Abstract
Random mutations were generated in the sequence for the 5′ untranslated region (5′UTR) of the Chlamydomonas reinhardtii chloroplast rps7 mRNA by PCR, the coding sequence for the mutant leaders fused upstream of the lacZ′ reporter in pUC18, and transformed into Escherichia coli, and white colonies were selected. Twelve single base pair changes were found at different positions in the rps7 5′UTR in 207 white colonies examined. Seven of the 12 mutant leaders allowed accumulation of abundant lacZ′ message. These mutant rps7 leaders were ligated into an aadA expression cassette and transformed into the chloroplast of C. reinhardtii and into E. coli. In vivo spectinomycin-resistant growth rates and in vitro aminoglycoside adenyltransferase enzyme activity varied considerably between different mutants but were remarkably similar for a given mutant expressed in the Chlamydomonas chloroplast and in E. coli. The variable effect of the mutants on aadA reporter expression and their complete abolition of lacZ′ reporter expression in E. coli suggests differences in the interaction between the 5′UTR of rps7 and aadA or lacZ′ coding regions. Several rps7 5′UTR mutations affected the predicted folding pattern of the 5′UTR by weakening the stability of stem structures. Site-directed secondary mutations generated to restore these structures in the second stem suppressed the loss of reporter activity caused by the original mutations. Additional site-directed mutations that were predicted to further strengthen (A-U→G-C) or weaken (G-C→A-U) the second stem of the rps7 leader both resulted in reduced reporter expression. This genetic evidence combined with differences between mutant and wild-type UV melting profiles and RNase T1 protection gel shifts further indicate that the predicted wild-type folding pattern in the 5′UTR is likely to play an essential role in translation initiation.
Translational regulation plays a major role in controlling the expression of chloroplast-encoded genes (7, 13, 18, 26, 29). While the translation machinery of the chloroplast shares many functional characteristics with that of its prokaryotic relatives (14, 16), several differences are beginning to emerge with respect to the well-defined Escherichia coli model (2, 11, 27, 38). Interactions between evolutionarily conserved Shine-Dalgarno (SD) sequences (GGAGG) found in the vast majority of genes in E. coli 7 ± 2 nucleotides (nt) upstream of the initiator AUG codon and anti-SD sequences near the 3′ end of the 16S rRNA (CCUCC) are known to be essential for translational initiation in this bacterium. Although the anti-SD sequences in the chloroplast 16S rRNAs are highly conserved, SD-like sequences in the leaders of chloroplast mRNAs vary significantly in location, size, and nucleotide composition or are absent altogether (3, 8, 31). Neither elimination of the variable putative SD sequences in chloroplast leaders by deletion mutagenesis (1, 19, 22, 32) or replacement mutagenesis (8, 21) nor insertion of canonical SD sequences (8) has significant effects on chloroplast gene expression, indicating that translation initiation in the chloroplast occurs largely in an SD-independent manner. Surprisingly, the same chloroplast reporter constructs lacking SD sequences are also expressed efficiently in E. coli and addition of canonical SD sequences to the leaders of these constructs only modestly enhances translation of these mRNAs by the bacterial protein synthesizing system (8).
In contrast to the majority of E. coli mRNAs, chloroplast mRNAs contain fairly long, AU-rich 5′ untranslated regions (5′UTRs) with little primary sequence conservation that appear to play a major role in the regulation of translation initiation (10, 13, 18). Analyses of representative chloroplast 5′UTRs, to determine likely secondary structure based on minimum energy models (42), predict that these leaders are highly folded, with several having large stem-loop structures directly upstream of the initiation codon (10, 18). Interactions between these structures and the translational apparatus of the chloroplast presumably allow for the regulation of translation initiation.
We have carried out a random PCR mutagenesis on the sequence for the 5′UTR of the chloroplast rps7 gene from Chlamydomonas reinhardtii that specifies a protein of the small subunit of the chloroplast ribosome. The population of rps7 mutant leaders was then fused to the E. coli lacZ′ coding sequence in a pUC18 plasmid, and bacterial transformants that were unable to express β-galactosidase on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates were identified as white colonies. Twelve of the 207 white colony mutants were found to have alterations in the coding sequence for the first 225 nt upstream of the initiation AUG codon of the rps7 5′UTR. Seven rps7 mutants that did not alter levels of lacZ′ mRNA accumulation affected expression of the aminoglycoside adenyltransferase (AAD) reporter protein to various degrees in both the C. reinhardtii chloroplast and in E. coli. The marked difference between the effect of a given mutant rps7 leader on translation of the lacZ′ and aadA reporter mRNAs in E. coli suggests that interactions between the coding sequences and the 5′UTR sequence play a role in the regulation of translation initiation. Three of the seven mutants altered the predicted structure of the second stem-loop upstream of the AUG initiation codon. Complementary nucleotide changes made in these mutants to reconstitute this predicted wild-type stem-loop structure restored spectinomycin-resistant growth and AAD enzyme activity. Additionally, site-directed mutants with base pair changes that strengthened or weakened the second predicted stem-loop of the rps7 5′UTR decreased growth and AAD enzyme activity in the C. reinhardtii chloroplast. These data strongly suggest that the secondary structure of this rps7 leader in vivo involves these predicted stem-loop structures that are essential for normal translational regulation.
Changes in the predicted secondary and tertiary structures of the mutant 5′UTRs were also supported by differences in the RNA melting-reannealing profiles and RNase T1 gel shift protection patterns from those of wild-type and suppressed mutant strains. The combined genetic and physical evidence supports the hypothesis that the wild-type rps7 leader sequence resulting in the predicted folding pattern for the second stem-loop is essential for normal function.
MATERIALS AND METHODS
Strains and culture conditions.
The C. reinhardtii atpB deletion mutant ac-u-c-2-21 (5) was used as the recipient for chloroplast transformation. Strain CC-3277 carried the chloroplast reporter construct rps7::aadA::rbcL with the wild-type rps7 leader (8). This strain and derived strains carrying rps7 leader mutations fused to the aadA reporter construct (with CC prefix) and plasmids with the mutant chimeric constructs (with P prefix) described in this paper are available from Elizabeth Harris (Chlamydomonas Genetics Center, Duke University, Durham, N.C.). C. reinhardtii and E. coli strains were grown and harvested for transformation and biochemical and molecular analyses as described previously (8, 15).
Mutagenesis of the rps7 chloroplast leader.
The chloroplast rps7::aadA::rbcL reporter gene (P-655) was constructed in pUC18 as described previously (8). To generate loss-of-function mutants, random mutagenesis was conducted in vitro on the 625-nt fragment from P-655 upstream of the translation start site (AUG) in the chloroplast rps7 construct from C. reinhardtii. This fragment was shown to contain the 266-nt 5′UTR by primer extension (data not shown) as well as the chloroplast promoter and an upstream transcriptional enhancer (8a). Mutagenic PCR mixtures (24, 41) included 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM ZnCl2, 62 μM deoxynucleoside triphosphates (dNTPs; a 20 μM concentration of each of three dNTPs and a 2 μM concentration of a fourth to generate a 10:1 nucleotide imbalance), 10 μl (0.1 ng/μl) of rps7 5′ leader template, 30 pmol of each of the primers, 10 μl of 2-mercaptoethanol, 10 μl of dimethyl sulfoxide, 5 U of Taq polymerase, and distilled water to 200 μl. Thirty PCR cycles of 94°C for 1 min, 59°C for 2 min, and 72°C for 3 min were carried out.
A population of the mutagenized 625-nt rps7 fragments was inserted between the lac promoter and the lacZ′ coding sequence by using XhoI and NcoI restriction sites in pUC18 and Ampr transformants selected in E. coli. Recombinant E. coli strains containing mutant rps7 fragments blocking expression of lacZ′ mRNA were identified as white colonies by an X-Gal plate assay (33). The first 225 nt of the 266-nt A-T-rich rps7 leader region from 207 white colony mutants were sequenced (A-T bases only) from double-stranded template by using the Sequenase II system (Amersham) to determine if specific alterations were introduced in this region. The 625-nt rps7 fragments from the 12 mutants identified were then sequenced by using appropriate primers and the Perkin-Elmer Dye Terminator Cycle Sequencing System with AmpliTaq DNA polymerase on an ABI Prism DNA sequencer. The 625-nt rps7 fragments containing mutations were reinserted into the chloroplast aadA expression vector (P-655), replacing the excised wild-type rps7 fragment as described previously (8).
Selected mutant rps7 leaders were subjected to site-directed PCR mutagenesis by using a Stratagene QuickChange site-directed mutagenesis kit. Primers for mutagenic amplification of the DNA fragments included the following: −36A→G (5′TCTATTTAACCGTTTAGTAAAAT3′), −36A→C (5′TCTATTTAACCCTTTAGTAAAAT3′), −92A→G (5′GTCCTATTAAGCCATCACATAAC3′), −65U→A (5′AATTGCTTATTAGGTATGAAAG3′), and −62U→A (5′GCTTATTTGGAATGAAAGTTTG3′). To generate the mutant weakening the second stem-loop structure, primers with nucleotide changes at −63G→A and −64G→A (5′CTAAAATTGCTTTAATATGAAAGTTTG3′) as well as at −91C→U and −90C→U (5′GTCCTATTAAATTATCACATAAC3′) were used. To create the mutant strengthening the second stem-loop, we employed primers to generate nucleotide changes at −83U→C and −82A→G (5′AACCATCACACGACTAAATTG3′) as well as at −69U→C and −68A→G (5′ACTAAAATTGCTCGTTTGGTATG3′). Mutant nucleotides are indicated in italics. The presence of the new mutations at position −36, of the second site mutations at positions −62, −65, and −92, and of the stem-weakening and -strengthening mutations in these rps7 fragments was verified by DNA sequence analysis with the Sequenase II system, and the site-directed mutant fragments were cloned into the aadA expression vector P-655 as described above.
To determine whether alterations of G-C base pairs in the first 225 nt of the rps7 leader or nucleotide changes elsewhere in the 400 nt upstream of the mutagenized fragment were blocking expression of the lacZ′ reporter in the remaining white colony mutants, we sequenced the entire 625-nt fragment in 20 additional randomly selected isolates by using the ABI sequencer.
Transformation of the aadA reporter constructs with mutant rps7 leaders into the C. reinhardtii chloroplast and into E. coli.
The nonphotosynthetic C. reinhardtii strain CC-373 was transformed by the biolistic method (4) with DNA from a chimeric pUC18 plasmid containing the wild-type atpB gene fused to aadA reporter constructs with mutant rps7 leaders (8). Photosynthetically competent transformants were selected on minimal HS medium as previously described (4, 5, 8). Homologous integration of the donor constructs at the atpB locus of the recipients’ chloroplast genome restored the function of the single-copy atpB gene. Transformants homoplasmic for the donor atpB gene were obtained after relatively few cell generations on HS medium (5). The linked reporter constructs, which integrate into the adjacent inverted repeat sequence, copy correct at high frequency (4). Continued selection of the wild-type rps7::aadA::rbcL chloroplast transformants on spectinomycin resulted in cells homoplasmic for two copies of this insert, whereas three or four rounds of subcloning in the absence of spectinomycin were necessary to obtain isolates homoplasmic for two copies of the insert with mutant leaders. Once homoplasmic isolates were obtained, they appeared to be stable. Only a single transformant with each mutant reporter construct was analyzed since previous studies demonstrated that insertion of the aadA reporter at this site in the chloroplast genome yielded independent transformants with no significant variation in AAD expression (8). E. coli cells (XL1-Blue) were transformed by electroporation with the same mutant reporter constructs, and transformants were selected on Luria-Bertani (LB) plates containing 100 μg of ampicillin per ml by using standard techniques (33). Chimeric constructs in E. coli were maintained as multicopy plasmids under ampicillin selection at 100 μg/ml. A single E. coli transformant was analyzed for each mutant leader since individual transformants of the chimeric reporter construct display minimal variability in expression (8).
Northern and Southern analysis of the transformants and primer extension.
DNA was isolated from transformed cells of E. coli (33) and C. reinhardtii (23) and digested with BamHI. Chloroplast DNA fragments were separated on 0.9% agarose gels, transferred to a Magna NT nylon membrane (MSI Scientific) with a Stratagene blot apparatus, and probed with the cloned chloroplast BamHI fragment 10 that covers the site of integration (8). RNA was extracted from E. coli and C. reinhardtii cells with 4 M guanidine isothiocyanate–25 mM sodium citrate (pH 7.0)–0.1 M 2-mercaptoethanol–0.05% Sarkosyl as described previously (6). RNA was separated on 1.1% agarose–2.2 M formaldehyde gels, blotted onto Magna NT membranes (MSI Scientific), and probed with an 0.81-kb aadA fragment or an 0.41-kb lacZ′ fragment from the respective coding sequences. For each DNA and RNA sample, nucleic acid concentrations were measured spectrophotometrically and standardized to ensure equal loading of lanes. Stringent hybridization and washing conditions were used on both the DNA and RNA blots. The probes were labeled with [α-32P]dATP (NEN) by using a random priming kit (Boehringer Mannheim). Blots were exposed to X-ray film (Kodak X OMAT-AR) at −70°C. RNA blots were also quantified with a Molecular Dynamics PhosphorImager. Primer extension analysis of the wild-type rps7 5′UTR was conducted to locate the transcription initiation site. These reactions employing a primer complementary to the amino-terminal end of the aadA coding sequence 5′-CGATCACCGCTTCCCTCAT-3′ and 10 U of avian myeloblastosis virus reverse transcriptase were run at 42°C for 60 min. Reactions were displayed on both a 5% polyacrylamide gel with DNA size standards and by running them next to the corresponding DNA sequence on a 6% polyacrylamide–8 M urea sequencing gel as described before (34).
Growth and enzyme activity assays.
Growth rates of 1.0-ml cultures of representative C. reinhardtii and E. coli rps7::aadA transformants with each rps7 5′UTR mutation were measured in 24-well microtiter plates (Falcon no. 3047) by the increase in A740 or A650, respectively, as described previously (8, 9). The AAD enzyme activity of these transformants was measured as the capacity to transfer α-32P-labeled ATP to spectinomycin by binding the spectinomycin breakdown product to chromatography paper (8, 12). β-Galactosidase activity was assayed for the original rps7::lacZ′ E. coli transformants by standard techniques (33).
Predicted RNA secondary structures.
RNA secondary structures were predicted by using the MFOLD algorithm available online (42a). Several wild-type rps7 structures were examined; they included the 625-nt upstream sequence that was mutagenized and is transcribed in E. coli, the 266-nt full-length 5′UTR, and the truncated 225-nt 5′UTR sequence. Additionally, the 266-nt 5′UTR sequence upstream of the aadA or the lacZ′ reporter sequence was examined for alteration of the predicted stem-loop structures by association with the downstream reporter RNA. In all of the constructs described above, the predicted wild-type secondary structure of the first five stem-loops corresponding to the 225 nt proximal to the AUG initiation codon (Fig. 1) are maintained. The MFOLD analyses were conducted at 25°C with 5% suboptimability in 1 M NaCl (default conditions). Additional predicted structures examined at 37°C displayed a similar global folding pattern with alterations in local stem-loop structures expected, given the increase in available free energy.
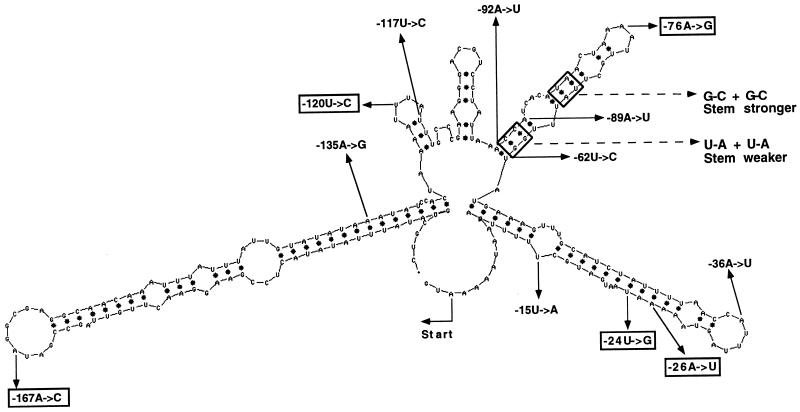
FIG. 1.
MFOLD prediction of partial secondary structure at 25°C of the first 225 nt proximal to the ATG initiation codon of the 5′UTR from the wild-type chloroplast rps7 gene of C. reinhardtii. The location and base pair changes of the 12 randomly induced mutations blocking expression of the lacZ′ reporter are indicated. Mutations that fail to accumulate normal levels of lacZ′ reporter mRNA are boxed, whereas those specifically affecting translation of lacZ′ mRNA are not boxed. The position of the site-directed mutants strengthening and weakening the second predicted stem-loop are indicated by boxes and dashed arrows.
UV melting-reannealing profile analyses.
RNA was synthesized in vitro from an SKII+ plasmid with the T7 promoter upstream of sequences corresponding to (i) the 266-nt wild-type rps7 5′UTR (P-833), (ii) three of the mutant 5′UTRs affecting the second predicted stem (−62U→C [P-848], −89A→U [P-849], and −92A→U [P-850]), (iii) three of the 5′UTRs with the corresponding suppressor double mutants (−62U→C and −92A→G [P-851], −89A→U and −65U→A [P-852], and −92A→U and −62U→A [P-853]), (iv) 5′UTRs with the three mutants at nucleotide position −36 (A→C [P-854]), A→G [P-855], and A→U [P-856]), and (v) 5′UTRs with the second stem-strengthening (double G-C [P-857]) and weakening (double A-U [P-858]) mutants. Each of the 20-ml reaction mixtures contained 4 mM each NTP, 25 mM MgCl2, 25 mM NaCl, 50 mM Tris-HCl (pH 8.1), 20 mM dithiothreitol, 50 nM T7 plasmid promoter template DNA, and 100 nM T7 RNA polymerase as described previously (22). Transcription reaction mixtures were extracted with phenol-chloroform and run over an 8 M urea–8% (wt/vol) polyacrylamide gel, excised, and electroeluted. The A260 was measured over a 20 to 80°C range at 2°C increments changing at 1°C/min on a Shimadzu TCC-260 spectrophotometer. Melting and reannealing profiles were obtained, and derivative values were determined to identify regions of varied hyperchromicity and changes in the temperature of the transition from primarily secondary structure to primarily tertiary structure unfolding (22).
RNase T1 protection gel shift assays.
Radiolabeled RNA leaders corresponding to the wild-type and mutant rps7 5′UTRs were synthesized in 20-μl reaction mixtures containing 1 μg of the linearized plasmid RNA synthesis templates as described above in 40 mM Tris-HCl (pH 7.5)–6 mM MgCl2–2 mM Spermidine (Sigma)–10 mM dithiothreitol–20 U of RNasin (Promega)–50 μCi of [α-32P]UTP (DuPont NEN)–12 μM nonradiolabeled UTP–an 0.3 mM concentration each of ATP, CTP, and GTP–20 U of T7 RNA polymerase for 1 h at 37°C. Five units of RNase-free DNase I (Sigma) was added, and the reaction mixtures were incubated for an additional 10 min at 37°C. RNAs were labeled to specific activities of 1 × 109 to 2 × 109 cpm/μg. The reaction mixtures were then extracted with phenol-chloroform and separated from unincorporated nucleotides on Sephadex G-25 spun columns as described previously (17).
Proteins used in the gel mobility retardation assays were obtained from a CC-400 (cell-wall-free) C. reinhardtii strain grown as described above and gently broken in a Yeda press (15). The preparation was enriched for chloroplasts by centrifugation over a 10 to 80% Percoll gradient (40), and nucleic acid binding proteins were separated by heparin-Actigel chromatography as described previously (17).
For the RNase T1 gel mobility shift assays, 50-μl reaction mixtures containing approximately 5 ng of the [32P]UTP-labeled RNA, 2.5 μg of yeast total RNA (to eliminate nonspecific binding), and 7 μg of the pooled heparin-Actigel protein extracts were incubated for 30 min at 25°C, treated with 10 U of RNase T1 (BRL) for 10 min, and electrophoresed on a 5% native polyacrylamide gel in 1× Tris-borate-EDTA TBE buffer. RNAs and protein-retarded RNA-protein bands were visualized by autoradiography as described before (17). RNA-protein binding competition experiments were conducted over a 0- to 100-fold range as described previously (17).
RESULTS
Isolation of random mutations in the chloroplast rps7 leader in E. coli.
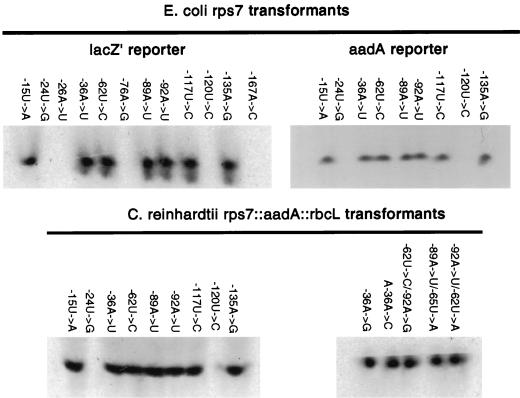
To identify regions of the 266-nt 5′UTR of the chloroplast rps7 gene from C. reinhardtii necessary for the initiation of translation, random PCR mutagenesis was conducted on the 625-nt wild-type fragment upstream of the AUG initiation codon. The population of amplified molecules was then purified on a 0.9% agarose gel and ligated into a pUC18 vector between the lac promoter and the lacZ′ coding sequence. These constructs were transformed into E. coli XL1-Blue competent cells, and ampicillin-resistant colonies were selected. Transformed isolates unable to produce β-galactosidase were identified as white colonies. Partial sequencing (A and T bases only) of the first 225 nt of the rps7 fragment upstream of the translation start site (greater than 70% A+T) from 207 of these white colonies identified 12 single base changes within the first 167 nt, each at a different position (Fig. 1). These changes were confirmed to be the only alterations present by determining the sequence of the entire 625-nt fragment mutagenized for each of the 12 mutants. A quantitative analysis of β-galactosidase activity revealed that the 12 white colony mutant strains had less than 6% of the enzyme activity level found in E. coli transformants with the wild-type rps7 5′UTR fused to lacZ′ (Table 1). RNA blot analysis demonstrated that 7 of the 12 mutants accumulated abundant rps7::lacZ′ mRNA (Fig. 2). These same seven mutants also accumulated abundant rps7::aadA mRNA (Fig. 2). Sequence analysis of the entire 625-nt rps7 fragment from 20 additional randomly selected white colony mutants with no A or T alterations in the first 225 nt revealed no further nucleotide changes.
TABLE 1.
Function of the mutant rps7::reporter constructs in E. coli
| Transformant | Nucleotide change |
lacZ′ expression
|
aadA expression
|
|||
|---|---|---|---|---|---|---|
| mRNAa | βGalb | mRNAa | Specr growthcd | AADde | ||
| P-780 | −15U→A | 77 | 4.9 ± 0.2 | 86 | 13 ± 2.4 | 11 ± 3.6 |
| P-781 | −24U→G | 1 | 2.6 ± 0.3 | 2 | NDf | ND |
| P-782 | −26A→U | 3 | 2.3 ± 0.2 | ND | ND | ND |
| P-783 | −36A→U | 90 | 5.5 ± 0.5 | 91 | 7 ± 2.1 | 5 ± 2.8 |
| P-784 | −62U→C | 98 | 4.2 ± 0.3 | 93 | 71 ± 1.8 | 66 ± 9.5 |
| P-785 | −76A→G | 1 | 2.2 ± 0.1 | ND | ND | ND |
| P-786 | −89A→U | 82 | 4.7 ± 0.3 | 94 | 33 ± 2.5 | 41 ± 6.9 |
| P-787 | −92A→U | 93 | 3.8 ± 0.4 | 96 | 21 ± 1.4 | 29 ± 6.0 |
| P-788 | −117U→C | 100 | 5.2 ± 0.5 | 99 | 72 ± 3.0 | 77 ± 11.1 |
| P-789 | −120U→C | 2 | 2.4 ± 0.2 | 1 | ND | ND |
| P-790 | −135A→G | 91 | 4.0 ± 0.4 | 100 | 92 ± 2.7 | 97 ± 8.8 |
| P-791 | −167A→C | 2 | 2.6 ± 0.3 | ND | ND | ND |
mRNA accumulation = percentage of mutant with highest value.
βGal activity = mean ± standard error of β-galactosidase activity as determined by spectrophotometric analysis of three independent cultures for whole-cell extracts adjusted to eliminate background levels (33).
Specr growth = mean ± standard error of the second derivative of the time to half-maximal inhibition (t1/2) for all concentrations of spectinomycin (8) for three independent cultures.
Values are standardized to those of E. coli transformants with the wild-type rps7 reporter construct as 100%.
AAD activity = mean ± standard error for three trials measured in microcuries per minute per microgram of S100 protein (8).
ND, not determined.
FIG. 2.
RNA blots of E. coli transformants and C. reinhardtii chloroplast transformants carrying the rps7::reporter constructs. E. coli blots were probed with a 0.41-kb lacZ′ and a 0.81-kb aadA fragment from the respective coding regions. C. reinhardtii blots were probed with the same 0.81-kb aadA fragment.
Functional analysis of the mutant rps7 leaders using the aadA reporter construct in chloroplast transformants of C. reinhardtii and in E. coli.
The DNA sequences encoding the seven mutant rps7 leaders that allowed normal or near-normal accumulation of lacZ′ mRNA in E. coli were subcloned into the C. reinhardtii chloroplast transformation cassette rps7::aadA::rbcL linked to a wild-type atpB gene, replacing the wild-type rps7 leader sequence. These chimeric plasmids were transformed into the C. reinhardtii recipient strain CC-373, an atpB partial deletion mutant, to complement the photosynthetic defect in this strain (4, 8). Homologous integration of the donor DNA fragment introduces the aadA reporter construct into one copy of the inverted repeat adjacent to the single-copy atpB gene, and the reporter sequence then copy corrects and segregates to homoplasmicity to yield progeny with one copy of the wild-type atpB gene and two copies of the rps7::aadA::rbcL construct. Homoplasmicity was confirmed by digesting total DNA from each subclone of the C. reinhardtii chloroplast transformants as well as the CC-373 recipient strain with BamHI and probing with the cloned chloroplast BamHI fragment 10 spanning this region (8). The rps7 leaders with the −24U→G and −120U→C mutations that failed to accumulate lacZ′ mRNA in E. coli were similarly ligated into the C. reinhardtii transformation cassette, transformed into the chloroplast, and subcloned to homoplasmicity. This set of nine mutant rps7::aadA constructs ligated into pUC18 by using XhoI and NcoI restriction sites was also transformed into E. coli for analysis of reporter gene expression.
Accumulation of the aadA reporter mRNA was analyzed for all nine mutant reporter constructs expressed in E. coli (Table 1) and in the chloroplast of C. reinhardtii (Table 2). Those mutant rps7 leaders that supported accumulation of the lacZ′ or aadA reporter mRNA in E. coli also permitted accumulation of aadA mRNA at high levels in the chloroplast. Similarly, the rps7 leader mutants with the −24U→G and −120U→C alterations that failed to accumulate lacZ′ mRNA in E. coli also failed to accumulate aadA mRNA in C. reinhardtii when fused to this reporter construct.
TABLE 2.
Function of the mutant rps7::aadA::rbcL reporter constructs in the chloroplast of C. reinhardtii
| Transformant | Nucleotide change |
aadA expression
|
||
|---|---|---|---|---|
| mRNAa | Specr growthbc | AAD activitycd | ||
| CC-3684 | −15U→A | 100 | 17 ± 1.9 | 19 ± 3.2 |
| CC-3685 | −24U→G | 2 | NDe | ND |
| CC-3686 | −36A→U | 88 | 13 ± 2.2 | 14 ± 4.3 |
| CC-3687 | −62U→C | 97 | 63 ± 2.0 | 58 ± 6.9 |
| CC-3688 | −89A→U | 94 | 26 ± 2.7 | 30 ± 3.4 |
| CC-3689 | −92A→U | 100 | 25 ± 4.6 | 23 ± 3.8 |
| CC-3690 | −117U→C | 94 | 70 ± 2.4 | 67 ± 7.2 |
| CC-3691 | −120U→C | 3 | ND | ND |
| CC-3692 | −135A→G | 93 | 98 ± 3.5 | 95 ± 7.1 |
mRNA accumulation = percentage of mutant with highest value.
Specr growth = mean ± standard error of the second derivative of the time to half-maximal inhibition (t1/2) for all concentrations of spectinomycin (8) for three independent cultures.
Values are standardized to those of C. reinhardtii chloroplast transformants with the wild-type rps7 reporter construct as 100%.
AAD activity = mean ± standard error for three trials measured in microcuries per minute per microgram of S100 protein (8).
ND, not determined.
Assay of aadA reporter activity.
The homoplasmic chloroplast transformants with mutant rps7 leaders fused to the aadA reporter were analyzed for their ability to express the AAD protein by two independent tests. In vivo growth rates for each of the mutant strains were determined with spectinomycin concentrations ranging from 0 to 500 μg/ml as described previously (8). We showed previously that spectinomycin-resistant growth was directly correlated with the level of AAD protein activity (8). The spectinomycin-resistant growth rates for chloroplast transformants with the seven rps7 leader mutants that accumulated normal levels of aadA mRNA varied from 13 to 98% that of the control strain with the wild-type rps7 leader (Table 2). The activity of the AAD reporter enzyme in the aadA transformants was measured in cell extracts (8, 12) as microcuries of 32P-labeled product bound and was found to be linear over the 40-min assay period up to 100 μg of S100 protein per ml. The in vitro enzyme activity of chloroplast transformants carrying the rps7 mutants was highly correlated with their respective spectinomycin-resistant growth rates (Table 2). In both assays, the −36A→U mutation was the most deleterious change and the −135A→G mutation was the least detrimental.
Each of the nine mutant rps7::aadA::rbcL constructs was also transformed into E. coli XL1-Blue cells. Comparable in vivo growth assays in spectinomycin at concentrations ranging from 0 to 100 μg/ml and in vitro enzyme activity assays (Table 1) were conducted as described before (8). A clear correlation between diminished spectinomycin-resistant growth and AAD enzyme activity for each of the rps7 leader mutants is apparent in the E. coli as well as the C. reinhardtii chloroplast transformants. In both organisms, the −36A→U mutant is the most impaired and the −135A→G mutant is the least affected compared to the wild type (compare Tables 1 and 2). We found that rps7 leader mutations that result in the apparent loss of lacZ′ expression in E. coli (white colonies on X-Gal plates, <6% of the enzyme activity of the construct with the wild-type rps7 leader) can permit widely varying levels of aadA reporter expression in both E. coli and the C. reinhardtii chloroplast (<10% to >90% of the aadA construct with the wild-type rps7 leader). Differences in interactions of the mutant rps7 leaders with the respective lacZ′ and aadA coding sequences in E. coli might explain these differences in reporter gene expression, either by causing alterations in the global folding of the mRNA or by promoting the differential ability of the mRNAs to recruit factors to the translation preinitiation complex.
Generation and analysis of site-directed alterations in the mutant rps7 leaders.
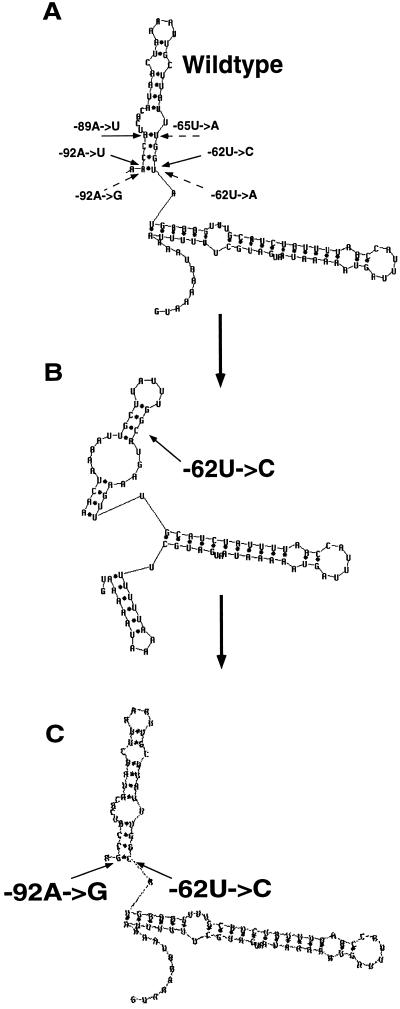
We used site-directed mutagenesis to determine whether nucleotide changes in four of the mutant rps7 leaders could restore their substantially reduced function. Second site −92A→G, −65U→A, and −62U→A alterations in the leader were tested for their ability to complement respectively the −62U→C, −89A→U, and −92A→U mutants that are predicted to reduce pairing in the second stem-loop (Fig. 3A). In each case, the alterations were designed to restore the putative wild-type rps7 secondary stem structure as is represented for the −62U→C mutation (Fig. 3B) and −92 A→G second site complementary nucleotide change (Fig. 3C). Since the −36A→U mutant falls in a predicted loop (Fig. 1), we generated −36A→G and −36A→C mutant constructs to examine the effects of all 4 nt at this position. Each of the three rps7::aadA::rbcL constructs containing both the original mutation and the complementary second site change, as well as the −36A→G and −36A→C alterations, was transformed into the chloroplast of C. reinhardtii and subcloned to homoplasmicity, and spectinomycin-resistant growth and AAD reporter enzyme activity were determined on individual representative isolates (Table 3). While −36A→G and −36A→C mutations failed to restore wild-type levels of aadA expression, the rps7 leader with the −36A→G alteration, a purine-for-purine replacement, is at least twice as competent at promoting translation as those with either the −36A→U or the −36A→C substitutions. Clearly, the wild-type sequence at nucleotide −36 is necessary for full expression of the rps7 leader.
FIG. 3.
MFOLD prediction of the partial secondary structure at 25°C of the first two stem-loop structures proximal to the AUG initiation codon of the rps7 5′UTR. (A) The wild-type rps7 5′UTR with three of the original random mutations, −62U→C, −89A→U, and −92A→U (solid arrows), and the complementary −92A→G, −65U→A, and −62U→A second site mutations (dashed arrows) designed to restore the second stem-loop structure are indicated. (B) The partial secondary structure of the rps7 5′UTR with the −62U→C mutation. (C) The partial secondary structure of the rps7 5′UTR with the −62U→C −92A→G double mutation. The other two single and double mutant combinations shown in panel A display similar alterations in the size of the first two predicted stem-loop structures as in the −62U→C mutant in panel B and the same restoration of the predicted wild-type secondary structure by the double mutant suppressor in panel C that displays normal reporter expression.
TABLE 3.
Function of second-site alterations of the mutant rps7 reporter constructs in the chloroplast of C. reinhardtii and in E. coli
| C. reinhardtii strain | E. coli transformant | Nucleotide change(s) |
C. reinhardtii
|
E. coli
|
|||
|---|---|---|---|---|---|---|---|
| aadA mRNAa | Specr growthbc | AADcd | βGale | AADcd | |||
| CC-3693 | P-792 | −36A→G | 83 | 32 ± 2.5 | 41 ± 4.5 | 4.7 ± 0.3 | 9 ± 1.2 |
| CC-3694 | P-793 | −36A→C | 100 | 8 ± 2.2 | 15 ± 3.3 | 4.1 ± 0.2 | 4 ± 0.8 |
| CC-3695 | P-794 | −62U→C, −92A→G | 96 | 79 ± 2.8 | 86 ± 8.7 | 76 ± 6.6 | 80 ± 6.7 |
| CC-3696 | P-795 | −89A→U, −65U→A | 89 | 104 ± 3.0 | 99 ± 9.2 | 92 ± 8.4 | 88 ± 5.5 |
| CC-3697 | P-796 | −92A→U, −62U→A | 90 | 93 ± 2.4 | 85 ± 8.9 | 87 ± 7.0 | 75 ± 9.4 |
mRNA accumulation = percentage of mutant with highest value.
Specr growth = mean ± standard error of the second derivative of the time to half-maximal inhibition (t1/2) for all concentrations of spectinomycin (8) for three independent cultures.
Values are standardized to those of C. reinhardtii chloroplast or E. coli transformants with the wild-type rps7 reporter construct as 100%.
AAD activity = mean ± standard error for three trials measured in microcuries per minute per microgram of S100 protein (8).
βGal activity = mean ± standard error of β-galactosidase activity as determined by spectrophotometric analysis of three independent cultures for whole-cell extracts adjusted to eliminate background levels (33).
The −92A→G and −62U→A second site mutations fall on opposite sides of the second extended stem structure predicted for the wild-type rps7 leader (Fig. 1). The presence of both mutations restores normal rps7 leader function, resulting in spectinomycin-resistant growth and AAD enzyme activity levels only moderately below those of the wild-type rps7 construct in the chloroplast aadA transformants (Table 3). The −65U→A second site mutation falls in the middle of the second predicted extended stem (Fig. 3A) and almost fully restores the loss of activity caused by the −89A→U alteration. The effects of secondary mutations were not examined in the −15U→A mutant, which falls in a predicted bulge, and the upstream −117U→C and −135A→G mutants that displayed aadA reporter expression closest to that of the wild type.
The same rps7 5′UTR constructs with the five site-directed mutations were also examined in E. coli by using both the aadA and lacZ′ reporters. A quantitative analysis of representative E. coli transformants with each of these mutations revealed that the alterations at the −36 position (−36A→G and −36A→C) failed to restore expression for either reporter (expression < 10% of transformants with the wild-type rps7 5′UTR for each reporter), whereas the three transformants carrying second site mutations designed to reform predicted stem-loop structures expressed both of the reporters at or near wild-type levels (Table 3).
Analysis of site-directed mutants designed to weaken or strengthen pairing in the predicted second stem-loop.
To determine the effects of mutations that either weaken or strengthen the second predicted stem without disrupting the predicted global folding pattern of the rps7 5′UTR, we generated two sets of site-directed mutants. We replaced two predicted A-U pairs with G-C pairs to make the stem stronger (A−68-U−82→G−68-C−82 and U−69-A−83→G−69-C−83) and replaced two predicted G-C pairs with A-U pairs to make the stem weaker (G−63-C−91→A−63-U−91 and G−64-C−90→A−64-U−90) (Fig. 1). 5′UTRs with both of these sets of alterations were placed upstream of the aadA reporter in P-655 and transformed into the chloroplast of C. reinhardtii. Homoplasmic transformants were analyzed by using the same growth and AAD enzyme activity assays described above. Either strengthening or weakening the stem beyond its predicted wild-type configuration had a deleterious effect on the translational activity of the 5′UTR. The stem-strengthening mutant with two adjacent (A-U→G-C) pairs created in the second stem-loop had a calculated increase in available free energy (ΔΔG = −4.8 kcal mol−1) and displayed spectinomycin-resistant growth at only 57% and AAD enzyme activity at only 61% of that of the wild type. The stem-weakening mutant with two adjacent (G-C→A-U) pairs had a calculated decrease in available free energy (ΔΔG = +3.9 kcal mol−1) and displayed spectinomycin-resistant growth at only 73% and AAD enzyme activity at only 63% of that of the wild type (Table 4).
TABLE 4.
Effect of mutations predicted to strengthen or weaken the second stem of the rps7 5′UTR on the expression of the aadA reporter constructs in the C. reinhardtii chloroplast
| C. reinhardtii strain | E. coli plasmid | Changes in paired bases in second stem |
C. reinhardtii chloroplast
|
|
|---|---|---|---|---|
| Specr growthab | AADbc | |||
| CC-3721 | P-802 | G−63-C−91→A−63-U−91 | 73 ± 1.9 | 63 ± 7.0 |
| G−64-C−90→A−64-U−90 | ||||
| CC-3722 | P-803 | A−68-U−82→G−68-C−82 | 57 ± 2.6 | 61 ± 8.3 |
| U−69-A−83→G−69-C−83 | ||||
Specr growth = mean ± standard error of the second derivative of the time to half-maximal inhibition (t1/2) for all concentrations of spectinomycin (8) for three independent cultures.
Values are standardized to those of C. reinhardtii chloroplast with the wild-type rps7 reporter construct as 100%.
AAD activity = mean ± standard error for three trials measured in microcuries per minute per microgram of S100 protein (8).
Melting-reannealing profile analyses of rps7 5′UTRs from the wild type, mutants, and designed suppressors.
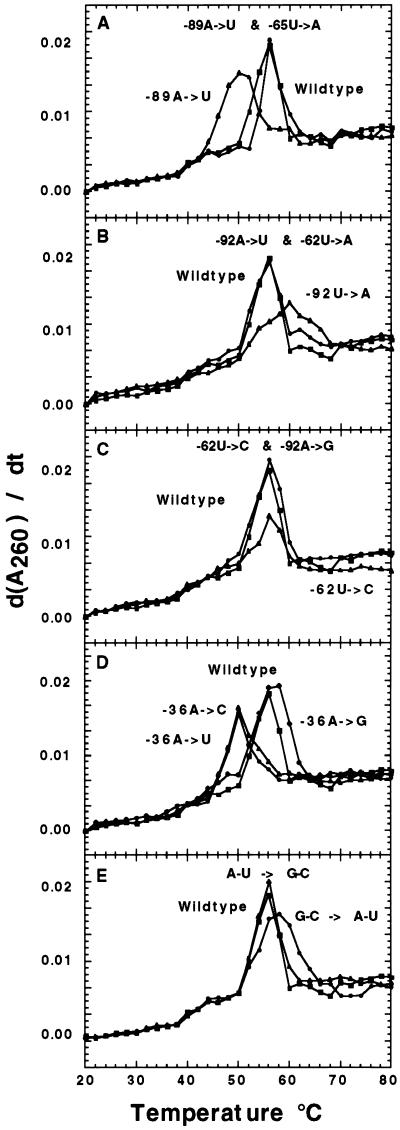
Melting and reannealing profiles of the wild-type and 11 mutant 266-nt rps7 5′UTR RNAs were examined for both the hyperchromicity (peak of the derivative curve) and the temperature at which the transition from predominantly tertiary to predominantly secondary structure melting occurs. Changes in these parameters are indicative of differences in the global folding structure between the wild-type and the mutant RNAs (22). Recovery of the wild-type melting-reannealing profile in the double mutant suppressors that were designed to restore predicted stem structures would provide further evidence that the predicted secondary structure is valid. Our melting-reannealing data indicate an alteration of wild-type secondary and tertiary folding in each of the original mutants and a near-wild-type recovery in the designed second-site suppressors (Fig. 4). The −89A→U mutant melting profile in Fig. 4A displays a shift in the derivative curve indicative of the transition from tertiary to secondary melting at 42°C instead of 50°C, suggesting a less-stable tertiary structure. The hyperchromicity is also decreased, indicating a difference in either the individual secondary structure elements or a difference in their interactions. The temperature of transition for the −92A→U and −62U→C mutants is similar to that of the wild type, but the hyperchromicity is again greatly reduced (Fig. 4B and C). The −36 substitution mutants fall in two structural populations (A or G and C or U) in terms of the temperature of transition from predominantly tertiary to predominantly secondary melting (Fig. 4D).
FIG. 4.
First derivative UV melting profiles of the wild-type and mutant C. reinhardtii chloroplast rps7 5′UTR RNAs. Each of the RNAs was transcribed in vitro as detailed in Materials and Methods. The representative melting profiles at A260 were carried out in 10 mM HEPES buffer (pH 7.5) containing 5 mM MgSO4 and 100 mM NH4Cl. Melting profiles for each genotype measured at A280 in the pH 7.5 buffer and at A260 and A280 in 10 mM morpholineethanesulfonic acid buffer (pH 5.5) containing 5 mM MgSO4 and 100 mM NH4Cl were similar. Reannealing profiles under the conditions described above for the wild-type and 11 mutant rps7 5′UTR RNAs were essentially superimposable with the melting profiles. (A to C) Symbols: ■, wild type; ▴, the original mutant; ●, the double mutant with the second site suppressor. (D) Symbols: ■, wild type; ●, −36 A→G mutant; ▴, −36A→C mutant; ⧫, −36A→U mutant. (E) Symbols: ■, wild type; ●, the G−63-C−91→A−63-U−91 and G−64-C−90→A−64-U−90 stem-weakened mutant; ▴, the A−68-U−82→G−68-C−82 and U−69-A−83→G−69-C−83 stem-strengthened mutant.
Analysis of the UV melting-reannealing profiles from the mutants predicted to strengthen or weaken the second stem demonstrates an apparent conservation of the overall tertiary structure, as indicated by the consistent temperature of transition from secondary to tertiary unfolding at 50°C. Differences in the hyperchromicity of the mutants are consistent with the expected strengthening or weakening of secondary-structure elements in the predicted second-stem structure. The derivative peak is increased for the stem-strengthening mutant with adjacent (A-U→G-C) changes, indicating increased overall secondary structure, and the derivative peak is decreased and broadened for the stem-weakening mutant with adjacent (G-C→A-U) changes, indicating decreased secondary structure (Fig. 4E). Unfortunately, the large size of the RNA leaders makes determining any specific secondary structures from the melting profiles impossible since interactions between the folding of stem-loops generates 2n + 1 (n = the number of stem-loops) possible interactions.
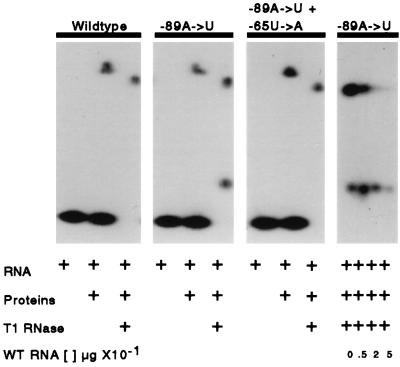
RNase T1 protection gel mobility shift assays.
5′UTRs from the wild type, the −89A→U mutant, and the −89A→U −65U→A double mutant were incubated with heparin-Actigel-purified chloroplast proteins from C. reinhardtii and run on native 5% polyacrylamide gels with and without T1 RNase digestion (Fig. 5). The assay examined the migration of the 5′UTR RNA alone, RNA plus enriched proteins, and RNA plus enriched proteins plus T1 RNase. A single up-shifted band is seen in the wild-type, the −89A→U mutant, and the −89A→U −65U→A double mutant lanes when combined with the enriched proteins. When T1 RNase is added, a single shifted-protected band migrating slightly faster is visible in the wild type and the double mutant. In contrast, two shifted-protected bands are seen in the −89A→U mutant, one in the same position as that in the wild type and the double mutant and one migrating much more rapidly through the gel. An assay with labeled mutant RNA and cold wild-type RNA in the presence of T1 RNase revealed that the higher of the two shifted-protected bands in the mutant was competed off at a lower RNA concentration than the lower band.
FIG. 5.
An RNase T1 protection gel mobility shift assay was conducted on a 5% polyacrylamide native gel with mutant and wild-type forms of the rps7 5′UTR and chloroplast-enriched nucleic acid binding proteins. The 266-nt RNA from the wild type, the −89A→U mutant, and the −89A→U −65U→A double mutant with the second site suppressor were analyzed. The presence of RNAs, proteins, and T1 RNase in each lane is indicated. The labeled mutant 5′UTR RNA (5 ng) was also competed with increasing concentrations (over a 0- to 100-fold range) of unlabeled wild-type 5′UTR RNA.
DISCUSSION
cis-acting mutations in the 5′UTR of the chloroplast rps7 leader that block expression of the lacZ′ reporter gene in E. coli (<6% of the β-galactosidase activity produced by the wild-type rps7 leader) show the same wide range of the aadA reporter activity (10% to >90% of the wild-type rps7 leader value) when examined in either E. coli or in the chloroplast of C. reinhardtii. One possible explanation for the variance between the expression levels of the lacZ′ and aadA reporters is feedback regulation by their protein products, as has been previously observed in other systems (27). Association of a 5′UTR and its protein product might block or promote the initiation of translation, as has been observed in the regulation of E. coli ribosomal protein expression (38). This seems an unlikely possibility since neither of the reporter proteins is known to have nucleic acid binding activity. A more likely possibility is that some structure or sequence within the coding region of the mRNA participates in the formation of the active element in translation initiation. This element might be located either within the coding region itself, as has been demonstrated with chloramphenicol acetyltransferase (28), or else involve interactions between the 5′UTR and the coding sequence to yield a conformation required for translation initiation. Our finding that these leader mutations have similar effects on gene expression both in E. coli and the chloroplast of C. reinhardtii suggests either that the sequences affected are important recognition regions in translation initiation in both organisms or that they present binding sites for trans-acting factors required for initiation common to both.
The 12 individual nucleotide alterations found in the 266-nt rps7 5′UTR among 207 white colony mutants screened map at different sites within the first 167 nt upstream of the initiation codon, but the 7 which alter mRNA translation are restricted to the first 135 nt. Since these mutations were identified by sequencing only A and T residues in the first 225 nt of the 625-nt mutagenized fragment which is 70% A+T rich, we recognized that our screen might have missed changes in other important residues. However, when we sequenced the entire 625-nt fragment from 20 of the remaining 195 white colony mutants, we found no additional changes. This localization of mutants affecting translation to the first 135 nt of the 266-nt 5′UTR suggests that structures proximal to the initiation codon may be most crucial for translation initiation (Fig. 1). While the entire 625-nt fragment is transcribed in E. coli from the endogenous plasmid promoter, no mutations were isolated in the upstream 400-nt region that folds independently as determined by using the MFOLD algorithm, suggesting that this remaining upstream sequence may not function in translation.
Our analysis of changes in the predicted folding pattern generated by the 225-nt rps7 leader mutations reveals the following. (i) Each of the five mutations that prevent accumulation of the reporter mRNA in E. coli and C. reinhardtii chloroplasts either falls in or creates a predicted loop structure (Fig. 1). This might indicate sensitivity of this region of the mRNA in the mutants to RNase activity. (ii) The three mutations localized to the predicted second stem-loop affect the overall size of the first two stem-loop structures and reduce the level of aadA and lacZ′ reporter expression. These nucleotide alterations can be suppressed by complementary mutations also in the second stem-loop that reconstitute the predicted wild-type folding pattern of the first two stem-loop structures (Fig. 3A), supporting the validity of the original predicted folding pattern. (iii) The free energy of each of the mutant rps7 5′UTR folding patterns is fairly similar, differing by less than 4%, arguing that gross thermodynamic changes do not account for the observed differences in translation of the reporter mRNAs.
Several recent studies provide insights into the possible mechanisms responsible for translational regulation of chloroplast gene expression (8, 17, 20, 35, 37). Since SD-like sequences in chloroplast mRNAs appear to be dispensable for translation initiation and other primary sequence motifs known to be involved in translation initiation in E. coli such as the downstream box are also absent, other structures within the long, AU-rich chloroplast leaders likely interact with trans-acting factors to facilitate association of the mRNA with the small subunit of the chloroplast ribosome. The highly ordered secondary structures predicted for the 5′UTRs from the majority of chloroplast mRNAs suggest that they may play a major role in controlling translation initiation.
Studies on a diverse set of chloroplast-encoded genes have identified higher-order RNA structure as an essential determinant in translational control. Multiple putative secondary structures have been detected by RNA mapping techniques in the relatively short (74-nt) 5′UTR from the chloroplast atpH mRNA of Euglena gracilis (1). Many of these structures mask the supposed ribosome binding site on the mRNA thought to be necessary for association with the ribosome. Deletion analysis has identified several small (3- to 8-nt) cis elements, distinct from SD sequences, in the chloroplast psbA mRNA of tobacco plants that associate specifically with trans-acting factors or with the ribosome to facilitate translation of a downstream reporter in an in vitro system (19). Two of these cis elements were proposed to bind cooperatively to the 16S rRNA and thereby position a third element for association of the message with specific trans factors. Deletion analysis has also identified a putative stem-loop structure in the 5′UTR of psbA mRNA in C. reinhardtii that is reported to modulate expression of the downstream D1 coding region in chloroplast transformants (25). However, since mutants lacking the loop sequence reduce accumulation of both the D1 protein and psbA mRNA, one cannot determine whether the region deleted is involved directly in translational regulation (29). In a similar study, two putative binding domains for a postulated initiation complex were identified in the 362-nt 5′UTR of petD mRNA from C. reinhardtii, 210 to 161 and 51 to 33 nt upstream of the initiation codon (32). These domains are thought to form a specific stem-loop structure and to facilitate the association of cis elements with trans factors or to associate directly with the ribosome. The ability of the 5′UTR to assume the correct higher-order structure appears to be crucial for initiation in each of these systems.
A detailed examination of the determinants of translation initiation in the mRNA of the C. reinhardtii chloroplast-encoded psbC gene has been carried out. Chloroplast mutations in C. reinhardtii that perturb translation of the psbC mRNA have been shown to alter the inverted repeat sequence coding for a predicted 97-nt stem-loop in the 5′UTR of this gene (30). The nonphotosynthetic mutant FuD34 enhances the base pairing and elevates the stability of this predicted stem structure by insertion of two adjacent T residues and deletion of a C residue 5 nt away. In contrast, the F34sul mutant, which suppresses a nuclear mutant F34 that blocks translation of the psbC mRNA, weakens the predicted stem by a T→A change. The behavior of these two cis mutants suggests that decreasing the free energy of this extended stem beyond that of the wild-type configuration reduces translation whereas increasing its free energy may alleviate the need for a nucleus-encoded trans-acting protein for translation initiation. Deletion analysis of the extended stem-loop structure of the psbC 5′UTR fused to an aadA reporter clearly indicates that specific interactions between this stem-loop structure and nucleus-encoded trans factors are critical for normal translation initiation of the downstream coding region (39, 40). Two models proposing that the secondary structure of the 5′UTR plays a major role in determining the level of protein expression from chloroplast-encoded genes both invoke trans factor association with wild-type stem-loop structures in the 5′UTR as a prerequisite for translation initiation on chloroplast ribosomes (39).
The foregoing results with the psbC leader mutations are consistent with our observations regarding the functional significance of the predicted stems in the rps7 leader if one assumes that the stability of the wild-type rps7 5′UTR is optimal. We have examined this directly by altering the stability of the predicted second stem-loop by replacing two adjacent A-U pairs with G-C pairs, making the stem more stable, and also by replacing two adjacent G-C pairs with A-U pairs making the stem less stable. Both the increase and the decrease of the local free energy of the predicted stem generated by these changes decrease the translational activity by about one-third, suggesting that the wild-type second stem-loop structure is necessary for optimal translation.
The folding, or unfolding, of RNAs can be visualized as occurring in two phases. Base pairing occurs to produce complex secondary structures, including stems, loops, mismatches, and bulges. Computer algorithms can reasonably predict these structures, which can be tested by using site-directed mutagenesis (42). This is followed by formation of tertiary structure in which loop-loop or other long-range interactions occur. Such tertiary interactions might occur between the unpaired components of the secondary structure, but these interactions will not necessarily produce the most thermodynamically stable final RNA structure since disassociation or recombination of some secondary structure elements leading to a more stable tertiary structure may occur. Thus, the algorithms that predict secondary structure may fail to reveal the true configuration of the RNA even at the secondary structure level despite their clear energy-minimizing parameters (42).
Our analyses of UV melting-reannealing profiles of the rps7 5′UTRs reveal a loss of the wild-type folding in each of the second stem mutants. Restoration of the wild-type structure was found in the double mutants with complementary second site alterations designed to restore base pairing in the predicted second stem-loop. Hyperchromicity is decreased in all three mutants, and in the −89A→U mutant, the temperature at which the transition from tertiary to secondary structure unfolding occurs is significantly diminished, indicating a much less stable tertiary folding pattern (22, 36). Analyses of rps7 5′UTRs with each of the nucleotides at position −36 indicate two distinct types of folding patterns, with the wild type and the −36A→G mutant each more stable in tertiary structure than the −36A→U or the −36A→C mutant. The increased hyperchromicity of the A-U→G-C mutant with the strengthened stem and the decreased hyperchromicity and broader peak of the G-C→A-U mutant with the weakened stem support the predicted increase and decrease of the local free energy of the second stem-loop in the rps7 5′UTR. In all of the cases described above, disruption of the normal melting-reannealing profile in the mutants is consistent with the predicted changes in the secondary structure of the mutant leaders and with the decrease of their biological activities.
Data from the gel mobility shift RNase T1 protection assay conducted on the wild-type, the −89A→U mutant, and the −89A→U −65U→A double mutant suppressor are consistent with the changes in folding indicated by the genetic and melting-reannealing profile analyses (Fig. 5). In the absence of RNase T1, the wild-type, mutant, and double mutant suppressor RNA samples show only a single up-shifted protein-protected band. However, when RNase T1 is added, the wild-type and double mutant suppressor display a single slightly lower up-shifted band protected by the protein, whereas the −89A→U mutant RNA sample shows a second up-shifted band that migrates much more rapidly through the native gel. Differences in protein association with the folded mutant in the wild-type or double mutant suppressor 5′UTR RNA could explain this difference in T1 sensitivity. An alteration in the binding of a specific protein or several proteins protecting the RNA might be responsible, assuming that the rps7 5′UTR can exist in two folding conformations. A change from 99% binding in one RNA conformation and 1% binding in a second in wild-type or the double mutant suppressor conformation (lower shift not visible in Fig. 5) to 50% binding in each conformation in the −89A→U mutant might occur. Competition experiments between the wild-type and −89A→U mutant RNAs for binding with the enriched nucleic acid binding proteins indicate that the complex in the higher band is likely the same in both the wild type and the mutant (Fig. 5). In contrast, the RNA in the lower band seen in the mutant may be binding a different set of proteins or may bind a subset of the same proteins in a different conformation with a stronger affinity and hence may have less of its structure protected from RNase digestion.
The restoration of a near-wild-type UV melting-reannealing profile in each of the double mutant suppressors and restoration of the gel mobility shift protection pattern in the −89A→U −65U→A double mutant is consistent with the observed restoration of normal growth rate and enzyme activity and strongly supports the biological relevance of the predicted folding pattern for the first two stem-loop domains of the rps7 leader.
Our generation of rps7 5′UTR mutants that display diminished spectinomycin-resistant growth when placed upstream of the aadA reporter will make possible a suppressor screen in both E. coli and in the chloroplast of C. reinhardtii to identify other relevant cis-acting sequences as well as genes encoding trans-acting proteins.
ACKNOWLEDGMENTS
We thank C. Hauser and M. Been for their assistance with both methodology and data interpretation.
This work was supported by NIH grant GM-19427.
REFERENCES
- 1.Betts L, Spremulli L L. Analysis of the role of Shine-Dalgarno sequence and mRNA secondary structure on the efficiency of translation initiation in the Euglena gracillis chloroplast atpH mRNA. J Biol Chem. 1994;269:26456–26463. [PubMed] [Google Scholar]
- 2.Blattner F R, Plunket G, Bloh C A, Perna N T, Burland M R, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bonham-Smith P C, Bourque D P. Translation of chloroplast-encoded mRNA: potential initiation and termination signals. Nucleic Acids Res. 1989;17:2057–2080. doi: 10.1093/nar/17.5.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boynton J E, Gillham N W. Chloroplast transformation in Chlamydomonas. Methods Enzymol. 1993;217:510–536. doi: 10.1016/0076-6879(93)17087-l. [DOI] [PubMed] [Google Scholar]
- 5.Boynton J E, Gillham N W, Harris E H, Hosler J P, Johnson A M, Jones A R, Randolph-Anderson B L, Robertson D, Klein T M, Shark K B, Sanford J C. Chloroplast transformation in C. reinhardtii with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi H. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fargo D C, Zhang M, Gillham N W, Boynton J E. Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplast or in Escherichia coli. Mol Gen Genet. 1998;257:271–282. doi: 10.1007/s004380050648. [DOI] [PubMed] [Google Scholar]
- 8a.Fargo, D. C., et al. Unpublished data.
- 9.Förster B, Heifetz P B, Lardans A, Boynton J E, Gillham N W. Herbicide resistance and growth of D1 Ala251 mutants in Chlamydomonas. Z Naturforsch. 1997;52c:654–664. [Google Scholar]
- 10.Gillham N W, Boynton J E, Hauser C R. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- 11.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 12.Goldschmidt-Clermont M. Transgenic expression of amino-glycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4090. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- 14.Gray M W. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:223–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 15.Harris E H. The Chlamydomonas sourcebook. San Diego, Calif: Academic Press, Inc.; 1989. [Google Scholar]
- 16.Harris E H, Boynton J E, Gillham N W. Chloroplast ribosomes and protein synthesis. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser C R, Gillham N W, Boynton J E. Translational regulation of chloroplast genes: proteins binding to the 5′ UTRs of chloroplast mRNAs in Chlamydomonas reinhardtii. J Biol Chem. 1996;271:1486–1497. doi: 10.1074/jbc.271.3.1486. [DOI] [PubMed] [Google Scholar]
- 18.Hauser C R, Gillham N W, Boynton J E. Regulation of chloroplast translation. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. Molecular biology of Chlamydomonas: chloroplasts and mitochondria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 19.Hirose T, Sugiura M. cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system for tobacco chloroplasts. EMBO J. 1996;15:1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Mullet J E. Ribosome binding sites on chloroplast rbcL and psbA mRNAs and the light-induced initiation of D1 translation. Plant Mol Biol. 1994;25:437–448. doi: 10.1007/BF00043872. [DOI] [PubMed] [Google Scholar]
- 21.Koo J S, Spremulli L L. Analysis of the translation initiation region of the Euglena gracilis chloroplast ribulose-bisphosphate carboxylase/oxygenase (rbcL) messenger RNA. J Biol Chem. 1994;269:7494–7500. [PubMed] [Google Scholar]
- 22.Laing L G, Draper D E. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J Mol Biol. 1994;237:560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- 23.Lers A, Heifetz P B, Boynton J E, Gillham N W, Osmond C B. The carboxyl-terminal extension of the D1 protein of photosystem II is not required for optimal photosynthetic performance under CO2− and light saturated growth conditions. J Biol Chem. 1992;267:17494–17497. [PubMed] [Google Scholar]
- 24.Leung D W, Chen E, Goedde D U. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 25.Mayfield S P, Cohen A, Danon A, Yohn C B. Translation of the psbA mRNA of Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′ untranslated region. J Cell Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayfield S P, Yohn C B, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- 27.McCarthy J E G, Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 28.Odjakova M, Golshani A, Ivanov G, Haidar M A, Ivanov I. The low level expression of chloramphenicol acetyltransferase (CAT) mRNA in Escherichia coli is not dependent on either Shine-Dalgarno or the downstream boxes in the cat gene. Microbiol Res. 1998;153:173–178. doi: 10.1016/S0944-5013(98)80037-2. [DOI] [PubMed] [Google Scholar]
- 29.Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 30.Rochaix J-D, Kuchka M, Mayfield S, Schirmer-Rahie M, Girard-Bascou J, Bennoun P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 1989;8:1013–1021. doi: 10.1002/j.1460-2075.1989.tb03468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruf M, Kössel K. Occurrence and spacing of ribosome recognition sites in mRNAs of chloroplasts from higher plants. FEBS Lett. 1988;240:41–44. [Google Scholar]
- 32.Sakamoto W, Chen X, Kindle K L, Stern D B. Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6f complex. Plant J. 1994;6:503–512. doi: 10.1046/j.1365-313x.1994.6040503.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Shapira M, Lers A, Heifetz P B, Irihimovitz V, Osmond C B, Gillham N W, Boynton J E. Differential regulation of chloroplast gene expression in Chlamydomonas reinhardtii during photoacclimation: light stress transiently suppresses synthesis of the Rubisco protein while enhancing synthesis of the PS II D1 protein. Plant Mol Biol. 1997;33:1001–1011. doi: 10.1023/a:1005814800641. [DOI] [PubMed] [Google Scholar]
- 35.Stampacchia O, Girard-Bascou J, Zanasco J-L, Bennoun P, Rochaix J-D. A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner M, Cech T. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(Ile) of the purple bacterium Azoarcus. RNA. 1996;1:74–83. [PMC free article] [PubMed] [Google Scholar]
- 37.Yohn C B, Cohen A, Danon A, Mayfield S P. Altered mRNA binding activity and decreased translational initiation of the chloroplast psbA mRNA. Mol Cell Biol. 1996;16:3560–3566. doi: 10.1128/mcb.16.7.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zengel J M, Lindahl L. Diverse mechanisms for regulation ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acids Res. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 39.Zerges W, Girard-Bascou J, Rochaix J-D. Translation of the chloroplast psbC mRNA is controlled by the interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol Cell Biol. 1997;17:3440–3448. doi: 10.1128/mcb.17.6.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerges W, Rochaix J-D. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol Cell Biol. 1994;14:5268–5277. doi: 10.1128/mcb.14.8.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y H, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 42a.Zuker, M. University of Washington. 1995–1999, copyright date. [Online.] http://mfold1.wustl.edu/mfold/rna/form1.cgi. [17 August 1999, last date accessed].