Abstract
Template activating factor I (TAF-I) was originally identified as a host factor required for DNA replication and transcription of adenovirus genome complexed with viral basic proteins. Purified TAF-I was shown to bind to core histones and stimulate transcription from nucleosomal templates. Human TAF-I consists of two acidic proteins, TAF-Iα and TAF-Iβ, which differ from each other only in their amino-terminal regions. Here, we report that TAF-I decondenses demembraned Xenopus sperm chromatin. Human TAF-Iβ has a chromatin decondensation activity comparable to that of NAP-I, another histone binding protein, whereas TAF-Iα has only a weak activity. Analysis of molecular mechanisms underlying the chromatin decondensation by TAF-I revealed that TAF-I interacts directly with sperm basic proteins. Deletion of the TAF-I carboxyl-terminal acidic region abolishes the decondensation activity. Interestingly, the acidic region itself is not sufficient for decondensation, since an amino acid substitution mutant in the dimerization domain of TAF-I which has the intact acidic region does not support chromatin decondensation. We detected the β form of TAF-I in Xenopus oocytes and eggs by immunoblotting, and the cloning of its cDNA led us to conclude that Xenopus TAF-Iβ also decondenses sperm chromatin. These results suggest that TAF-I plays a role in remodeling higher-order chromatin structure as well as nucleosomal structure through direct interaction with chromatin basic proteins.
Structural change of chromatin in eukaryotic cells has an impact on biological processes such as gene expression, replication, and maintenance (57, 59). Disruption and reformation of nuclear architecture involve global remodeling of chromatin organization in mitotic and meiotic cell cycle (29, 47). Thus, chromatin remodeling has been one of the hot topics in molecular and cell biology in recent years (3, 12, 22, 52, 53, 55).
Chromatin consists of a repeating unit, nucleosome, in which about 200 bp of DNA wrap around a core histone octamer (H2A, H2B, H3, H4)2. Mixing of histones with DNA at physiological ionic strength generally results in precipitation of histone-DNA aggregates. It is therefore thought that nucleosome assembly-remodeling factors are required to mediate nucleosome assembly under physiological conditions (22). These factors are classified into two classes at least. One includes so-called histone chaperones, proteins which recruit and/or deposit histones to DNA. Nucleoplasmin is the first identified histone chaperone; it was originally identified as an abundant nucleosome assembly factor in oocytes of Xenopus laevis (31). It is reported that in Xenopus egg extracts, core histones are complexed with nucleoplasmin and a pair of other histone chaperones, N1 and N2 (10, 28). From somatic cells, other nucleosome assembly factors, such as chromosome assembly factor I (CAF-I) and nucleosome assembly protein I (NAP-I), have been identified by using DNA replication-dependent or -independent nucleosome assembly systems (18, 50). Recent progress in our understanding of chromatin remodeling from considerable experimental efforts has identified the second class of nucleosome assembly-remodeling factors, i.e., chromatin remodeling factors dependent on ATP hydrolysis (22, 53, 55). One of these ATP-dependent chromatin remodeling factors, ACF, can act with NAP-I or CAF-I to mediate the formation of periodic nucleosome arrays (20).
Dynamic remodeling in chromatin structure takes place during early development, represented by sperm chromatin decondensation and pronuclei formation upon fertilization (47). During spermatogenesis in Xenopus, subtypes of somatic histones are replaced with sperm-specific basic proteins. Xenopus sperm chromatin contains all four histones, but the amounts of H2A and H2B are considerably less than those of H3 and H4 (11, 23, 45). Demembraned Xenopus sperm chromatin is decondensed by incubation with Xenopus egg extracts in cell-free systems (11, 12, 23, 34, 42, 46). In the past decade, demembraned Xenopus sperm chromatin has been used extensively to study functions of histone binding proteins (7, 21, 24, 42, 45, 46). Again first studied was nucleoplasmin, which is most likely the actual player to decondense sperm chromatin in Xenopus eggs (42, 46). Depletion of nucleoplasmin from egg extracts results in a much lower rate of chromatin decondensation than that in the mock-depleted control extracts (46). Recently, Drosophila embryo extracts used to study the nucleosome assembly and fractionation of the extracts led to the identification of factors that decondense Xenopus sperm chromatin (6, 7, 19–21, 24). Thus, it is thought that factors which had been found originally as nucleosome assembly factors can be involved in remodeling of chromatin structure.
In the course of study to establish a cell-free adenovirus DNA replication system with a viral DNA template complexed with viral basic proteins (adenovirus core), we identified and purified a host factor designated template activating factor I (TAF-I) (35). Subsequently, we showed that TAF-I is also able to stimulate transcription from the adenovirus core but not from the naked adenovirus DNA as a template, suggesting that TAF-I functions as a remodeling factor of the structure of the adenovirus core (36). TAF-I was purified as a fraction containing 41- and 39-kDa proteins from uninfected HeLa cell extracts (35). cDNA cloning of human TAF-I revealed that the 39-kDa protein TAF-Iβ is encoded by a previously identified gene called SET, which is fused to the CAN/NUP214 gene in a case of acute undifferentiated leukemia (39, 56). The 41-kDa protein, TAF-Iα, differs from TAF-Iβ only at its amino-terminal region. TAF-Iα and TAF-Iβ are highly acidic proteins with their pIs of 4.23 and 4.12, respectively. TAF-I has structural homology with NAP-I, and it was shown that TAF-I facilitates assembly of nucleosomes in the supercoiling assay, in which TAF-I introduces negative supercoiling in circular DNA when incubated with histones and DNA topoisomerase I (25, 39). We previously showed that NAP-I is also able to stimulate DNA replication and transcription of adenovirus core, suggesting a functional similarity between TAF-I and NAP-I (25, 39). Importantly, it has been indicated that TAF-I remodels the structure of the chromatin reconstituted with a DNA fragment and purified histones and stimulates transcription from it (44).
In this paper, we studied the function of TAF-I in decondensation of Xenopus sperm chromatin. We found that recombinant human TAF-I mediates the decondensation of sperm chromatin by releasing sperm basic proteins. Immunoblotting revealed the existence of the Xenopus homologue of TAF-Iβ in oocyte nuclei and in the eggs. We cloned its cDNA and found that Xenopus TAF-I is active in sperm chromatin decondensation. Domain analyses of TAF-I suggested that the acidic region and the region possibly required for the structural integrity are essential for chromatin decondensation. The results presented here extend our knowledge on the structure-function relationships of histone-binding chromatin remodeling proteins.
MATERIALS AND METHODS
Preparation of proteins.
Recombinant human TAF-I (hTAF-I) and its deletion mutants with a six-histidine tag at their amino termini were prepared as described previously (39). To generate hTAF-IβPME, DNA fragments were prepared by PCR-mediated amplification with a cDNA clone as a template and oligonucleotide primers. TAF-I cDNA was separated into two parts, parts 1 and 2, in the middle of the region to be mutated. Each DNA fragment corresponding to two parts was amplified by PCR with oligonucleotide primers as follows: 5′-GGCAGCCATATGTCGGCGCCGGCGGCCAAAGTC-3′ and 5′-CATTAGATCTGTCTTCTTCATTTTGTTCTTCATCAATGTGTTC-3′ as an amino-terminal primer and a carboxyl-terminal primer, respectively, for part 1 of βPME; 5′-GACAGATCTAATGAACAAGAAAGTGAGGAGATTTTG-3′ and 5′-CGCGGATCCTTAGTCATCTTCTCCTTCATC-3′ as an amino-terminal primer and a carboxyl-terminal primer, respectively, for part 2 of βPME. Amplified DNA fragments for part 1 was treated with Klenow fragment (Takara Shuzo, Kusatsu, Japan) to blunt the ends and with polynucleotide kinase (TOYOBO, Osaka, Japan) to phosphorylate the 5′ ends and cloned into the EcoRV site of plasmid pBluescript (Stratagene). The resultant plasmid was designated pBluescript-TAF-IβPME. Amplified DNA fragments for part 2 were digested with BamHI and BglII and cloned into BamHI-BglII sites of pBluescript-TAF-IβPME. A DNA fragment generated by digestion of the plasmid with both NdeI and BamHI were cloned into NdeI-BamHI sites of plasmid pET14b (Novagen). To obtain recombinant TAF-I protein, Escherichia coli BL21 (DE3) was transformed with each plasmid. Recombinant TAF-I was overexpressed by the addition of isopropyl-β-d-thiogalactopyranoside to the bacterial culture (800 ml). Bacterial cells were sonicated in 30 ml of sonication buffer consisting of 20 mM Tris HCl (pH 8.0) and 100 mM NaCl. The cell lysate was centrifuged at 12,000 × g for 10 min, and the supernatant was applied to a Talon metal affinity column (Clontech). Six-histidine-tagged TAF-I proteins were eluted with sonication buffer containing 100 mM imidazole. hTAF-IβΔN1 and hTAF-IβΔC3 were further purified by the isolation from a polyacrylamide gel and subjected to the denature-renature protocol (36). For preparation of glutathione S-transferase (GST)–hTAF-Iβ and GST-AR, DNA fragments were obtained by PCR using the TAF-Iβ cDNA clone as a template with the following primers: 5′-GGCGCGGATCCATGTCGGCGCAGGCGGCCAAAGTC-3′ for GST-hTAF-Iβ and 5′-CGCGGATCCGATGATGAAGAAGGAGAAGGAG-3′ for GST-AR as amino-terminal primers, and 5′-CCGGAATTCCTTAGTCATCTTCTCCTTCATC-3′ as a common carboxyl-terminal primer. The PCR products were digested with BamHI and EcoRI and cloned into BamHI-EcoRI sites of plasmid pGEX2TK (Amersham Pharmacia Biotech) for GST–hTAF-Iβ or pGEX4T-3 for GST-AR. GST fusion recombinant proteins were overexpressed in the E. coli BL21 (DE3) strain as described above. Bacterial cells were suspended in phosphate-buffered saline (PBS) and sonicated. The soluble fractions were applied to a glutathione-Sepharose 4B column (Amersham Pharmacia Biotech), and the GST fusion proteins were eluted with 10 mM reduced glutathione in 50 mM Tris HCl (pH 8.0) as described in the manufacturer’s instructions.
HeLa TAF-I represents the fraction eluted from MonoQ column chromatography (35). Recombinant yeast and mouse NAP-I were prepared as described previously (15, 39). Core histones were purified from HeLa cells as described before (49). Protein concentration was determined as described with reagents from Bio-Rad by using bovine serum albumin (BSA) as the standard (5).
Preparation of extracts.
To prepare the oocyte lysates used for detection of Xenopus TAF-I, defolliculated oocytes were prepared by treating frog ovaries with collagenase, and healthy stage VI oocytes (300 μl) were homogenized in 1 ml of 90 mM HEPES (pH 7.5)–70 mM KCl–5% sucrose–1 mM dithiothreitol (DTT). The homogenate was centrifuged in a microtube at 12,000 × g rpm for 10 min at 4°C, and the supernatant was used as the oocyte lysate. To examine the localization of proteins, nuclei were isolated from oocytes manually. Nuclear or cytoplasmic fractions from 10 oocytes were pooled and analyzed by immunoblotting as described below.
Fractionated interphase egg extracts from Xenopus were prepared as described previously (51). To obtain heat-labile fraction, the egg extracts were diluted 10-fold with 20 mM Tris HCl (pH 7.5) containing 0.25 mM phenylmethylsulfonyl fluoride (PMSF) and heated at 80°C for 10 min. After being chilled on ice, the extracts were centrifuged in a microtube at 12,000 × g for 10 min at 4°C. The soluble fraction (heat-stable fraction) was removed, and the pellet (heat-labile fraction) was solubilized in a volume of the original egg extracts by adding 6 M guanidine HCl, 10 mM Tris HCl (pH 7.5), 0.2 M KCl, 0.1 mM EDTA, 5% glycerol, and 0.1 mM DTT. The heat-labile fraction was renatured by dialysis against 10 mM Tris HCl (pH 7.5)–0.1 mM EDTA–10% glycerol–0.1 mM DTT–0.25 mM PMSF (36). TAF-I was depleted from the renatured heat-labile fraction by using anti-TAF-Iβ monoclonal antibody coupled to protein G Sepharose. Fifty microliters of the fraction was mixed with 10 μl of antibody-coupled Sepharose beads for 1 h at 4°C, and the mixture was then centrifuged at 2,000 × g for 2 min. The resultant supernatant was mixed with another 10 μl of antibody-coupled Sepharose beads for 1 h and centrifuged to obtain the TAF-I-depleted fraction. For immunoprecipitation, oocyte extracts were prepared as described for the egg extracts (51). The egg or oocyte extracts (1.5 ml) or the recombinant hTAF-Iβ protein (30 μg) was mixed for 1 h at 4°C with 20 μl of protein G Sepharose beads which had been coupled with anti-TAF-Iβ antibody or control mouse immunoglobulin G (IgG; Chemicon). The beads were washed extensively with 50 mM Tris HCl (pH 7.5)–1 mM EDTA–150 mM NaCl–0.1% NP-40–1 mM PMSF. Proteins were released from the beads into 40 μl of 10 mM N-cyclohexyl-3-aminopropanesulfonic acid (pH 11), followed by the addition of one-tenth volume of 1 M Tris HCl (pH 7.5).
Decondensation of Xenopus sperm chromatin.
Demembraned Xenopus sperm chromatin was prepared as described previously (51). In a standard assay, the demembraned sperm chromatin was incubated at a final concentration of 5 × 103 sperm/μl with TAF-I at room temperature in 10 μl of reaction mixture containing 8 mM HEPES (pH 7.5), 8 mM KCl, 2 mM MgCl2, 200 mM sucrose, 3.3 mM ATP, 33 mM creatine phosphate, 0.33 mg of phosphocreatine kinase per ml, and 0.8 mM DTT. After incubation, a 2-μl aliquot of the reaction mixture was added to 5 μl of PBS containing 10 μg of Hoechst 33258 per ml, 50% glycerol, and 7.4% formaldehyde on a slide glass. The DNA stained with the dye was visualized under a fluorescent microscope (Olympus). To analyze the chromatin-bound and released proteins during chromatin decondensation by TAF-I, sperm chromatin (1.5 × 106) was incubated with 150 μg of GST or 250 μg of GST–hTAF-Iβ at room temperature for 60 min under the conditions for the decondensation assay. The mixture was centrifuged at 12,000 × g for 10 min to separate the chromatin from the released proteins. The resulting supernatants were incubated with 20 μl of glutathione-Sepharose 4B by gentle agitation at 4°C for 60 min. Protein complexes bound to the Sepharose beads were precipitated by centrifugation and washed extensively with buffer A (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, 1 mM DTT) containing 50 mM NaCl and then with buffer A containing 200 mM NaCl. Proteins were eluted with buffer A containing 1 M NaCl and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The chromatin was resuspended in buffer A containing 50 mM NaCl, and then HCl was added at a final concentration of 0.5 N. The mixture was incubated on ice for 10 min, and insoluble proteins were removed by centrifugation for 10 min. The HCl-soluble proteins (basic proteins) were concentrated by precipitation with trichloroacetic acid and analyzed by SDS-PAGE.
Chemical cross-linking analysis.
Dimer formation of TAF-I was examined by chemical cross-linking of TAF-I proteins. One hundred nanograms of TAF-I was cross-linked at room temperature for 30 min with 0.05% glutaraldehyde in a buffer containing 30 mM HEPES (pH 7.9), 0.5 mM EDTA, 50 mM NaCl, and 10% glycerol. The reaction was stopped by adding the sample buffer for SDS-PAGE. Samples were then heated at 98°C for 2 min and separated by SDS–7.5% PAGE.
Restriction enzyme sensitivity assay.
Adenovirus core (30 ng) was incubated with TAF-I at 30°C for 30 min in a buffer containing 12.5 mM MgCl2, 60 mM KCl, 20 mM NaCl, 12 mM HEPES (pH 7.9), 1 mg of BSA per ml, and 8% glycerol and then digested at 30°C for 2 min with 1 unit of PvuII. DNA was purified and separated in a 1% agarose gel. DNA was transferred to a nylon membrane and visualized by hybridization with radiolabelled DNA spanning nucleotide positions 455 to 628 of the adenovirus genome around the E1A promoter region (25, 44).
cDNA cloning of Xenopus TAF-I cDNAs.
A Xenopus oocyte cDNA library was constructed in the λZIPLOX vector by using SuperScript Lambda System (Life Technologies Inc.). A degenerated oligonucleotide set of 5′-GARAARGARCARCARGARGC-3′ (R = G or A) encoding amino acids EKEQQEA and 5′-CCYTCYTCRTCRTCCATRTC-3′ (R = G or A, Y = T or C) encoding amino acids DMDDEEG was used for reverse transcriptase-mediated PCR using Xenopus oocyte mRNA. Nucleotide sequence analysis of amplified DNA cloned into a plasmid vector revealed that the amino acid sequence encoded by the cloned DNA showed high homology with human TAF-I. Using this DNA fragment, the Xenopus oocyte cDNA library was screened. Positive clones were plaque-purified twice, and the cDNA clones were recovered in plasmids by using in vivo excision as described in the manufacturer’s protocol and sequenced. To construct Xenopus TAF-Iβ1 and TAF-Iβ2 expression plasmids, PCR was performed with the cDNA clones as templates with primer sets of 5′-GGCAGCCATATGTCGGCGCCGGCGGCCA-3′ for xTAF-Iβ1 or 5′-GGCAGCCATATGTCGGCGCCCAAAGTCAGTAAAAAG-3′ for xTAF-Iβ2 as amino-terminal primers and 5′-AGCCCGCTCGAGATCATCTTCACCTTCGTCTTCC-3′ as a common carboxyl-terminal primer. The PCR products were digested with NdeI and XhoI and inserted into NdeI-XhoI sites of pET24b (Novagen).
Immunoblot analysis.
Cell-free transcription-coupled translation in rabbit reticulocyte lysate was performed with 1 μg of expression plasmids for xTAF-Iβ1 or xTAF-Iβ2 (see above) in a 50-μl reaction mixture with the TNT T7 Quick Coupled Transcription/Translation System (Promega). The cell-free translation products, Xenopus extracts, and purified proteins were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The following procedures were performed at room temperature. The membrane was blocked with 10% BSA in PBS plus 0.2% Tween 20 for 1 h, followed by incubation with first antibodies in PBS containing 5% BSA and 0.2% Tween 20 for 1 h. After washing with three changes of PBS containing 0.3% Triton X-100, the membrane was incubated with a secondary antibody, goat anti-mouse IgG antibody conjugated to horseradish peroxidase (Promega), in PBS containing 1% BSA and 0.2% Triton X-100 for 1 h. The membrane was washed as described above, and the signals were detected with ECL Western blotting detection reagent (Amersham Pharmacia Biotech).
Nucleotide sequence accession number.
The complete nucleotide sequences of xTAF-Iβ1 and xTAF-Iβ2 cDNA obtained in this study will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession no. AB022691 and AB022692, respectively.
RESULTS
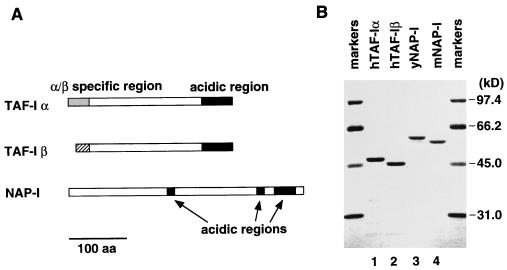
TAF-I, NAP-I, and nucleoplasmin share the acidic region in their carboxyl-terminal regions (13, 15, 39). The acidic region of nucleoplasmin has been suggested to be important for its interaction with histones (13). As shown in Fig. 1A, TAF-I has an acidic region with glutamic and aspartic acid stretches located at its carboxyl-terminal region (also see Fig. 7 for amino acid sequence). The carboxyl-terminal third of NAP-I is also acidic, consisting of tripartite highly acidic regions (Fig. 1A). This structural similarity prompted us to examine whether TAF-I can act as a chromatin remodeling factor in a sperm chromatin decondensation assay. We then found that this is indeed the case.
FIG. 1.
Structure of TAF-I and NAP-I. (A) Schematic diagrams of TAF-Iα, TAF-Iβ, and NAP-I. TAF-Iα- and TAF-Iβ-specific regions are indicated by gray and hatched boxes, respectively. The TAF-I acidic region and the NAP-I tripartite acidic regions are shown by black boxes. Bar, 100 amino acids. (B) Recombinant proteins used in this study. Purified recombinant human TAF-Iα and TAF-Iβ and yeast and mouse NAP-I proteins (yNAP-I and mNAP-I, respectively) were analyzed by SDS–10% PAGE and stained with Coomassie brilliant blue. The sizes of the molecular mass markers (Bio-Rad) are shown on the right.
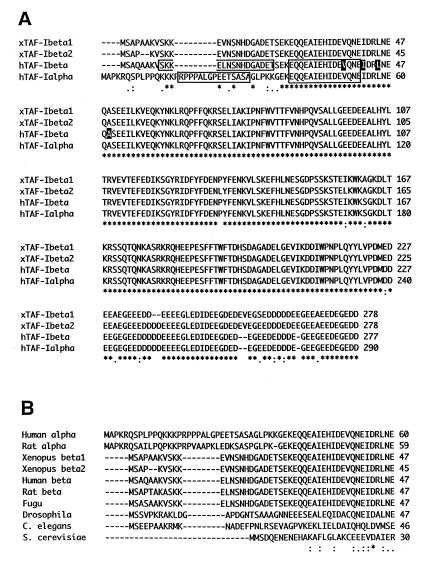
FIG. 7.
Sequence comparison of TAF-I. (A) The complete amino acid sequences of Xenopus TAF-Iβ1 and TAF-Iβ2, human TAF-Iβ (accession no. M93651), and TAF-Iα (D45198) aligned by use of a CLUSTALW program. Amino acids identical in all four sequences are indicated by asterisks, conserved substitutions are indicated by colons, and semiconserved substitutions are indicated by periods at the bottom of the alignment. The four amino acids mutated in hTAF-IβPME are highlighted with black shading. The peptide sequence to produce monoclonal antibodies against TAF-I (α and β common, EQQEAIEHIDEVQNE; α-specific, RPPPALGPEETSASA; β-specific, SKKELNSNHDGADET) are boxed. (B) Amino acid sequence alignment of amino-terminal regions of TAF-I/SET from different organisms. Sequences are aligned from human, rat (accession no. S68589 and S68987), Xenopus, puffer fish (fugu fish; AF007219), Drosophila (U30470), C. elegans (Z54236), and yeast (Z71522) TAF-I/SET. Only the amino-terminal portion of the alignment is shown.
TAF-Iβ, but not TAF-Iα, efficiently decondenses Xenopus sperm chromatin.
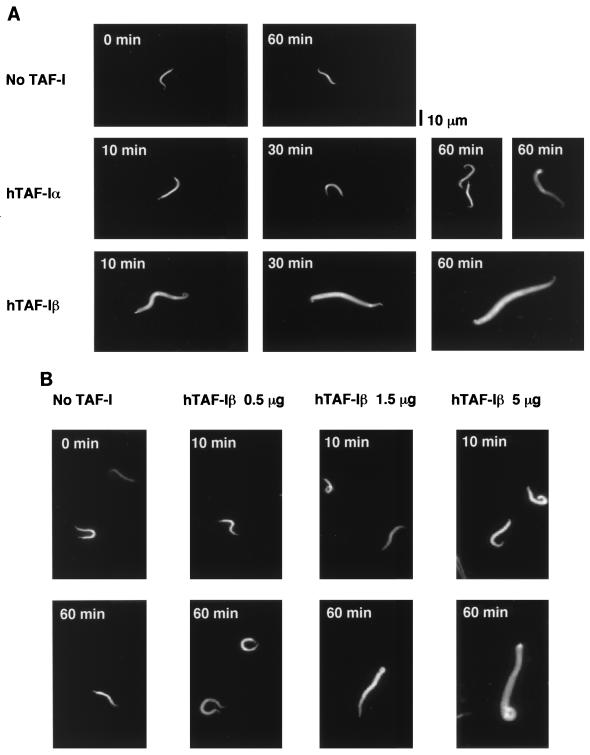
Demembraned Xenopus sperm chromatin was incubated with recombinant human TAF-Iα and TAF-Iβ (Fig. 1B), and the chromosomal DNA was stained with Hoechst 33258 stain and visualized under a fluorescent microscope. After incubation with TAF-Iβ for 60 min, sperm chromatin was fully decondensed (Fig. 2A). Although TAF-Iα shares all the amino acid sequence with TAF-Iβ except its amino-terminal specific region (Fig. 1A), it showed a very weak decondensation activity. In samples taken after incubation for 10 and 30 min with TAF-Iα, we did not observe any decondensed sperm chromatin. At 60 min, we found about 1% of sperm chromatin decondensed; its size was slightly larger than that of the condensed chromatin but not as large as that of the decondensed chromatin with TAF-Iβ (Fig. 2A and data not shown).
FIG. 2.
Decondensation of Xenopus sperm chromatin by TAF-Iα and TAF-Iβ. (A) Time course of decondensation by TAF-Iα and TAF-Iβ. Sperm chromatin (5 × 104 sperm) was incubated without (top panels) or with 5 μg of recombinant hTAF-Iα (middle panels) or hTAF-Iβ (lower panels) as described in Materials and Methods. At the indicated times, aliquots were mixed with fixation buffer containing Hoechst 33258 stain and the chromosomal DNA was visualized under a fluorescent microscope. For the sample from the 60-min incubation with hTAF-Iα, two panels are shown to represent a weak decondensation activity (see text for details). Bar, 10 μm. (B) Dose response of hTAF-Iβ in chromatin decondensation. Sperm chromatin (5 × 104 sperm) was incubated with 0, 0.5, 1.5, and 5 μg of hTAF-Iβ for 10 and 60 min.
To examine the amount of TAF-I required for chromatin decondensation, we titrated hTAF-Iβ in the decondensation assay with a constant number of sperm chromatin. Samples were aliquoted at 10 and 60 min, and the chromosomal DNA was visualized (Fig. 2B). With 0.5 μg of TAF-Iβ, we did not observe any significant decondensation, whereas the incubation with 1.5 μg of TAF-Iβ showed chromatin decondensation to a limited extent. To observe fully decondensed chromatin, a 60-min incubation with 5 μg of TAF-Iβ was required. The decondensed chromatin under this condition was of a size comparable to that prepared by incubation with interphase extracts of Xenopus eggs (see Fig. 6C). ATP and the ATP regeneration system were included in our decondensation assay, and they stimulated the reaction. However, it is possible that they act as monovalent cations since under higher ionic strength the stimulatory effect of them was not observed. The time course of decondensation by TAF-I is slower than that by egg extracts or purified nucleoplasmin (46). Although we have not examined the effect of a higher amount of TAF-Iβ on the time course and extent of decondensation, the amount of TAF-Iβ used here (5 μg) is comparable to that of purified nucleoplasmin (7 μg) in the experiments by Philpott et al. (46).
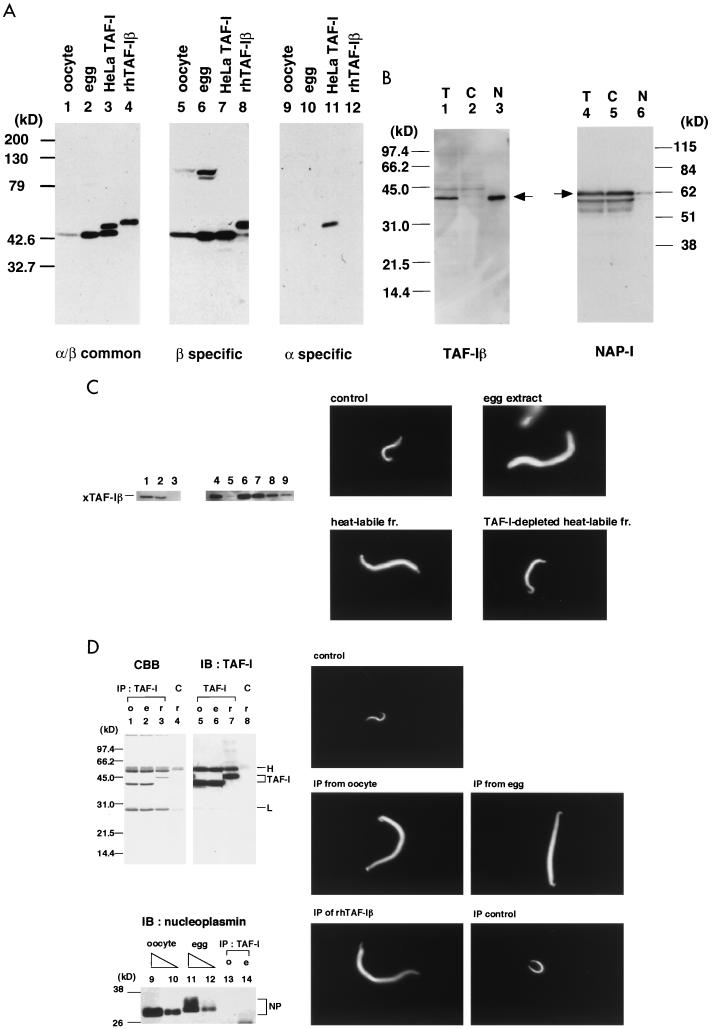
FIG. 6.
Existence of TAF-Iβ, but not TAF-Iα, in Xenopus oocytes and eggs. (A) Detection of TAF-Iβ in Xenopus oocytes and eggs. Xenopus oocyte lysates (30 μg of protein; lanes 1, 5, and 9), egg extracts (30 μg of protein; lanes 2, 6, and 10), purified HeLa TAF-I (lanes 3, 7, and 11), and recombinant human TAF-Iβ (40 ng; lanes 4, 8, and 12) were analyzed by SDS–10% PAGE and transferred to a polyvinylidene difluoride membrane. TAF-I was detected with monoclonal antibodies which recognize human TAF-Iα and TAF-Iβ common region (clone KM1725 in reference 40) (lanes 1 to 4), TAF-Iβ-specific region (clone KM1720) (lanes 5 to 8), or TAF-Iα-specific region (clone KM1712) (lanes 9 to 12) by immunoblotting (see Fig. 7 for the epitope of each antibody). It should be noted that recombinant hTAF-Iβ has a six-histidine tag at its amino terminus, which might have resulted in its lower mobility than that of the native hTAF-Iβ (compare a band in lane 4 and the lower band in lane 3). Molecular masses of marker proteins are shown on the left. (B) Localization of TAF-Iβ and NAP-I in oocytes. A Xenopus oocyte was separated into nucleus and cytoplasm. Lysates of total oocyte (T), nuclear (N), and cytoplasmic fractions (C) were analyzed by immunoblotting with anti-TAF-Iβ antibody (lanes 1 to 3). Immunoblotting with anti-NAP-I antibody of the same oocyte fractionation is also shown (lanes 4 to 6). The positions of TAF-I and NAP-I are indicated by arrows. (C) Chromatin decondensation activity of TAF-Iβ in heat-labile fraction of egg extracts. Xenopus egg extracts were heated at 80°C for 10 min and separated into heat-stable and heat-labile fractions. The heat-labile fraction was subjected to the denature-renature protocol, and TAF-I was depleted from the renatured heat-labile fraction. Left panels show immunoblotting analysis using anti-TAF-Iβ antibody of egg extracts (lane 1), heat-labile fraction solubilized in guanidine-containing buffer (lane 2), heat-stable fraction (lane 3), mock-depleted renatured heat-labile fraction (lane 4), TAF-I depleted renatured heat-labile fraction (lane 5), and sequential dilutions of renatured heat-labile fraction (lane 6, 100%; lane 7, 30%; lane 8, 10%; lane 9, 3%). Right panels show chromatin decondensation with these fractions. Sperm chromatin (104 sperm) was incubated for 10 min with egg extracts, mock-depleted renatured heat-labile fraction, and TAF-I-depleted renatured heat-labile fraction. The chromosomal DNA was stained with Hoechst 33258 stain and visualized under a fluorescent microscope. (D) Sperm chromatin decondensation by TAF-Iβ immunopurified from oocyte and egg extracts. The oocyte and egg extracts and recombinant hTAF-Iβ were subjected to immunoprecipitation with anti-TAF-Iβ antibody or control mouse IgG as described in Materials and Methods. The immunoprecipitates of oocyte extracts (labeled “o”) (lanes 1, 5, and 13), egg extracts (labeled “e”) (lanes 2, 6, and 14), and recombinant hTAF-Iβ (labeled “r”) (lanes 3 and 7) with anti-TAF-Iβ antibody or the immunoprecipitates of recombinant hTAF-Iβ with control mouse IgG (lanes 4 and 8) were analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue (lanes 1 to 4) and by immunoblotting with anti-TAF-Iβ antibody (lanes 5 to 8) and with anti-nucleoplasmin monoclonal antibody (lanes 13 and 14). Two microliters (lanes 9 and 11) and 0.6 microliter (lanes 10 and 12) of oocyte (lanes 9 and 10) and egg (lanes 11 and 12) extracts were also analyzed by immunoblotting with antinucleoplasmin antibody. Positions of TAF-Iβ, IgG heavy chain (H) and light chain (L), and nucleoplasmin (NP) are indicated. It is important to note that the egg nucleoplasmin has lower mobility than the oocyte nucleoplasmin due to the hyperphosphorylation (32, 43, 48). The proteins released at pH 11 from the protein G beads were assayed for mediating the decondensation of sperm chromatin (2 × 104) for 60 min. The panel labelled “control” shows the sperm chromatin incubated with no added protein. The panel labelled “IP control” shows the sperm chromatin incubated with the proteins released after the immunoprecipitation of recombinant hTAF-Iβ with control mouse IgG.
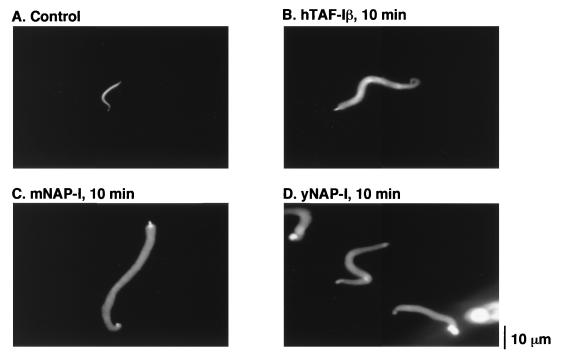
We next tested whether NAP-I, another histone binding protein, can decondense Xenopus sperm chromatin. NAP-I homologues have been identified in many organisms, including humans, mice, Xenopus and Drosophila species, and S. cerevisiae (15). NAP-I proteins from mice and yeast were shown to be capable of replacing TAF-I functionally in DNA replication and transcription of the adenovirus core (25, 39). Yeast and mouse recombinant NAP-I proteins were overexpressed in E. coli and purified, and then 5 μg of each protein was incubated with sperm chromatin under the conditions used for the decondensation. In a 10-min incubation, sperm chromatin was decondensed efficiently by both yeast and mouse NAP-I proteins as well as by TAF-Iβ (Fig. 3). These results are consistent with those of Ito et al., who have shown that Drosophila NAP-I decondenses Xenopus sperm chromatin (21).
FIG. 3.
Chromatin decondensation by NAP-I. Sperm chromatin (5 × 104 sperm) was incubated with 5 μg of hTAF-Iβ (B), mouse NAP-I (C), and yeast NAP-I (D) for 10 min. The chromosomal DNA was visualized as described in Materials and Methods. Sperm chromatin that was not incubated with any proteins is shown in panel A.
Domains of TAF-I required for the chromatin decondensation.
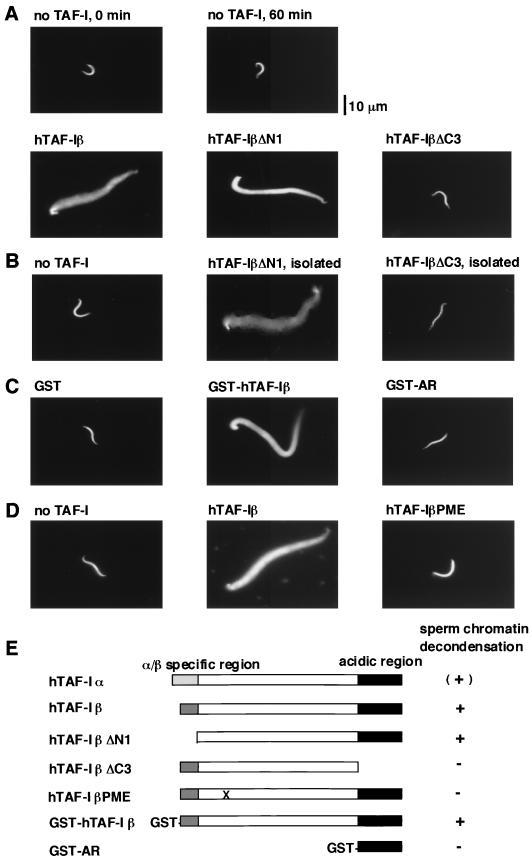
Having established that acidic proteins TAF-I and NAP-I decondense Xenopus sperm chromatin, we wished to determine the domain which is required for this activity by using recombinant human TAF-I derivatives. We compared the activities of TAF-Iα and TAF-Iβ and found that the chromatin decondensation caused by TAF-Iα was to a very limited extent in contrast to the efficient decondensation by TAF-Iβ (Fig. 2A). In the first of the domain analyses, shown in Fig. 4, we used deletion mutants of hTAF-Iβ. hTAF-IβΔN1, which lacks the amino-terminal β-specific domain, represents the α/β common regions (Fig. 4E). hTAF-IβΔN1 binds to histones as efficiently as full-length hTAF-Iβ (44). Sperm chromatin incubated with this mutant TAF-I was decondensed as efficiently as that with full-length hTAF-Iβ. Given that TAF-Iα differs from TAF-Iβ only at its amino-terminal region, these results suggest that the α-specific region has a negative effect on the chromatin decondensation activity. In marked contrast, hTAF-IβΔC3 [previously termed TAF-Iβ(1-225) in reference 39] lacking the carboxyl-terminal acidic domain did not show any decondensation activity (Fig. 4A).
FIG. 4.
Domains required for chromatin decondensation. (A to D) Sperm chromatin was incubated with 5 μg of hTAF-Iβ and its derivatives for 60 min. The chromosomal DNA was stained with Hoechst 33258 stain and visualized under a fluorescent microscope. In panel B, the gel-isolated hTAF-IβΔN1 and hTAF-IβΔC3 were used (see Materials and Methods). (E) The results of domain analysis for sperm chromatin decondensation are summarized. The decondensation activity of each hTAF-I derivative is shown to the right of its schematic diagram. + and −, active and inactive in the decondensation assay, respectively. hTAF-Iα has a very weak activity, indicated by (+). Point mutations in hTAF-IβPME are shown by X in the schematic diagram.
Polyanions such as polyglutamic acids can mediate chromatin decondensation as well as nucleosome assembly (46). To exclude the possibility that proteins or RNA contaminating in the recombinant TAF-I preparations mediate the chromatin decondensation, we made use of gel isolation of six-histidine-tagged TAF-I after the metal affinity column chromatography, followed by renaturation of the protein (36). The gel-purified proteins gave the same results with the column-purified proteins in the decondensation assay (compare Fig. 4B and A), indicating that the decondensation activity examined here is mediated by TAF-I itself.
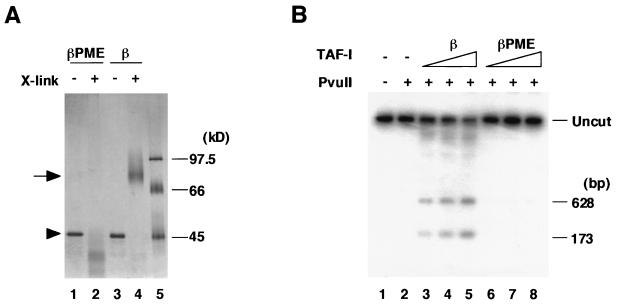
The results described above indicate the importance of the carboxyl-terminal acidic region. It was possible that interaction between the acidic region of TAF-I and sperm basic proteins would be sufficient for the chromatin decondensation. To further examine the role of the acidic region, we prepared GST fusion proteins containing the full-length hTAF-Iβ (GST–hTAF-Iβ) or the acidic region alone (GST-AR). When GST itself or GST-AR was employed in the decondensation assay, no change in sperm chromatin was observed (Fig. 4C). GST–hTAF-Iβ was fully active in the decondensation assay, indicating that the GST moiety is not inhibitory. We concluded that the carboxyl-terminal acidic region is essential but not sufficient for the decondensation. This raised a possibility that some structural integrity mediated through a region(s) other than the acidic region of TAF-Iβ is necessary for the decondensation activity. In solution, TAF-I exists and functions as an oligomer, possibly a dimer (35). Recent results with deletion mutant TAF-I proteins have shown that the amino-terminal portion of 40 amino acids common to both TAF-Iα and TAF-Iβ is required for dimerization (38a). Based on this information, we prepared hTAF-IβPME, a mutant TAF-Iβ protein in which four amino acids possibly involved in hydrophobic intermolecular interactions are replaced with the hydrophilic amino acids (V38E, I42E, L45S, and A49E) (see Fig. 7). Cross-linking by glutaraldehyde allowed us to detect the dimeric form of hTAF-Iβ, while hTAF-IβPME completely lost the dimerization capability (Fig. 5A). hTAF-Iβ has been shown to make adenovirus core accessible to restriction enzymes (25). hTAF-IβPME was inactive in this assay as well as in the adenovirus core DNA replication assay (Fig. 5B) (38a). These observations led us to test the ability of hTAF-IβPME to decondense sperm chromatin (Fig. 4D). hTAF-IβPME was found inactive in the decondensation assay, suggesting that the dimer formation of TAF-I is a prerequisite for chromatin decondensation. In summary, we have found at least three domains of TAF-I which regulate its chromatin decondensation activity, i.e., a negative effect by α-specific region and essential roles of the acidic region and the dimerization region.
FIG. 5.
Dimerization of TAF-I. (A) Chemical cross-linking of hTAF-I proteins. One hundred nanograms of hTAF-IβPME (lanes 1 and 2) or hTAF-Iβ (lanes 3 and 4) was cross-linked with (lanes 2 and 4) or without (lanes 1 and 3) 0.05% glutaraldehyde. Samples were analyzed by SDS–7.5% PAGE, and proteins were detected by silver staining. Lane 5 shows marker proteins. Monomer and dimer are indicated by an arrowhead and an arrow, respectively. The cross-linked product migrating faster than the monomer appears to be a compact form of TAF-I due to intramolecular cross-linking. (B) Restriction enzyme sensitivity assay. Adenovirus core (30 ng) was incubated at 30°C for 30 min without (lanes 1 and 2) or with 100 ng (lanes 3 and 6), 200 ng (lanes 4 and 7), or 400 ng (lanes 5 and 8) of hTAF-Iβ (lanes 3 to 5) and hTAF-IβPME (lanes 6 to 8) and then digested with PvuII. Lane 1 shows the undigested DNA. DNA was purified and separated by electrophoresis in a 1% agarose gel. DNA fragments around the E1A promoter region were detected by Southern blotting. One-hundred-seventy-three- and 628-bp-long fragments were shown by the probe after partial digestion with PvuII.
TAF-I in the Xenopus oocytes and eggs.
So far we have used recombinant hTAF-I in the decondensation assay of Xenopus sperm chromatin. Next we looked for TAF-I in Xenopus oocytes and eggs. We have prepared monoclonal antibodies against either hTAF-Iα- or hTAF-Iβ-specific peptides as well as a monoclonal antibody that recognizes the α/β common region (40). Immunoblotting was employed to detect the Xenopus protein(s) by using these antibodies (Fig. 6A). With α/β common and β-specific antibodies, we found a Xenopus protein which comigrated with hTAF-Iβ purified from HeLa cells. TAF-Iα-specific antibody detected no protein in the Xenopus oocyte and egg extracts. These results suggest the existence of the β form of TAF-I in Xenopus oocytes and eggs. We next examined the localization of the Xenopus TAF-I protein in the oocytes. Nuclei were isolated from the oocytes manually under a microscope, and the resultant nuclear and cytoplasmic fractions were used for immunoblotting with anti-TAF-Iβ antibody (Fig. 6B). The signal detected with anti-TAF-Iβ antibody was found only in the oocyte nucleus. Although TAF-I was originally purified from HeLa cytoplasmic fractions, immunocytochemical analysis indicated its accumulation in the nuclei of somatic cells (35, 40). Previously, NAP-I homologues were detected in Xenopus oocytes (15). We then performed an immunoblotting of the same oocyte fractions with anti-NAP-I monoclonal antibody (17) and found the cytoplasmic localization of Xenopus NAP-I (Fig. 6B).
We then wished to examine whether Xenopus TAF-I in the egg extracts functions in sperm chromatin decondensation (Fig. 6C). In this regard, it is established that nucleoplasmin plays a pivotal role in sperm chromatin decondensation in the egg extracts (42, 46). In preliminary experiments, depletion of TAF-I from the egg extracts did not show any apparent effects on sperm chromatin decondensation. We then took advantage of the knowledge that nucleoplasmin is heat stable and remains soluble after heat treatment at 80°C for 10 min (31). Thus, Xenopus egg extracts were heated and separated into heat-stable and heat-labile fractions by centrifugation. TAF-I was detected predominantly in the heat-labile fraction (Fig. 6C, lane 2). The heat-labile fraction was renatured and examined for the decondensation activity. Incubation of sperm chromatin with the heat-labile fraction showed modest decondensation (Fig. 6C). Quantitative analysis revealed that the heat-labile fraction contained less than 10% of activity of the original egg extracts and the heat-stable fraction (data not shown). Since TAF-I was fractionated into the heat-labile fraction, we tried to deplete TAF-I from this fraction by using anti-TAF-Iβ antibody coupled to protein G-Sepharose. After two consecutive incubations with the antibody-coupled beads, the heat-labile fraction was depleted of >97% of TAF-I (Fig. 6C, lanes 5 to 9). Sperm chromatin incubated with the TAF-I-depleted heat-labile fraction showed very little decondensation.
Since the experiments described above utilized the TAF-I fraction that had been denatured and renatured, we cannot rule out the possibility that TAF-I in the egg extracts would not be active in sperm chromatin decondensation possibly because of its association with other proteins. As an alternative approach to address whether native TAF-I functions in the sperm chromatin decondensation, we performed immunopurification of TAF-I from the extracts under mild conditions (Fig. 6D). The immunoprecipitates from the egg or oocyte extracts with anti-TAF-Iβ antibody contained TAF-I as the predominant component, based on the SDS-PAGE and immunoblotting analysis. The proteins released from the protein G beads mediated the decondensation of sperm chromatin to the same extent with recombinant hTAF-Iβ. Immunoblotting with antinucleoplasmin antibody revealed the absence of nucleoplasmin in the immunoprecipitates with anti-TAF-Iβ antibody. These data strongly suggest that Xenopus TAF-I in the egg extracts is active in the chromatin decondensation.
cDNA cloning of xTAF-I and its characterization.
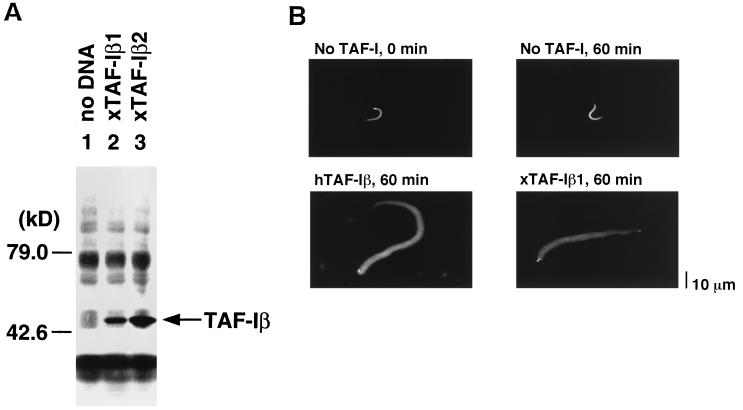
To obtain direct evidence that the Xenopus TAF-I indeed acts as a chromatin decondensation factor, we decided to clone its cDNA. Using degenerated oligonucleotides encoding amino acid sequences of hTAF-I, a DNA fragment from Xenopus mRNA whose sequence showed high homology with hTAF-I was amplified. This DNA fragment was used to screen the Xenopus oocyte cDNA library. We have obtained several positive clones, and two of them, which are designated xTAF-Iβ1 and xTAF-Iβ2, were completely sequenced (Fig. 7). Alignment of amino acid sequence deduced from the cloned cDNAs shows 96% identity between xTAF-Iβ1 and xTAF-Iβ2 and 93 to 94% identity between xTAF-Iβ and hTAF-Iβ. Since there was one amino acid difference between the xTAF-Iβ sequence and the peptide sequence which was used to raise the monoclonal antibody against hTAF-Iβ (Fig. 7A), we performed cell-free translation (Fig. 8A). The products of the cell-free transcription-coupled translation using xTAF-Iβ cDNAs were specifically recognized by our anti-TAF-Iβ antibody. These results confirmed that we have cloned xTAF-Iβ cDNAs which actually encode the protein(s) detected by the immunoblotting in oocyte and egg extracts (Fig. 6). We then purified recombinant xTAF-Iβ and used it in the chromatin decondensation assay (Fig. 8B). In a 60-min incubation with xTAF-Iβ1, sperm chromatin was decondensed as efficiently as with hTAF-Iβ.
FIG. 8.
Analysis of recombinant Xenopus TAF-Iβ. (A) Immunoblotting of cell-free translation products. Transcription-coupled translation in rabbit reticulocyte lysate was performed with no DNA (lane 1), expression plasmids of xTAF-Iβ1 (lane 2), and xTAF-Iβ2 (lane 3). Aliquots were subjected to immunoblotting with anti-TAF-Iβ antibody. (B) Decondensation activity of xTAF-I. Sperm chromatin (5 × 104 sperm) was incubated with 5 μg of hTAF-Iβ or xTAF-Iβ1 for 60 min.
TAF-I directly interacts with sperm basic proteins.
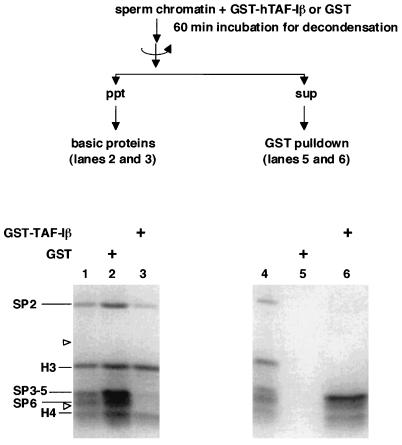
To examine protein composition of the chromatin decondensed by TAF-I, we analyzed chromatin-bound basic proteins by SDS-PAGE (Fig. 9). Before incubation with TAF-I, sperm chromatin contains sperm-specific proteins SP2 to SP6 in addition to histones H3 and H4 and small amounts of H2A and H2B. SP3 to SP5 comigrate towards each other under this condition (23). Almost all SP3 to SP6 and some of SP2 were removed from the chromatin by incubation with GST–hTAF-Iβ (Fig. 9, lane 3). These results indicate that TAF-I mediates the decondensation of Xenopus sperm chromatin by releasing sperm-specific basic proteins. We performed a GST pulldown assay to test whether TAF-I interacts directly with core histones and/or sperm proteins upon chromatin decondensation. Protein complexes formed in the decondensation reaction were precipitated with GST–hTAF-Iβ. SP3 to SP5 and SP6 were coprecipitated with GST–hTAF-Iβ (Fig. 9, lane 6). In this experiment, we detected faint bands corresponding to SP2 and histone H4. These results demonstrate that TAF-I decondenses the sperm chromatin by interacting directly with sperm-specific basic proteins and releasing them from the chromatin.
FIG. 9.
Interaction of TAF-I with chromatin basic proteins. Sperm chromatin was incubated with GST (lanes 2 and 5) or GST–hTAF-Iβ (lanes 3 and 6) under the conditions for chromatin decondensation for 60 min. The chromatin was precipitated by centrifugation, and chromatin-bound basic proteins were analyzed by SDS–15% PAGE (lanes 2 and 3). Proteins released during chromatin decondensation were subjected to a GST pulldown assay. Proteins eluted from the glutathione-Sepharose beads at 1 M NaCl were analyzed (lanes 5 and 6). Total proteins of sperm chromatin were electrophoresed in parallel (lane 1 and 4). The gel was stained with Coomassie brilliant blue. The position of sperm-specific basic proteins (SP2 to SP6) and core histones (H3 and H4) are indicated. Arrowheads on the left of the gel show the positions of marker proteins of 21.5 and 14.4 kDa.
DISCUSSION
In this paper, we have shown that histone-binding acidic protein TAF-I, which was originally identified in somatic cells, has an activity to mediate the decondensation of Xenopus sperm chromatin dependent on its acidic region. We found the β form of TAF-I in Xenopus egg and oocyte extracts. Although it remains to be examined whether xTAF-I indeed functions in sperm chromatin decondensation in eggs, we found that both xTAF-Iβ in the oocyte and egg extracts and its recombinant form are active in the chromatin decondensation assay. The extent of chromatin decondensation by purified TAF-I is comparable to that performed by egg extracts or by purified nucleoplasmin, but the time course of decondensation by TAF-I was slow (Fig. 2) (45, 46). Philpott et al. (46) observed decondensation of sperm chromatin in their nucleoplasmin-depleted egg extracts that occurred more slowly than that in the mock-depleted extracts and suggested the existence of other decondensation factors. Our observation supports this notion in that TAF-I present in egg extracts could have a redundant function to decondense sperm chromatin at a lower rate than that of nucleoplasmin.
Molecular mechanism of chromatin decondensation by TAF-I.
We have shown that the sperm chromatin incubated with TAF-I has lost most of its sperm-specific basic proteins upon chromatin decondensation. The similar change in the protein composition of chromatin occurs when sperm chromatin was incubated with egg extracts or purified nucleoplasmin (45). We also showed the direct interaction of TAF-I with those released basic proteins in solution. These results suggest that TAF-I decondenses the sperm chromatin by interacting with sperm-specific basic proteins on the chromatin and releasing them from the chromatin.
Previous experiments suggested that TAF-I interacts with histones H3 and H4 but not with H2A and H2B in isolation, while all four core histones were complexed with TAF-I when they were incubated with TAF-I (36a, 44). Among Xenopus sperm-specific basic proteins, SP3 to SP6 have a high arginine content, while SP2 has a relatively high lysine content (23). Our results indicate that TAF-I interacts strongly with SP3 to SP6 but not, if any, with SP2 (Fig. 9). If one considers the fact that histones H3 and H4 have high arginine contents, it can be concluded that TAF-I may interact with arginine-rich basic proteins. The carboxyl-terminal acidic region of TAF-I is required for the interaction with histones (44). Obviously, however, TAF-I is not acting as a simple acidic polypeptide. Our domain analyses of TAF-I in sperm chromatin decondensation indicated that the acidic region is necessary but insufficient for decondensation. The PAIR COIL algorithm (4) predicts that TAF-I contains a putative coiled-coil region between amino acid positions 21 and 65 in hTAF-Iβ. The coiled-coil structure is thought to be formed through the intermolecular hydrophobic interaction. Point mutations that impair the dimerization of TAF-I abolished the decondensation activity, suggesting the requirement of the dimerization of TAF-I. These considerations remind us of nucleoplasmin, which forms pentamer. It has been suggested that interaction between nucleoplasmin and histones requires the carboxyl-terminal acidic region of nucleoplasmin acting together as “five fingered grab” (13).
The amino-terminal α-specific region also regulates the activity of TAF-I to decondense the sperm chromatin. TAF-Iα showed much weaker activity than the α/β common region. We have also shown that TAF-Iα is less active in stimulating DNA replication of adenovirus core (39). While HeLa TAF-I consists of almost equal amounts of TAF-Iα and TAF-Iβ, no TAF-Iα was detected in Xenopus oocytes and eggs (Fig. 6), suggesting the absence of the α form of TAF-I in Xenopus early development. Consistent with this, we failed to clone the α form of TAF-I from the Xenopus oocyte cDNA library. Although a database search revealed the presence of homologues of human TAF-I/SET (39) in rat (27), puffer fish, Drosophila (26), Caenorhabditis elegans, and yeast, TAF-Iα/SETα was cloned only from humans and rats (Fig. 7B). It is therefore possible that TAF-Iα has an unknown regulatory role in mammalian cells.
It was observed that the adenoviral core proteins were not released from the adenovirus core upon incubation with TAF-I (44). Instead, incubation of the adenovirus core with TAF-I allows the viral DNA accessibility to restriction endonucleases, suggesting that TAF-I induces a structural change in the adenovirus core (references 25 and 44 and this study). The results presented in this paper indicate that in the case of Xenopus sperm chromatin, TAF-I releases sperm basic proteins through direct interaction with them. TAF-I interacts with histones H3 and H4 in solution, but TAF-I releases SP3 to SP6 rather than histones in the decondensation reaction. However, it is currently unknown whether TAF-I remodels the sperm chromatin structure by interacting with core histones on sperm chromatin in addition to releasing sperm basic proteins. An understanding of the exact nature of interaction of TAF-I with histones and other basic proteins should await the structural analysis of TAF-I, although we have shown here the role of both the acidic region and dimerization.
TAF-I and other histone chaperones.
Xenopus oocytes contain a large stockpile of proteins and mRNAs that will be used upon oocyte maturation and fertilization to midblastula transition, when zygotic transcription is repressed (9). The stored proteins in the oocytes include core histones to package newly synthesized DNA in the embryos (58). There are two complexes in the oocytes which contain histones and specific histone binding proteins: one consists of histones H2A and H2B and nucleoplasmin, and the other complex contains H3 and H4 and N1 (10, 28). Nucleoplasmin was found to be increasingly associated with maternal mRNA upon oocyte maturation (37). It remains to be examined whether xTAF-I is bound to core histones in the oocytes and eggs. Nucleoplasmin and N1 and N2 are the most abundant acidic proteins in the oocyte nucleus (30, 38). A homology to a bipartite nuclear localization signal consisting of two runs of basic amino acids separated by a spacer region, which was originally identified in nucleoplasmin, is found in hTAF-I and xTAF-I sequences (KRSSQTQNKASRKR) (41). Indeed, TAF-I was found in nuclear fractions of somatic cells and Xenopus oocytes (references 1 and 40 and this study). Our preliminary estimation indicates that 40 to 60 ng of TAF-I is present in one egg (36a). Assuming that one Xenopus egg contains 250 ng of nucleoplasmin and about 50 ng of TAF-Iβ, it is thought that the contribution of xTAF-Iβ to sperm chromatin decondensation in the eggs would be small. Indeed, we showed that depletion of TAF-I after the heat fractionation of the egg extracts resulted in the significant decrease of chromatin decondensation activity. We also proved that xTAF-I has a function in chromatin decondensation by isolating its cDNA and using the recombinant protein in the decondensation assay.
It is reported that TAF-Iβ/SET is a phosphoprotein (1). We have neither tested whether TAF-Iβ in Xenopus oocytes and eggs is phosphorylated nor detected a change in the mobility of TAF-I in oocyte and egg extracts in SDS-PAGE (Fig. 6A). Nucleoplasmin was shown to be highly phosphorylated in the eggs, and the egg nucleoplasmin is more active in sperm chromatin decondensation and nucleosome assembly than oocyte nucleoplasmin is (32, 43, 48) (Fig. 6D). Phosphorylation of acidic histone chaperones will result in the additional negative charges or induce conformational changes on these proteins, either of which might lead to a stronger interaction with basic proteins. It is of interest to examine quantitatively if native TAF-Iβ in Xenopus eggs would be more active than recombinant TAF-Iβ used in this study.
Histone binding proteins in Drosophila embryo extracts have been identified to decondense Xenopus sperm chromatin. Two heat-stable factors, p22/CRP1 and DF31/CRP2, were purified based on the decondensation activity (7, 8, 24). Drosophila NAP-I was also shown to decondense Xenopus sperm chromatin (21). Here we showed that Xenopus sperm chromatin decondensation can be mediated by yeast and mouse NAP-I proteins. NAP-I and TAF-I have high homology to each other and have some structural and functional similarities, including being acidic proteins with the carboxyl-terminal highly acidic regions; having histone binding activity (see below); nucleosome assembly activity demonstrated by the DNA supercoiling assay; the activity to stimulate DNA replication and transcription of the adenovirus core; and interacting with cyclin B in yeast and Xenopus egg extracts (15, 25, 26, 39). Interestingly, the carboxyl-terminal long acidic tail among the tripartite acidic regions of NAP-I is dispensable to form a complex with histones (14). It should be noted that NAP-I has higher affinity to histones H2A and H2B than to H3 and H4, although it can introduce negative supercoiling of DNA with H3 and H4 and DNA topoisomerase I (14, 16). In contrast, TAF-I interacts with H3 and H4 at a much higher affinity than with H2A and H2B (36a, 44). It is possible that TAF-I and NAP-I have separate roles as histone chaperones to associate with H3 and H4 and with H2A and H2B, respectively, in somatic cells, just like N1 and nucleoplasmin in Xenopus oocytes. However, it was also found that NAP-I is predominantly cytoplasmic whereas TAF-I is nuclear in somatic cells as well as in the oocytes (references 1, 26, and 40 and this study). Ito et al. reported cell cycle-specific nuclear localization of NAP-I in Drosophila embryos (19). Nuclear envelope breaks down upon oocyte maturation in Xenopus, and soluble components in the nucleus will be mixed with cytoplasmic materials. It is speculated that NAP-I and TAF-I have redundant functions with each other and/or with nucleoplasmin in chromatin decondensation.
Cellular functions of TAF-I.
We have originally identified TAF-I as an acidic host factor for DNA replication and transcription of the adenovirus core and proposed its possible function as a chromatin disassembly factor (35). Recently it was shown that TAF-I stimulates transcription from nucleosomal templates (44). TAF-I was also found to facilitate nucleosome assembly in the supercoiling assay (25), suggesting that TAF-I can act as a chromatin assembly or remodeling factor. NAP-I can be involved in promoter-specific chromatin remodeling to activate transcription from nucleosomal templates in conjunction with ATP-utilizing chromatin assembly factor, ACF (20). ACF also functions in assembly of regularly spaced nucleosomes together with CAF-I as well as with NAP-I. Considering the structural and functional similarity between TAF-I and NAP-I, one can speculate that TAF-I cooperates as a nucleosome assembly or remodeling protein with ACF or other chromatin remodeling factors. In the cell-free transcription system, TAF-I activated transcription of the adenovirus core from the E1A promoter but not from the major late promoter (36). Taken together, TAF-I can induce a local change in chromatin or chromatin-like structure as well as global chromatin remodeling such as sperm chromatin decondensation.
TAF-I/SET was identified as a protein that associates with class II human histocompatibility leukocyte antigen, with HRX, the human homologue of Drosophila Trithorax protein, and with protein phosphatase 2A (2, 54). The interaction with HRX might recruit TAF-I to specific chromosomal regions. It was reported that TAF-I inhibits protein phosphatase 2A (33, 47a), which is involved in multiple steps of cell cycle regulation. In addition, NAP-I and TAF-I/SET bind to cyclin B in Xenopus egg extracts and in yeast (26). The biological significance of these protein-protein interactions is to be elucidated. These reports, however, raise the possibility that TAF-I also plays an important role in regulatory pathways other than chromatin remodeling. It is of interest to examine the regulation of TAF-I activity during cell cycle. Future experiments on TAF-I in the Xenopus system and in somatic cells will explore these issues.
ACKNOWLEDGMENTS
We thank Yukio Ishimi for providing anti-NAP-I antibody and Yoshihiro Yoneda and Naoko Imamoto for anti-nucleoplasmin antibody. We are also grateful to Fumio Hanaoka for support at the initial stage of this work, Takeshi Mizuno for help with the fluorescence microscopy, and Keita Ohsumi for useful discussions.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan, by a grant for a Biodesign Research Program from RIKEN, and by a grant from the Naito Foundation.
REFERENCES
- 1.Adachi Y, Pavlakis G N, Copeland T D. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994;269:2258–2262. [PubMed] [Google Scholar]
- 2.Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. doi: 10.1074/jbc.272.45.28407. [DOI] [PubMed] [Google Scholar]
- 3.Almouzni G, Wolffe A P. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp Cell Res. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 4.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bulger M, Ito T, Kamakaka R T, Kadonaga J T. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc Natl Acad Sci USA. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crevel G, Cotterill S. DF31, a sperm decondensation factor from Drosophila melanogaster: purification and characterization. EMBO J. 1995;14:1711–1717. doi: 10.1002/j.1460-2075.1995.tb07160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crevel G, Huikeshoven H, Cotterill S, Simon M, Wall J, Philpott A, Laskey R A, McConnell M, Fisher P A, Berrios M. Molecular and cellular characterization of CRP1, a Drosophila chromatin decondensation protein. J Struct Biol. 1997;118:9–22. doi: 10.1006/jsbi.1996.3836. [DOI] [PubMed] [Google Scholar]
- 9.Davidson E H. Gene activity in early development. 3rd ed. Orlando, Fla: Academic Press, Inc.; 1986. [Google Scholar]
- 10.Dilworth S M, Black S J, Laskey R A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrov S, Dasso M C, Wolffe A P. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126:591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrov S, Wolffe A P. Chromatin and nuclear assembly: experimental approaches towards the reconstitution of transcriptionally active and silent states. Biochim Biophys Acta. 1995;1260:1–13. doi: 10.1016/0167-4781(94)00182-3. [DOI] [PubMed] [Google Scholar]
- 13.Dingwall C, Dilworth S M, Black S J, Kearsey S E, Cox L S, Laskey R A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem. 1992;267:20980–20986. [PubMed] [Google Scholar]
- 15.Ishimi Y, Kikuchi A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem. 1991;266:7025–7029. [PubMed] [Google Scholar]
- 16.Ishimi Y, Kojima M, Yamada M, Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur J Biochem. 1987;162:19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 17.Ishimi Y, Sato W, Kojima M, Sugasawa K, Hanaoka F, Yamada M. Rapid purification of nucleosome assembly protein (AP-I) and production of monoclonal antibodies against it. Cell Struct Funct. 1985;10:373–382. doi: 10.1247/csf.10.373. [DOI] [PubMed] [Google Scholar]
- 18.Ishimi Y, Yasuda H, Hirosumi J, Hanaoka F, Yamada M. A protein which facilitates assembly of nucleosome-like structures in vitro in mammalian cells. J Biochem (Tokyo) 1983;94:735–744. doi: 10.1093/oxfordjournals.jbchem.a134414. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Bulger M, Kobayashi R, Kadonaga J T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 21.Ito T, Tyler J K, Bulger M, Kobayashi R, Kadonaga J T. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J Biol Chem. 1996;271:25041–25048. doi: 10.1074/jbc.271.40.25041. [DOI] [PubMed] [Google Scholar]
- 22.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri C, Ohsumi K. Remodeling of sperm chromatin induced in egg extracts of amphibians. Int J Dev Biol. 1994;38:209–216. [PubMed] [Google Scholar]
- 24.Kawasaki K, Philpott A, Avilion A A, Berrios M, Fisher P A. Chromatin decondensation in Drosophila embryo extracts. J Biol Chem. 1994;269:10169–10176. [PubMed] [Google Scholar]
- 25.Kawase H, Okuwaki M, Miyaji M, Ohba R, Handa H, Ishimi Y, Fujii-Nakata T, Kikuchi A, Nagata K. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells. 1996;1:1045–1056. doi: 10.1046/j.1365-2443.1996.d01-223.x. [DOI] [PubMed] [Google Scholar]
- 26.Kellogg D R, Kikuchi A, Fujii-Nakata T, Turck C W, Murray A W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim E-G, Choi M E, Ballermann B J. Spatially restricted expression of set mRNA in developing rat kidney. Am J Physiol. 1994;266:F155–F161. doi: 10.1152/ajprenal.1994.266.1.F155. [DOI] [PubMed] [Google Scholar]
- 28.Kleinschmidt J A, Fortkamp E, Krohne G, Zentgraf H, Franke W W. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 29.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 30.Krohne G, Franke W W. Immunological identification and localization of the predominant nuclear protein of the amphibian oocyte nucleus. Proc Natl Acad Sci USA. 1980;77:1034–1038. doi: 10.1073/pnas.77.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskey R A, Honda B M, Mills A D, Finch J T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 32.Leno G H, Mills A D, Philpott A, Laskey R A. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J Biol Chem. 1996;271:7253–7256. doi: 10.1074/jbc.271.13.7253. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 34.Lohka M J, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto K, Nagata K, Ui M, Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J Biol Chem. 1993;268:10582–10587. [PubMed] [Google Scholar]
- 36.Matsumoto K, Okuwaki M, Kawase H, Handa H, Hanaoka F, Nagata K. Stimulation of DNA transcription by the replication factor from the adenovirus genome in a chromatin-like structure. J Biol Chem. 1995;270:9645–9650. doi: 10.1074/jbc.270.16.9645. [DOI] [PubMed] [Google Scholar]
- 36a.Matsumoto, K. Unpublished observations.
- 37.Meric F, Matsumoto K, Wolffe A P. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation. A role for the chaperone nucleoplasmin. J Biol Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- 38.Mills A D, Laskey R A, Black P, De Robertis E M. An acidic protein which assembles nucleosomes in vitro is the most abundant protein in Xenopus oocyte nuclei. J Mol Biol. 1980;139:561–568. doi: 10.1016/0022-2836(80)90148-5. [DOI] [PubMed] [Google Scholar]
- 38a.Miyaji-Yamaguchi M, Okuwaki M, Nagata K. Coiled-coil structure-mediated dimerization of Template Activating Factor-I is critical for its chromatin remodeling activity. J Mol Biol. 1999;290:547–557. doi: 10.1006/jmbi.1999.2898. [DOI] [PubMed] [Google Scholar]
- 39.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagata K, Saito S, Okuwaki M, Kawase H, Furuya A, Kusano A, Hanai N, Okuda A, Kikuchi A. Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res. 1998;240:274–281. doi: 10.1006/excr.1997.3930. [DOI] [PubMed] [Google Scholar]
- 41.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 42.Ohsumi K, Katagiri C. Characterization of the ooplasmic factor inducing decondensation of and protamine removal from toad sperm nuclei: involvement of nucleoplasmin. Dev Biol. 1991;148:295–305. doi: 10.1016/0012-1606(91)90338-4. [DOI] [PubMed] [Google Scholar]
- 43.Ohsumi K, Shimada A, Okumura E, Kishimoto T, Katagiri C. Dependence of removal of sperm-specific proteins from Xenopus sperm nuclei on the phosphorylation state of nucleoplasmin. Dev Growth Differ. 1995;37:329–336. doi: 10.1046/j.1440-169X.1995.t01-1-00011.x. [DOI] [PubMed] [Google Scholar]
- 44.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J Biol Chem. 1998;273:34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 45.Philpott A, Leno G H. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- 46.Philpott A, Leno G H, Laskey R A. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- 47.Poccia D. Remodeling of nucleoproteins during gametogenesis, fertilization, and early development. Int Rev Cytol. 1986;105:1–65. doi: 10.1016/s0074-7696(08)61061-x. [DOI] [PubMed] [Google Scholar]
- 47a.Saito S, Miyaji-Yamaguchi M, Shimoyama T, Nagata K. Functional domains of Template Activating Factor-I as a protein phosphatase 2A inhibitor. Biochem Biophys Res Commun. 1999;259:471–475. doi: 10.1006/bbrc.1999.0790. [DOI] [PubMed] [Google Scholar]
- 48.Sealy L, Cotten M, Chalkley R. Xenopus nucleoplasmin: egg vs. oocyte. Biochemistry. 1986;25:3064–3072. doi: 10.1021/bi00358a049. [DOI] [PubMed] [Google Scholar]
- 49.Simon R H, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 51.Smythe C, Newport J W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- 52.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 53.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 54.Vaesen M, Barnikol-Watanabe S, Gotz H, Awni L A, Cole T, Zimmermann B, Kratzin H D, Hilschmann N. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol Chem Hoppe-Seyler. 1994;375:113–126. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- 55.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 56.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolffe A P. Chromatin: structure and function. 3rd ed. London, United Kingdom: Academic Press, Ltd.; 1998. [Google Scholar]
- 58.Woodland H R, Adamson E D. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev Biol. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- 59.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]