Abstract
The nucleolus has long been known as a functionally highly specialized subnuclear compartment where synthesis, posttranscriptional modification, and processing of cytoplasmic rRNAs take place. In this study, we demonstrate that the nucleolus contains all the trans-acting factors that are responsible for the accurate and efficient synthesis of the eight 2′-O-methylated nucleotides and three pseudouridine residues carried by the mammalian U6 spliceosomal small nuclear RNA. Factors mediating the formation of pseudouridine residues in the U3 small nucleolar RNA are also present and functionally active in the nucleolus. For selection of the correct target nucleotides in the U6 and U3 RNAs, the nucleolar 2′-O-methylation and pseudouridylation factors rely on short sequences located around the target nucleotide to be modified. This observation further underscores a recently proposed role for small nucleolar guide RNAs in the 2′-O-methylation of the U6 spliceosomal RNA (K. T. Tycowski, Z.-H. You, P. J. Graham, and J. A. Steitz, Mol. Cell 2:629–638, 1998). We demonstrate that a novel 2′-O-methylated nucleotide can be generated in the yeast U6 RNA by use of an artificial 2′-O-methylation small nucleolar guide RNA. We also show that a short fragment of the 5.8S rRNA, when expressed as part of the human U6 RNA, is faithfully 2′-O-methylated and pseudouridylated. These results are most consistent with a trafficking pathway in which the U6 spliceosomal RNA cycles through the nucleolus to undergo nucleolar RNA-directed modifications.
In eukaryotes, most stable cellular RNAs undergo extensive posttranscriptional nucleoside modifications (5, 41, 62). For tRNAs, rRNAs, and small nuclear RNAs (snRNAs), about 90 different modified nucleotides have been identified (39). Despite the great structural diversity of modified residues, methylation of the backbone ribose at the 2′-hydroxyl position and conversion of uridine residues into pseudouridine residues represent the most abundant RNA modifications. The versatile hydrogen-bonding capacity of pseudouridines and the hydrophobic nature of 2′-O-methyl groups can modulate the three-dimensional structure of the RNA or fine-tune its interactions with other RNAs or proteins (36, 41, 55).
In tRNAs, modified nucleosides are important determinants of the specificity and efficiency of both aminoacylation and codon recognition (5, 81). The actual role of the large number of 2′-O-methyl groups and pseudouridines found in rRNAs and snRNAs is unknown. However, the fact that these modifications cluster around the functionally essential regions of these RNAs suggests that they contribute to RNA function (1, 41, 62). Consistent with this notion, a 2′-O-methyl group at G2251 in yeast mitochondrial 21S RNA has been found to be essential for the production of functional 50S ribosomal subunits (71). The importance of modifications in spliceosomal snRNAs has recently been underscored by the findings that in vitro-synthesized U2 snRNAs failed to reconstitute pre-mRNA splicing both in a U2-depleted HeLa cell splicing extract (66) and in Xenopus oocytes (57, 82).
A cell follows different strategies to accomplish the accurate synthesis of modified nucleosides in tRNAs, rRNAs, and snRNAs. The formation of pseudouridines and 2′-O-methylated nucleosides in tRNAs is catalyzed by protein enzymes which recognize the sequence and/or structure of the target site (3, 10, 38, 70). In 18S, 5.8S, and 28S rRNAs, the selection of more than 200 2′-O-methylation and pseudouridylation sites that occupy diverse sequence and structural environments is mediated by small nucleolar guide RNAs (snoRNAs). For each modification site, transient base-pairing interactions between a specific snoRNA and the target rRNA sequence occur. Methylation snoRNAs form 10- to 21-bp perfect double helices with rRNAs that are immediately followed by the conserved D or D′ box sequence motifs of the snoRNAs. In the snoRNA-rRNA double helix, the fifth ribosomal nucleotide upstream of the D or D′ box represents the target nucleotide for the methyltransfer reaction (11, 31, 32). Pseudouridylation snoRNAs take part in two short interactions with rRNA sequences that precede and follow the target uridine residue. Normally, in the pseudouridylation snoRNA-rRNA interaction, the substrate uridine is located 14 nucleotides (nt) upstream of the ACA or H box motif of the snoRNA (8, 16, 52). A pseudouridine synthase and a methyltransferase enzyme, most probably the Nap57p/Cbf5p (34) and fibrillarin/Nop1 (53, 75) proteins, that are directly or indirectly bound to the H/ACA or C/D box elements of the snoRNAs catalyze the nucleoside modification reaction. Therefore, the formation of numerous ribosomal pseudouridines and 2′-O-methylated nucleotides most likely is catalyzed by a single pseudouridine synthase and a methyltransferase enzyme.
Less is known about the generation of pseudouridines and 2′-O-methylated residues in spliceosomal snRNAs. In vitro pseudouridylation experiments suggest that multiple pseudouridine synthase activities direct the pseudouridylation of the polymerase II-synthesized U1, U2, and U5 snRNAs (59, 60). Since maturation of these snRNAs proceeds via a cytoplasmic phase (20, 47, 58), it remains unclear whether their modification occurs in the nucleoplasm or the cytoplasm. The U6 snRNA, an RNA polymerase III product (12, 21), represents the most conserved and most extensively modified spliceosomal RNA (62). Microinjection experiments indicated that U6 does not leave the nucleus of Xenopus oocytes, suggesting that its maturation occurs in the nucleus (80). Unexpectedly, it has recently been found that 2′-O-methylation of U6 snRNA at C-77 and perhaps at A-47 is directed by C and D box-containing methylation snoRNAs (79).
The nucleolus has long been considered a subnuclear compartment that is devoted to the maturation of cytoplasmic rRNAs (18). The formation of mature rRNAs, including 2′-O-ribose methylation, pseudouridylation, and nucleolytic processing of the newly synthesized precursor rRNA is assisted by many snoRNAs (48, 72, 73, 77). Here, we demonstrate that factors directing the correct posttranscriptional modification of mammalian U6 snRNA at the eight known 2′-O-methylation and three pseudouridylation sites are present and functionally active in the nucleolus.
MATERIALS AND METHODS
General procedures and oligos.
Unless otherwise noted, all cloning and nucleic acid manipulation procedures were performed according to standard laboratory protocols (65). The identity of all constructs was confirmed by sequence analyses. The oligodeoxynucleotides (oligos) used in this study and their sequences are as follows: 1, CTAGTACTAAAATTGGAACGATACAGAGAA; 2, TCGATTCTCTGTATCGTTCCAATTTTAGTA; 3, CTAGAGAAGATTAGCATGGCCC; 4, TCGAGGGCCATGCTAATCTTCT; 5, CTAGATGGCCCCTGCGCAAGGATGACA; 6, TCGATGTCATCCTTGCGCAGGGGCCAT; 7, CTAGAGATGACACGCAAATTCGTGAAGCGC; 8, TCGAGCGCTTCACGAATTTGCGTGTCATCT; 9, CTAGAGTGTAGTATCTGTTCTTATCAGC; 10, TCGAGCTGATAAGAACAGATACTACACT; 11, CTAGAAAGACTATACTTTCAGGGATCAC; 12, TCGAGTGATCCCTGAAAGTATAGTCTTT; 13, CTAGATTAATGTGAATTGCAGGACACATGACTAGTC; 14, TCGAGACTAGTCATGTGTCCTGCAATTCACATTAAT; 15XhoI, TTTCTCGAGCCCCAGTGGAAAGACGCGCAG; 16SspI, CCCAATATTGGAACGCTTCACGAATTTGCG; 17SpeI, TTTACTAGTAATATTTTTACATCAGGTTG; 18SacI, TTTGAGCTCTGGTAAACCGTGCACCGGCG; 19 ATTAATGTGAATTGCAGGACACAGA; 20, CTAGTCTGTGTCCTGCAATTCACATTAAT; 21SacI, CTCAATATTATGTGCTGCCGAAGCG; 22SpeI, CTCACTAGTCATACTAAAATTGGAACGATACA; 23XhoI, TTCTCGAGTAGCTGGGACTACAGACGG; 24BclI, TTTTGATCACTATAGAAATGATCCCTG; 25SpeI, TTTACTAGTGTTACTAGAGAAGTTTCTC; 26KpnI, TTTGGTACCTTTCTCGCGACATTGCCAAGC; 27, GATCATTAATGTGAATTGCAGGACACATGA; 28, CTAGTCATGTGTCCTGCAATTCACATTAAT; 29, GATCCTTAATGTGAATTGCAGGACACATGACTAGTG; 30, GATCCACTAGTCATGTGTCCTGCAATTCACATTAAG; 31KpnI, TTTGGTACCTGGTGCATCAGTTTGGTCAATTTGATTAAAATGTCATCA; 32XhoI, ATACTCGAGTGTGCAGATGATGTAAAAG; 33, CCAGCTCAAGATCGTAATAT; 34, GTTATTACATCATTTGA; 35XbaI, TGCTCTAGAGTGCTCGCTTCGGCAGC; 36XhoI, CCGCTCGAGAAAATATGGAACGCTTCAC; 37XbaI, TGCTCTAGATCCCAATGATGAGTTGCCATGC; 38XhoI, CCGCTCGAGACCCCTCAGATCTTCATGTGAG; 39, AAAATATTACTAGTCTGTG; 40, ATTTTAGTATGACTAGTCTGTG; 41, GTGGACGGAGCAACTAGTCATG; 42, TTCTCTAGTAACACTAGTCATG; 43, TTCTCAGGATCCACTAGTCATG; 44, CCAGTGATTTTTTTCTCCATTTTAGC; 45, GTCTTCAAAGTTCTCATTTG; and 46, ACTGCTGATCATCTCTGTATTG.
Construction of plasmids and transfection of mammalian cells.
To generate the pW-U6-1 expression construct, appropriate oligos (oligos 1 and 2) were annealed and inserted into the XbaI and XhoI sites of recombinant plasmid pW(Xb/Xh) carrying the mouse ribosomal minigene (16, 19). The same strategy was used to obtain pW-U6-2 (oligos 3 and 4), pW-U6-3 (oligos 5 and 6), pW-U6-4 (oligos 7 and 8), pW-U2 (oligos 9 and 10), pW-U3 (oligos 11 and 12), and pW-5.8S (oligos 13 and 14). Transfection of mouse L929 (American Type Culture Collection [ATCC] CCL1) cells was achieved by use of the DEAE-dextran method (67).
To construct pGL/U6-5.8S(3′), two contiguous fragments of the human U6 gene (33) (GenBank accession no. M14486) from positions −328 to +100 and from positions +101 to +199 were PCR amplified with Vent DNA polymerase (New England BioLabs), human genomic DNA as a template, and oligo primers 15XhoI/16SspI and 17SpeI/18SacI, respectively. The amplified fragments were digested with appropriate endonucleases and joined in a quadrimolecular ligation reaction in the presence of the XhoI- and SacI-digested pGL2 promoter vector (Promega) and annealed oligos 19 and 20, which represented a fragment of the human 5.8S rRNA gene from positions 62 to 84 and formed SspI- and SpeI-compatible termini. The same approach was used to construct pGL/U6-5.8S(5′) and pGL/U3-5.8S. For amplification of the 5′ and 3′ halves of the human U6 gene from positions −328 to +22 and from positions +23 to +199, oligos 15XhoI/21SacI and 22SpeI/18KpnI were used as specific primers, respectively. The amplified fragments were digested and connected via ligation of annealed oligos 19 and 20 to the PCR-introduced SspI and SpeI sites. The 5′ and 3′ halves of the human U3 gene (83) (GenBank accession no. X14945) from positions −478 to +33 and from positions +34 to +313 were PCR amplified with oligos 23XhoI/24BclI and 25SpeI/26KpnI, respectively. Annealed oligos 27 and 28 carrying 5.8S-specific tag sequences were inserted between the PCR-introduced BclI and SpeI sites of the U3 gene fragments. The resulting 5.8S-tagged U6 and U3 genes were cloned into the XhoI/SacI and XhoI/KpnI sites of the pGL2 promoter vector, respectively. To generate pG-5.8S, annealed oligos 29 and 30 were inserted into the BamHI site of the pG expression construct (29). Transfection of COS-7 (ATCC CRL 1651) cells was performed with DOTAP {N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate} transfection reagent (Boehringer) according to the manufacturer’s instructions.
Cell fractionation.
Fractionation of transfected simian COS-7 and mouse cells as well as human HeLa cells was performed as described earlier (27, 78).
Construction of plasmids for transformation of yeast cells.
The pFL45/ACT/U24 yeast expression construct has already been described (32). To generate pFL45/ACT/U24-6, the coding region of the human U24 snoRNA gene was PCR amplified with oligos 31KpnI and 32XhoI as 3′ and 5′ end-specific primers, respectively. The resulting U24-6 fragment was inserted into the XhoI and KpnI sites of the pFL45/ACT expression construct (32). Transformation of the yeast ΔU24 strain (a trp1 Δ his3 Δ ura3,52 lys2,801 ade2,101 URA3::U24) (31) was performed by the lithium acetate transformation procedure (22).
RNA analysis.
Total RNAs from human HeLa, mouse L929, and simian COS-7 cells (17) and yeast cells (76) were isolated by guanidinium thiocyanate–phenol-chloroform extraction. For Northern analysis, 10 μg of yeast cellular RNAs was separated on a 6% sequencing gel, electroblotted onto a Hybond-N nylon membrane, and probed with a mixture of 5′-end-labeled oligos complementary to the yeast snR36 (oligo 33) and human U24 (oligo 34) snoRNAs. RNase A and T1 mapping was performed as described previously (17). Generation of a sequence-specific antisense RNA probe for human U3 snoRNA has been reported elsewhere (15, 27). To obtain RNA probes for the U6-5.8S(5′) and U6-5.8S(3′) RNAs, the XhoI/SacI fragments of the pGL/U6-5.8S(5′) and pGL/U6-5.8S(3′) constructs were inserted into the same sites of pBluescript KS (Stratagene). The resulting pU6-5.8S(5′) and pU6-5.8S(3′) plasmids were linearized with XhoI and used as templates for the synthesis of antisense RNA probes by use of T7 RNA polymerase. To generate probes for the human U6 and mgU6-53 RNAs, the coding regions of the U6 (oligos 35XbaI and 36XhoI) and mgU5-53 (oligos 37Xbal and 38XhoI) snRNAs were PCR amplified with human genomic DNA as a template. The resulting DNA fragments were inserted into the XbaI/XhoI sites of pBluescript KS, linearized with XhoI, and transcribed by use of T3 RNA polymerase. To generate an antisense probe for the W-U6-1 mouse minigene transcript, the PstI/EcoRI fragment of pW-U6-1 encompassing the full-length ribosomal minigene was subcloned into the same sites of pBluescript KS, linearized with HindIII, and transcribed by use of T7 RNA polymerase.
Mapping of 2′-O-methylated nucleotides and pseudouridine residues.
Detection of 2′-O-ribose-methylated nucleotides and pseudouridine residues was performed by primer extension analyses as described by Maden et al. (42) and Bakin and Ofengand (2), respectively. Terminally labeled oligos 39 [for U6-5.8S(5′) RNA], 40 [for U6-5.8S(3′) RNA], 41 [for human U2 snRNA], 42 (for U3-5.8S RNA), 43 (for G-5.8S RNA), 44 (for mouse ribosomal minigene transcripts), 45 (for yeast 25S rRNA), and 46 (for yeast U6 snRNA) complementary to the appropriate target RNAs were used as primers for reverse transcription. The extended DNA products were analyzed on 6% sequencing gels.
RESULTS
A putative guide snoRNA for 2′-O-methylation of human U6 snRNA at Am53.
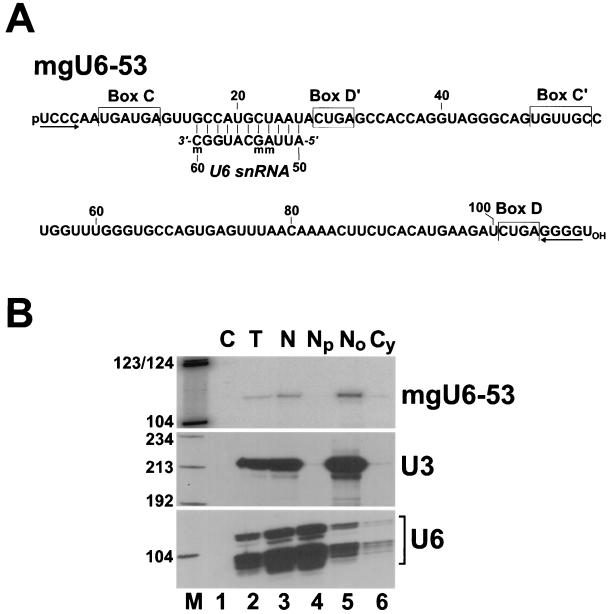
During characterization of a cDNA library of human snoRNAs (31), we identified a 109-nt novel small RNA (Fig. 1A). Cell fractionation experiments demonstrated that the newly identified RNA, like the authentic U3 snoRNA (78), copurifies with the nucleolar fraction of human HeLa cells (Fig. 1B, upper panel, lane 5) and is absent from the nucleoplasmic fraction, where the U6 spliceosomal snRNA accumulates (lane 4). The new RNA features all of the structural elements, the box C, C′, D, and D′ motifs and a short 5′-, 3′-terminal helix, that are essential for the expression, nucleolar localization, and function of 2′-O-methylation snoRNAs (31, 32, 37, 64). However, the novel putative 2′-O-methylation snoRNA lacks a significant sequence complementarity to rRNA sequences, indicating that it cannot function in rRNA methylation.
FIG. 1.
Human mgU6-53 is a novel C and D box-containing snoRNA that possesses sequence complementarity to U6 spliceosomal RNA. (A) Nucleotide sequence of mgU6-53 snoRNA. The conserved sequence box motifs of 2′-O-methylation snoRNAs are indicated. Sequences potentially involved in the formation of a 5′-, 3′-terminal helix are indicated by inverted arrows. Sequences of the human U6 snRNA that are complementary to the mgU6-53 snoRNA sequence are shown. Nucleotides carrying 2′-O-methyl groups are indicated (m). The nucleotide sequence of mgU6-53 has been deposited in the EMBL database under accession no. AJ243222. (B) Subcellular localization of mgU6-53 snoRNA. RNA samples (200 ng) extracted either from human HeLa cells (T) or from nuclear (N), nucleoplasmic (Np), nucleolar (No), and cytoplasmic (Cy) fractions of HeLa cells were mapped by RNase A and T1 protection by use of sequence-specific antisense RNA probes as indicated on the right. Lane C represents a control mapping with Escherichia coli tRNA. Lane M, size markers in nucleotides (a mixture of HaeIII- and TaqI-digested pBR322).
Recently, it was shown that 2′-O-methylation of the Cm77 residue of human U6 spliceosomal snRNA, upon injection into Xenopus oocytes, is dependent on a C and D box-containing methylation snoRNA called mgU6-77 (methylation guide for U6 RNA at position 77) (79). Another C and D box-containing snoRNA (mgU6-47) was implicated in the synthesis of the 2′-O-methylated Am47 residue in U6 snRNA (79). A closer examination of our new C and D box-containing snoRNA revealed that it carries an 11-nt sequence that is perfectly complementary to the human U6 snRNA from positions 50 to 60 (Fig. 1A). A putative base-pairing interaction between the snoRNA and the U6 snRNA places the D′ box of the snoRNA 5 bp upstream of the A53 residue that is known to be 2′-O-methylated in mammalian U6 snRNAs (62). We therefore concluded that the novel RNA, now termed the mgU6-53 snoRNA, represents a new member of the group of C and D box-containing snoRNAs and is likely to direct the 2′-O-methylation of U6 snRNA.
Factors directing 2′-O-methylation and pseudouridylation of U6 and U3 snRNAs are functional in the nucleolus.
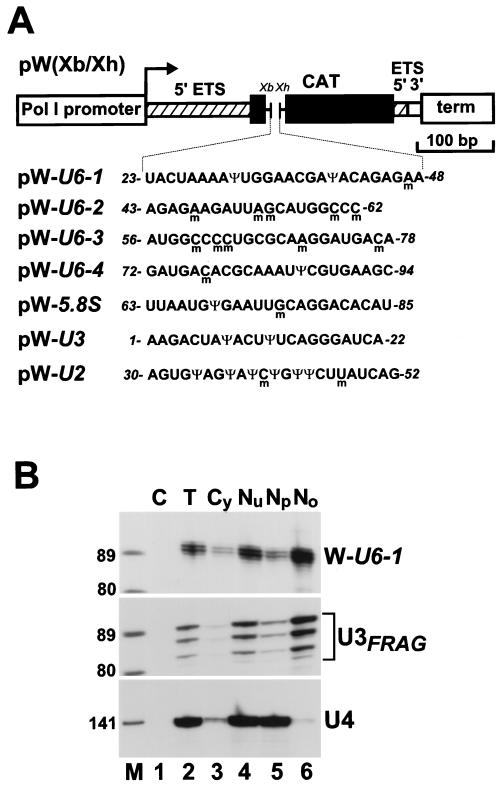
Implication of the mgU6-47, mgU6-77, and mgU6-53 snoRNAs in 2′-O-methylation of the U6 spliceosomal snRNA raises the possibility that posttranscriptional modification of the U6 RNA takes place in the nucleolus. We therefore tested whether factors supporting the 2′-O-methylation and pseudouridylation of U6 snRNA are present in the nucleolus. Short overlapping fragments of the rat U6 snRNA, U6-1 to U6-4 (Fig. 2A), encompassing its eight 2′-O-methylation (m) and three pseudouridylation (Ψ) sites (62), were inserted into the pW(Xb/Xh) mouse ribosomal minigene (19). As controls, fragments of the human 5.8S rRNA (from U63 to U85), U2 snRNA (from A30 to G52), and U3 snoRNA (from A1 to A22) were inserted into the pW(Xb/Xh) minigene. Each fragment contained residues that are 2′-O-methylated and/or pseudouridylated in wild-type RNAs (Fig. 2A) (41, 62). Upon transfection into mouse cells, the polymerase I-directed transcription of the ribosomal minigene occurs in the nucleolus (18, 50), and the resulting RNA transcript accumulates in the nucleolus (16, 19). Indeed, cell fractionation experiments followed by RNase A and T1 mapping and phosphorimager quantification showed that between 90 and 96% of the minigene transcripts studied in our experiments localized to the nucleolus of transfected mouse cells. As a representative example, the intracellular distribution of the W-U6-1 transcript is shown in Fig. 2B. The small amount of W-U6-1 RNA detected in the nucleoplasmic fraction derived most likely from cross-contaminating nucleoli, since the nucleolar U3 RNA was also detectable in this fraction (Fig. 2B, lane 5).
FIG. 2.
Nucleolar expression of the mouse ribosomal minigene tagged with human U6, U2, U3, and 5.8S RNA-specific sequences. (A) Schematic structure of the pW(Xb/Xh) mouse ribosomal minigene construct. pW(Xb/Xh) contains the mouse polymerase I (Pol I) promoter and terminator (term), some 5′ (hatched boxes) and 3′ (open box) external transcribed spacer (ETS) sequences, and a fragment of the chloramphenicol acetyltransferase (CAT) gene that carries the XbaI (Xb) and XhoI (Xh) restriction sites. The pW-U6-1, pW-U6-2, pW-U6-3, pW-U6-4, pW-U5.8S, pW-U3, and pW-U2 constructs were created by insertion of appropriate synthetic DNA fragments into the XbaI and XhoI sites of pW(Xb/Xh). Nucleotides which are 2′-O-methylated (m) or pseudouridylated (Ψ) in rat U6, U3, and U2 snRNAs and human 5.8S rRNA are indicated. (B) Subcellular localization of the W-U6-1 ribosomal minigene transcript. Upon transfection of the pW-U6-1 minigene into mouse cells, RNA samples (200 ng) extracted either from total cells (T) or from cytoplasmic (Cy), nuclear (Nu), nucleoplasmic (Np), and nucleolar (No) fractions were analyzed by RNase A and T1 protection by use of antisense RNA probes specific to the W-U6-1 transcript (upper panel), U3 snoRNA (middle panel), or U4 snRNA (lower panel). Lane C represents a control mapping with E. coli tRNA. Lane M, size markers in nucleotides.
The state of 2′-O-methylation and pseudouridylation of the minigene transcripts was observed by primer extension analyses with a 32P-labeled deoxyoligonucleotide primer complementary to the minigene RNA downstream of the insertion site. In the presence of a low concentration of deoxynucleoside triphosphates, (0.004 mM), reverse transcriptase stops 1 nt before and/or at the 2′-O-methylated nucleotide (31, 32, 42). Pseudouridine residues were visualized by primer extension analysis with CMC [N-cyclohexyl-N′-β-(4-methyl-morpholinium)ethylcarbodiimide p-tosylate]-treated RNAs (2). CMC reacts irreversibly with N3 of pseudouridine and stops reverse transcriptase 1 nt before the pseudouridylation site.
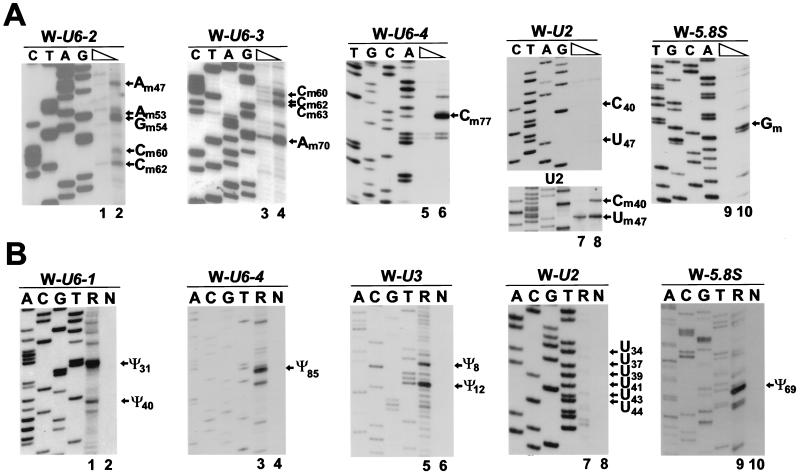
As expected (16), the 5.8S rRNA sequences in the nucleolar W-5.8S transcript were faithfully 2′-O-methylated at G75 (Fig. 3A, lane 10) and pseudouridylated at U69 (Fig. 3B, lane 9). More interestingly, the U6-specific sequences expressed in the nucleolus were accurately 2′-O-methylated at A47, A53, G54 (Fig. 3B, lane 2), C60, C62 (lanes 2 and 4), C63, A70 (lane 4), and C77 (lane 6) as well as pseudouridylated at U31, U40 (lane 1), and U85 (lane 3). The same results were obtained when 2′-O-methylation of the W-U6-3 RNA and pseudouridylation of the W-U6-1 RNA were monitored with RNA samples obtained from the nucleolar fraction of transfected mouse cells (data not shown). Both pseudouridine residues of the U3 snoRNA (Ψ8 and Ψ12) (62) were accurately formed in the nucleolar W-U3 transcript (Fig. 3B, lane 5). However, neither ribose methylation nor pseudouridine formation was observed in the U2-specific tag sequences (Fig. 3A, upper panel, lane 8, and Fig. 3B, lane 7). To ensure that the lack of stop signals in the primer extension mapping of 2′-O-methyl groups in the W-U2 transcript was not due to the special sequence context of the U2 RNA, we performed control mapping with the human U2 snRNA (Fig. 3A, lower panel, lanes 7 and 8). No RNA modification was detected when another fragment of the U2 RNA (from A48 to U70) carrying two known pseudouridylation sites (Ψ54 and Ψ58) and one 2′-O-methylation site (Am61) was tested (data not shown). Likewise, primer extension analyses of the W-U6-1 and W-U6-3 transcripts revealed no stop signals that could have indicated the formation of the base-methylated m6A43 and m2G72 nucleotides that are present in the wild-type U6 snRNA (62) (Fig. 3A, lanes 3 and 4, and Fig. 3B, lane 1).
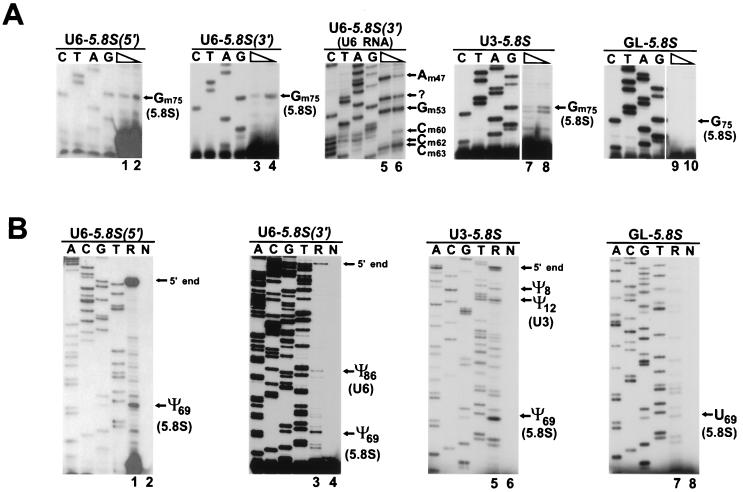
FIG. 3.
Primer extension mapping of modified nucleotides in mouse ribosomal minigene transcripts. (A) Mapping of 2′-O-methylated nucleotides. RNAs isolated from mouse cells transfected with the pW-U6-2, pW-U6-3, pW-U6-4, pW-U2, or pW-U5.8S construct were analyzed by primer extension in the presence of a high (1 mM) or a low (0.004 mM) concentration of deoxynucleoside triphosphates. Panel U2 shows mapping of 2′-O-methyl groups in the wild-type human U2 snRNA. Lanes C, T, A, and G represent sequencing ladders. The extension products were fractionated on 6% sequencing gels. (B) Mapping of pseudouridines. RNAs obtained from mouse cells transfected (R) or not transfected (N) with the pW-U6-1, pW-U6-4, pW-U3, pW-U2, or pW-U5.8S expression construct were subjected to CMC-alkali treatment. The modified pseudouridine-CMC residues were visualized by primer extension.
These results demonstrate that trans-acting factors directing the site-specific 2′-O-methylation and pseudouridylation of the U6 spliceosomal snRNA and pseudouridylation of the U3 snoRNA are present and are functionally active within the nucleolus. The fact that short fragments of the U6 snRNA and the U3 snoRNA are faithfully methylated and/or pseudouridylated in the nucleolus lends further support to the assumption that modification of these snRNAs is directed by snoRNAs. The snoRNA-directed synthesis of 2′-O-methylated nucleotides requires a 10-bp interaction between the snoRNA and the substrate RNA (11, 31). Since the target nucleotide is located in the middle of this interaction, the synthesis of Cm60 and Cm62 residues lacking U6-specific 3′-flanking sequences in the W-U6-2 transcript (Fig. 3A, lane 2) was unexpected. However, we noticed that minigene sequences following the U6-2 sequences are capable of extending a base-pairing interaction with a putative snoRNA up to 12 bp with two mismatches (GGCCCctcgaga; the authentic U6 sequences are shown in uppercase letters).
5.8S rRNA-specific tag sequences carried by U6 RNA are faithfully modified.
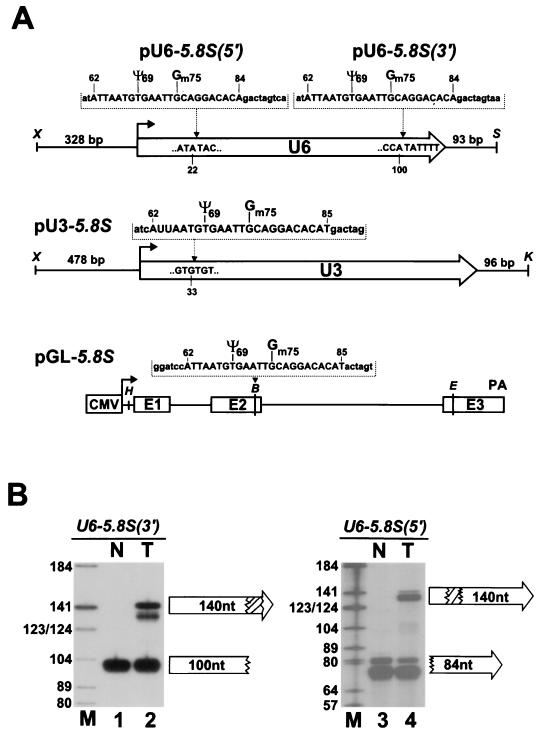
In view of the above results, it seems possible that the U6 snRNA cycles through the nucleolus for snoRNA-mediated nucleotide modification. The snoRNA-directed synthesis of ribose-methylated nucleotides and pseudouridine residues in the 18S, 5.8S, and 28S rRNAs occurs within the nucleolus shortly after or during the synthesis of precursor rRNA (13, 41). In accordance with this fact, rRNA methylation and pseudouridylation snoRNAs show an exclusive nucleolar localization (48). To assess whether the U6 snRNA can establish an interaction with the rRNA modification snoRNAs, a short fragment of the human 5.8S rRNA containing the pseudouridylated U69 and 2′-O-methylated G75 residues (41) was inserted into the 5′- or 3′-terminal part of the coding region of the human U6 snRNA gene (33) (Fig. 4A). The same 5.8S tag was inserted into the human U3 snoRNA gene (83) as well as into the second exon of the human β-globin gene (29). Upon transfection of the 5.8S-tagged genes into simian COS-7 cells, RNase A and T1 mapping performed with sequence-specific antisense probes revealed that the chimeric U6-5.8S(5′) and U6-5.8S(3′) RNAs (Fig. 4B, lanes 2 and 4) and U3-5.8S and GL-5.8S RNAs (data not shown) were correctly and efficiently expressed. The observed heterogeneity of the U6-5.8S(5′)- and U6-5.8S(3′)-specific protected fragments likely represents RNase mapping artifacts, since fragments protected by the 3′-terminal part of the endogenous U6 snRNA also appeared as a doublet (Fig. 4B, lane 3) and primer extension revealed a unique 5′ terminus for both U6-5.8S(5′) and U6-5.8S(3′) (Fig. 5B, lanes 1 and 3).
FIG. 4.
Expression of human U6 snRNA, U3 snoRNA, and β-globin mRNA tagged with 5.8S-specific sequences. (A) Schematic structures of the expression constructs used for the transfection of COS-7 cells. The coding regions of human U6 and U3 snRNA genes are represented by open arrows. The cytomegalovirus promoter (CMV), three exons (E1 to E3), and the polyadenylation region (PA) of the human β-globin gene are indicated for pGL-5.8S. The inserted tag sequences and their positions as well as relevant restriction sites (B, BamHI; E, EcoRI; H, HindIII; K, KpnI; S, SacI; X, XhoI) are indicated above the constructs. Nucleotides representing authentic 5.8S rRNA sequences are in uppercase letters. Nucleotides introduced to facilitate cloning are in lowercase letters. The U69 and G75 residues that are pseudouridylated or 2′-O-methylated in human 5.8S rRNA are marked. (B) Accumulation of U6-5.8S(5′) and U6-5.8S(3′) RNAs in COS-7 cells. RNAs extracted from transfected (T) and nontransfected (N) cells were analyzed by RNase A and T1 mapping by use of antisense RNA probes specific for either the U6-5.8S(5′) or the U6-5.8S(3′) RNA. The protected fragments were separated on a 6% sequencing gel. Lane M, size markers.
FIG. 5.
Primer extension mapping of modified nucleotides. (A) Mapping of 2′-O-methyl groups. RNAs obtained from COS-7 cells transfected with the pU6-5.8S(5′), pU6-5.8S(3′), pU3-5.8S, or pGL-5.8S expression construct were annealed with specific 32P-labeled primers and extended with avian myeloblastosis virus reverse transcriptase in the presence of 1 mM (lanes 1, 3, 5, 7, and 9) or 0.004 mM (lanes 2, 4, 6, 8, and 10) deoxynucleoside triphosphates. The origin of the stop signal at A51 in the U6 snRNA is unknown (lanes 5 and 6). (B) Mapping of pseudouridine residues. RNAs isolated from COS-7 cells transfected (R) or not transfected (N) with the pU6-5.8S(5′), pU6-5.8S(3′), pU3-5.8S, or pGL-5.8S construct were treated with CMC and analyzed by primer extension. For other details, see the legend to Fig. 3.
Primer extension mapping of 2′-O-methylated nucleotides in the U6-5.8S(5′) and U6-5.8S(3′) RNAs in the presence of 0.004 mM deoxynucleoside triphosphates (Fig. 5A, lanes 2 and 4) resulted in stop signals at the G75 residue, indicating that the 5.8S tag sequence is correctly methylated in both chimeric U6-5.8S RNAs. Mapping of pseudouridines revealed that the U69 residue was converted into pseudouridine in both U6-5.8S(5′) and U6-5.8S(3′) (Fig. 5B, lanes 1 and 3). Moreover, mapping of the 5′-terminal region of the U6-5.8S(3′) RNA showed that 2′-O-methylated nucleotides and pseudouridines found in the rat U6 snRNA (62) were also present in the chimeric U6-5.8S(3′) RNA (Fig. 5A, lanes 5 and 6, Fig. 5B, lane 4, and data not shown), indicating that not only the 5.8S but also the U6-specific sequences were correctly modified in the chimeric U6-5.8S(3′) RNA. Likewise, when expressed as part of the U3-5.8S RNA, the 5.8S tag sequences were correctly 2′-O-methylated at G75 (Fig. 5A, lane 8) and pseudouridylated at U69 (Fig. 5B, lane 5). Pseudouridines found in the rat U3 snoRNA (Ψ8 and Ψ12) were also readily detectable in the chimeric U3-5.8S RNA (Fig. 5B, lane 5). However, in marked contrast to the findings for the U6-5.8S and U3-5.8S RNAs, no 2′-O-methyl group and no pseudouridine residue were detected in the 5.8S tag sequences carried by the cytoplasmic β-globin mRNA (Fig. 5A, lane 10, and Fig. 5B, lane 7).
These results demonstrate that the chimeric U6-5.8S and U3-5.8S RNAs, when expressed in simian cells, undergo correct posttranscriptional modifications. This finding indicates that these RNAs are able to establish physical contacts with rRNA methylation and pseudouridylation snoRNAs as well as with all the factors that are responsible for the site-specific 2′-O-methylation and pseudouridylation of U6 and U3 RNAs.
Targeted 2′-O-methylation of yeast U6 snRNA is directed by an artificial snoRNA.
By use of artificial 2′-O-methylation snoRNAs that carry properly designed rRNA recognition motifs, novel 2′-O-methylation sites can be generated in eukaryotic rRNAs (11, 14, 30, 31). We tested whether site-specific 2′-O-methylation of the yeast Saccharomyces cerevisiae U6 snRNA can be achieved by an artificial snoRNA that carries an antisense element complementary to the U6 snRNA.
When expressed in yeast cells, the human U24 snoRNA can direct 2′-O-methylation of the 25S rRNA at two positions (32). The upstream antisense element (UAE) directs the methylation of C1450, and the downstream antisense element (DAE) selects the C1436 residue (Fig. 6A). The DAE of human U24 was replaced with sequences complementary to the yeast U6 snRNA from U27 to A40. The resulting U24-6 snoRNA is predicted to direct the 2′-O-methylation of residue G31 in yeast U6 RNA in addition to residue C1450 in 25S rRNA. DNA fragments encoding the mutant U24-6 and wild-type U24 snoRNAs were inserted into the intron region of the yeast alcohol dehydrogenase gene and transformed into a yeast strain that lacks a functional U24 locus (ΔU24) (31, 32). Northern analysis showed that the U24 and U24 snoRNAs were correctly expressed in yeast (Fig. 6B).
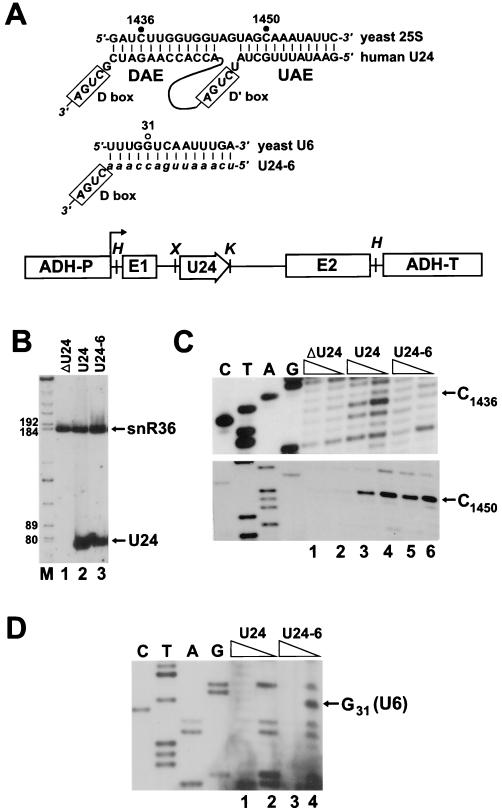
FIG. 6.
Site-specific 2′-O-ribose methylation of yeast U6 snRNA directed by an artificial snoRNA. (A) Potential base-pairing interactions formed between human U24 snoRNA and yeast 25S rRNA or U24-6 snoRNA and yeast U6 snRNA. Nucleotides known or expected to be 2′-O-methylated are indicated by closed or open circles, respectively. DNA fragments encoding U24 or U24-6 snoRNAs were inserted into the intron of the yeast actin gene that had been placed under the control of the promoter (ADH-P) and terminator (ADH-T) of the yeast alcohol dehydrogenase gene. The relevant restriction sites are shown (H, HindIII; X, XhoI; K, KpnI). (B) Northern blot analysis of human U24 and U24-6 snoRNAs. RNAs isolated from control ΔU24 yeast cells (lane 1) and ΔU24 cells transformed with the pFL45/ACT expression construct carrying either the U24 (lane 2) or the U24-6 (lane 3) snoRNA gene were fractionated on a 6% sequencing gel and probed with a mixture of labeled oligos specific for U24 and snR36 snoRNAs. Lane M, size markers. (C) Mapping of ribose-methylated nucleotides in yeast 25S rRNA. RNAs obtained from the ΔU24 strain (lanes 1 and 2) and from cells expressing either the U24 (lanes 3 and 4) or the U24-6 (lanes 5 and 6) snoRNA were analyzed by primer extension in the presence of 1 or 0.004 mM deoxynucleoside triphosphates. Lanes C, T, A, and G represent sequencing ladders. (D) Primer extension mapping of ribose-methylated nucleotides in yeast U6 snRNA. Oligo primers specific for the yeast U6 RNA were annealed to RNAs obtained from the yeast ΔU24 strain expressing the human U24 (lanes 1 and 2) or U24-6 (lanes 3 and 4) snoRNA. Primer extension was performed in the presence of 1 mM (lanes 1 and 3) or 0.004 mM (lanes 2 and 4) deoxynucleoside triphosphates.
The 2′-O-methylation pattern of the 25S rRNA (Fig. 6C) and the U6 snRNA (Fig. 6D) was tested by primer extension analyses. The U24-6 snoRNA (Fig. 6C, upper panel, lane 6), in contrast to the wild-type U24 snoRNA (lane 4), did not support the DAE-dependent methylation of 25S rRNA at C1436. The U24-6 snoRNA, like the wild-type U24 snoRNA, directed the UAE-dependent methylation of the C1450 residue (Fig. 6C, lower panel). More interestingly, in the ΔU24 yeast strain expressing the U24-6 snoRNA, the U6 RNA was 2′-O-methylated at the G31 position (Fig. 6D, lane 4), demonstrating that 2′-O-methylation snoRNAs can direct the site-specific methylation of the U6 spliceosomal snRNA. These results also show that the UAE and DAE of a methylation snoRNA can independently direct 2′-O-methylation of an snRNA and an rRNA (see also reference 79).
DISCUSSION
The rat U6 spliceosomal snRNA carries eight ribose- and two base-methylated nucleosides and three pseudouridines (62). Many of these modified nucleosides are present in plant U6 snRNA as well (28), indicating that they play an important and phylogenetically conserved role in the assembly and/or function of the U6 snRNP. This study has focused on the molecular mechanism that is responsible for the generation of modified nucleosides in U6 snRNA.
Factors directing site-specific modification of U6 snRNA and U3 snoRNA.
We have demonstrated that short fragments of the U6 snRNA embedded in mouse ribosomal minigene transcripts are efficiently and accurately 2′-O-methylated and pseudouridylated within the nucleolus of mouse cells (Fig. 3). Two major conclusions can be drawn from these observations. First, recognition of the correct 2′-O-methylation and pseudouridylation sites in U6 snRNA relies on short nucleotide sequences located around the target site. Second, trans-acting factors directing the modification of U6 snRNA at all known 2′-O-methylation and pseudouridylation sites are present and are functionally active in the nucleolus.
After injection of in vitro-transcribed human U6 snRNA into Xenopus oocytes, the synthesis of the Cm77 2′-O-methylated nucleotide in human U6 snRNA depends on the presence of a C and D box-containing methylation snoRNA (79). Mammalian cells contain at least two additional C and D box-containing snoRNAs with the potential to direct the 2′-O-methylation of U6 snRNA at Am47 (79) and Am53 (Fig. 1). These observations, coupled with the conclusion that the recognition of all known 2′-O-methylation sites of U6 RNA depends on nucleotide sequences located around the actual target site, rather than the secondary structure of the RNA, strongly support the notion that the 2′-O-methylation of U6 snRNA is directed exclusively by snoRNAs. We have demonstrated that trans-acting factors directing the synthesis of all pseudouridine residues in the U6 (Ψ31, Ψ40, and Ψ85) and U3 (Ψ8 and Ψ12) RNAs are also present in the nucleolus (Fig. 3B). Since these pseudouridylation factors recognize the nucleotide sequences around the substrate uridines of the U6 and U3 RNAs, we can anticipate that the pseudouridylation of these snRNAs may turn out to be a guide RNA-mediated process.
Mammalian U6 snRNAs also contain two base-methylated nucleotides, an N-6-methyladenosine (m6A43) and a 2-methylguanosine (m2G72) (62). No base methylation was detected at A43 or G72 in short fragments of the U6 snRNA that were expressed in the nucleolus (Fig. 3). This finding lends further support to the idea that base methylation of the U6 snRNA is dependent on the three-dimensional structure of the RNA (69) and may take place in the nucleoplasm. Similarly, short U2 snRNA-specific sequences are neither 2′-O-methylated nor pseudouridylated in the nucleolus (Fig. 3), suggesting that factors directing the modification of U2 snRNA are not present in the nucleolus and/or they recognize the three-dimensional structure of the U2 snRNA. Indeed, a yeast tRNA pseudouridine synthase enzyme, Pus1p, is also responsible for the synthesis of the Ψ44 residue in yeast U2 snRNA (44). The nucleoplasmic localization of Pus1p (70) and pseudouridylation analyses of in vitro-synthesized U2 RNAs that were microinjected into the nucleoplasm or cytoplasm of Xenopus oocytes (82) suggest that the pseudouridylation of U2 snRNA takes place in the nucleoplasm.
Does modification of U6 snRNA occur in the nucleolus?
The nucleus is highly compartmentalized, and most nuclear processes can be linked to distinct subnuclear structure (35, 74). The nucleolus, the most extensively studied subnuclear organelle, has long been known as the site of the biogenesis of cytoplasmic ribosomes (18). Recently, several lines of evidence suggest that the nucleolus has more diverse functions than earlier anticipated. In yeast, the early processing of some precursor tRNAs has been found to occur within the nucleolus (4). A fraction of mammalian telomerase (49), signal recognition particle (63), and RNase P (24) RNAs has been shown to be present in the nucleolus. Microinjection of in vitro-synthesized signal recognition particle (23), RNase MRP (25), and RNase P (24) RNAs into the nucleoplasm of mammalian cells is followed by transient localization of these RNAs to the nucleolus. These findings suggest that in addition to the biogenesis of cytoplasmic ribosomes, the nucleolus also functions in the processing and/or export of some stable small RNAs (61).
Based on biochemical criteria, the mgU6-47, mgU6-53, and mgU6-77 putative 2′-O-methylation guide RNAs for U6 RNA have been localized to the nucleolus (79) (Fig. 1B). These results, coupled with the observation that trans-acting factors directing the 2′-O-methylation and pseudouridylation of U6 and U3 snRNAs are functionally active in the nucleolus (Fig. 3), suggest that the nucleolus may function in the posttranscriptional modification of the U6 spliceosomal snRNA and the U3 snoRNA. Since mature U6 snRNA shows a steady-state nucleoplasmic localization, the notion that its posttranscriptional modification occurs in the nucleolus presupposes that U6 snRNA cycles through the nucleolus during its maturation. Demonstration that the wild-type U6 snRNA in yeast (Fig. 6) and the chimeric U6-5.8S RNA in mammalian cells (Fig. 5) can undergo snoRNA-directed 2′-O-methylation and pseudouridylation further supports this notion. Recently, it was shown that an in vitro-transcribed U6 snRNA, upon injection into the nucleoplasm of Xenopus oocytes, transiently accumulates in the dense fibrillar compartment of the nucleolus (51). The fibrillar compartment of the nucleolus also harbors the fibrillarin snoRNP protein that is associated with all 2′-O-methylation snoRNAs. This protein is most likely the methyltransferase enzyme that catalyzes the ribose methylation of the target nucleotide specified by the RNA component of the snoRNP particles (53, 75).
The above-mentioned study (51) also revealed that microinjected U6 snRNA localizes to other conserved subnuclear organelles, the coiled bodies, in addition to the nucleolus. The nucleolus and coiled bodies have several common antigens and appear to be remarkably related in both structure and function (6, 45, 74). Coiled bodies are attached to the nucleolar periphery and, under certain physiological conditions, are found within the nucleolus (40, 43, 54), suggesting that they either emerge from or fuse to the nucleolar structures (6, 45). An exciting feature of coiled bodies is that they contain all the major spliceosomal snRNPs, including the U6 snRNP (9, 46). Hence, coiled bodies have been implicated in the processing, modification, or export of spliceosomal snRNAs (6, 40, 45).
Although no H or ACA box pseudouridylation guide snoRNP has yet been detected in coiled bodies, the presence of some C and D box-containing snoRNPs has been well documented (26, 56, 64, 68). Since the C and D boxes have been identified as the key transport elements which target these RNAs to the nucleolus and coiled bodies (37, 64), all snoRNAs carrying these box motifs may be present in coiled bodies. Thus, snoRNA-directed 2′-O-methylation of U6 snRNA may take place in coiled bodies (6, 79). Coiled bodies also contain the putative 2′-O-methyltransferase (fibrillarin) and pseudouridine synthase (Nap57/Cbf5) enzymes that likely catalyze the U6 modification reactions. However, the putative guide RNAs directing U6 methylation are localized to the nucleolus and are practically absent from the nucleoplasm (79) (Fig. 1B). In marked contrast, a coiled body-specific nuclear protein, p80 coilin, was detected mainly, if not exclusively, in the nucleoplasmic fraction (data not shown). Therefore, a potential copurification of coiled bodies with nucleoli cannot account for the observed nucleolar localization of the U6 methylation snoRNAs. We cannot, however, exclude the possibility that coiled bodies contain a small but still sufficient amount of guide snoRNPs to conduct the modification of U6 snRNA. Likewise, the available data cannot unambiguously rule out the formal possibility that the modification of U6 snRNA occurs in the nucleoplasm. In this context, however, it is noteworthy that, despite our repeated efforts, artificial snoRNAs failed to direct the 2′-O-methylation of the human β-globin mRNA and precursor mRNA as well as the nucleoplasmic U19H RNA (7, 27).
In summary, the data presented in this paper are most consistent with the idea that the posttranscriptional modification of U6 spliceosomal snRNA and U3 snoRNA takes place in the nucleolus, although we acknowledge that other subnuclear compartments, such as coiled bodies, may also contribute to this process. Nevertheless, the fact that trans-acting factors directing the site-specific 2′-O-methylation and pseudouridylation of the U6 spliceosomal RNA and pseudouridylation of the U3 snoRNA are present and are functionally active in the nucleolus documents that the nucleolus, directly or indirectly, is involved in the biogenesis of some snRNAs and snoRNAs. These results further substantiate the emerging idea that the nucleolus is a multifunctional organelle that functions in the maturation and/or intracellular trafficking of different classes of cellular RNAs.
ACKNOWLEDGMENTS
Thanks are due to Y. de Préval for synthesis of oligodeoxynucleotides and to L. Poljak for critical reading of the manuscript.
B. E. Jády has been supported by the French Government and the Hungarian Research Foundation (OTKA, T 029042). This research was supported by the Centre National de la Recherche Scientifique and by grants from la Ligue Nationale Contre le Cancer, l’Association pour la Recherche sur le Cancer, and the Hungarian Research Foundation (OTKA, T 029042).
REFERENCES
- 1.Ares M, Jr, Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog Nucleic Acid Res. 1995;50:131–159. doi: 10.1016/s0079-6603(08)60813-2. [DOI] [PubMed] [Google Scholar]
- 2.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 3.Becker H F, Motorin Y, Planta R J, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björk G R. Biosynthesis and function of modified nucleosides. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 165–205. [Google Scholar]
- 6.Bohmann K, Ferreira J, Santama N, Weis K, Lamond A I. Molecular analysis of the coiled body. J Cell Sci. 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- 7.Bortolin M-L, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 8.Bortolin M-L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca M, Pepperkok R, Carvalho M T, Lamond A I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavaillé J, Chetouani F, Bachellerie J-P. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA. 1999;5:66–81. doi: 10.1017/s1355838299981475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 12.Dahlberg J E, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Berlin, Germany: Springer-Verlag KG; 1988. pp. 38–70. [Google Scholar]
- 13.Eichler D C, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:179–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 14.Fournier, M. J. Personal communication.
- 15.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 16.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 17.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 18.Hadjiolov A A. The nucleolus and ribosome biogenesis. In: Alfert M, Beermann W, Goldstein L, Porter K R, Sitte P, editors. Cell biology monographs. Vienna, Austria: Springer-Verlag; 1985. [Google Scholar]
- 19.Hadjiolova K V, Normann A, Cavaillé J, Soupène E, Mazan S, Hadjiolov A A, Bachellerie J-P. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol Cell Biol. 1994;14:4044–4056. doi: 10.1128/mcb.14.6.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamm J, Kazmaier M, Mattaj I W. In vitro assembly of U1 snRNPs. EMBO J. 1987;6:3479–3488. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S, Yamamoto K, editors. transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson M R, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson M R, Cao L-G, Taneja K, Singer R H, Wang Y-L, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson M R, Cao L-G, Wang Y-L, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez-Garcia L F, Segura-Valdez M L, Ochs R L, Rothblum L I, Hannan R, Spector D L. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss T, Marshallsay C, Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992;11:3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiss T, Antal M, Solymosy F. Plant small nuclear RNAs. II. U6 RNA and a 4.5SI-like RNA are present in plant nuclei. Nucleic Acids Res. 1987;15:543–560. doi: 10.1093/nar/15.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 30.Kiss-László, Z., and T. Kiss. Unpublished data.
- 31.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 32.Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel G R, Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988;2:196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- 34.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamond A I, Earnshow W C. Structure and function of the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 36.Lane B G, Ofengand J, Gray M W. Pseudouridine and O2-methylated nucleotides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie. 1995;77:7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- 37.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecointe F, Simos G, Sauer A, Hurt E C, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of Ψ38 and Ψ39 in tRNA anticodon loop. J Biol Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 39.Limbach P A, Crain P F, McCloskey J A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyon C E, Bohmann K, Sleeman J, Lamond A I. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- 41.Maden B E H. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 42.Maden B E H, Corbett M E, Heeney P A, Pugh K, Ajuh P M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 43.Malatesta M, Zancanaro C, Martin T E, Chan E K, Amalric F, Lührmann R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res. 1994;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- 44.Massenet S, Motorin Y, Lafontaine D L J, Hurt E C, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matera A G. Of coiled bodies, gems and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- 46.Matera A G, Ward D C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattaj I W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 48.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell J R, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moss T, Stefanovsky V Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 51.Narayanan, A., R. Terns, and M. Terns. Personal communication.
- 52.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 53.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 54.Ochs R L, Stein T W, Jr, Tan E M. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- 55.Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane B G. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. doi: 10.1139/o95-099. [DOI] [PubMed] [Google Scholar]
- 56.Olmedilla A, de Dios Alche J, Rodriguez-Garcia M I. Nucleolar evolution and coiled bodies during meiotic prophase in Olea europaea: differential localization of nucleic acids. Eur J Cell Biol. 1997;74:181–189. [PubMed] [Google Scholar]
- 57.Pan Z-Q, Prives C. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 1989;3:1887–1898. doi: 10.1101/gad.3.12a.1887. [DOI] [PubMed] [Google Scholar]
- 58.Parry H D, Scherly D, Mattaj I W. ’Snurpogenesis’: the transcription and assembly of U snRNP components. Trends Biochem Sci. 1989;14:15–19. [Google Scholar]
- 59.Patton J R. Multiple pseudouridine synthase activities for small nuclear RNAs. Biochem J. 1993;290:595–600. doi: 10.1042/bj2900595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patton J R. Formation of pseudouridine in U5 small nuclear RNA. Biochemistry. 1994;33:10423–10427. doi: 10.1021/bi00200a025. [DOI] [PubMed] [Google Scholar]
- 61.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reddy R, Busch H. Small nuclear RNAs: RNA sequences, structure, and modifications. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Berlin, Germany: Springer-Verlag KG; 1988. pp. 1–37. [Google Scholar]
- 63.Reddy R, Li W-Y, Henning D, Choi Y C, Nohga K, Busch H. Characterization and subcellular localization of 7-8S RNAs of Novikoff hepatoma. J Biol Chem. 1981;256:8452–8457. [PubMed] [Google Scholar]
- 64.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 66.Ségault V, Will C L, Sproat B S, Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selden R F. Transfection using DEAE-dextran. In: Ausubel F M, Brent R, Kingson R E, Moore D D, Seidman J G, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. pp. 9.2.1–9.2.4. [Google Scholar]
- 68.Shaw P J, Beven A F, Leader D J, Brown J W S. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- 69.Shimba S, Bokar J A, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995;23:2421–2426. doi: 10.1093/nar/23.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 71.Sirum-Connolly K, Mason T L. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 72.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 73.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA processing in eukaryotes. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA. Structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 469–490. [Google Scholar]
- 74.Spector D. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 75.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 76.Tollervey D, Mattaj I W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1987;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 78.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tycowski K T, You Z-H, Graham P J, Steitz J A. Modification of U6 spliceosomal RNA is guided by another small RNAs. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 80.Vankan P, McGuigan C, Mattaj I W. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990;9:3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 82.Yu Y-T, Shu M-D, Steitz J A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan Y, Reddy R. Genes for human U3 small nucleolar RNA contain highly conserved flanking sequences. Biochim Biophys Acta. 1989;1008:14–22. doi: 10.1016/0167-4781(89)90164-4. [DOI] [PubMed] [Google Scholar]