Abstract
The bcr-abl oncogene plays a critical role in the pathogenesis of chronic myelogenous leukemia (CML). The fusion of Bcr sequences to Abl constitutively activates the Abl protein tyrosine kinase. We have recently shown that expression of Bcr-Abl in bone marrow cells by retroviral transduction efficiently induces in mice a myeloproliferative disease resembling human CML and that Abl kinase activity is essential for Bcr-Abl to induce a CML-like myeloproliferative disease. However, it is not known if activation of the Abl kinase alone is sufficient to induce a myeloproliferative disease. In this study, we examined the role of the Abl SH3 domain of Bcr-Abl in induction of myeloproliferative disease and tested whether c-Abl activated by SH3 deletion can induce a CML-like disease. We found that Bcr-Abl with an Abl SH3 deletion still induced a CML-like disease in mice. In contrast, c-Abl activated by SH3 deletion induced only lymphoid malignancies in mice and did not stimulate the growth of myeloid colonies from 5-fluorouracil-treated bone marrow cells in vitro. These results indicate that Bcr sequences in Bcr-Abl play additional roles in inducing myeloproliferative disease beyond simply activating the Abl kinase domain and that functions of the Abl SH3 domain are either not required or redundant in Bcr-Abl-induced myeloproliferative disease. The results also suggest that the type of hematological neoplasm induced by an abl oncogene is influenced not only by what type of hematopoietic cells the oncogene is targeted into but also by the intrinsic oncogenic properties of the particular abl oncogene. In addition, we found that ΔSH3 c-Abl induced less activation of Akt and STAT5 than did Bcr-Abl, suggesting that activation of these pathways plays a critical role in inducing a CML-like disease.
The bcr-abl oncogene is produced when breakpoint cluster region gene (c-bcr) sequences on chromosome 22 are fused to c-abl sequences on chromosome 9 by a reciprocal translocation (reviewed in reference 46). It is found in 95% of the patients with chronic myelogenous leukemia (CML) and also in 20% of the adult patients and 2 to 5% of the pediatric patients with acute lymphoblastic leukemia (ALL) (reviewed in reference 32). Different kinds of bcr-abl oncogenes are found in leukemia patients, depending on the nature of the translocation and exactly how the bcr and abl sequences become spliced into a final bcr-abl mRNA. Often major bcr exons b2 and b3 become fused to abl exon 2 (a2) to produce Bcr-Abl/p210 (b2a2) and Bcr-Abl/p210 (b3a2), which contain, respectively, the first 902 and 927 amino acids of Bcr as well as all of c-Abl except for sequences from the first variable exon. Another common fusion combines minor bcr exon 1 with a2 to produce Bcr-Abl/p185 (e1a2), which contains the first 426 amino acids of Bcr and c-Abl lacking exon 1 sequences. Less frequently, fusions of other bcr and abl exons have been found in leukemia patients (32). Bcr-Abl/p210 is primarily associated with CML and is infrequently associated with ALL, while Bcr-Abl/p185 is usually associated with ALL and is rarely associated with CML.
The Bcr-Abl fusion proteins display an elevated Abl kinase activity. There is a wealth of published data that has revealed that many signaling pathways and cellular functions can be affected by Bcr-Abl and has defined the functional domains and properties of Bcr-Abl (reviewed in reference 40). Most of these data were obtained from experiments in in vitro systems or from studies of the properties of cells derived from leukemia patients at particular stages of disease. However, development of leukemia in vivo is a complex process, which involves both the effects of Bcr-Abl within its target cells and interactions of Bcr-Abl target cells with the rest of the in vivo environment. Although in vitro assays reveal the oncogenic potential of Bcr-Abl, they do not address the complex process of the pathogenesis of CML. Moreover, even in in vitro assays, it has been shown that the role of the functional properties of Bcr-Abl in transformation of cell lines can be dependent on the context of the assay system (7, 13). Therefore it remains unclear what roles are played in leukemogenesis in vivo by specific domains and properties of Bcr-Abl and by the pathways and cellular functions affected by Bcr-Abl.
A recently developed effective mouse model for CML (34, 54) provides an in vivo experimental system that allows direct comparison of the disease phenotypes that result from different abl oncogenes or from specifically mutated versions of bcr-abl. By providing information about disease latency and the phenotypes of the malignant cells that arise, the in vivo mouse model of CML provides a detailed picture of the end result of a complex disease process. It has been shown in many systems, and also in the mouse model of CML, that Abl kinase activity is essential for Bcr-Abl to transform cells and cause disease (54). However, it is still not known if an activated Abl kinase, when targeted into the correct hematopoietic cells, is sufficient to induce a myeloproliferative disease. In this study we tested whether an Abl kinase activated by deletion of the Abl SH3 domain, ΔSH3 c-Abl (12, 23), can induce a CML-like disease in the present CML model.
The Abl SH3 domain has been shown to bind to a number of proteins, including 3BP-1, 3BP-2, Abi-1, Abi-2, AAP-1, Ena, SHPTP-1, PAG, and Rin1 (1, 52; reviewed in reference 40). Thus, the Abl SH3 domain may play a positive role in cellular transformation and leukemogenesis, in the context of Bcr-Abl, through its binding proteins (1). To test this possibility, we compared the diseases induced in the CML model by Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3). Bcr-Abl/p210 lacking abl exon 2 sequences [Bcr-Abl/p210 (b3a3)] has been reported to occur as a rare variant in a few CML patients and ALL patients (32, 38). The deletion of abl exon 2 sequences in Bcr-Abl/p210 (b3a3) results in deletion of abl common exon sequences and of an essential part (the first beta strand and RT loop) of the Abl SH3 domain (see Fig. 1A) (15, 33). Here we demonstrate that Bcr-Abl/p210 (b3a3) induced a myeloproliferative disease in mice similar to the disease induced by Bcr-Abl/p210 (b3a2). However, we found that ΔSH3 c-Abl induced only lymphoid malignancy with a long latency in mice and did not stimulate the growth of myeloid colonies from 5-fluorouracil (5-FU)-treated bone marrow (BM) cells in vitro. These results indicate that the SH3 domain in Bcr-Abl is not essential for induction of a CML-like disease and that an activated Abl kinase alone is not sufficient to induce a CML-like disease in mice. We also found that the ability of ΔSH3 c-Abl to induce the phosphorylation of Akt in tumor cells, and to induce phosphorylation of STAT5 in some tumor cells and in NIH 3T3 cells, was significantly weaker than that of Bcr-Abl.
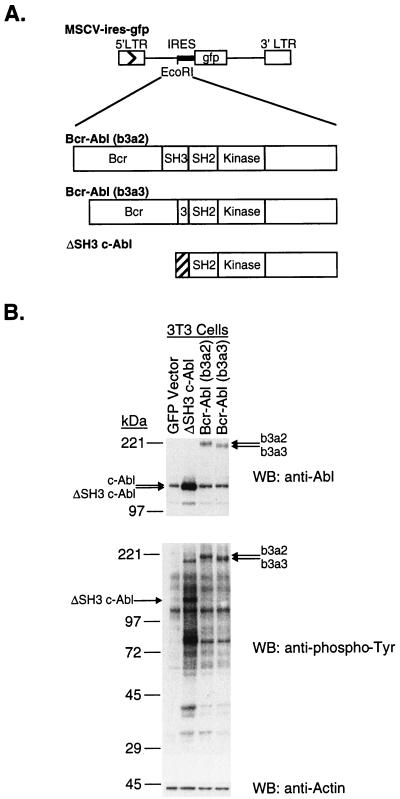
FIG. 1.
(A) Abl oncogenes transduced by retrovirus infection. cDNAs carrying bcr-abl (b3a2), bcr-abl (b3a3), and ΔSH3 c-abl were ligated into the EcoRI site of the retroviral vector MSCV-IRES-gfp. Retroviral vectors were transfected into the packaging cell line Bosc23, to create viral stocks. The hatched region of ΔSH3 c-abl encodes the amino-terminal sequence of c-Abl type IV. MCSV, murine stem cell virus vector; IRES, internal ribosome entry site; LTR, long terminal repeat. (B) Transient expression of Abl oncoproteins in NIH 3T3 cells. Viral stocks produced from Bosc23 cells with the GFP vector (MSCV-IRES-gfp) or GFP vector containing the indicated abl oncogene were normalized to equivalent titers and used to infect NIH 3T3 cells. Two days after infection, cell lysates were prepared, run on 6 to 15% polyacrylamide gradient gels, transferred to nitrocellulose filters, and probed with anti-Abl (Ab-3), antiphosphotyrosine (PY20), or antiactin (AC-40) antibodies, as indicated. WB, Western blot.
MATERIALS AND METHODS
DNA constructs.
To create a bcr-abl/p210 (b3a3) cDNA, sequences corresponding to abl exon 2 were deleted from the bcr-abl/p210 (b3a2) cDNA (9) by using the b3a2 cDNA as the template with a PCR-based strategy (19). First, two DNA fragments with an overlapping sequence were amplified by using 5′ primer A (5′-GAG AAG AGG GCG AAC AAG GGC AG-3′) and 3′ primer B (5′-GAG CTT TTC ACT TGA ACT CTG CTT AAA TC-3′) for fragment 1 and by using 5′ primer C (5′-CAG AGT TCA AGT GAA AAG CTC CGG GTC-3′) and 3′ primer D (5′-CGG AAT TCA TGA GAT ACT GGA TTC CTG GAA C-3′) for fragment 2. Then, these two overlapping fragments were purified by the QIAquick PCR Purification kit (Qiagen Inc., Chatsworth, Calif.) and were used together as the template in a PCR with 5′ primer A and 3′ primer D. Unique BsmI and Asp718 enzyme sites were used to combine the resulting b3a3 fragment with the rest of the bcr-abl/p210 cDNA and thus make a full-length bcr-abl/p210 (b3a3) cDNA. The portion of the bcr-abl/p210 (b3a3) cDNA that was produced by PCR amplification was verified to be correct by sequencing.
Construction of the retroviral vectors MSCV-IRES-gfp and MSCV-bcr-abl/p210 (b3a2)-IRES-gfp was previously described by Zhang and Ren (54). The bcr-abl/p210 (b3a3) cDNA described above and the ΔSH3 c-abl IV cDNA (also called ΔXB [23]) were cloned into the EcoRI site of MSCV-IRES-gfp to create MSCV-bcr-abl/p210 (b3a3)-IRES-gfp and MSCV-ΔSH3 c-abl-IRES-gfp, respectively (see Fig. 1A).
Retroviruses.
Retroviruses were produced by transfecting Bosc23 cells with retroviral vectors and tested on NIH 3T3 cells essentially as described previously (35). Bosc23 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS), 100 U of penicillin/ml, and 100 μg of streptomycin (Gibco BRL, Grand Island, N.Y.) per ml. Two days after transfection, the culture supernatant containing the retroviruses was collected and used to infect BM cells for the CML model and to infect NIH 3T3 cells for determination of a relative viral titer. NIH 3T3 cells were grown in DMEM with 10% donor calf serum (Gibco BRL), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. For titering, 105 NIH 3T3 cells were plated in 60-mm-diameter plates and infected the next day with 0.05 to 1 ml of viral culture supernatant for 4 h in a total volume made up to 2 ml with medium plus 8-μg/ml polybrene (Sigma, St. Louis, Mo.). Two days after infection, NIH 3T3 cells were tested for expression of green fluorescent protein (GFP) by flow cytometry and the relative virus titer was measured by calculating the percentage GFP-positive NIH 3T3 cells. The amount of viral culture supernatant used to infect NIH 3T3 cells was directly proportional to the percent GFP-positive cells in a range where up to 50% of the cells were GFP positive.
BM infection, transplantation, and in vitro soft agar colony assays.
As previously described (34, 54), BM cells from 5-FU-treated male donor BALB/cByJ mice (The Jackson Laboratory, Bar Harbor, Maine) were infected for 2 days in a mixture containing DMEM, 15% FCS, 5% conditioned medium of WEHI-3B cells, 30% viral culture supernatant, 3 μg of polybrene (Sigma) per ml, 2 mM l-glutamine (Gibco BRL), 100 μg of streptomycin/ml, 100 U of penicillin/ml, 0.25 μg of amphotericin B (Gibco BRL) per ml, 7 ng of interleukin (IL-3) (R&D Systems, Inc., Minneapolis, Minn.) per ml, 12 ng of IL-6 (R&D Systems, Inc.) per ml, and 56 ng of stem cell factor (R&D Systems, Inc.) per ml. After 1 day of infection the cells were collected, and the cells were infected for a second day in a freshly made retrovirus cocktail as described above. Then infected BM cells were washed and resuspended in phosphate-buffered saline (PBS) (Gibco BRL), and 4 × 105 BM cells were injected into the tail vein of each lethally irradiated (two times with 450 rads each time; 4 h between doses) female recipient BALB/cByJ mouse.
In vitro soft agar colony assays were performed as described previously (44), with modifications. In soft agar assay 1 (see Table 2), cells from the same common pool of 5-FU-treated BM cells as used for ΔSH3 c-Abl in vivo experiment 2 (see Fig. 4) were infected with bcr-abl/p210 (b3a2), bcr-abl/p210 (b3a3), or ΔSH3 c-abl viruses and then plated in soft agar. The titer of ΔSH3 c-abl virus was twice that of each of the bcr-abl/p210 viruses, while the titers of b3a2 and b3a3 viruses were equal. In soft agar assay 2 (see Table 2), viral culture supernatants were held at 4°C while samples of each were titered as described under “Retroviruses.” Viruses were normalized to give equivalent titers and used to infect 5-FU-treated BM cells by the 2-day infection protocol described above, and the infected cells were then plated in soft agar. For soft agar plating, infected BM cells were plated in 35-mm-diameter wells in DMEM, 20% FCS, 100 μg of streptomycin/ml, 100 U of penicillin/ml, 0.25 μg of amphotericin B/ml, 50 μM 2-mercaptoethanol (Sigma), and 0.3% Bacto-agar, on top of a layer of medium containing 0.6% Bacto-agar. Colonies were counted after 10 days. Because no colonies formed in the ΔSH3 c-Abl plates, the plates were incubated for 2 additional weeks.
TABLE 2.
Growth in soft agar of colonies from infected 5-FU-treated BM cellsa
| Type of abl oncogene in retrovirus | No. of colonies/105 infected cells (avg ± SD) in soft agar assay
|
|
|---|---|---|
| 1 | 2 | |
| bcr-abl (b3a2) | 22.2 ± 2.6 | 37.3 ± 9.3 |
| bcr-abl (b3a3) | 26.4 ± 4.9 | 26.0 ± 2.0 |
| ΔSH3 c-abl | <0.2 | <0.3 |
Infected 5-FU-treated BM cells (105) were plated per well in soft agar, at five wells for each type of virus-infected cells in assay 1 and three wells for each type in assay 2. Colonies showed morphologies typical of myeloid lineage. The lowest possible number was 0.2 in assay 1 and 0.3 in assay 2; < the lowest possible number means that no colonies were seen.
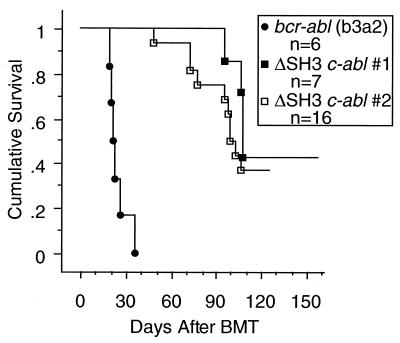
FIG. 4.
Survival of mice after receiving BMT of either bcr-abl (b3a2) or ΔSH3 c-abl virus-infected cells. Data from two independent experiments with ΔSH3 c-abl are presented separately. ΔSH3 c-abl 1 and bcr-abl (b3a2) mice received BMT at the same time, with cells for retrovirus infection coming from a common pool of donor BM cells. In a second experiment, cells from a common pool of donor BM cells were used for ΔSH3 c-abl 2 BMT and also for in vitro soft agar colony assay 1 (Table 2).
Transformation of cell lines in vitro.
32D cells (2 × 106) were infected with titer-matched viruses in a total volume of 2 ml of DMEM–10% FCS–10% conditioned medium of WEHI-3B cells–8-μg/ml polybrene for 8 h. After infection, the cells were collected and resuspended in 10 ml of DMEM–10% FCS–10% conditioned medium of WEHI-3B cells. Two days after infection, the cells were collected, washed, and resuspended in 50 ml of DMEM–10% FCS without any source of IL-3. Factor-independent populations were considered to be established when the medium began to be acidified, the cell concentration ranged from 1 × 106/ml to 2 × 106/ml, and flow cytometry showed that a majority of the cells were GFP positive and excluded propidium iodide.
Transformation of NIH 3T3 cells was quantified by formation on soft agar of NIH 3T3 cell colonies in the same 0.3% agar medium as used for soft agar BM cell colony assays, without 2-mercaptoethanol. Identical plates of NIH 3T3 cells were infected as described under “Retroviruses,” and 2 days later the percent of initially infected cells was determined by measuring the percentage GFP-positive cells in duplicate infection plates. Dilutions of infected cells were plated in triplicate in soft agar, to give 103 to 105 cells per well. The number of colonies formed per 104 GFP-positive cells plated was calculated. To observe the morphological transformation of NIH 3T3 cells in foci, infected plates of cells were allowed to become confluent and the media were changed every 3 days. Foci formed 7 to 10 days after infection.
Analysis of disease phenotype in mice.
Beginning 2 weeks after BM transplantation (BMT), mice were monitored by measurement of peripheral blood leukocyte (WBC) counts and staining of blood smears with Hema 3 (Fisher Scientific, Pittsburgh, Pa.). When the peripheral WBC counts became elevated, immunophenotype and GFP expression were measured by flow cytometry. For the ΔSH3 c-Abl mice, which did not develop highly elevated peripheral WBC counts, randomly selected mice were checked for the presence of GFP-positive peripheral blood cells by flow cytometry. After death or sacrifice due to moribund condition, the mice were examined for tumors or internal abnormalities and samples of tumors and tissues were removed.
Statistical analysis of survival curve data was performed with Survival Tools for StatView 4.5 (Abacus Concepts, Inc., Berkeley, Calif.) by using the Kaplan-Meier survival analysis and Mantel-Cox (log-rank) test functions.
Flow cytometry.
Standard protocols for antibody staining of cell surface proteins were as described in reference 6. Cells were treated with ACK (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA [pH 7.3]) to lyse red blood cells, resuspended in staining buffer (Hanks’ balanced salt solution, 5% fetal bovine serum, 0.1% sodium azide), and blocked with anti-mouse CD16-CD32 (2.4G2; Pharmingen, San Diego, Calif.). The following antibody reagents (all from Pharmingen; antibody clone numbers are given in parentheses) were used: phycoerythrin-conjugated Mac-1 (M1/70), Thy-1.2 (53-2.1), B220 (RA3-6B2), Ter-119, CD4 (H129.19), and heat-stable antigen (M1/69); biotin-conjugated CD8a (53-6.7), CD43 (S7), BP-1 (6C3/BP-1 ag), and immunoglobulin M (IgM) (R6-60.2), streptavidin-Cy-Chrome, and streptavidin-allophycocyanin. Flow cytometry measurements were made on a FACSCalibur machine, and data were analyzed with CellQuest software (Becton Dickinson, San Jose, Calif.).
Cell lysates and immunoblotting.
Peripheral blood, pleural effusion, spleen, and thymus cells were treated with ACK to lyse erythrocytes, washed with PBS, and resuspended in PBS at a concentration of 20 × 106 cells/ml. Total cell lysates were prepared by adding an equal volume of 2× Laemmli sample buffer to each cell suspension, heating at 100°C for 10 min, and pelleting debris by centrifugation. Equal volumes of these lysates were run on 6 to 15% polyacrylamide gradient gels and transferred to nitrocellulose filters (Schleicher and Schuell, Keene, N.H.).
Before lysates from transiently infected NIH 3T3 cells were made, viral culture supernatants were held at 4°C while samples of each were titered as described under “Retroviruses.” Viruses were normalized to give equivalent titers, and several 60-mm-diameter plates of NIH 3T3 cells were infected with each type of virus, as described under “Retroviruses.” Two days later the percentage GFP-positive cells in plates infected with each type of virus was measured. At the same time, cells from triplicate plates for each type of virus were combined and washed in PBS containing 1 mM sodium orthovanadate and 10 mM sodium fluoride. The cells were resuspended and held for 15 min at 4°C in lysis buffer (1% Triton X-100, 50 mM HEPES [pH 7.4], 150 mM NaCl, 10% glycerol, 1 mM EGTA, 1.5 mM MgCl2, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 1× Complete Protease Inhibitor Cocktail ([Boehringer Mannheim, Indianapolis, Ind.]) and then centrifuged to remove debris. Total protein concentrations were measured with Coomassie Protein Assay Reagent (Pierce, Rockford, Ill.). Equal amounts of total proteins of all lysates were run on 6 to 15% polyacrylamide gradient gels and transferred to nitrocellulose filters.
Protein blots were probed with the following primary antibodies: anti-Abl (Ab-3; Oncogene Research Products, Cambridge, Mass.); antiphosphotyrosine (PY20; prepared in our lab); antiactin (AC-40; Sigma); anti-Akt, anti-phospho-Akt (Ser473), anti-p44/42 mitogen-activated protein (MAP) kinases (Erk1 and Erk2), and anti-phospho-p44/42 (Thr202/Tyr204) MAP kinases (Erk1 and Erk2; New England Biolabs, Beverly, Mass.); anti-phospho-STAT5A/B (Y694/Y699) (8-5-2; Upstate Biotechnology, Lake Placid, N.Y.); and anti-STAT5 (G-2), anti-JNK2 (D-2), and anti-phospho-JNK (G-7; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The secondary antibodies used were horseradish peroxidase-labeled goat anti-mouse IgG or goat anti-rabbit IgG (Southern Biotechnology Associates, Inc., Birmingham, Ala.). Blots were probed with a phospho-specific antibody, stripped according to the instructions of the manufacturer (Amersham, Arlington Heights, Ill.), and reprobed with the corresponding non-phospho-specific antibody. Identical blots were probed in parallel, so each blot was stripped and reprobed only one time.
RESULTS
Bcr-Abl/p210 with a deletion in the Abl SH3 domain induces a CML-like myeloproliferative disease in mice.
To compare the expression levels of Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) proteins in a single type of cell with minimal opportunity for selection to occur, we measured the amounts of Bcr-Abl proteins in NIH 3T3 cells 2 days after they were infected with titer-matched viruses (Fig. 1). Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) proteins were expressed to the same level. There was no major difference in the patterns of tyrosine-phophorylated protein bands for lysates from cells expressing either Bcr-Abl/p210 (b3a2) or Bcr-Abl/p210 (b3a3).
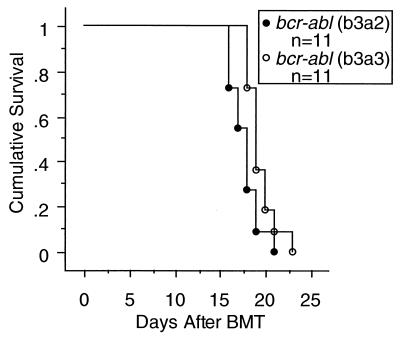
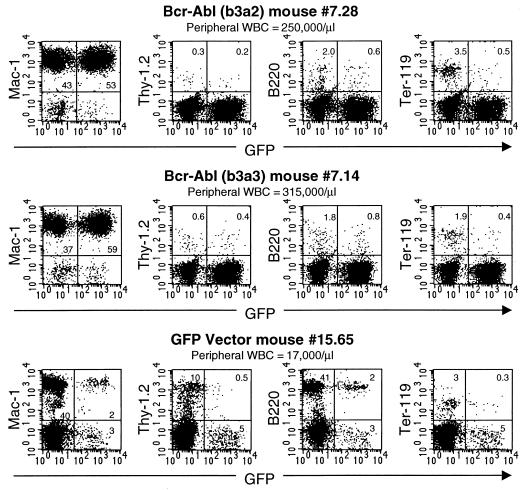
To make a direct comparison of bcr-abl/p210 (b3a2) and bcr-abl/p210 (b3a3) in vivo, a common pool of BM cells was collected from 5-FU-treated donor mice and infected with titer-matched bcr-abl/p210 (b3a2) or bcr-abl/p210 (b3a3) retroviruses (Fig. 1A). For each kind of virus, infected BM cells were injected into 15 irradiated recipient mice. To compare the speed of disease development, 11 randomly chosen mice from each group were monitored by evaluating peripheral blood counts and smears. Both groups of mice developed peripheral blood WBC counts of >200,000/μl (WBCs were seen on blood smears to be mostly granulocytes) and died within 4 weeks (Fig. 2). Mice in both groups had hepatomegaly, splenomegaly, and pulmonary hemorrhages. These are the same as the general features previously described for the Bcr-Abl-induced CML-like myeloproliferative disease in the mouse model of CML (34, 54). There was a small, but consistent, increase in survival time following BMT for mice receiving bcr-abl/p210 (b3a3) (Fig. 2) (Mantel-Cox [log-rank] test, χ2 = 4.692, P < 0.030). This increased survival paralleled a delay of the increase in peripheral blood WBC counts in the bcr-abl/p210 (b3a3) mice as compared to the increase in the bcr-abl/p210 (b3a2) mice (data not shown). Analysis of the four remaining mice from each group included immunophenotyping of peripheral blood cells by flow cytometry. For comparison, it can be seen that mice that received GFP vector-infected BM cells to rescue them from lethal irradiation remained healthy, with normal peripheral blood WBC counts, and that GFP-positive cells were present as a proportional fraction of the normal blood cell types (Fig. 3, mouse 15.65). In contrast, mice in both the Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) BMT groups were seen to have elevated peripheral WBC counts due to massive expansion of the numbers of Mac-1-positive (myeloid) cells, with the counts of both GFP-positive and -negative cells expanded, as previously described (Fig. 3, mice 7.28 and 7.14) (54). No difference between the peripheral blood profiles of diseased mice in the Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) groups was revealed by flow cytometry (Fig. 3).
FIG. 2.
Survival of mice after receiving BMT of either bcr-abl (b3a2) or bcr-abl (b3a3) virus-infected cells. Curves were generated by Kaplan-Meier survival analysis. A Mantel-Cox (log-rank) test of these two survival curves yielded χ2 = 4.692, P = 0.030.
FIG. 3.
Immunophenotypes of peripheral blood from representative Bcr-Abl (b3a2) and Bcr-Abl (b3a3) mice with CML-like myeloproliferative disease and from a healthy GFP vector mouse. Peripheral blood samples were stained with the indicated antibodies and analyzed by flow cytometry for expression of cell surface markers and GFP.
ΔSH3 c-Abl induces lymphoid malignancy with pleural effusion in mice.
Since the Abl SH3 domain was shown not to be essential in the context of Bcr-Abl/p210 for induction of myeloproliferative disease in the CML model, we tested what kind of disease, if any, can be induced by a c-Abl kinase activated by an SH3 deletion (ΔSH3 c-Abl, Fig. 1A). It has been shown that c-Abl with a deleted SH3 domain, without the addition of sequences from any other genes, has an activated Abl kinase domain, and can transform fibroblast and hematopoietic cell lines as well as primary lymphoid cells from normal BM (12, 23). Therefore, by comparing ΔSH3 c-Abl to Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) in the mouse CML model, we can assess if Bcr sequences play additional roles in Bcr-Abl-induced myeloproliferative disease beyond simply activating the Abl tyrosine kinase domain.
The protein expression level of ΔSH3 c-Abl from our retroviral vector construct was compared to the protein expression levels of our Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) constructs in NIH 3T3 cells, 2 days after the cells were infected with titer-matched viruses (Fig. 1). While Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) proteins were expressed to about the same level, ΔSH3 c-Abl protein was expressed to a much higher level. There were no major differences in the patterns of tyrosine-phosphorylated protein bands for lysates from cells expressing any of these three Abl oncoproteins. Consistent with the higher level of expression of ΔSH3 c-Abl, there was an increased overall amount of tyrosine-phosphorylated proteins in the ΔSH3 c-Abl lysate compared to those in the Bcr-Abl/p210 lysates.
When ΔSH3 c-Abl was tested in the murine CML model, a striking difference between the diseases induced by ΔSH3 c-Abl and Bcr-Abl quickly became apparent. Compared to Bcr-Abl, ΔSH3 c-Abl induced in mice disease with a greatly extended latency and a greatly decreased efficiency (Fig. 4 and Table 1). When mice received Bcr-Abl/p210 BMT, 100% of them succumbed to a fatal myeloproliferative disease in 3 to 4 weeks. However, in ΔSH3 c-Abl mice, disease usually developed after more than 3 months (Fig. 4 and Table 1). Furthermore, only 4 of 7 mice in ΔSH3 c-Abl experiment 1 developed disease within 22 weeks after BMT, and 10 of 16 mice in ΔSH3 c-Abl experiment 2 developed disease within 18 weeks after BMT (Fig. 4 and Table 1). It should be noted that in these experiments the ΔSH3 c-abl viral titers were actually twice those of the bcr-abl/p210 viruses.
TABLE 1.
Features of disease in ΔSH3 c-Abl BMT and Bcr-Abl BMT mice
| Mouse | Latencya (no. of days) | Pleural effusionb | Thymic lymphomac | Phenotype of GFP+ cellsd |
|---|---|---|---|---|
| 7.28 [Bcr-Abl (b3a2)] | 17 | No | No | Mac-1+ |
| 7.14 [Bcr-Abl (b3a3)] | 17 | No | No | Mac-1+ |
| ΔSH3 c-Abl | ||||
| 6.27 | 48 | Yes | No | B220+ |
| 6.16 | 72 | Yes | No | ND |
| 6.30 | 72 | Yes | Yes | ND |
| 6.25 | 78 | Yes | No | B220+ |
| 5.2 | 95 | Yese | Yes | ND |
| 6.22 | 95 | Yese | Yes | 15% Thy-1.2+/CD4+/CD8+, 85% Thy-1.2+/CD4+/CD8− |
| 6.23 | 98 | Yes | No | ND |
| 6.20 | 99 | Yes | Yes | Thy-1.2+/CD4+/CD8+ |
| 6.31f | 99 | No | No | B220+/CD43+/HSA+/BP-1+/sIgM− |
| 6.28f | 103 | No | No | B220+/CD43+/HSA+/BP-1+/sIgM− |
| 5.4 | 106 | Yes | Yes | Thy-1.2+/CD4+/CD8+ |
| 6.21 | 106 | Yes | Yes | ND |
| 5.3 | 108 | Yes | Yes | Thy-1.2+/CD4+/CD8+ |
| 5.6 | 108 | Yes | Yes | 60% Thy-1.2+/CD4+/CD8+, 40% Thy-1.2+/CD4+/CD8− |
Latency is the time after BMT for mice to die or become moribund.
Pleural effusion is 0.5 to 1 ml of unclotted, bloody fluid in the chest cavity, containing erythrocytes and many GFP-positive (GFP+) lymphoblastic cells.
Thymic lymphoma was diagnosed by the presence of a greatly enlarged thymus (0.3 to 0.6 g).
Phenotypes of GFP+ cells were determined by flow cytometry. ND, not determined.
Pleural effusion appeared less bloody, with fewer erythrocytes and WBCs.
Developed rear leg paralysis and elevated peripheral WBCs with >50% GFP+ lymphoblastic cells. No tumors, pleural effusion, or lung hemorrhages were seen.
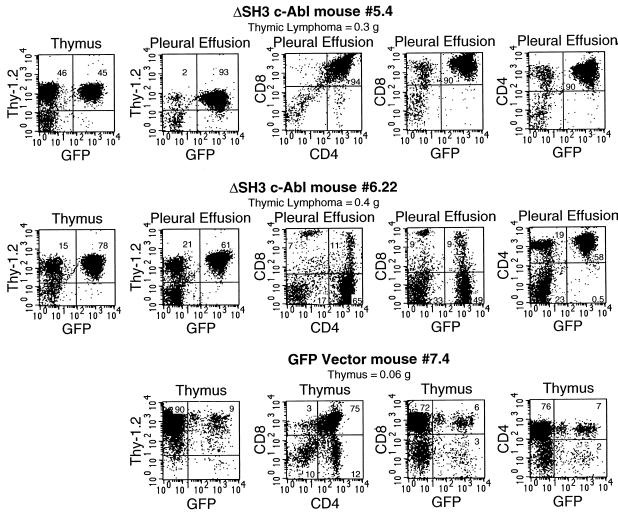
Unlike the Bcr-Abl BMT mice, most ΔSH3 c-Abl BMT mice did not develop elevated peripheral WBC counts. Therefore, disease was first detected by observation of the symptoms of cachexia, abnormal gait, and labored breathing. Pleural effusion, the likely cause of death, and lung involvement were found in 12 of the 14 ΔSH3 c-Abl mice that developed disease and are described in detail below. The marked hepatomegaly and splenomegaly seen in Bcr-Abl BMT mice were not seen in ΔSH3 c-Abl BMT mice. Detailed analysis revealed that among the 14 ΔSH3 c-Abl mice that developed disease, 8 developed thymic lymphoma and pleural effusion (Table 1). As shown in Fig. 5, in each mouse a large number of GFP-positive, Thy-1.2-positive cells were present in the greatly enlarged thymus as well as in the pleural effusion. Further analysis showed that in some mice the GFP-positive tumor cells were all CD4+ CD8+ cells, while in other mice there was a mixed population of CD4+ CD8− cells and CD4+ CD8+ cells (Fig. 5 and Table 1). It should be noted that in the latter case among the GFP-positive CD4+ CD8+ cells, there was a range from low to high level of expression of CD8 rather than a single distinct level of CD8 (Fig. 5, mouse 6.22). This could reflect tumor cells at different stages of T-cell development. Thymus cells from mice in the GFP vector BMT group were mostly CD4+ CD8+ cells, with smaller populations of doubly negative and singly positive cells present and CD4 and CD8 expression occurring at distinct levels (Fig. 5, mouse 7.4).
FIG. 5.
Immunophenotypes of cells from two ΔSH3 c-Abl mice with thymic lymphoma and pleural effusion. Thymus and pleural effusion cells were stained with the indicated antibodies and analyzed by flow cytometry for expression of cell surface markers and GFP. For comparison, thymus cells from a healthy GFP vector mouse were also analyzed.
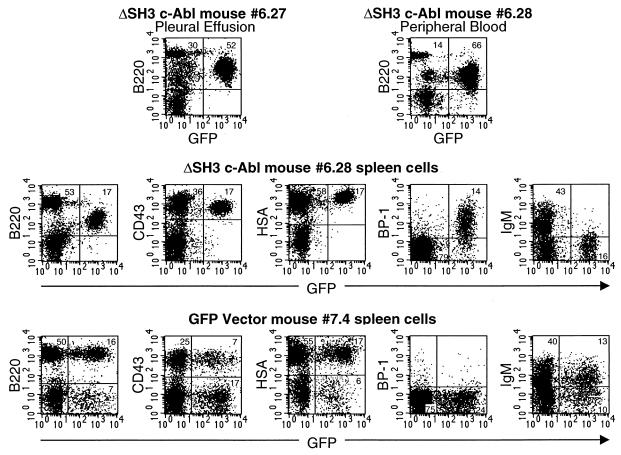
The second major disease phenotype seen in ΔSH3 c-Abl mice was distinguished by the lack of thymic involvement (Table 1). In these six mice the tumor cells were of the B-lymphoid lineage (Fig. 6). All but two of these mice also had pleural effusion and lung involvement (Table 1). The two mice without pleural effusion (mice 6.28 and 6.31) developed rear leg paralysis and elevated peripheral WBC counts (55,000/μl and 97,000/μl, respectively) and were not found to have lung hemorrhages or visible tumors. The peripheral blood of these two mice contained more than 50% GFP-positive B-lymphoblastic cells (Fig. 6). In Fig. 6 examples of flow cytometry data from diseased ΔSH3 c-Abl mice without thymic involvement with (mouse 6.27) or without (mouse 6.28) pleural effusion are shown. Mouse 6.27 had a large number of B220-positive, GFP-positive cells in the pleural effusion, while mouse 6.28 had B220-positive, GFP-positive cells in peripheral blood and spleen. Further immunophenotypic analysis was performed on mouse 6.28 and mouse 6.31 to determine the developmental stage of the B-lymphoblastic cells (Table 1). The GFP-positive cells were shown to be in the early B-cell developmental stage previously designated fraction C (Fig. 6) (these cells are positive for B220, CD43, HSA, and BP-1 and negative for sIgM) (16). Consistent with this developmental stage, the GFP-positive cells in ΔSH3 mice with B-lymphoid disease were B220low (Fig. 6). By contrast, the GFP-positive spleen cells from GFP vector BMT mice paralleled the normal GFP− cells. In particular, note that many, but not all, GFP-positive spleen cells were B220high and had surface IgM expression, while there was no significant BP-1-positive population (Fig. 6, mouse 7.4). This is characteristic of the more mature B cells that are normally found outside of the BM in nonleukemic mice.
FIG. 6.
Immunophenotypes of cells from two diseased ΔSH3 c-Abl mice without thymic involvement. Mouse 6.27 had pleural effusion, while mouse 6.28 had hind limb paralysis and no pleural effusion. Pleural effusion, peripheral blood, and spleen cells were stained with the indicated antibodies and analyzed by flow cytometry for expression of cell surface markers and GFP. For comparison, spleen cells from a healthy GFP vector mouse were also analyzed.
As mentioned above, 12 of the 14 diseased ΔSH3 c-Abl mice had pleural effusion (Table 1). The pleural effusion in these mice was usually 0.5 to 1 ml of unclotted, bloody fluid filling the chest cavity and containing erythrocytes and a high concentration of GFP-positive lymphoblastic cells. The lungs of these mice did appear to have some hemorrhages and when tested by flow cytometry were found to contain a large number of GFP-positive lymphoblastic cells (data not shown). However, this phenotype was very different from the pulmonary hemorrhages seen in Bcr-Abl/p210 mice (34, 54). In Bcr-Abl/p210 mice the lungs contained many blood clots that appeared to be evidence of extensive bleeding followed by clotting, but the hemorrhages and blood were contained within the lungs. In contrast, the pleural effusion in ΔSH3 c-Abl mice was outside of the lungs, filling the chest cavity, and the clotting within the lungs was not as extensive, nor was there clotting of the pleural effusion itself.
ΔSH3 c-Abl does not stimulate the growth of myeloid colonies in vitro.
The lack of any myeloid disease in ΔSH3 c-Abl mice could result if the Abl kinase activated by SH3 deletion, in the absence of additional sequences from other genes, is unable to transform myeloid lineage cells, but can still transform lymphoid cells. Alternatively, the end result we saw, i.e., purely lymphoid disease, could be caused by an in vivo selection process that favors the expansion of ΔSH3 c-Abl infected lymphoid cells over the expansion of ΔSH3 c-Abl infected myeloid cells. In this case there should not be any intrinsic defect in the ability of ΔSH3 c-Abl to transform myeloid lineage cells.
To test if ΔSH3 c-Abl has an intrinsic deficiency in transforming myeloid lineage cells, we performed an in vitro soft agar colony assay (Table 2). Under the conditions used in this experiment the growth of myeloid, but not lymphoid, cell colonies was stimulated by Bcr-Abl (data not shown). Both kinds of Bcr-Abl/p210 induced equivalent numbers of myeloid cell colonies (Table 2). However, no colonies at all were seen to grow from the ΔSH3 c-Abl-infected cells, even though the ΔSH3 c-Abl plates were allowed to incubate long after the myeloid cell colonies were visible in the other plates.
Although our ΔSH3 c-Abl retroviral construct did not induce myeloid cell colonies to grow from 5-FU-treated BM cells in vitro, it does have transforming activity in fibroblast and hematopoietic cell lines. When equal numbers of 32D cells were infected with titer-matched retroviruses, factor-independent GFP-positive cell populations grew to a high density 7 days after infection with the Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) viruses and 14 days after infection with the ΔSH3 c-Abl virus. In addition, we compared the transforming potentials of ΔSH3 c-Abl and Bcr-Abl in NIH 3T3 cells. Earlier, Bcr-Abl was found not to transform NIH 3T3 cells (8). Later it was found that there are sublines of NIH 3T3 cells that are permissive and nonpermissive for transformation by abl oncogenes (41, 42). Our subline of NIH 3T3 cells was originally isolated from a pool of NIH 3T3 cells by M. Kamps in D. Baltimore’s laboratory and was found to be highly susceptible to Bcr-Abl transformation (unpublished results). Using this cell line we found (i) that ΔSH3 c-Abl induced foci of cells with morphological changes similar to those seen in v-Abl foci and more drastic than the morphological changes seen in Bcr-Abl/p210 induced foci; and (ii) that ΔSH3 c-Abl, Bcr-Abl (b3a2), and Bcr-Abl (b3a3) induced 35 ± 6, 197 ± 69, and 159 ± 76 NIH 3T3 cell colonies/104 infected cells (average ± standard deviation) in soft agar.
Expression of Bcr-Abl/p210 (b3a2), Bcr-Abl/p210 (b3a3), and ΔSH3 c-Abl, total tyrosine phosphorylation profiles, and activation of important signaling pathways in tumor cells.
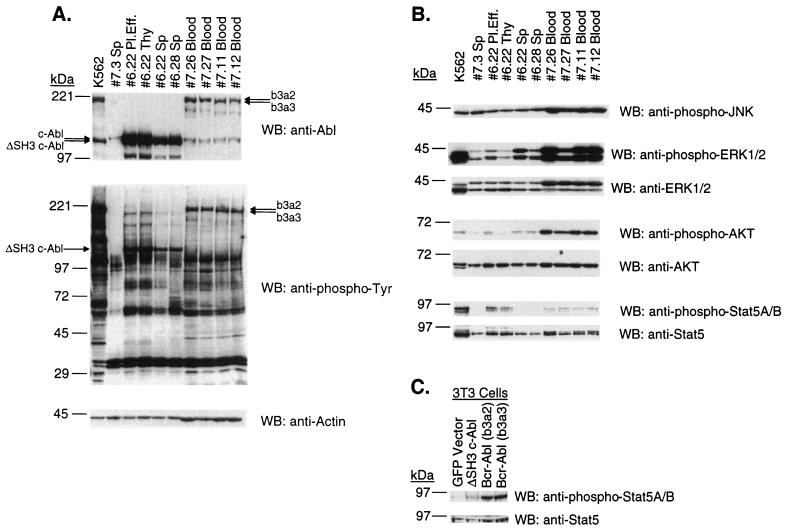
The protein expression levels of Bcr-Abl/p210 (b3a2), Bcr-Abl/p210 (b3a3), and ΔSH3 c-Abl, total tyrosine phosphorylation profiles, and activation status of important signaling pathways were measured by Western blotting of lysates of tumor cells from diseased mice. It is important to note that for some of these comparisons protein expression was measured in different types of cells, since mice with different disease phenotypes have expanded different kinds of tumor cells. In these blots (Fig. 7A and B), K562 cell line lysate was included as a positive control and lysate of spleen cells from GFP vector BMT mouse 7.3 was included as a negative control. Lysates of peripheral blood cells from two of the Bcr-Abl/p210 (b3a2) mice (mouse 7.26 and mouse 7.27) and from two of the Bcr-Abl/p210 (b3a3) mice (mouse 7.11 and mouse 7.12) that had developed a CML-like myeloproliferative disease were probed. Greater than 90% of the cells in these peripheral blood samples were myeloid cells, and more than half of the myeloid cells were GFP-positive cells (Fig. 3). Also probed in these blots were lysates of pleural effusion, thymic lymphoma, and spleen cells from a ΔSH3 c-Abl mouse that developed a T-cell disease (mouse 6.22, Fig. 5 and Table 1). These samples from the different tissues of mouse 6.22 contained 61, 78, and 24% GFP-positive, Thy-1.2-positive cells, respectively (Fig. 5 and data not shown). Lysate from spleen cells of a ΔSH3 c-Abl mouse that developed a B-cell disease (mouse 6.28, Fig. 6 and Table 1) containing 17% GFP-positive, B220-positive B-lymphoblastic cells was also included in these blots.
FIG. 7.
(A) Expression of Abl proteins and total tyrosine phosphorylation profiles in tumor cells. Lysates from equal numbers of cells, and 10 μg of lysate from K562 cells, were run on 6 to 15% polyacrylamide gradient gels, transferred to nitrocellulose filters, and probed with anti-Abl (Ab-3), antiphosphotyrosine (PY20), or antiactin (AC-40) antibodies, as indicated. Samples 7.26 and 7.27 [Bcr-Abl/p210 (b3a2)] and samples 7.11 and 7.12 [Bcr-Abl/p210 (b3a2)] are peripheral WBC lysates from mice that had developed a CML-like myeloproliferative disease. Pleural effusion (Pl.Eff.), spleen (Sp), and thymic lymphoma (Thy) lysate samples 6.22 are from a ΔSH3 c-Abl mouse that developed a T-cell leukemia and lymphoma. Spleen lysate 6.28 is from a ΔSH3 c-Abl mouse that developed a B-cell leukemia. Spleen lysate 7.3 is from a GFP vector mouse. (B) Activation of signaling pathways in tumor cells. The same lysates as probed in panel A were probed with anti-phospho-JNK (G-7), anti-phospho-p44/42 (Thr202/Tyr204) MAP kinases (Erk1 and Erk2), anti-p44/42 MAP kinases (Erk1 and Erk2), anti-phospho-Akt (Ser473), anti-Akt, anti-phospho-STAT5A/B (Y694/Y699) (8-5-2), and anti-STAT5 (G-2). (C) Activation of STAT5 in NIH 3T3 cells. Two days after infection with the indicated titer-matched virus, lysates were prepared from NIH 3T3 cells, run on 6 to 15% polyacrylamide gradient gels, transferred to nitrocellulose filters, and probed with anti-phospho-STAT5A/B (Y694/Y699) (8-5-2), and anti-STAT5 (G-2). WB, Western blot.
Each lane contains lysate from an equal number of cells, except where 10 μg of K562 total cell lysate was loaded. Comparison with the amount of actin in each lane, as a loading control (Fig. 7A), revealed that lysates from equal numbers of cells of the same type (lymphoid or myeloid) contained about the same amount of protein but that myeloid cells yielded more protein than an equal number of lymphoid cells.
The anti-Abl blot (Fig. 7A) shows that ΔSH3 c-Abl protein was expressed to a much higher level in the lymphoid tumor cells compared to the levels of Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) proteins found in myeloid tumor cells in the mouse CML model. As expected, the spleen cell lysates that contained lower percentages GFP-positive lymphoblastic cells (from mouse 6.22 Sp and mouse 6.28 Sp) contained less ΔSH3 c-Abl protein.
Total tyrosine phosphorylation profiles (Fig. 7A) showed a similar overall amount of tyrosine-phosphorylated proteins in peripheral blood lysates from mice with the CML-like disease and in pleural effusion and thymic lymphoma lysates from a mouse with T-cell disease. The decreased overall amount of tyrosine-phosphorylated proteins in spleen cell lysates could simply reflect the smaller fraction of ΔSH3 c-Abl-expressing cells in the spleen. Bcr-Abl/p210- and ΔSH3 c-Abl-expressing tumor cells have most major tyrosine-phosphorylated protein bands in common, most notably bands of approximately 120, 62, and 39 kDa. These proteins are likely to be Cbl, p62Dok, and Crk/Crkl, respectively, since they have been shown to be major tyrosine-phosphorylated proteins in Bcr-Abl-transformed cells (reviewed in references 40 and 46). Interestingly, p39 appears to be tyrosine phosphorylated to a greater extent in myeloid cells expressing Bcr-Abl/p210 than in lymphoid cells expressing ΔSH3 c-Abl. In contrast to the tyrosine phosphorylation in NIH 3T3 cells (Fig. 1B), Bcr-Abl/p210 proteins appear to phosphorylate proteins more efficiently than ΔSH3 c-Abl in hematopoietic cells, if the relative expression levels of the oncoproteins is considered (Fig. 7A).
To test if there were differences in the abilities of Bcr-Abl/p210 (b3a2), Bcr-Abl/p210 (b3a3), and ΔSH3 c-Abl to activate major signaling pathways in the tumor cells that expanded in vivo, the tumor cell lysates were probed with antibodies that recognize activation-specific phosphorylated sites of the signaling proteins JNK1, JNK2, Erk1, Erk2, Akt, STAT5A, and STAT5B (see the Materials and Methods section). Then the amounts of the activated forms of these signaling proteins were compared among the tumor cell lysates and were also compared to the total amounts of the proteins present, as revealed by antibodies that recognize both activated and nonactivated forms of the proteins. We chose to examine activation of these particular signaling molecules because they participate in signaling pathways previously shown to be activated by Bcr-Abl (JNK, Akt, and STAT5) and in a major pathway downstream of Ras (Erk) (reviewed in references 40 and 46).
Figure 7B shows the results for these proteins. No difference in the amounts of activated phospho-JNK was detected among the lysate from K562 cells, spleen cell lysate from GFP vector mouse 7.3, and tumor cell lysate from Bcr-Abl/p210 or ΔSH3 c-Abl mice. The phospho-JNK antibody recognized a 46-kDa form (p46) of JNK in these lysates, while the JNK2 antibody recognized mainly the p54 form and also weakly the p46 forms of JNK in the cell lysates (data not shown). This result indicates that Abl oncoproteins selectively activate p46 JNK. All of the lysates from cells that express Bcr-Abl/p210 contained significant amounts of activated phospho-Erk. Lysates from pleural effusion and spleen cells from the ΔSH3 c-Abl-expressing mouse 6.22 and from spleen cells from the ΔSH3 c-Abl-expressing mouse contained greater amounts of activated phospho-Erk than the spleen cell lysate from the GFP vector-expressing mouse 7.3. This suggests that the Ras-MAP kinase pathway is activated in the tumor cells of ΔSH3 c-Abl-expressing mice. All Bcr-Abl/p210-expressing myeloid tumor cells contained a large proportion of activated phospho-Akt. Increased amounts of activated phospho-Akt were also detected in pleural effusion and spleen cells from the ΔSH3 c-Abl-expressing mouse 6.22 and in spleen cells from mouse 6.28; however, this activation appeared to be much weaker than that in Bcr-Abl/p210-expressing tumor cells. Myeloid tumor cells expressing Bcr-Abl/p210 all contained activated phospho-STAT5, as did pleural effusion and thymic lymphoma cells from the ΔSH3 c-Abl-expressing mouse 6.22. However, no activated STAT5 was detected in tumor cells from the spleens of ΔSH3 c-Abl-expressing mice, even though expression of ΔSH3 c-Abl and activation of Erk and Akt were readily detected in these cells (Fig. 7A and B). The antibody used to detect the total content of both activated and nonactivated STAT5, G-2, recognized several prominent bands in the lysates. We identified the specific STAT5 bands in our blots by their position just below the 97-kDa marker and by precisely aligning the anti-STAT5 blot to the anti-phospho-STAT5A/B blot; only these STAT5 bands are shown in Fig. 7B and C.
Transient expression of Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) in NIH 3T3 cells induces much greater activation of STAT5 than does expression of ΔSH3 c-Abl.
The complexity of examining signaling from Abl oncoproteins in tumor cells that arise in vivo can be appreciated by noticing that lysates from spleen cells of the ΔSH3 c-Abl-expressing mouse (mouse 6.22) did not have detectable amounts of activated phospho-STAT5. Moreover, comparing different kinds of tumor cells that have been obtained from different in vivo environments greatly complicates the analysis of signaling pathways in these cells. Therefore, to test if there is a difference in the abilities of Bcr-Abl/p210 (b3a2), Bcr-Abl/p210 (b3a3), and ΔSH3 c-Abl to activate STAT5 in a single type of cell, under equivalent conditions, we measured the amounts of activated phospho-STAT5 in NIH 3T3 cells transiently expressing Bcr-Abl/p210 (b3a2), Bcr-Abl/p210 (b3a3), and ΔSH3 c-Abl (Fig. 7C). Under these conditions, Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) both induced significant activation of STAT5 as compared to GFP vector-infected NIH 3T3 cells. A much weaker activation of STAT5 was detected in NIH 3T3 cells expressing ΔSH3 c-Abl.
DISCUSSION
We have used a mouse model of CML to examine the role of the Abl SH3 domain of Bcr-Abl/p210 in induction of myeloproliferative disease and to test if an activated Abl kinase that is not fused to any other protein is sufficient to induce a CML-like disease. Both Bcr-Abl/p210 (b3a2) and Bcr-Abl/p210 (b3a3) induced a CML-like myeloproliferative disease in the mouse model. These results demonstrated that the Abl SH3 domain, in the context of Bcr-Abl/p210, is not absolutely essential for efficient induction of a myeloproliferative disease. We also found that ΔSH3 c-Abl, which has an activated Abl kinase domain and can transform fibroblast and hematopoietic cell lines, induced only lymphoid malignancies with a long latency in vivo under the conditions used for study of the mouse model of CML. We found that the amount of phosphorylation of Akt in tumor cells of ΔSH3 c-Abl-expressing mice was significantly less than that in tumor cells of Bcr-Abl-expressing mice, and that the ability of ΔSH3 c-Abl to induce phosphorylation of STAT5 in both tumor cells and NIH 3T3 cells was significantly weaker than that of Bcr-Abl. Together, our results indicate that the Bcr sequences in Bcr-Abl play additional roles in inducing myeloproliferative disease beyond simply activating the Abl kinase domain and suggest that activation of the Akt and STAT5 pathways plays a critical role in inducing a CML-like disease. The results also demonstrated the importance of delineating the molecular mechanisms of CML by using the in vivo model.
The importance of Bcr sequences in Bcr-Abl oncogenic potential has been well documented in the in vitro transforming assays (reviewed in references 40 and 46; see also reference 31). However, the role of Bcr sequences in determining disease specificity remains unclear. For example, it was shown that both Bcr-Abl and v-Abl can induce formation of lymphoid and myeloid cell colonies (25). For a mouse model of CML used earlier, there is a controversy as to whether v-Abl can induce the same myeloproliferative disorder as Bcr-Abl (24, 47). For the first time in a much more efficient mouse model (where Bcr-Abl induces exclusively a CML-like disease), we found that the presence of Bcr sequences is essential for causing myeloproliferative disease in vivo.
The Bcr region of Bcr-Abl/p210 contains multiple functional domains and/or motifs, including a coiled-coil oligomerization domain, a serine-threonine kinase domain, a pleckstrin homology domain, a Dbl/CDC24 guanine-nucleotide exchange factor homology domain, several serine-threonine and tyrosine phosphorylation sites, and binding sites for the Abl SH2 domain and Grb2, Grb10, and 14-3-3 proteins (2, 26–29, 36, 37, 39, 40, 43, 53). The coiled-coil oligomerization domain at the amino terminus of Bcr has been shown to play an important role in activation of the Abl kinase domain, association of Bcr-Abl with actin filaments, and in vitro cellular transformation by Bcr-Abl (29, 30). In the oncoprotein Tel-Abl, part of the Ets transcription factor family member Tel is fused to abl exon 2. Although the Bcr and Tel sequences are very different, they have been shown to have at least one common property. A helix-loop-helix domain in Tel mediates oligomerization of Tel-Abl and is required for activation of the Abl kinase domain, localization of Tel-Abl along actin filaments, and in vitro transformation of cells by Tel-Abl (14). These comparisons of Bcr-Abl and Tel-Abl raise the possibility that the only role of Bcr sequences in Bcr-Abl-induced leukemia is to activate Abl functions by oligomerizing Abl. Since an activated Abl kinase alone is shown here not to be sufficient to induce a CML-like disease, the role of functions of the coiled-coil domain beyond the activation of the Abl kinase requires further investigation in the mouse model of CML.
Another important motif in Bcr is the major Grb2 binding site, tyrosine 177 (37, 39). It is believed that the Grb2 binding leads to activation of the Ras pathway. Mutation of the tyrosine residue (Y177F) has been shown to block fibroblast transformation by Bcr-Abl but not to affect induction of factor independence in hematopoietic cell lines (7, 13). We have recently found that the Y177F mutant of Bcr-Abl/p210 had a greatly reduced ability to induce myeloproliferative disease (50a). Consistently, coexpression of wild-type Bcr-Abl with a dominant-negative form of Ras resulted predominantly in lymphoid disease in the mouse model of CML (3). These results suggest that activation of Ras in Bcr-Abl target cells is an important signaling event for Bcr-Abl to induce a myeloproliferative disease.
However, ΔSH3 c-Abl differs from the Y177F mutant of Bcr-Abl at least in that it can transform fibroblasts in vitro. In this study we found that Erk is activated in tumor cells expressing ΔSH3 c-Abl, including leukemic T cells in pleural effusion and T- or B-leukemic cells in the spleen, to levels similar to those in myeloid tumor cells expressing Bcr-Abl. This result suggests that the Ras-MAP kinase pathway is activated in hematological tumors of both ΔSH3 c-Abl- and Bcr-Abl-expressing mice directly or indirectly by the abl oncogenes and that ΔSH3 c-Abl may lack the combination of additional signaling properties provided by Bcr sequences that are required for myeloid cell expansion in vivo. The lack of ability of ΔSH3 c-Abl to induce a myeloproliferative disease appears not to be due to a lower level of expression of ΔSH3 c-Abl protein nor to a drastic reduction in the overall tyrosine phosphorylation of cellular proteins compared to Bcr-Abl (Fig. 1B and Fig. 7A). Interestingly, we found much less activated Akt in tumor cells of ΔSH3 c-Abl-expressing mice than in tumor cells of Bcr-Abl-expressing mice. One possible cause of this difference in activation of Akt could be the different in vivo environments of the myeloid and lymphoid cells. Notably, the phosphorylation of Akt in K562 cells, a Bcr-Abl-positive erythroblast cell line, was also lower than that in Bcr-Abl-induced myeloid tumor cells (Fig. 7B). Alternatively, this result may indicate that ΔSH3 c-Abl is a weaker activator of the Akt pathway than is Bcr-Abl. In addition, we found that activation of STAT5 in tumor cells of ΔSH3 c-Abl-expressing mouse spleens was much weaker than that in Bcr-Abl-expressing myeloid tumor cells. However, ΔSH3 c-Abl-expressing tumor cells from thymic lymphoma and pleural effusion did contain activated STAT5. These results suggest that activation of STAT5 in ΔSH3 c-Abl-expressing tumor cells may also depend on the extracellular microenvironment. In a single cell type, NIH 3T3, under the same conditions, we did find that ΔSH3 c-Abl was a much weaker activator of STAT5 compared to Bcr-Abl. The inability of ΔSH3 c-Abl to fully activate the Akt and/or STAT5 pathways may underlie the defect of ΔSH3 c-Abl in stimulating the growth of BM cell colonies in vitro and in inducing a CML-like disease in vivo. In other words, these pathways may play an important role in inducing myeloproliferative disease. Consistent with this idea, it has been reported for a different mouse model that a dominant-negative mutant of Akt inhibited Bcr-Abl-mediated leukemogenesis (49). Also, STAT5 has been found to be activated in a variety of Bcr-Abl-expressing cells (4, 5, 22, 48). Whether differences in activation of these, or other, signaling pathways can account for the difference between Bcr-Abl/p210 and ΔSH3 c-Abl in inducing myeloproliferative disease will be the subject of further study.
Our results also showed that the b3a3 version of Bcr-Abl/p210 induced myeloproliferative disease with a small delay in the onset of disease compared to the b3a2 version. This result suggests that the Abl SH3 domain may help to increase the oncogenic potential of Bcr-Abl. The protein Rin1 has been reported to increase transformation of cells when coexpressed with Bcr-Abl, dependent on the binding of Rin1 to the Abl SH2 and SH3 domains (1). Rin1 may be a type of Abl SH3 ligand that is a substrate and effector of Bcr-Abl that enhances oncogenic signaling. Therefore, one possible explanation for the slightly increased survival of Bcr-Abl/p210 (b3a3)-expressing mice could be disruption of Abl SH3 binding to host cell ligands such as Rin1. Another factor that may have influenced the delayed disease onset and increased survival of these mice is a possible effect of the SH3 deletion on adhesion and homing properties of infected BM cells. Decreased homing to the spleen and bone marrow was reported for 32Dc13 clones made factor independent by ΔSH3 Bcr-Abl and for G418-selected BM cells that had been infected with ΔSH3 Bcr-Abl virus, as compared to cells with wild-type Bcr-Abl (50). Nonetheless, our results showed that Bcr-Abl/p210 (b3a3) can still efficiently induce a CML-like disease in mice. This result is consistent with the finding of Bcr-Abl/p210 (b3a3) in association with human leukemias, including CML. Together these data indicate that the function of the Abl SH3 domain, in the context of Bcr-Abl/p210, is either redundant or not essential for induction of a myeloproliferative disease.
Different abl oncogenes were originally found to be predominantly associated with different kinds of hematological malignancies (32, 45). In several earlier mouse models, Bcr-Abl expression induced a variety of hematological neoplasms, including ALL, pre-B-cell lymphoma, T-cell leukemia and/or lymphoma, macrophage, erythroid, and mast cell tumors, myelomonocytic leukemia, and, with a low efficiency, myeloproliferative disease (9–11, 18, 20, 21, 24, 51). The nature of the hematological neoplasms induced by Bcr-Abl has been shown to be influenced by the genetic background of the mouse strains, as well as by infection conditions or the promoters used in transgenic animals (10, 11, 17, 18, 20, 21, 51). In this study we have demonstrated that different types of activated Abl kinases differ in their abilities to induce myeloid or lymphoid cell neoplasms. Our results suggest that the type of hematopoietic cell neoplasm induced by an activated Abl kinase is influenced not only by what type of hematopoietic cells the oncogene is targeted to but also by the intrinsic oncogenic properties of the particular type of activated Abl kinase.
ACKNOWLEDGMENTS
We thank Ben Hentel for help in analyzing mice and with flow cytometry.
This work was supported by National Cancer Institute grant CA68008 (to R.R.). R.R. is a recipient of the Leukemia Society of America Scholar Award.
REFERENCES
- 1.Afar D E, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte O N, Colicelli J. Regulation of the oncogenic activity of BCR-ABL by a tightly bound substrate protein RIN1. Immunity. 1997;6:773–782. doi: 10.1016/s1074-7613(00)80452-5. [DOI] [PubMed] [Google Scholar]
- 2.Bai R Y, Jahn T, Schrem S, Munzert G, Weidner K M, Wang J Y, Duyster J. The SH2-containing adapter protein GRB10 interacts with BCR-ABL. Oncogene. 1998;17:941–948. doi: 10.1038/sj.onc.1202024. [DOI] [PubMed] [Google Scholar]
- 3.Baum K, Hao X, Ren R. Inhibition of the Ras pathway by a dominant negative Ras suppresses Bcr-Abl induced myeloid but not lymphoid leukemia. Blood. 1998;92:94. [Google Scholar]
- 4.Carlesso N, Frank D A, Griffin J D. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chai S K, Nichols G L, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159:4720–4728. [PubMed] [Google Scholar]
- 6.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley and Sons, Inc.; 1996. [Google Scholar]
- 7.Cortez D, Kadlec L, Pendergast A M. Structural and signaling requirements for BCR-ABL-mediated transformation and inhibition of apoptosis. Mol Cell Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daley G Q, McLaughlin J, Witte O N, Baltimore D. The CML-specific P210 bcr/abl protein, unlike v-abl, does not transform NIH/3T3 fibroblasts. Science. 1987;237:532–535. doi: 10.1126/science.2440107. [DOI] [PubMed] [Google Scholar]
- 9.Daley G Q, Van Etten R A, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 10.Elefanty A G, Cory S. Hematologic disease induced in BALB/c mice by a bcr-abl retrovirus is influenced by the infection conditions. Mol Cell Biol. 1992;12:1755–1763. doi: 10.1128/mcb.12.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elefanty A G, Hariharan I K, Cory S. bcr-abl, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. EMBO J. 1990;9:1069–1078. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz W M, Berger P, Wang J Y. Deletion of an N-terminal regulatory domain of the c-abl tyrosine kinase activates its oncogenic potential. EMBO J. 1989;8:137–147. doi: 10.1002/j.1460-2075.1989.tb03358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goga A, McLaughlin J, Afar D E, Saffran D C, Witte O N. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 14.Golub T R, Goga A, Barker G F, Afar D E H, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosser Y Q, Zheng J, Overduin M, Mayer B J, Cowburn D. The solution structure of Abl SH3, and its relationship to SH2 in the SH(32) construct. Structure. 1995;3:1075–1086. doi: 10.1016/s0969-2126(01)00243-x. [DOI] [PubMed] [Google Scholar]
- 16.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hariharan I K, Harris A W, Crawford M, Abud H, Webb E, Cory S, Adams J M. A bcr-v-abl oncogene induces lymphomas in transgenic mice. Mol Cell Biol. 1989;9:2798–2805. doi: 10.1128/mcb.9.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale P K, Groffen J. Acute leukemia in bcr/abl transgenic mice. Nature. 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Honda H, Fujii T, Takatoku M, Mano H, Witte O N, Yazaki Y, Hirai H. Expression of p210bcr-abl by metallothionein promoter induced T-cell leukemia in transgenic mice. Blood. 1995;85:2853–2861. [PubMed] [Google Scholar]
- 21.Honda H, Oda H, Suzuki T, Takahashi T, Witte O N, Ozawa K, Ishikawa T, Yazaki Y, Hirai H. Development of acute lymphoblastic leukemia and myeloproliferative disorder in transgenic mice expressing p210bcr/abl: a novel transgenic model for human Ph1-positive leukemias. Blood. 1998;91:2067–2075. [PubMed] [Google Scholar]
- 22.Ilaria R L, Jr, Van Etten R A. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 23.Jackson P, Baltimore D. N-terminal mutations activate the leukemogenic potential of the myristoylated form of c-abl. EMBO J. 1989;8:449–456. doi: 10.1002/j.1460-2075.1989.tb03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelliher M A, McLaughlin J, Witte O N, Rosenberg N. Induction of a chronic myelogenous leukemia-like syndrome in mice with v-abl and BCR/ABL. Proc Natl Acad Sci USA. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelliher M A, Weckstein D J, Knott A G, Wortis H H, Rosenberg N. ABL oncogenes directly stimulate two distinct target cells in bone marrow from 5-fluorouracil-treated mice. Oncogene. 1993;8:1249–1256. [PubMed] [Google Scholar]
- 26.Liu J, Campbell M, Guo J Q, Lu D, Xian Y M, Andersson B S, Arlinghaus R B. BCR-ABL tyrosine kinase is autophosphorylated or transphosphorylates P160 BCR on tyrosine predominantly within the first BCR exon. Oncogene. 1993;8:101–109. [PubMed] [Google Scholar]
- 27.Liu J, Wu Y, Ma G Z, Lu D, Haataja L, Heisterkamp N, Groffen J, Arlinghaus R B. Inhibition of Bcr serine kinase by tyrosine phosphorylation. Mol Cell Biol. 1996;16:998–1005. doi: 10.1128/mcb.16.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maru Y, Witte O N. The Bcr gene encodes a novel serine/threonine kinase activity within a single exon. Cell. 1991;67:459–468. doi: 10.1016/0092-8674(91)90521-y. [DOI] [PubMed] [Google Scholar]
- 29.McWhirter J R, Galasso D L, Wang J Y. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McWhirter J R, Wang J Y. Activation of tyrosine kinase and microfilament-binding functions of c-abl by bcr sequences in bcr/abl fusion proteins. Mol Cell Biol. 1991;11:1553–1565. doi: 10.1128/mcb.11.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWhirter J R, Wang J Y. Effect of Bcr sequences on the cellular function of the Bcr-Abl oncoprotein. Oncogene. 1997;15:1625–1634. doi: 10.1038/sj.onc.1201342. [DOI] [PubMed] [Google Scholar]
- 32.Melo J V. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- 33.Musacchio A, Saraste M, Wilmanns M. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat Struct Biol. 1994;1:546–551. doi: 10.1038/nsb0894-546. [DOI] [PubMed] [Google Scholar]
- 34.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pendergast A M, Bronson R, Aster J C, Scott M L, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 35.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pendergast A M, Muller A J, Havlik M H, Maru Y, Witte O N. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991;66:161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- 37.Pendergast A M, Quilliam L A, Cripe L D, Bassing C H, Dai Z, Li N, Batzer A, Rabun K M, Der C J, Schlessinger J, Gishizky M L. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the Grb-2 adapter protein. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- 38.Polak J, Zemanova Z, Michalova K, Klamova H, Cermak J, Haskovec C. A new case of chronic myeloid leukemia (CML) in myeloid blast crisis with an atypical (b3/a3) junction of the BCR/ABL gene. Leukemia. 1998;12:250. doi: 10.1038/sj.leu.2400913. [DOI] [PubMed] [Google Scholar]
- 39.Puil L, Liu J, Gish G, Mbamalu G, Bowtell D, Pelicci P G, Arlinghaus R, Pawson T. Bcr-Abl oncoproteins bind directly to activators of the Ras signalling pathway. EMBO J. 1994;13:764–773. doi: 10.1002/j.1460-2075.1994.tb06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raitano A B, Whang Y E, Sawyers C L. Signal transduction by wild-type and leukemogenic Abl proteins. Biochim Biophys Acta. 1997;1333:F201–F216. doi: 10.1016/s0304-419x(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 41.Renshaw M W, Kipreos E T, Albrecht M R, Wang J Y. Oncogenic v-Abl tyrosine kinase can inhibit or stimulate growth, depending on the cell context. EMBO J. 1992;11:3941–3951. doi: 10.1002/j.1460-2075.1992.tb05488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renshaw M W, McWhirter J R, Wang J Y. The human leukemia oncogene bcr-abl abrogates the anchorage requirement but not the growth factor requirement for proliferation. Mol Cell Biol. 1995;15:1286–1293. doi: 10.1128/mcb.15.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuther G W, Fu H, Cripe L D, Collier R J, Pendergast A M. Association of the protein kinases c-Bcr and Bcr-Abl with proteins of the 14-3-3 family. Science. 1994;266:129–133. doi: 10.1126/science.7939633. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg N, Baltimore D. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J Exp Med. 1976;143:1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg N, Witte O N. The viral and cellular forms of the Abelson (abl) oncogene. Adv Virus Res. 1988;35:39–81. doi: 10.1016/s0065-3527(08)60708-3. [DOI] [PubMed] [Google Scholar]
- 46.Sawyers C L. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 47.Scott M L, Van Etten R A, Daley G Q, Baltimore D. v-abl causes hematopoietic disease distinct from that caused by bcr-abl. Proc Natl Acad Sci USA. 1991;88:6506–6510. doi: 10.1073/pnas.88.15.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers C L. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 49.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi J K, Trotta R, Wlodarski P, Perrotti D, Chan T O, Wasik M A, Tsichlis P N, Calabretta B. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skorski T, Nieborowska-Skorska M, Wlodarski P, Wasik M, Trotta R, Kanakaraj P, Salomoni P, Antonyak M, Martinez R, Majewski M, Wong A, Perussia B, Calabretta B. The SH3 domain contributes to BCR/ABL-dependent leukemogenesis in vivo: role in adhesion, invasion, and homing. Blood. 1998;91:406–418. [PubMed] [Google Scholar]
- 50a.Subrahmanyam, R., and R. Ren. Unpublished data.
- 51.Voncken J W, Kaartinen V, Pattengale P K, Germeraad W T, Groffen J, Heisterkamp N. BCR/ABL P210 and P190 cause distinct leukemia in transgenic mice. Blood. 1995;86:4603–4611. [PubMed] [Google Scholar]
- 52.Wen S T, Van Etten R A. The PAG gene product, a stress-induced protein with antioxidant properties, is an Abl SH3-binding protein and a physiological inhibitor of c-Abl tyrosine kinase activity. Genes Dev. 1997;11:2456–2467. doi: 10.1101/gad.11.19.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Liu J, Arlinghaus R B. Requirement of two specific tyrosine residues for the catalytic activity of Bcr serine/threonine kinase. Oncogene. 1998;16:141–146. doi: 10.1038/sj.onc.1201524. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]