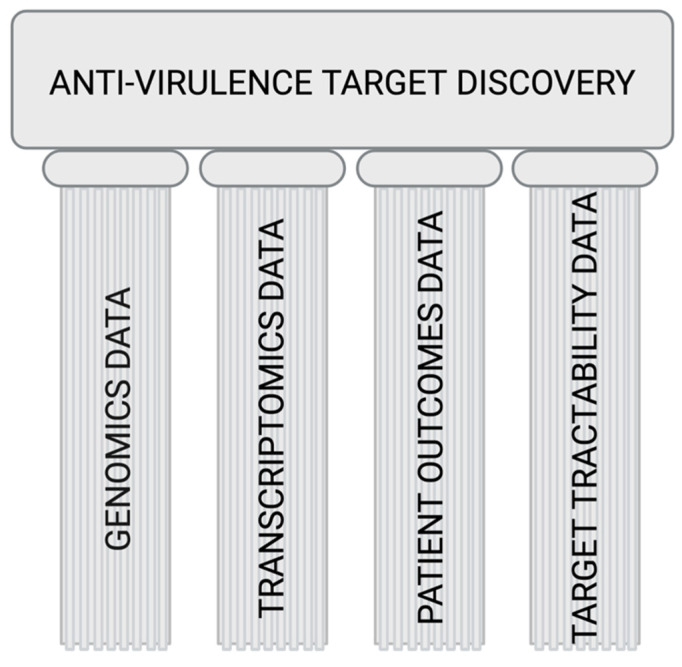
Overall, it is clear that appropriately curated and analyzed data will help us identify the best way(s) to fight intracellular infections by bringing forth good drug targets. A platform that rationally helps identify good drug targets must include, at the very least, these cogs (Figure 2).
Figure 2.
Pillars of target discovery: the essential infrastructure needed in the public domain to help facilitate and advance rational, guided efforts in anti-virulence drug discovery. The selection of targets will require an understanding of how the bacterial genome dictates virulence signaling via the transcriptome and contributes to patient outcomes. Only after understanding the chemical tractability of those targets will we be able to rationally select drug targets and focus our efforts. We envision a comprehensive platform that provides data to connect all these factors, allowing a rational approach towards target selection for the development of anti-virulence agents.
2.3. Current Models of Studying Virulence
A very important factor behind the successes in antimicrobial drug discovery is the availability of broth microdilution as a phenotypic assay. Antimicrobial susceptibility measurement using broth microdilution is such a successful model that it has been standardized and used uniformly across the globe. It represents exactly what it needs to represent—the growth of the bacterium in the presence of an antimicrobial. Furthermore, reagents are inexpensive, and procedures are easy. In comparison, current models to study virulence are much more difficult and expensive [60].
Part of the problem is that there is no widely accepted, standardized method to measure virulence itself. This is mostly because virulence is an agglomerate of multiple phenotypes, unlike growth. Furthermore, standardized molecular biology techniques (e.g., RT-qPCR, Western blots, etc.) are not useful because of how virulence manifests itself—different virulence factors are often involved, and each must be measured in unique ways [40,61,62].
It is also difficult to study intracellular bacteria because the technique necessitates more expensive and challenging mammalian cell culture methods. There has been some advancement in this regard. A method has been proposed for rapid and reliable isolation of intracellular bacteria during urinary tract infections, involving rapid and inexpensive procedures [63]. However, multiple intracellular bacteria (Supplementary Table S1) manifest distinct infections of multiple tissues, requiring several highly specific models to study each infection. More generalizable methods will certainly be helpful, if they are able to adequately simulate multiple infections.
Relatively inexpensive in vitro models do exist to simulate biofilm, and they are valuable for their tractability and reproducibility. For example, U.S. CDC approved biofilm reactors are cost-effective, high-throughput methods for assessing biofilm formation; however, this method also shows attachment of the biofilm to host cell epithelia, an action not always observed in in vivo infections [64,65]. Complicating matters, antimicrobial susceptibility and bacterial growth rates may be different within and outside biofilm, and even between aggregated, surface-bound biofilm versus suspended biofilm, which can be difficult to measure in a reproducible manner. This situation is even more complicated with intracellular bacteria because it is hard to separate those surviving inside intracellular biofilms, or exiting intracellular biofilm but remaining within the cell, or perhaps even breaking out of the host cell completely. The effectiveness of an anti-virulence agent against biofilm would require accurate and precise quantitation of all these outcomes because these could lead to different clinical end points. While biofilms grown in the laboratory do not necessarily represent biofilms grown in vivo, it is always possible to follow up on the results from in vitro model with an ex vivo or in vivo model.
Animal models are ideal for studying intracellular bacteria, but they are expensive. It has always been difficult to balance factors such as inoculum dosage; high doses may be lethal while low doses result in a swift immune response [66]. Furthermore, while vertebrate models are more similar to humans, there are ethical concerns over use, and therefore must be used only when absolutely required. Invertebrate models have been introduced (e.g., Caenorhabditis elegans and Drosophila melanogaster [67,68,69,70,71]), but we are unsure if permeation of chemicals resembles human cells. Theoretically, unless similarities in permeation are well established, it is possible to find chemicals that eliminate intracellular bacteria in these models that do not reach intracellular bacteria in human cells. The major advantage of using vertebrate animals, of course, is the presence of a fully formed immune system. Just as humans, rats and mice have both innate and adaptive branches of the immune system, while invertebrates such as C. elegans and D. melanogaster do not. Invertebrates also typically do not provide large amounts of tissue for analysis [65,66]. The silkworm provides a larger invertebrate model with a simpler genome to identify genes contributing to virulence and larger tissue samples for use in virulence assays [72]. However, a simpler genome also likely means bigger differences when compared to humans.
Ex Vivo models are an alternative to animal models. Human tissues can be grown in a test tube with fewer ethical concerns, and drugs can be tested. This method permits control over experimental conditions in which the environmental aspects of bacterial cell survival and virulence can be assessed. However, even these are not cheap or easy, and the lack of an immune response coupled with a severe lack in standardization prevent large-scale expansion of this method in modelling pathogenesis [66]. Furthermore, tissue culture is not the same as humans. For example, the immune system is absent. Are these as effective as an animal model? Not likely.
In our opinion, we need new, in vitro models capable of simulating the intracellular environment. In Vitro models may never capture the complexity of a host immune response, but if we are able to simulate the stresses undergone by pathogens in an intracellular environment, we may be able to identify anti-virulence agents at the same rate as antimicrobials were discovered using broth microdilution methods. The key difference between current in vitro models and these new models should be a clear and precise demonstration of phenotypic and transcriptomic similarities. These could even be prepared separately for different cell types, but they will need to be lucid enough to run in a standardized manner, akin to antimicrobial susceptibility assays by broth microdilution.
3. Perspective
In the face of rising antibiotic resistance, anti-virulence therapies are a hopeful prospect in combating persistent infections caused by intracellular bacteria. From a drug discovery perspective, three changes from status quo are needed to truly advance the field:
Find better targets.
Develop cheaper, more effective models (maybe in vitro, for instance broth microdilution).
Understanding chemical space that is best to permeate both human and bacterial cells.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091172/s1. Section SI: Intracellular Bacteria Are Particularly Challenging to Treat [2,3,4,5,6,7,11,12,13,14,15,73,74,75,76,77,78]; Section SII: Virulence Factors Are Important Enablers of Intracellular Pathogen Survival [7,16,17,18,19,79,80,81,82,83,84,85,86,87,88]; Section SIII: Examples of Proteomics Being Used to Understand Bacterial Virulence [89,90,91]; Table S1: A list of clinically important intracellular bacteria, including clinical conditions and therapy options [12,13,14,15,92,93,94,95,96,97].
Author Contributions
A.S. conceived the document. A.N.T. and T.J.C. identified data sources. A.N.T., T.J.C., and A.S. interpreted the data, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by High Point University startup funds to A.S. and T.J.C., and a High Point University Natural Sciences Fellowship to A.N.T.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization The Top 10 Causes of Death. [(accessed on 22 January 2021)]. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- 2.Rowley J., Hoorn S.V., Korenromp E., Low N., Unemo M., Abu-Raddad L.J., Chico R.M., Smolak A., Newman L., Gottlieb S., et al. Chlamydia, Gonorrhoea, Trichomoniasis and Syphilis: Global Prevalence and Incidence Estimates, 2016. Bull. World Health Organ. 2019;97:548–562P. doi: 10.2471/BLT.18.228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millet J.P., Orcau A., de Olalla P.G., Casals M., Rius C., Cayla J.A. Tuberculosis Recurrence and Its Associated Risk Factors among Successfully Treated Patients. J. Epidemiol. Community Health. 2009;63:799–804. doi: 10.1136/jech.2008.077560. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline. [(accessed on 22 January 2021)]. Available online: https://www.who.int/publications/i/item/9789240000193.
- 5.Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. [(accessed on 22 January 2021)]; Available online: https://www.cdc.gov/drugresistance/biggest-threats.html.
- 6.Moulder J.W. Comparative Biology of Intracellular Parasitism. Microbiol. Rev. 1985;49:298–337. doi: 10.1128/mr.49.3.298-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakur A., Mikkelsen H., Jungersen G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019;2019:1356540. doi: 10.1155/2019/1356540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassini A., Högberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the Eu and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R., Eggleston K., Rotimi V., Zeckhauser R.J. Antibiotic Resistance as a Global Threat: Evidence from China, Kuwait and the United States. Glob. Health. 2006;2:6. doi: 10.1186/1744-8603-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar A., Garneau-Tsodikova S. Resisting Resistance: Gearing up for War. MedChemCommun. 2019;10:1512–1516. doi: 10.1039/C9MD00330D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley C.L., Iaccarino J.M., Lange C., Cambau E., Wallace R.J., Andrejak C., Bottger E.C., Brozek J., Griffith D.E., Guglielmetti L., et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official Ats/Ers/Escmid/Idsa Clinical Practice Guideline: Executive Summary. Clin. Infect. Dis. 2020;71:e1–e36. doi: 10.1093/cid/ciaa241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nahid P., Dorman S.E., Alipanah N., Barry P.M., Brozek J.L., Cattamanchi A., Chaisson L.H., Chaisson R.E., Daley C.L., Grzemska M., et al. Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical Practice Guidelines: Treatment of Drug-Susceptible Tuberculosis. Clin. Infect. Dis. 2016;63:e147–e195. doi: 10.1093/cid/ciw376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tunkel A.R., Hartman B.J., Kaplan S.L., Kaufman B.A., Roos K.L., Scheld W.M., Whitley R.J. Practice Guidelines for the Management of Bacterial Meningitis. Clin. Infect. Dis. 2004;39:1267–1284. doi: 10.1086/425368. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert D.N., Chambers H.F., Saag M.S., Pavia A.T., Black D., Boucher H.W., Freedman D.O., Kim K., Schwartz B.S., editors. The Sanford Guide to Antimicrobial Therapy 2020. 50th ed. Antimicrobial Therapy, Inc.; Sperryville, VA, USA: 2020. [Google Scholar]
- 15.Griffith D.E., Aksamit T., Brown-Elliott B.A., Catanzaro A., Daley C., Gordin F., Holland S.M., Horsburgh R., Huitt G., Iademarco M.F., et al. An Official Ats/Idsa Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Am. J. Respir. Crit. Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 16.Anderson G.G., Palermo J.J., Schilling J.D., Roth R., Heuser J., Hultgren S.J. Intracellular Bacterial Biofilm-Like Pods in Urinary Tract Infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 17.Justice S.S., Hung C., Theriot J.A., Fletcher D.A., Anderson G.G., Footer M.J., Hultgren S.J. Differentiation and Developmental Pathways of Uropathogenic Escherichia Coli in Urinary Tract Pathogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y.S., Bastidas R.J., Saka H.A., Carpenter V.K., Richards K.L., Plano G.V., Valdivia R.H. The Chlamydia Trachomatis Type Iii Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including Tepp, a Tyrosine-Phosphorylated Protein Required for the Recruitment of Crki-Ii to Nascent Inclusions and Innate Immune Signaling. PLoS Pathog. 2014;10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawana K., Quayle A.J., Ficarra M., Ibana J.A., Shen L., Kawana Y., Yang H., Marrero L., Yavagal S., Greene S.J., et al. Cd1d Degradation in Chlamydia Trachomatis-Infected Epithelial Cells Is the Result of Both Cellular and Chlamydial Proteasomal Activity. J. Biol. Chem. 2007;282:7368–7375. doi: 10.1074/jbc.M610754200. [DOI] [PubMed] [Google Scholar]
- 20.Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for Bad Bugs: Confronting the Challenges of Antibacterial Discovery. Nat. Rev. Drug. Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 21.Chan P.F., Holmes D.J., Payne D.J. Finding the Gems Using Genomic Discovery: Antibacterial Drug Discovery Strategies—The Successes and the Challenges. Drug Discov. Today Ther. Strateg. 2004;1:519–527. doi: 10.1016/j.ddstr.2004.11.003. [DOI] [Google Scholar]
- 22.Brown E.D., Wright G.D. Antibacterial Drug Discovery in the Resistance Era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea R., Moser H.E. Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski C.A. Drug-Like Properties and the Causes of Poor Solubility and Poor Permeability. J. Pharmacol. Toxicol. Methods. 2001;44:235–249. doi: 10.1016/S1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 25.Ebejer J.P., Charlton M.H., Finn P.W. Are the Physicochemical Properties of Antibacterial Compounds Really Different from Other Drugs? J. Cheminform. 2016;8:30. doi: 10.1186/s13321-016-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar A. Enabling Design of Screening Libraries for Antibiotic Discovery by Modeling Chembl Data. Eur. J. Pharm. Sci. 2019;143:105166. doi: 10.1016/j.ejps.2019.105166. [DOI] [PubMed] [Google Scholar]
- 27.Parker E.N., Drown B.S., Geddes E.J., Lee H.Y., Ismail N., Lau G.W., Hergenrother P.J. Implementation of Permeation Rules Leads to a Fabi Inhibitor with Activity against Gram-Negative Pathogens. Nat. Microbiol. 2020;5:67–75. doi: 10.1038/s41564-019-0604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter M.F., Drown B.S., Riley A.P., Garcia A., Shirai T., Svec R.L., Hergenrother P.J. Predictive Compound Accumulation Rules Yield a Broad-Spectrum Antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper S.J., Krishnamoorthy G., Wolloscheck D., Walker J.K., Rybenkov V.V., Parks J.M., Zgurskaya H.I. Molecular Properties That Define the Activities of Antibiotics in Escherichia Coli and Pseudomonas Aeruginosa. ACS Infect. Dis. 2018;4:1223–1234. doi: 10.1021/acsinfecdis.8b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnamoorthy G., Leus I.V., Weeks J.W., Wolloscheck D., Rybenkov V.V., Zgurskaya H.I. Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. mBio. 2017;8:e01172-17. doi: 10.1128/mBio.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamoorthy G., Wolloscheck D., Weeks J.W., Croft C., Rybenkov V.V., Zgurskaya H.I. Breaking the Permeability Barrier of Escherichia Coli by Controlled Hyperporination of the Outer Membrane. Antimicrob. Agents Chemother. 2016;60:7372–7381. doi: 10.1128/AAC.01882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansbach R.A., Leus I.V., Mehla J., Lopez C.A., Walker J.K., Rybenkov V.V., Hengartner N.W., Zgurskaya H.I., Gnanakaran S. Machine Learning Algorithm Identifies an Antibiotic Vocabulary for Permeating Gram-Negative Bacteria. J. Chem. Inf. Model. 2020;60:2838–2847. doi: 10.1021/acs.jcim.0c00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha P., Sikdar S., Krishnamoorthy G., Zgurskaya H.I., Rybenkov V.V. Drug Permeation against Efflux by Two Transporters. ACS Infect. Dis. 2020;6:747–758. doi: 10.1021/acsinfecdis.9b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergalli J., Atzori A., Pajovic J., Dumont E., Malloci G., Masi M., Vargiu A.V., Winterhalter M., Refregiers M., Ruggerone P., et al. The Challenge of Intracellular Antibiotic Accumulation, a Function of Fluoroquinolone Influx versus Bacterial Efflux. Commun. Biol. 2020;3:198. doi: 10.1038/s42003-020-0929-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westfall D.A., Krishnamoorthy G., Wolloscheck D., Sarkar R., Zgurskaya H.I., Rybenkov V.V. Bifurcation Kinetics of Drug Uptake by Gram-Negative Bacteria. PLoS ONE. 2017;12:e0184671. doi: 10.1371/journal.pone.0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zgurskaya H.I., Rybenkov V.V. Permeability Barriers of Gram-Negative Pathogens. Ann. N. Y. Acad. Sci. 2020;1459:5–18. doi: 10.1111/nyas.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Zahed S.S., French S., Farha M.A., Kumar G., Brown E.D. Physicochemical and Structural Parameters Contributing to the Antibacterial Activity and Efflux Susceptibility of Small Molecule Inhibitors of Escherichia Coli. Antimicrob. Agents Chemother. 2021;65:e01925-20. doi: 10.1128/AAC.01925-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tommasi R., Iyer R., Miller A.A. Antibacterial Drug Discovery: Some Assembly Required. ACS Infect. Dis. 2018;4:686–695. doi: 10.1021/acsinfecdis.8b00027. [DOI] [PubMed] [Google Scholar]
- 39.Casadevall A., Pirofski L.A. Host-Pathogen Interactions: Redefining the Basic Concepts of Virulence and Pathogenicity. Infect. Immun. 1999;67:3703–3713. doi: 10.1128/IAI.67.8.3703-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassenaar T.M., Gaastra W. Bacterial Virulence: Can We Draw the Line? FEMS Microbiol. Lett. 2001;201:1–7. doi: 10.1111/j.1574-6968.2001.tb10724.x. [DOI] [PubMed] [Google Scholar]
- 41.Gu J., Wang Y., Lilburn T. A Comparative Genomics, Network-Based Approach to Understanding Virulence in Vibrio Cholerae. J. Bacteriol. 2009;191:6262–6272. doi: 10.1128/JB.00475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. Vfdb: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L., Xiong Z., Sun L., Yang J., Jin Q. Vfdb 2012 Update: Toward the Genetic Diversity and Molecular Evolution of Bacterial Virulence Factors. Nucleic Acids Res. 2012;40:D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou C.E., Smith J., Lam M., Zemla A., Dyer M.D., Slezak T. Mvirdb—A Microbial Database of Protein Toxins, Virulence Factors and Antibiotic Resistance Genes for Bio-Defence Applications. Nucleic Acids Res. 2007;35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng L.-L., Li Y.-X., Ding J., Guo X.-K., Feng K.-Y., Wang Y.-J., Hu L.-L., Cai Y.-D., Hao P., Chou K.-C. A Comparison of Computational Methods for Identifying Virulence Factors. PLoS ONE. 2012;7:e42517. doi: 10.1371/journal.pone.0042517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng A.C., Coleman R.G., Smyth K.T., Cao Q., Soulard P., Caffrey D.R., Salzberg A.C., Huang E.S. Structure-Based Maximal Affinity Model Predicts Small-Molecule Druggability. Nat. Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 47.Desaphy J., Azdimousa K., Kellenberger E., Rognan D. Comparison and Druggability Prediction of Protein-Ligand Binding Sites from Pharmacophore-Annotated Cavity Shapes. J. Chem. Inf. Model. 2012;52:2287–2299. doi: 10.1021/ci300184x. [DOI] [PubMed] [Google Scholar]
- 48.Fauman E.B., Rai B.K., Huang E.S. Structure-Based Druggability Assessment--Identifying Suitable Targets for Small Molecule Therapeutics. Curr. Opin. Chem. Biol. 2011;15:463–468. doi: 10.1016/j.cbpa.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 49.Hajduk P.J., Huth J.R., Fesik S.W. Druggability Indices for Protein Targets Derived from Nmr-Based Screening Data. J. Med. Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 50.Halgren T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. J. Chem. Inf. Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 51.Henrich S., Salo-Ahen O.M., Huang B., Rippmann F.F., Cruciani G., Wade R.C. Computational Approaches to Identifying and Characterizing Protein Binding Sites for Ligand Design. J. Mol. Recognit. 2010;23:209–219. doi: 10.1002/jmr.984. [DOI] [PubMed] [Google Scholar]
- 52.Keller T.H., Pichota A., Yin Z. A Practical View of ‘Druggability’. Curr. Opin. Chem. Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 53.Schmidtke P., Barril X. Understanding and Predicting Druggability. A High-Throughput Method for Detection of Drug Binding Sites. J. Med. Chem. 2010;53:5858–5867. doi: 10.1021/jm100574m. [DOI] [PubMed] [Google Scholar]
- 54.Seco J., Luque F.J., Barril X. Binding Site Detection and Druggability Index from First Principles. J. Med. Chem. 2009;52:2363–2371. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- 55.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. Combining Global and Local Measures for Structure-Based Druggability Predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 56.Volkamer A., Rarey M. Exploiting Structural Information for Drug-Target Assessment. Future Med. Chem. 2014;6:319–331. doi: 10.4155/fmc.14.3. [DOI] [PubMed] [Google Scholar]
- 57.Krasowski A., Muthas D., Sarkar A., Schmitt S., Brenk R. Drugpred: A Structure-Based Approach to Predict Protein Druggability Developed Using an Extensive Nonredundant Data Set. J. Chem. Inf. Model. 2011;51:2829–2842. doi: 10.1021/ci200266d. [DOI] [PubMed] [Google Scholar]
- 58.Sarkar A., Brenk R. To Hit or Not to Hit, That Is the Question—Genome-Wide Structure-Based Druggability Predictions for Pseudomonas Aeruginosa Proteins. PLoS ONE. 2015;10:e0137279. doi: 10.1371/journal.pone.0137279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thurlow L.R., Joshi G.S., Richardson A.R. Virulence Strategies of the Dominant Usa300 Lineage of Community-Associated Methicillin-Resistant Staphylococcus Aureus (Ca-Mrsa) FEMS Immunol. Med. Microbiol. 2012;65:5–22. doi: 10.1111/j.1574-695X.2012.00937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schumacher A., Vranken T., Malhotra A., Arts J.J.C., Habibovic P. In Vitro Antimicrobial Susceptibility Testing Methods: Agar Dilution to 3d Tissue-Engineered Models. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:187–208. doi: 10.1007/s10096-017-3089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang S.L., Mekalanos J.J., Holden D.W. In Vivo Genetic Analysis of Bacterial Virulence. Annu. Rev. Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 62.Totsika M. Benefits and Challenges of Antivirulence Antimicrobials at the Dawn of the Post-Antibiotic Era. Curr. Med. Chem. 2016;6:30–37. doi: 10.2174/2210303106666160506120057. [DOI] [Google Scholar]
- 63.Duraiswamy S., Chee J.L.Y., Chen S., Yang E., Lees K., Chen S.L. Purification of Intracellular Bacterial Communities during Experimental Urinary Tract Infection Reveals an Abundant and Viable Bacterial Reservoir. Infect. Immun. 2018;86:e00740-17. doi: 10.1128/IAI.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alhede M., Kragh K.N., Qvortrup K., Allesen-Holm M., van Gennip M., Christensen L.D., Jensen P.Ø., Nielsen A.K., Parsek M., Wozniak D., et al. Phenotypes of Non-Attached Pseudomonas Aeruginosa Aggregates Resemble Surface Attached Biofilm. PLoS ONE. 2011;6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts A.E., Kragh K.N., Bjarnsholt T., Diggle S.P. The Limitations of in Vitro Experimentation in Understanding Biofilms and Chronic Infection. J. Mol. Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Shi D., Mi G., Wang M., Webster T. In Vitro and Ex Vivo Systems at the Forefront of Infection Modeling and Drug Discovery. Biomaterials. 2018;198:228–249. doi: 10.1016/j.biomaterials.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Begun J., Gaiani J.M., Rohde H., Mack D., Calderwood S.B., Ausubel F.M., Sifri C.D. Staphylococcal Biofilm Exopolysaccharide Protects against Caenorhabditis Elegans Immune Defenses. PLoS Pathog. 2007;3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sifri C.D., Baresch-Bernal A., Calderwood S.B., von Eiff C. Virulence of Staphylococcus Aureus Small Colony Variants in the Caenorhabditis Elegans Infection Model. Infect. Immun. 2006;74:1091–1096. doi: 10.1128/IAI.74.2.1091-1096.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sifri C.D., Begun J., Ausubel F.M. The Worm Has Turned--Microbial Virulence Modeled in Caenorhabditis Elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Sifri C.D., Begun J., Ausubel F.M., Calderwood S.B. Caenorhabditis Elegans as a Model Host for Staphylococcus Aureus Pathogenesis Caenorhabditis Elegans as a Model Host for Staphylococcus Aureus Pathogenesis. Infect. Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.García-Lara J., Needham A.J., Foster S.J. Invertebrates as Animal Models for Staphylococcus Aureus Pathogenesis: A Window into Host–Pathogen Interaction. FEMS Immunol. Med. Microbiol. 2005;43:311–323. doi: 10.1016/j.femsim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Miyazaki S., Matsumoto Y., Sekimizu K., Kaito C. Evaluation of Staphylococcus Aureus Virulence Factors Using a Silkworm Model. FEMS Microbiol. Lett. 2012;326:116–124. doi: 10.1111/j.1574-6968.2011.02439.x. [DOI] [PubMed] [Google Scholar]
- 73.Hoffmann S., Maculloch B., Batz M. Economic Burden of Major Foodborne Illnesses Acquired in the United States, Eib-140. Economic Research Service U.S. Department of Agriculture; Washington, DC, USA: 2015. [Google Scholar]
- 74.WHO . Estimates of the Global Burden of Foodborne Diseases. World Health Organization; Geneva, Switzerland: 2015. 2007–2015, Foodborne Disease Burden Epidemiology Reference Group. [Google Scholar]
- 75.World Health Organization . Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections-2008. World Health Organization; Geneva, Switzerland: 2012. [Google Scholar]
- 76.Simmering J.E., Tang F., Cavanaugh J.E., Polgreen L.A., Polgreen P.M. The Increase in Hospitalizations for Urinary Tract Infections and the Associated Costs in the United States, 1998–2011. Open Forum Infect. Dis. 2017;4:ofw281. doi: 10.1093/ofid/ofw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein E.Y., Jiang W., Mojica N., Tseng K.K., McNeill R., Cosgrove S.E., Perl T.M. National Costs Associated with Methicillin-Susceptible and Methicillin-Resistant Staphylococcus Aureus Hospitalizations in the United States, 2010–2014. Clin. Infect. Dis. 2019;68:22–28. doi: 10.1093/cid/ciy399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fisher R.A., Gollan B., Helaine S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017;15:453. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 79.Ray K., Marteyn B., Sansonetti P.J., Tang C.M. Life on the Inside: The Intracellular Lifestyle of Cytosolic Bacteria. Nat. Rev. Microbiol. 2009;9:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 80.Chai Q., Wang L., Liu C.H., Ge B. New Insights into the Evasion of Host Innate Immunity by Mycobacterium Tuberculosis. Cell Mol. Immunol. 2020;17:901–913. doi: 10.1038/s41423-020-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robino L., Scavone P., Araujo L., Algorta G., Zunino P., Pírez M.C., Vignoli R. Intracellular Bacteria in the Pathogenesis of Escherichia Coli Urinary Tract Infection in Children. Clin. Infect. Dis. 2014;75:e158–e164. doi: 10.1093/cid/ciu634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freitag N.E., Port G.C., Miner M.D. Listeria Monocytogenes—From Saprophyte to Intracellular Pathogen. Nat. Rev. Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagamatsu K., Hannan T.J., Guest R.L., Kostakioti M., Hadjifrangiskou M., Binkley J., Dodson K., Raivio T.L., Hultgren S.J. Dysregulation of Escherichia Coli A-Hemolysin Expression Alters the Course of Acute and Persistent Urinary Tract Infection. Proc. Natl. Acad. Sci. USA. 2015;75:E871–E880. doi: 10.1073/pnas.1500374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carlson J.H., Whitmire W.M., Crane D.D., Wicke L., Virtaneva K., Sturdevant D.E., Kupko J.J., 3rd, Porcella S.F., Martinez-Orengo N., Heinzen R.A., et al. The Chlamydia Trachomatis Plasmid Is a Transcriptional Regulator of Chromosomal Genes and a Virulence Factor. Infect. Immun. 2008;76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heuer D., Lipinski A.R., Machuy N., Karlas A., Wehrens A., Siedler F., Brinkmann V., Meyer T.F. Chlamydia Causes Fragmentation of the Golgi Compartment to Ensure Reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 86.Tullius M.V., Harth G., Horwitz M.A. Glutamine Synthetase Glna1 Is Essential for Growth of Mycobacterium Tuberculosis in Human Thp-1 Macrophages and Guinea Pigs. Infect. Immun. 2003;71:3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loisel-Meyer S., de Bagüés M.P.J., Bassères E., Dornand J., Köhler S., Liautard J.-P., Jubier-Maurin V. Requirement of Nord for Brucella Suis Virulence in a Murine Model of in Vitro and in Vivo Infection. Infect. Immun. 2006;75:1973–1976. doi: 10.1128/IAI.74.3.1973-1976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ammendola S., Pasquali P., Pistoia C., Petrucci P., Petrarca P., Rotilio G., Battistoni A. High-Affinity Zn2+ Uptake System Znuabc Is Required for Bacterial Zinc Homeostasis in Intracellular Environments and Contributes to the Virulence of Salmonella Enterica. Infect. Immun. 2007;75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vitoriano I., Saraiva-Pava K.D., Rocha-Gonçalves A., Santos A., Lopes A.I., Oleastro M., Roxo-Rosa M. Ulcerogenic Helicobacter Pylori Strains Isolated from Children: A Contribution to Get Insight into the Virulence of the Bacteria. PLoS ONE. 2011;6:e26265. doi: 10.1371/journal.pone.0026265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mirrashidi K.M., Elwell C.A., Verschueren E., Johnson J.R., Frando A., von Dollen J., Rosenberg O., Gulbahce N., Jang G., Johnson T., et al. Global Mapping of the Inc-Human Interactome Reveals That Retromer Restricts Chlamydia Infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Purschke F.G., Hiller E., Trick I., Rupp S. Flexible Survival Strategies of Pseudomonas Aeruginosa in Biofilms Result in Increased Fitness Compared with Candida Albicans. Mol. Cell. Proteom. MCP. 2012;12:1652–1669. doi: 10.1074/mcp.M112.017673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chey W.D., Leontiadis G.I., Howden C.W., Moss S.F. Acg Clinical Guideline: Treatment of Helicobacter Pylori Infection. Am. J. Gastroenterol. 2017;112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 93.Metlay J.P., Waterer G.W., Long A.C., Anzueto A., Brozek J., Crothers K., Cooley L.A., Dean N.C., Fine M.J., Flanders S.A., et al. Diagnosis and Treatment of Adults with Community-Acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care Med. 2019;200:e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fournier P.E., Thuny F., Richet H., Lepidi H., Casalta J.P., Arzouni J.P., Maurin M., Celard M., Mainardi J.L., Caus T., et al. Comprehensive Diagnostic Strategy for Blood Culture-Negative Endocarditis: A Prospective Study of 819 New Cases. Clin. Infect. Dis. 2010;51:131–140. doi: 10.1086/653675. [DOI] [PubMed] [Google Scholar]
- 95.St Cyr S., Barbee L., Workowski K.A., Bachmann L.H., Pham C., Schlanger K., Torrone E., Weinstock H., Kersh E.N., Thorpe P. Update to Cdc’s Treatment Guidelines for Gonococcal Infection, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;50:1911–1916. doi: 10.15585/mmwr.mm6950a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baddour L.M., Wilson W.R., Bayer A.S., Fowler V.G., Jr., Tleyjeh I.M., Rybak M.J., Barsic B., Lockhart P.B., Gewitz M.H., Levison M.E., et al. Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications: A Scientific Statement for Healthcare Professionals from the American Heart Association. Circulation. 2015;15:1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 97.Osmon D.R., Berbari E.F., Berendt A.R., Lew D., Zimmerli W., Steckelberg J.M., Rao N., Hanssen A., Wilson W.R. Diagnosis and Management of Prosthetic Joint Infection: Clinical Practice Guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 2013;56:e1–e25. doi: 10.1093/cid/cis803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.