Abstract
The splicing of mammalian mRNA precursors requires both protein phosphorylation and dephosphorylation, likely involving modification of members of the SR protein family of splicing factors. Several kinases have been identified that can phosphorylate SR proteins in vitro, and transfection assays have provided evidence that at least one of these, Clk/Sty, can modulate splicing in vivo. But evidence that a specific kinase can directly affect the splicing activity of SR proteins has been lacking. Here, by using purified recombinant Clk/Sty, a catalytically inactive mutant, and individual SR proteins, we show that Clk/Sty directly affects the activity of SR proteins, but not other essential splicing factors, in reconstituted splicing assays. We also provide evidence that both hyper- and hypophosphorylation inhibit SR protein splicing activity, repressing constitutive splicing and switching alternative splice site selection. These findings indicate that Clk/Sty directly and specifically influences the activity of SR protein splicing factors and, importantly, show that both under- and overphosphorylation of SR proteins can modulate splicing.
SR proteins constitute a family of essential pre-mRNA splicing factors that are highly conserved throughout metazoa (reviewed in references 14, 26, and 42). They have been shown to function in vitro as essential factors required for all splices, as concentration-dependent regulators of alternative splice site selection, and as activators-inhibitors of splicing as components of complexes assembled on splicing enhancer-silencer elements in the pre-mRNA. Although most studies have employed in vitro assays, individual SR proteins have been shown to be capable of modulating alternative splice site selection when overexpressed in transient transfection assays (2, 36, 45). One SR protein, B52/SRp55, is required for proper development in Drosophila (33, 34), and another, ASF/SF2, is essential for the viability of a cultured cell line (46), and in vivo depletion of the protein can affect alternative splicing (47). SR proteins contain one or two N-terminal RNP-type RNA binding domains (RBDs) and C-terminal regions enriched in repeated arginine-serine dipeptides (RS domains). The RBDs function in binding the pre-mRNA (1, 39, 51), while RS domains have been implicated in protein-protein interactions (24, 48). SR proteins are extensively phosphorylated (35).
Reversible protein phosphorylation plays an important role in pre-mRNA splicing, and SR proteins are likely to be key targets. Initial in vitro studies, employing phosphatases and their inhibitors, provided evidence that protein phosphorylation, possibly of SR proteins, is required for spliceosome assembly (28); that dephosphorylation is necessary at a subsequent step (27, 40); and that phosphatase treatment of nuclear extract can influence alternative splice site selection (5). More recent experiments have begun to suggest possible molecular mechanisms for these effects. Phosphorylation of serines within RS domains can influence RNA binding by SR proteins by preventing strong sequence nonspecific interactions with RNA (38, 49). Such phosphorylation also enhances the RS-domain-mediated protein-protein interaction with the U1 snRNP 70-kDa protein (44, 49), which is likely important for 5′ splice site recognition by U1 snRNP (22, 24). At the same time, phosphorylation can reduce interactions among certain SR proteins (50), although how this contributes to function is not yet known. SR protein phosphorylation and dephosphorylation are required for splicing in vitro (4, 49, 50).
Several protein kinases have been described that can phosphorylate SR protein RS domains (reviewed in references 14 and 26). The two most extensively studied are SRPK1 (18) and Clk/Sty (8, 11), and these appear to be prototypes of larger families of differentially expressed kinases (13, 20, 31, 32, 44). SRPK1 may modulate SR protein phosphorylation during the cell cycle, and addition of the purified enzyme to in vitro reactions has been shown to inhibit splicing (18, 19). Clk/Sty contains an N-terminal region enriched in RS dipeptides and was found to interact in yeast two-hybrid assays with several SR proteins, including ASF/SF2, via this domain (8). A catalytically inactive version of Clk/Sty colocalizes in nuclear speckles with endogenous splicing factors when expressed by transient transfection, whereas overexpression of the wild-type kinase causes redistribution of splicing factors into a more diffuse pattern (8). Excess SRPK1 likewise causes a redistribution of splicing factors when introduced into permeabilized cells (18). Together, these results indicate that protein kinases that phosphorylate SR proteins in vitro (9) can affect the localization of splicing factors when overexpressed in vivo. Supporting the functional significance of these observations, transient overexpression of Clk/Sty has been shown to influence alternative splicing of a cotransfected reporter transcript (12). In Drosophila, mutations in a Clk/Sty-like kinase, Doa, can influence phosphorylation of an SR protein and affect alternative splicing (10). However, there is no evidence that any of these kinases (or for that matter any kinase) directly influences splicing by phosphorylating a specific target protein(s) and changing its activity in splicing. Indeed, transiently overexpressed Clk/Sty was recently shown to result in the accumulation of ASF/SF2 in the cytoplasm, leading to the suggestion that previously observed effects on splicing might be at least in part indirect, resulting from mislocalization of splicing factors (3).
In this study, we provide evidence that Clk/Sty directly targets SR proteins, specifically ASF/SF2 and SC35, and show that both hyper- and hypophosphorylation can reduce SR protein activity in constitutive, activated, and alternative splicing in vitro. Hypophosphorylation and splicing inhibition can be induced by a dominant-negative, kinase-inactive Clk/Sty mutant, but only when de novo SR protein phosphorylation is required. Hyperphosphorylation of SR proteins by Clk/Sty also reduces their activity in splicing. In both cases, we provide evidence, using reconstituted splicing assays, that the effects on splicing are due specifically to alterations in SR protein phosphorylation and not due to modification of other required factors in the complex splicing machinery. We discuss the implications of these results with respect to mechanisms by which SR protein activity, and hence splicing, can be controlled.
MATERIALS AND METHODS
Recombinant proteins.
His-tagged ASF/SF2 was expressed in JM101, Escherichia coli-produced ASF/SF2 (ecASF), recombinant baculovirus-infected SF9 cells, or baculovirus-produced ASF/SF9 (bvASF) cells and purified using Ni2+ chromatography essentially as described (16, 17). Glutathione S-transferase (GST)-Clk/Sty (Clk) and GST-ClkK190R (ClkR) were also expressed in JM101 and were purified by using glutathione agarose as described (8, 11). SC35 from SF9 cells was purified by ammonium sulfate and MgCl2 precipitation (39, 51).
In vitro splicing.
Template DNAs were linearized with appropriate restriction enzymes before transcription with SP6 RNA polymerase, and RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE). Splicing reactions with HeLa nuclear extract or S100 were performed essentially as described previously (17, 50). The final concentrations of components during the splicing reactions were 12 mM HEPES-KOH (pH 7.9), 20 mM creatine phosphate, 0 to 42 mM (NH)4SO4, 20 to 60 mM KCl, 2.1 to 3.2 mM MgCl2, 0.12 mM EDTA, 0.5 mM dithiothreitol, 2.6% polyvinal alcohol, 2 U of RNasin, and 6 to 10% glycerol. (Salt concentrations were optimized separately for each pre-mRNA.) Preincubations (20 min), where indicated, were carried out in the absence of substrate which, together with the indicated protein(s), was added at time zero. Splicing reactions were routinely 90 min, in a final volume of 25 μl. For splicing assays employing S100 extracts, the amounts of ecASF, bvASF, or SC35 indicated in the figure legends were added to S100 extracts along with the indicated amounts of Clk or ClkR, in final reaction volumes of 25 μl. For the mixing experiment with tat pre-mRNA, 100 ng of ecASF and/or 200 ng of SC35 was added to the S100 extracts as described (50). On completion of splicing reactions, mixtures were treated with proteinase K (50 μg/ml), extracted with phenol-chloroform, and precipitated with ethanol. RNA was fractionated by denaturing PAGE, and splicing products were visualized by autoradiography. Where indicated, splicing efficiencies were quantitated by densitometry.
Western blot analysis.
Proteins in splicing assays were diluted 10-fold with water and precipitated with 10% trichloroacetic acid for 60 min on ice. Pellets were washed with acetone before resuspending in sodium dodecyl sulfate (SDS) gel sample buffer. Proteins were fractionated by SDS–10% PAGE and then transferred to nitrocellulose. Blots were probed with monoclonal antibody (MAb) 104 as described (49) and were visualized using a chemiluminescence kit (Amersham).
RESULTS
Clk/Sty and ClkK190R can both inhibit in vitro splicing.
To begin to address the question of whether Clk/Sty phosphorylation of SR proteins directly affects their activity in splicing, we first wished to determine whether the kinase has any effect on constitutive splicing when added to in vitro reactions. In order to accomplish this, we purified from E. coli Clk and, initially as a control, the catalytically inactive mutant ClkR (8, 11). Figure 1, lanes 1 and 2, shows a silver-stained gel of the two purified proteins. Note the reduced mobility of the wild-type derivative, reflecting the expected autophosphorylation. Figure 1 also shows two versions of the purified recombinant SR protein ASF/SF2 used in these studies. The difference in mobility was due entirely to the phosphorylation of the protein expressed in SF9 cells (lane 3), but not of the E. coli-expressed protein (lane 4), as judged by phosphatase treatment of the purified proteins (results not shown). Clk, but not ClkR, efficiently phosphorylated recombinant ASF/SF2 (results not shown).
FIG. 1.
SDS-PAGE analysis of recombinant proteins. Five milligrams each of Clk (lane 1), ClkR (lane 2), ecASF (lane 3), and bvASF (lane 4) were fractionated by SDS–10% PAGE and were stained with Coomassie blue R-250. The species indicated by the asterisk reflects an N-terminal truncation consisting of GST plus an approximate 10-kDa segment of Clk/Sty which becomes phosphorylated in the wild-type but not the mutant sample. The fragment is unlikely to have affected activity in our assays, as its presence was variable and not correlated with activity, and E. coli- and baculovirus-expressed Clks behaved similarly, although the latter lacks the truncated fragment (unpublished data). Lane M contains marker proteins (masses are indicated on the left in kilodaltons).
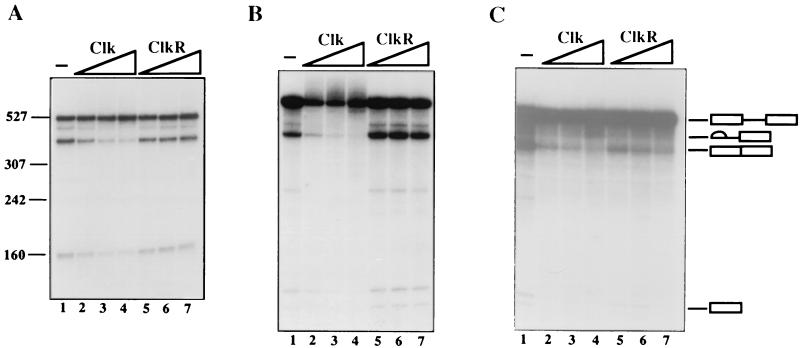
As a first test of the ability of wild-type Clk/Sty to influence splicing, Clk was added to nuclear extract (NE), and its effect on splicing of a human β-globin pre-mRNA was determined. Figure 2A shows that increasing amounts of Clk inhibited splicing (lanes 1 to 4; >4-fold inhibition). This finding is similar to a report with purified SRPK1 (18), although our results extend this study by showing that inhibition requires kinase activity, as ClkR was without effect (Fig. 2A, lanes 5 to 7). Although these results are consistent with inhibition resulting from SR protein phosphorylation, it is also possible that the phosphorylation of other splicing factors was responsible. To begin to address this, we performed similar experiments, except using S100 extract supplemented with recombinant ASF/SF2. When bvASF was used (Fig. 2B), the results were similar to those obtained with NE, with one significant quantitative exception: inhibition induced by Clk (lanes 1 to 4; >10-fold inhibition) was more robust than that observed in NE (compare Fig. 2A and B, lanes 1 to 4). The results obtained when ecASF was used to activate splicing (Fig. 2C) were, however, distinct in an important and unexpected way: although Clk again inhibited the reaction (lanes 1 to 4), in this case ClkR also inhibited splicing, and nearly as strongly as did the wild-type kinase (compare lanes 2 to 4 [∼4.5-fold inhibition] with lanes 5 to 7 [∼3-fold inhibition]). These results indicate that both wild-type Clk and catalytically inactive ClkR were able to inhibit constitutive splicing, but the latter only when an unphosphorylated SR protein was used to activate splicing.
FIG. 2.
Effects of Clk and ClkR on constitutive splicing. (A) Effect of Clk and ClkR on human β-globin pre-mRNA splicing in nuclear extracts. Lane 1, nuclear extract alone. Increasing amounts (50, 100, and 200 ng) of Clk (lanes 2 to 4) and ClkR (lanes 5 to 7) were added to nuclear extracts, and splicing was allowed to proceed for 90 min. RNAs were purified and fractionated by denaturing PAGE. (B) Effects of Clk and ClkR of β-globin splicing in S100 complemented with bvASF (300 ng). Lane 1, S100 plus bvASF only. Increasing amounts (25, 50, and 100 ng) of Clk (lanes 2 to 4) or ClkR (lanes 5 to 7) were added to reaction mixtures. The apparent loss of pre-mRNA in lanes 2 to 4 reflects poly(A) addition to the 3′ end of precursor, which resulted in upward smearing of the RNA during electrophoresis. (C) Effect of Clk and ClkR on β-globin splicing in S100 extracts complemented with ecASF (500 ng). Lane 1, S100 plus ecASF only; lanes 2 to 4 and 5 to 7, increasing amounts (50, 100, and 200 ng) of Clk and ClkR, respectively. The positions of the pre-mRNA, spliced product, and intermediates are depicted by symbols on the right.
Clk inhibits splicing by specifically targeting SR proteins.
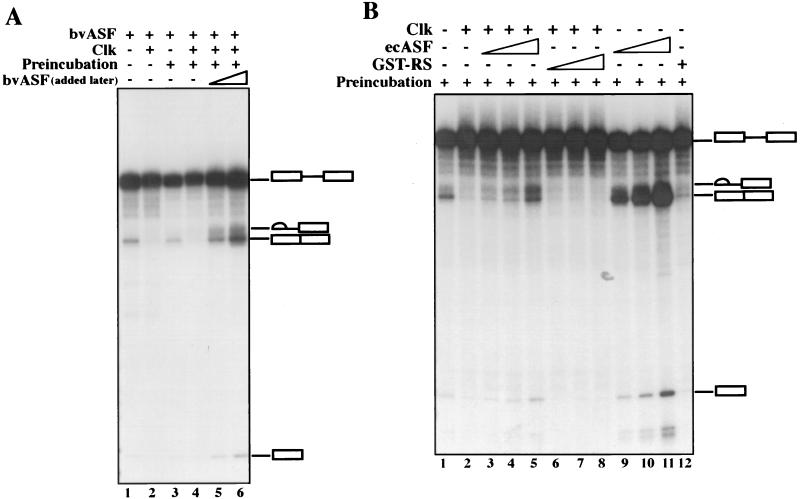
Given that the only difference between the conditions under which ClkR was without effect (Fig. 2B) and inhibitory (Fig. 2C) was the source of ASF/SF2, the inhibition of splicing by ClkR must have been due to an effect specifically on ASF/SF2. However, because Clk inhibited splicing under all conditions tested, it was still possible that inhibition was due to modification of another factor. To provide evidence that SR proteins, and specifically ASF/SF2, are indeed the target of Clk, we performed an experiment similar to that shown in Fig. 2B, except incorporating a preincubation-rescue procedure. Specifically, bvASF, Clk, and S100 were preincubated for 20 min, at which time the pre-mRNA and additional bvASF were added. The results (Fig. 3A) again show significant inhibition by Clk, which was not affected by the preincubation period per se (compare lanes 1 and 2 with lanes 3 and 4; note that the preincubation did slightly reduce splicing efficiency). Remarkably, addition of bvASF with the pre-mRNA not only restored processing but allowed splicing at levels significantly higher than those observed in the absence of Clk or preincubation (lanes 5 and 6). This was especially striking, because the amounts of bvASF added after the preincubation (50 or 100 ng) were less than the amount added to all samples at time zero (200 ng), and splicing was, in fact, greater than that observed when 300 ng was added at time zero (results not shown). Although we do not know how the preincubation primes the extract to enhance splicing (it likely reflects titration of the limiting amount of Clk [50 ng] by the bvASF), these results indicate that no component was inactivated by Clk during the preincubation, and small amounts of active bvASF can fully rescue splicing. To extend these findings, we repeated the experiment using ecASF instead of bvASF (Fig. 3B). The results obtained were very similar to those described above, as the recombinant bacterial protein was also able to efficiently restore Clk-inhibited splicing (lanes 1 to 5). When the same amounts of ecASF were added to reaction mixtures preincubated in the absence of Clk, significantly higher levels of splicing were detected (compare lanes 3 to 5 with lanes 9 to 11), reflective of the inhibitory effect of the added Clk on ASF/SF2 function.
FIG. 3.
Both bvASF and ecASF can rescue Clk-mediated inhibition of constitutive splicing. (A) Effect of preincubation of Clk on bvASF and rescue of splicing. bvASF (200 ng) was preincubated (lanes 3 to 6) in the presence (lanes 4 to 6) or absence (lane 3) of 50 ng of Clk for 20 min in the absence of pre-mRNA. 50 or 100 ng of bvASF was added subsequently (lanes 5 and 6) along with the pre-mRNA (lanes 3 to 6), and splicing continued for 90 min. Samples in lanes 1 and 2 lacked or contained Clk, respectively, but were not preincubated. (B) Effect of preincubation of Clk on ecASF. ecASF (200 ng) was preincubated in the presence (lanes 2 to 8) or absence (lanes 1 and 9 to 12) of Clk. Subsequently, 50, 100, and 200 ng of ecASF (lanes 3 to 5 and 9 to 11) or GST-RS (lanes 6 to 8) were added along with pre-mRNA. Lane 12 contained 200 ng of GST-RS. The positions of the pre-mRNA, spliced product, and intermediates are indicated.
It was conceivable that the addition of ecASF to reaction mixtures preincubated with Clk restored splicing not by providing SR protein activity, but instead by serving as a “sink” for kinase activity, such that Clk phosphorylated ecASF instead of another putative target. To test this, we added GST-RS, which contains only the ASF/SF2 RS domain, instead of ecASF. GST-RS is phosphorylated by Clk/Sty just as efficiently as the full-length protein (8, 49) but is not active in splicing. Figure 3B shows that GST-RS was completely unable to restore Clk-inhibited splicing (compare lane 2 with lanes 5 to 8). Note that GST-RS added in the absence of Clk had an inhibitory effect, although even at the highest concentration tested, splicing was readily detectable (compare lanes 1 and 12). Taken together, our results indicate that addition of Clk to in vitro splicing reactions inhibits splicing by interfering with the activity of SR proteins such as ASF/SF2 and not with any other required factor.
Hyper- and hypophosphorylation of SR proteins is induced by Clk/Sty derivatives.
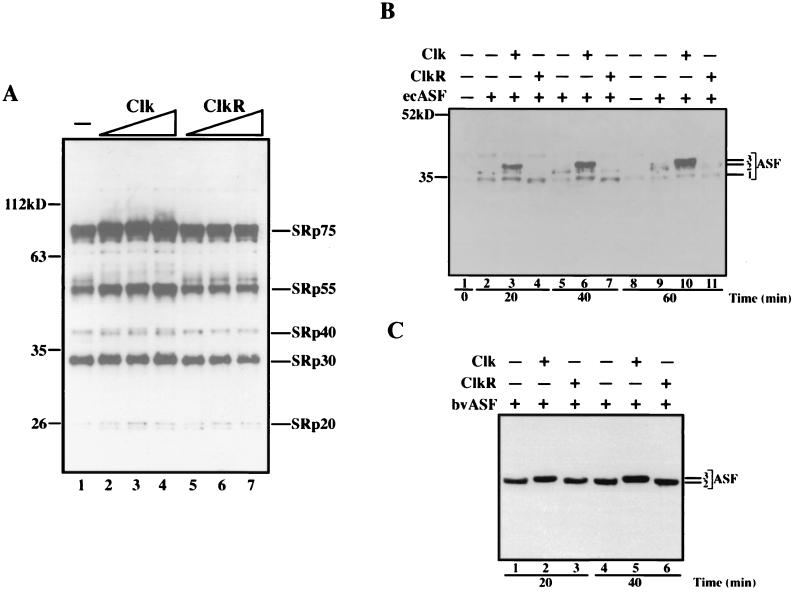
The effects on splicing described above are consistent with inhibition of SR protein activity by either hyperphosphorylation (induced by Clk) or hypophosphorylation (induced by ClkR). While it is known that purified recombinant Clk can phosphorylate purified ASF/SF2, there is no evidence that this can occur in the complex background of a splicing reaction, and there is no data at all suggesting that ClkR can have the dominant-negative effect required to induce the hypophosphorylation we propose. We therefore examined the phosphorylation status of SR proteins in NE and supplemented S100 in the presence of Clk or ClkR. Figure 4A displays a Western blot with MAb104 of NE incubated under splicing conditions (i.e., as in Fig. 2A) with increasing amounts of Clk (lanes 2 to 4) or ClkR (lanes 5 to 7). MAb104 detects a conserved phosphoepitope in the RS domains of all SR proteins (35) and thus can detect changes in phosphorylation quantitatively (via changes in intensity) and qualitatively (via changes in mobility). By one or both of these criteria, Clk caused an increase in phosphorylation of all the classical SR proteins, including SRp75, SRp55, SRp40, SRp30 (which includes ASF/SF2 and SC35), and SRp20. These concentration-dependent increases in phosphorylation thus correlate with the concentration-dependent inhibition of splicing observed in Fig. 2. In contrast, ClkR had almost no detectable effect on SR protein phosphorylation (a slight decrease in intensity was detected at the highest concentration), consistent with its similar lack of effect on splicing in NE.
FIG. 4.
Effects of Clk and ClkR on SR protein phosphorylation in splicing-competent extracts. (A) Western blot analysis of endogenous SR proteins in nuclear extracts. Aliquots of splicing reactions as in Fig. 2A were resolved by SDS-PAGE and subjected to Western blotting using MAb104. Lane 1, nuclear extract alone; lanes 2 to 4 and 5 to 7, increasing amounts of Clk and ClkR, respectively. The identities of specific SR proteins are indicated on the right, and masses (in kilodaltons) are indicated on the left. (B) Western blot analysis of S100 extracts containing ecASF. Aliquots of splicing reactions terminated at the indicated times were fractionated by SDS-PAGE and were analyzed by Western blotting as in panel A. Lanes 1 and 8, S100 extract alone; lanes 2, 5, and 9, S100 extracts plus 300 ng of ecASF; lanes 3, 6, and 10 and 4, 7, and 11, S100 plus 300 ng of ecASF and 100 ng of either Clk or ClkR, respectively. The positions of the three forms of ASF detected are indicated (see text). (C) Western blot analysis of S100 extracts containing bvASF. Conditions were exactly as in panel B, except that bvASF replaced ecASF and was present in all samples. Lanes 1 and 4 contain no added kinase, lanes 2 and 5 contain Clk, and lanes 3 and 6 contain ClkR. The positions of the two forms of ASF detected (see text) are indicated.
We next examined the phosphorylation status of ecASF added to S100 extract in the presence of Clk or ClkR. Recall that under these conditions both kinase derivatives inhibited splicing of β-globin pre-mRNA (Fig. 2C), and we wished to determine if this reflected altered phosphorylation. Figure 4B displays a Western blot (with MAb104) of a splicing time course supplemented with no added kinase (lanes 1, 2, 5, 8, and 9) or identical amounts of Clk (lanes 3, 6, and 10) or ClkR (lanes 4, 7, and 11). In the absence of exogenously added kinase, phosphorylated ecASF was readily detected at the 20 min time point, giving rise to two closely spaced MAb104-reactive species (labeled forms 1 and 2). At the 40 and 60 min time points, the amount of reactive material did not increase significantly, but the relative levels of the lower mobility (and presumably more phosphorylated) form 2 were markedly enhanced. In the presence of Clk, evidence of hyperphosphorylation was clearly evident. Not only was reactivity with MAb104 substantially increased, but an additional, lower-mobility species (labeled form 3) was also observed at all time points, increasing to the most abundant form after 60 min. In contrast, ClkR induced the opposite effects. Although MAb104 reactivity was only slightly reduced (at the 60 min time point; compare lanes 9 and 11), at all time points, and especially at the first two, the ratio of the low-mobility form 2 to the high-mobility form 1 was significantly decreased (compare lanes 2 and 4, 5 and 7, and 9 and 11), and no form 3 was detected. Thus, the data indicate that both Clk and ClkR can influence ASF/SF2 phosphorylation, and this correlates with their ability to inhibit splicing under the same conditions.
When the effects of Clk and ClkR on bvASF in S100 were examined (Fig. 4C), the results were again entirely consistent with the effects of the two kinase derivates on splicing (Fig. 2B). In the absence of Clk or ClkR, only a single MAb104-reactive species, equivalent to ecASF form 2, was detected, and it did not change significantly as the splicing reaction proceeded (lanes 1 and 4, 20 and 40 min time points). Clk quantitatively converted bvASF to the hyperphosphorylated form 3 (lanes 2 and 5). This complete conversion is consistent with the very strong inhibition of splicing caused by Clk in the presence of bvASF. It is likely that bvASF is more completely hyperphosphorylated by Clk than is ecASF, simply because bvASF is already extensively phosphorylated. ClkR, in contrast, did not detectably affect the phosphorylation status of bvASF (lanes 3 and 6), consistent with its lack of effect on slicing.
Clk and ClkR block ASF/SF2-activated splicing of tat pre-mRNA.
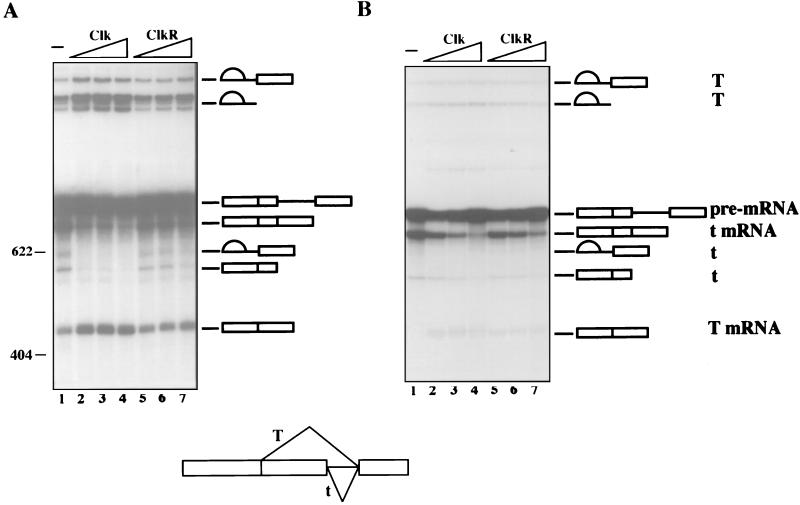
To determine whether the effects on splicing described above could be extended to different types of mRNA precursors, we analyzed splicing of several additional pre-mRNAs in the presence of increasing amounts of Clk or ClkR. In the experiment shown in Fig. 5, the behavior of a human immunodeficiency virus tat pre-mRNA was examined. This RNA is distinctive in that it displays an unusual dependence on ASF/SF2. In vitro, tat RNA is spliced very poorly in NE unless the extract is supplemented with ASF/SF2 (25, 50), and it can also be committed to splicing by preincubation with ASF/SF2, but not with several other SR proteins (6, 15). Paradoxically, tat RNA splicing in vivo is stimulated when ASF/SF2 is genetically depleted (47). Thus it appears that ASF/SF2 can function as either an activator or repressor of tat splicing, depending on the experimental conditions. Both of these activities likely depend on a complex interplay between splicing enhancers and silencers in the 3′ exon (reference 37 and references therein). To test the effects of Clk and ClkR on ASF/SF2-activated tat splicing, we added increasing amounts of each kinase derivative to S100-based splicing reactions containing either ecASF or bvASF. The results were remarkably similar to those obtained with β-globin pre-mRNA: in the presence of (unphosphorylated) ecASF, both kinases inhibited splicing, with wild-type Clk marginally more effective than the mutant derivative (Fig. 5A). When (phosphorylated) bvASF was used (Fig. 5B), Clk again inhibited splicing, but ClkR was without significant effect. The data provide evidence that both hyper- and hypophosphorylation can interfere with the ability of ASF/SF2 to activate tat splicing.
FIG. 5.
Inhibition of ASF/SF2-activated human immunodeficiency virus tat splicing by Clk and ClkR. (A) Effect of Clk and ClkR on tat pre-mRNA splicing in S100 extracts complemented with ecASF (400 ng). Lane 1, ecASF alone; lanes 2 to 4 and 5 to 7, increasing amounts (50, 100, and 200 ng) of Clk and ClkR, respectively. (B) Effect of Clk and ClkR on S100 extracts complemented with bvASF (250 ng). Lane 1, bvASF alone; lanes 2 to 4 and 5 to 7, increasing amounts (25, 50, 100 ng) of Clk and ClkR, respectively. (C) Effect of ClkR on ecASF-mediated activation of splicing in the presence of SC35. Lanes 1 and 3 to 5, 100 ng of ecASF; lanes 2 to 5, 200 ng of SC35; lanes 4 and 5, 100 and 200 ng of ClkR, respectively. The positions of the pre-mRNA, spliced product, and intermediates are shown. ∗ indicates an artifactual cleavage product unrelated to splicing.
We also employed the tat pre-mRNA and a previously described assay (50) to address another question: is phosphorylation required for the sequence-specific (as opposed to the general or constitutive) function of an SR protein? Although, as discussed above, SR protein phosphorylation is necessary for general splicing, whether it is also required for sequence-specific (or activated) splicing has not been addressed. The possibility that there may be distinct phosphorylation requirements is raised by our recent discovery that SR protein dephosphorylation, while necessary for general splicing, is not required for sequence-specific activation function (50). Part of the evidence for this came from use of the ASF/SF2-dependent tat pre-mRNA, which we showed could be efficiently spliced in the presence of SC35 plus thiophosphorylated ASF/SF2. Given that the modified ASF/SF2 derivative, which cannot be dephosphorylated, is by itself inactive on all substrates tested (4, 50), this provided evidence that ASF/SF2 dephosphorylation is not required for splicing activation when another SR protein (SC35) provides the general function. To extend this analysis to phosphorylation, we examined tat splicing in S100 plus SC35 (from baculovirus), a limiting amount of ecASF, and increasing concentrations of ClkR, to induce ecASF hypophosphorylation (as with bvASF, SC35 was not affected by ClkR; see below). As expected, SC35 cooperated with ecASF to give efficient splicing (Fig. 5C, compare lanes 1 to 3). However, this splicing showed at least the same sensitivity to ClkR (lanes 4 to 5) as did splicing activated by ecASF alone (Fig. 5A). These results confirm that ecASF is inhibited by ClkR and also provide evidence that ASF/SF2 phosphorylation is required for its activation as well as its general functions.
Hyper- and hypophosphorylation of SR proteins influences alternative splicing.
A well-established property of SR proteins is their ability to influence selection of alternative splice sites in a concentration-dependent manner (reviewed in references 14 and 26). For example, increasing the concentration of ASF/SF2 favors usage of the most downstream 5′ splice site in mRNAs containing alternative 5′ splice sites, while decreasing its concentration (e.g., by genetic depletion in vivo) (47) increases splicing from more upstream splice sites. If Clk and ClkR indeed exert their effects on splicing by inhibiting SR protein function, then addition of the kinase derivatives to reaction mixtures containing such alternatively spliced pre-mRNAs should, under the appropriate conditions, shift splicing in favor of the upstream 5′ splice site. We tested this prediction in different ways with two different pre-mRNAs, simian virus 40 (SV40) early and adenovirus E1a.
The SV40 RNA was first spliced in NE without supplementation (Fig. 6A, lane 1), with Clk (lanes 2 and 3), or with ClkR (lanes 4 and 5). The effects of both kinase derivatives were largely in keeping with expectation. Clk significantly reduced splicing from the downstream 5′ splice site (which produces small t mRNA) while simultaneously increasing use of the upstream 5′ splice site, leading to increased synthesis of large T mRNA. In contrast, ClkR had, at most, minor effects and these only at the highest concentration tested. This is consistent with the very limited effects of ClkR on both β-globin splicing and SR protein phosphorylation in NE (see above). To extend these results, we examined SV40 splicing in S100 plus ecASF in the presence or absence of Clk or ClkR (Fig. 6B). Sufficient ecASF was added so that nearly all splicing was from the downstream (small t) 5′ splice site in the absence of added kinase (lane 1). Strikingly, under these conditions both Clk (lanes 2 to 4) and ClkR (lanes 5 to 7) reduced small t splicing while simultaneously enhancing large T splicing. These results suggest that the switch in SV40 splicing was due to reduced ASF/SF2 activity, which can be caused by either hyper- or hypophosphorylation, and are consistent with the effects observed with the β-globin and tat pre-mRNAs.
FIG. 6.
Clk and ClkR modulate SV40 pre-mRNA alternative splicing. (A) Effect of Clk and ClkR on SV40 pre-mRNA splicing in nuclear extract. Lane 1, nuclear extract alone; lanes 2 to 4 and 5 to 7, increasing amounts (50, 100, and 200 ng) of Clk and ClkR, respectively. (B) Effect of Clk and ClkR on SV40 pre-mRNA splicing in S100 extracts complemented with ecASF (750 ng). Lane 1, ecASF only; lanes 2 to 4 and 5 to 7, increasing amounts (50, 100, and 200 ng) of Clk and ClkR, respectively. Positions of pre-mRNA, spliced products, and intermediates are shown. A schematic of the pre-mRNA is shown at the bottom.
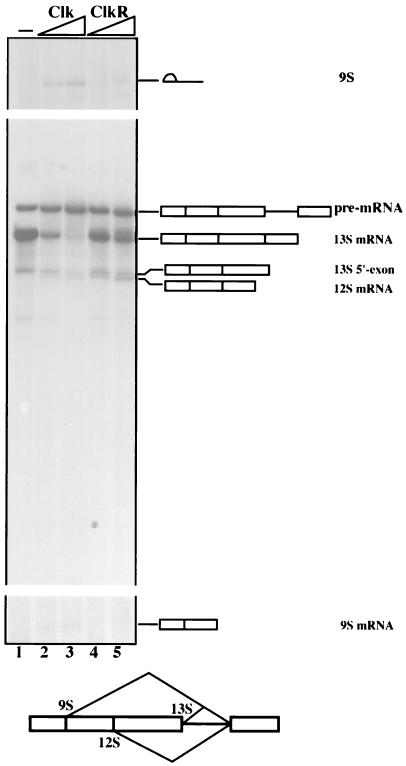
The above results are significant in part because they provide the first direct evidence that changes in SR protein phosphorylation can influence alternative splicing. We therefore wished to determine whether this effect could be extended to an additional pre-mRNA and to another SR protein. To this end, we used the adenovirus E1a pre-mRNA, which contains three 5′ splice sites and is known to be responsive to SR proteins in vitro (21), and SC35 (prepared from baculovirus) instead of ASF/SF2 in splicing reactions performed with S100 supplemented with Clk or ClkR (Fig. 7). In the absence of added kinase, nearly all splicing was from the downstream-most 5′ splice site, to generate 13S mRNA, with a very small amount of splicing from the intermediate 12S mRNA 5′ splice site. Addition of Clk (lanes 2 and 3) greatly reduced 13S splicing and weakly activated 12S, but significantly enhanced splicing from the far upstream 9S 5′ splice site (because of the small size of the mRNA, the lariat intron is the most readily detected product). ClkR, in contrast, had no significant effect on E1a splicing, with the exception that the very small increase in 12S splicing was still observed (lanes 4 and 5). Overall, however, the behavior of SC35 and bvASF was similar, suggesting that ASF/SF2 and SC35 respond similarly to Clk/Sty-induced changes in phosphorylation status. Furthermore, these results together provide direct evidence that changes in SR protein phosphorylation can be a mechanism for modulating alternative splicing.
FIG. 7.
Clk and ClkR modulate adenovirus E1a pre-mRNA alternative splicing in S100 extracts complemented with SC35. All samples contained E1a pre-mRNA and baculovirus-produced SC35 (500 ng). Lane 1, SC35 only; lanes 2 and 3 and 4 and 5, increasing amounts (100 and 200 ng) of Clk and ClkR, respectively. Positions of pre-mRNA, spliced products, and intermediates are shown. A schematic of the pre-mRNA is shown at the bottom.
DISCUSSION
The experiments described above have provided evidence that Clk/Sty kinase directly targets SR proteins and influences their activity in splicing. There are almost certainly additional proteins that are phosphorylated during the splicing cycle (43), but our data suggests that they are not functionally significant targets of Clk/Sty, at least not to the extent that their activity is affected in the preincubation assays we employed here. Our experiments have also shown that hyper- and hypophosphorylation both affect SR protein function, in each case reducing their activity in splicing, with constitutive, activated, and alternative splicing all being similarly affected. While the molecular interactions affected in the two situations may well be distinct, our findings indicate that the level of SR protein phosphorylation must be precisely maintained to provide optimum activity. We discuss these and other implications of our data below.
Clk/Sty has previously been shown to be capable of phosphorylating SR proteins in vitro (8, 9) and to influence alternative splicing in transient transfection assays (12). So in one sense it is not unexpected that Clk can influence the splicing activity of SR proteins in vitro. However, in the initial two-hybrid screen that suggested an SR protein-Clk/Sty interaction, a number of other potential targets, including hnRNP proteins, were also detected (8). Furthermore, a number of proteins can be phosphorylated by Clk in vitro, and the specificity of the kinase appears not to be limited strictly to RS dipeptide repeats (8, 9). Finally, transiently expressed Clk/Sty can alter the subcellular localization of SR proteins, which had suggested that the effects on splicing may be indirect (3). Thus our data are significant because they indicate that SR proteins are authentic, functionally important targets of Clk/Sty.
Overexpression of Clk/Sty by transient transfection (12) resulted in changes in splicing of two reporter transcripts, derived from Ela and Clk/Sty itself, similar to those reported here, consistent with decreased SR protein activity. In the case of the Clk/Sty pre-mRNA, this reflected decreased inclusion of an alternative exon, which would result in synthesis of a truncated protein, likely part of an autoregulatory mechanism. The effect of ClkR overexpression was also tested with the Clk/Sty pre-mRNA, and, surprisingly, the mutant kinase resulted in enhanced inclusion of the alternative exon, the opposite effect of wild-type Clk/Sty. If this change in splicing was also due to alterations in SR protein activity, it would seem to indicate that expression of the mutant kinase increased their activity. This contrasts with our in vitro results in which the mutant either had no effect or decreased activity, depending on the assay conditions. Although there are numerous explanations for these differences, one is that the in vivo results reflect a specialized mechanism related to the putative autoregulatory pathway, such that the splicing machinery directly responds to decreased Clk/Sty activity as opposed to altered SR protein activity.
Clk/Sty and other Clk family members contain an N-terminal domain enriched in RS dipeptides, which is essential for the yeast two-hybrid interaction with SR proteins (8). Given the known function of RS domains in protein-protein interactions, could it be that a protein-protein interaction between Clk or ClkR and SR proteins, as opposed to phosphorylation changes, was responsible for the effects we observed? While such interactions may indeed contribute to Clk/Sty function (12), they are unlikely to have played a significant role in our experiments. In NE, Clk, but not ClkR, inhibited splicing. However, when the ability of these two GST proteins to interact with SR proteins in the NE was determined using “pull-down” assays, binding was observed only with ClkR, and this binding was very weak (unpublished data). Thus the inhibitory effect of Clk on splicing can only have been due to increased phosphorylation and not to protein-protein interaction, consistent with the SR protein hyperphosphorylation observed. ClkR is also unlikely to work solely by protein-protein interaction, as the mutant kinase derivative interacted with phosphorylated SR proteins, including bvASF (unpublished data), but did not inhibit their activity; only unphosphorylated SR proteins (i.e., ecASF) were inhibited. Thus ClkR most likely functioned by a dominant-negative mechanism, blocking the activity of endogenous kinases, which is consistent with the observed hypophosphorylation.
It is remarkable that both over- and underphosphorylation have the same effect on SR protein activity in splicing. Based on the known effects of phosphorylation on SR protein interactions, we propose specific, distinct mechanisms that lead in each case to reduced activity. It has already been shown that SR protein underphosphorylation can inhibit splicing. Xiao and Manley (49) showed that addition of nonspecific RNA to splicing reactions, together with ecASF, prevented both phosphorylation and splicing, and Cao et al. (4) used a related approach to reach the same conclusion. Given that unphosphorylated RS domains greatly enhance nonspecific RNA binding by SR proteins (38, 49), it could be that sequence-nonspecific interactions with RNA sequester SR proteins and interfere with function, as seen in the experiments just described, in the data presented here (where hypophosphorylation was induced by the dominant-negative ClkR), and possibly in vivo (see below). Additionally, underphosphorylation could interfere with required protein-protein interactions, such as the one between ASF/SF2 and U1 snRNP, which likely helps define the 5′ splice site (24, 49).
Our data provides the first direct evidence that SR protein hyperphosphorylation can interfere with the proteins’ activity in splicing. One explanation for this again stems from the fact that phosphorylation enhances the interaction between ASF/SF2 and U1 snRNP (49). It is possible that this interaction becomes too strong if the SR protein is hyperphosphorylated, which could then prevent or interfere with the dissociation of U1 snRNP from the 5′ splice that must occur during the splicing cycle. Alternatively, under these conditions, the SR protein-U1 snRNP interaction might be so strong that complexes form independently of RNA, sequestering SR proteins and/or U1 snRNP. Indeed, evidence that this can happen in vivo is discussed below. Finally, SR protein hyperphosphorylation per se may not be inhibitory, but it could prevent dephosphorylation of the proteins, known to be required for constitutive splicing (4, 50). This could reflect either a direct competition between the excess Clk and the relevant phosphatase, or simply that the hyperphosphorylated SR protein is too heavily modified to be dephosphorylated effectively.
It is intriguing that treatments designed to induce either hypo- or hyperphosphorylation of SR proteins in vivo can have similar effects on the proteins’ subnuclear localization, analogous to our demonstration that both modifications can similarly affect activity in vitro. Misteli and Spector (30) found that the addition of protein phosphatase 1 to permeabilized cells caused hypophosphorylation of SR proteins and, unexpectedly, resulted in a diffuse nuclear distribution of SR proteins, similar to that observed when SRPK1 or Clk/Sty was overexpressed (8, 18). Furthermore, both kinase and phosphatase inhibitors were shown to alter the dynamic distribution of a GFP-ASF/SF2 fusion protein in living cells (29). Although the molecular interactions influenced by phosphorylation that affect SR protein subnuclear localization are unknown, it is conceivable, perhaps even likely, that they involve, at least in part, the same factors responsible for the effects on splicing observed in vitro.
It is likely that cells use both hyper- and hypophosphorylation of SR proteins to control gene expression, and there are already several apparent examples. Early studies with MAb104, even before the identity of the antigen was known, showed enhanced reactivity in the nuclei of M-phase cells relative to interphase cells (35). Subsequently, it was shown that SR proteins isolated from 32P-labeled HeLa cells arrested in M phase were more extensively phosphorylated than were SR proteins from cells arrested at G1/S phase (18). Coupled with our data that hyperphosphorylated SR proteins are repressed, these findings suggest that mRNA splicing is inhibited by SR protein hyperphosphorylation during M phase. This is in keeping with the silencing of gene expression that occurs during mitosis, previously shown to involve inhibition by phosphorylation of the transcription (reviewed in reference 17a) and polyadenylation (7) machineries. In Jurkat T cells induced to undergo apoptosis, but not in normal cells, a stable complex containing U1 snRNP and several apparently hyperphosphorylated SR proteins can be isolated (41). Details of what activates formation of this complex and the functional significance of the complex are lacking, but its existence is intriguing, especially in light of our suggestion that such a complex might form following SR protein hyperphosphorylation. This could result in the sequestration of SR proteins and contribute to changes in splicing patterns during apoptosis. SR proteins have also recently been reported to be hyperphosphorylated, and inactive, in early embryos of the nematode Ascaris lumbricoides (35a). Concurrent with activation of zygotic gene expression, the proteins become partially dephosphorylated and functional in in vitro splicing assays, again consistent with the view that hyperphosphorylation inhibits SR protein activity. Finally, it has been reported that SR proteins become hypophosphorylated during adenovirus infection and that SR proteins isolated from infected cells display reduced activity in splicing assays (23). Although it is unclear if the reduced activity was due solely to hypophosphorylation, as the proteins should be rapidly phosphorylated and activated upon addition to splicing extracts (Fig. 4B and reference 49), these results nonetheless provide evidence that hypophosphorylation can be a physiologically significant way of modulating SR protein activity and hence splicing, at least during viral infection.
In summary, our results have established that SR proteins are functionally significant targets of Clk/Sty kinase. Our data has also shown that SR protein hyper- and hypophosphorylation have similar inhibitory effects on splicing activity, influencing constitutive, activated, and alternative splicing. These findings provide a firm foundation for the hypothesis that the Clk kinases, four of which are now known (32), participate in tissue- and/or cell-specific control of SR protein activity and that changes in SR protein modification, as well as abundance (52), can contribute to the regulation of alternative splicing.
ACKNOWLEDGMENTS
We are grateful to K. G. K. Murthy for valuable discussions, R. Gattoni and J. Stevenin (CNRS, France) for the ASF baculovirus, D. Reifman for help with preliminary experiments, H. Shi for technical assistance, and M. Riley and I. Boluk for help with the manuscript.
This work was supported by NIH grant GM48259 to J.L.M. T.P. was supported by the National Cancer Institute of Canada and is a Distinguished Scientist of the Medical Research Council of Canada.
REFERENCES
- 1.Cáceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cáceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 3.Cáceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;1:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao W, Jamison S F, Garcia-Blanco M A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;12:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinali B, Cohen P, Lamond A I. Protein phosphatase PP1 can modulate 5′-splice site selection in a HeLa splicing extract. FEBS Lett. 1994;352:276–280. doi: 10.1016/0014-5793(94)00973-2. [DOI] [PubMed] [Google Scholar]
- 6.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;8:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgan D F, Murthy K G, Prives C, Manley J L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 8.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intracellular distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 9.Colwill K, Feng L L, Yeahkley J M, Gish G D, Caceres J F, Pawson T, Fu X-D. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine-arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 10.Du C, McGuffin M E, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 1998;6:741–750. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- 11.Duncan P I, Howell B W, Marius R M, Drmanic S, Douville E M, Bell J C. Alternative splicing of STY, a nuclear dual specificity kinase. J Biol Chem. 1995;270:21524–21531. doi: 10.1074/jbc.270.37.21524. [DOI] [PubMed] [Google Scholar]
- 12.Duncan P I, Stojdl D F, Marius R M, Bell J C. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;10:5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan P I, Stojdl D F, Marius R M, Scheit K H, Bell J C. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;2:300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- 14.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 15.Fu X D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;65:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 16.Gallego M E, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;7:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 17a.Gottesfeld J M, Forbes D J. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 18.Gui J G, Lane W S, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 19.Gui J G, Tronchere H, Chandler S E, Fu X-D. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanes J, von der Kammer H, Klaudiny J, Scheit K H. Characterization by cDNA cloning of two new human protein kinases: evidence by sequence comparison of a new family of mammalian protein kinases. J Mol Biol. 1994;244:665–672. doi: 10.1006/jmbi.1994.1763. [DOI] [PubMed] [Google Scholar]
- 21.Harper J E, Manley J L. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;1:19–29. [PMC free article] [PubMed] [Google Scholar]
- 22.Jamison S F, Pasman Z, Wang J, Will C, Luhrmann R, Manley J L, Garcia-Blanco M A. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR protein. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 24.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 25.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;7:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 26.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 27.Mermoud J E, Cohen P T W, Lamond A I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermoud J E, Cohen P T W, Lamond A I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:3679–3688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misteli T, Caceres J F, Spector D L. The dynamics of a pre-mRNA splicing factor. Nature. 1997;387:523–526. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 30.Misteli T, Spector D L. Serine/threonine phosphatase I modulates the subnuclear distribution of pre-mRNA splicing factor. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayler O, Schnorrer F, Stamm S, Ullrich A. The cellular localization of the murine serine/arginine-rich protein kinase CLK2 is regulated by serine 141 autophosphorylation. J Biol Chem. 1998;51:34341–34348. doi: 10.1074/jbc.273.51.34341. [DOI] [PubMed] [Google Scholar]
- 32.Nayler O, Stamm S, Ullrich A. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem J. 1997;326:693–700. doi: 10.1042/bj3260693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng X, Mount S M. Genetic enhancement of RNA processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ring H Z, Lis J T. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth M B, Murthy C, Gall J G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Sanford J R, Bruzik J P. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si Z H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 Ta exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;9:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tacke R, Chen Y, Manley J L. Sequence specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazi J, Daugeron M C, Cathala G, Brunel C, Jeanteur P. Adenosine phosphorothioates (ATPαS and ATPτS) differentially affect the two steps of mammalian pre-mRNA splicing. J Biol Chem. 1992;267:4322–4326. [PubMed] [Google Scholar]
- 41.Utz P J, Hottelet M, van Venrooij W J, Anderson P. Association of phosphorylated serine/arginine (SR) splicing factors with the U1-small ribonucleoprotein (snRNP) autoantigen complex accompanies apoptotic cell death. J Exp Med. 1998;4:547–560. doi: 10.1084/jem.187.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–302. [PubMed] [Google Scholar]
- 43.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;10:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H Y, Lin W, Dyck J A, Yeakley J M, Songyang Z, Cantley L C, Fu X D. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;4:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Xiao S H, Manley J L. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;14:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 49.Xiao S-H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 50.Xiao S H, Manley J L. Phosphorylation-dephosphorylation differentially affects activities of splicing factors ASF/SF2. EMBO J. 1998;21:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;5:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 52.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 53.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]