Abstract
Simple Summary
Gram-negative bacteria are common causes of urinary tract infections (UTIs), some of which can resist treatment by antibiotics, including carbapenems, which are last resort treatment options. This study aimed to report the resistance of some Gram-negative bacteria causing complicated UTIs to carbapenems at two important hospitals in Cairo, Egypt, and to determine the possible transfer of this resistance to other bacterial species. The collected bacteria were tested for antibiotic resistance and detection of the genes responsible for this resistance. A total of 256 Gram-negative bacterial clinical isolates were collected, 65 (25.4%) of which showed carbapenem resistance. The detected carbapenem resistance genes were blaOXA-48, blaVIM, blaKPC, and blaNDM genes. The blaOXA-48, among other genes, was successfully transferred to a previously susceptible bacteria, making it resistant. The study concluded that the rate of carbapenem resistance among Gram-negative bacteria causing UTIs in Cairo, Egypt is relatively high and can be transferred among bacterial hosts.
Abstract
Gram-negative bacteria are common causes of urinary tract infections (UTIs). Such pathogens can acquire genes encoding multiple mechanisms of antimicrobial resistance, including carbapenem resistance. The aim of this study was to detect the carbapenemase-producing ability of some Gram-negative bacterial isolates from urine specimens of patients suffering from complicated UTIs at two vital tertiary care hospitals in Cairo, Egypt; to determine the prevalence of carbapenemase genes among plasmid-bearing isolates; and explore the possibility of horizontal gene transfer to other bacterial species. The collected isolates were subjected to antimicrobial susceptibility testing, phenotypic analysis of carbapenemase production, and molecular detection of plasmid-borne carbapenemase genes, then the extracted plasmids were transformed into competent E. coli DH5α. A total of 256 Gram-negative bacterial clinical isolates were collected, 65 (25.4%) isolates showed carbapenem resistance of which 36 (55.4%) were carbapenemase-producers, and of these 31 (47.7%) harbored plasmids. The extracted plasmids were used as templates for PCR amplification of blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP carbapenemase genes. The blaOXA-48 gene was detected in 24 (77.4%) of the tested isolates while blaVIM gene was detected in 8 (25.8%), both blaKPC and blaNDM genes were co-present in 1 (3.2%) isolate. Plasmids carrying the blaOXA-48 gene from 4 K. pneumoniae clinical isolates were successfully transformed into competent E. coli DH5α. The transformants were carbapenemase-producers and acquired resistance to some of the tested antimicrobial agents as compared to untransformed E. coli DH5α. The study concluded that the rate of carbapenem resistance among Gram-negative bacterial uropathogens in Cairo, Egypt is relatively high and can be transferred horizontally to other bacterial host(s).
Keywords: antimicrobial resistance, carbapenemase genes, urinary tract infections, plasmids, Gram-negative bacteria, carbapenem-resistant Enterobacteriaceae
1. Introduction
Urinary tract infections (UTIs) represent one of the most widespread bacterial infections that require antimicrobial treatment [1,2]. They are caused by uropathogens that are capable of colonization in the naturally sterile urinary tract [3]. UTIs can either be uncomplicated or complicated, the difference being that uncomplicated UTIs affect individuals that have no functional or anatomical abnormalities of the urinary tract, while complicated UTIs (cUTIs) are associated with anatomically abnormal urinary tracts and/or the presence of one or more risk factors complicating the infection. These risk factors include urolithiasis, renal insufficiency, urinary obstruction, draining devices and indwelling catheters, voiding dysfunction, surgery of the urinary tract, compromised immune system, diabetes mellitus, the male gender, and pregnancy [4,5]. Antimicrobial resistance and subsequent treatment failure are common in cUTI [6].
The emergence of antimicrobial resistance by bacteria which produce extended spectrum β-lactamase enzymes (ESBLs) has limited the effectiveness of β-lactams used to treat UTIs [7]. In addition to this, co-resistance among uropathogens to β-lactams, aminoglycosides, and fluoroquinolones further limits the available treatment options [8]. As ESBL-producers spread throughout healthcare facilities, carbapenems such as imipenem/cilastatin, meropenem, doripenem, and ertapenem were used more frequently, and were regarded as the last resort treatment for infections caused by ESBL-producers and multidrug-resistant (MDR) pathogens [9,10]. Unfortunately, the increased use of carbapenems has led to the emergence of carbapenem resistance (CR) in Gram-negative bacteria (GNB), such as Enterobacterales, Pseudomonas spp., and Acinetobacter spp., as well as the emergence of pathogens carrying up to three different carbapenemase genes. These CR pathogens are capable of spreading in the hospital setting and in the community [11,12]. The rapid spread of CR has posed a global public health crisis owing to the lack of novel antimicrobials that could be used as an alternative last resort treatment [13,14].
There are three major mechanisms of CR: overexpression of efflux pumps, porin-mediated resistance, and enzyme-mediated resistance [15,16]. The latter is due to the production of carbapenemases, which are β-lactamase enzymes capable of hydrolyzing β-lactam antimicrobials and carbapenems. Based on the Ambler classification system, carbapenemases belong to three classes of β-lactamases according to their chemical structures and substrate specificity: classes A, B, and D [15]. Classes A and D possess a serine residue at the active site that facilitates β-lactam ring opening; they are therefore called serine β-lactamases (SBLs) [17]. Class B comprises metallo-β-lactamases (MBLs), the active site of which contains zinc ions, hence their name [18].
Most carbapenemase-producers (CPs) are MDR pathogens carrying multiple resistance determinants to other antimicrobial agents [19]. Carbapenemase-mediated resistance is of great concern, as these enzymes are encoded by plasmid-borne genes which can be transferred horizontally to other bacterial species [10]. The most common carbapenemases, based on carbapenem hydrolysis and geographical dissemination of outbreaks, are Klebsiella pneumoniae carbapenemases (KPC), New Delhi metallo-β-lactamase (NDM), imipenem-resistant Pseudomonas-type carbapenemases (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and oxacillinase (OXA-48-like) types. They are encoded by blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48 genes, respectively [20,21]. Enterobacterales are extensively reported as CPs, especially Klebsiella pneumoniae and Escherichia coli. However, carbapenemase genes and their mobile genetic elements (MGEs) are not limited to these species [22]. CR, especially due to carbapenemase production, is already widespread and well-reported in some parts of the world including Europe, South America, and Asia, while the situation is not well-reported in other parts such as Africa [14].
Our aim was to detect the carbapenemase-producing ability of GNB isolates recovered from urine specimens of patients suffering from cUTIs at two vital tertiary care hospitals in Cairo, Egypt, to detect the prevalence of carbapenemase genes among plasmid-bearing isolates, and to explore the possibility of horizontal gene transfer to other bacterial host(s).
2. Materials and Methods
2.1. Collection of Clinical Isolates
Starting November 2019 to November 2020, 256 Gram-negative clinical bacterial isolates were collected from the microbiology laboratories of El-Demerdash and Kasr Al-Ainy Tertiary Care Hospitals (each with approximately 3200 beds), Cairo, Egypt. According to the hospitals’ records, all isolates were non-duplicates and were recovered from urine specimens of patients suffering from cUTIs. Written and oral informed consents were obtained from the patients or their legal guardians after explaining the purpose of the study. The inclusion criteria were: admission to the above-mentioned hospitals during the study period; patients having colony count of uropathogens >105 cfu/mL in urine sample, indicating significant bacteriuria; patients with risk factors of cUTIs (urolithiasis, renal insufficiency, urinary obstruction, indwelling catheters, voiding dysfunction, surgery of the urinary tract, diabetes mellitus); and consent of the patient or legal guardian. Patients with uncomplicated UTIs were excluded from the study. The study complied with the principles laid out in the Declaration of Helsinki and was approved by the ethics committee of Faculty of Pharmacy Ain Shams University (ENREC-ASU-2019-98).
The identification of isolates was based on their microscopic, morphologic, and biochemical characteristics as stated in Bergey’s manual of determinative bacteriology [23]. The bacterial identification was confirmed by comparing the results to the hospitals’ records. Identification of carbapenem-resistant isolates was further confirmed by using the commercially available API® 20E identification kit (BioMérieux® SA, Marcy l’Etoile, France).
2.2. Antimicrobial Susceptibility of the Collected Isolates
The Kirby–Bauer disk diffusion test was performed on all collected isolates [24] using 4 carbapenem disks including imipenem (10 μg), meropenem (10 μg), ertapenem (10 μg), and doripenem (10 μg), followed by measuring the inhibition zone diameters, then interpreting the susceptibility by referring to Clinical and Laboratory Standards Institute (CLSI) guidelines 2020 [25]. Isolates that showed resistance to at least one of the above-mentioned carbapenems were considered CR-GNB, and were selected for further studying. They were each tested against a panel of 13 additional disks of antimicrobials commonly used to treat UTIs, including amoxicillin/clavulanic acid (30 μg), ampicillin/sulbactam (20 μg), cefoxitin (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefepime (30 μg), amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), trimethoprim/sulfamethoxazole (25 μg), nitrofurantoin (300 μg), and fosfomycin (200 μg). The reference strain E. coli ATCC® 25922 was used as control.
2.3. Phenotypic Detection of CPs
2.3.1. Blue-Carba Test
This test was validated for the detection of CPs directly from bacterial cultures of Enterobacterales, Pseudomonas, and Acinetobacter species. It was carried out as described by Pires et al. [26]. The reference strain E. coli ATCC® 25922 was used as a negative control.
2.3.2. Modified Carbapenem Inactivation Method (mCIM)
This test was used for the detection of carbapenemases in Enterobacterales and P. aeruginosa, as recommended by the CLSI guidelines 2020 [25]. A meropenem disk was submerged in a 2 mL TSB suspension of the tested isolate and incubated at 37 °C for 4 h. The disk was then transferred to the center of a Mueller–Hinton agar plate previously inoculated with E. coli ATCC® 25922. Following overnight incubation, the inhibition zone around the disk was measured. Isolates showing a 6–15 mm zone diameter or the presence of pinpoint colonies within a 16–18 mm zone were considered CPs [25].
2.3.3. EDTA-Modified Carbapenem Inactivation Method (eCIM)
The eCIM test was used together with mCIM to detect the production of MBLs in Enterobacterales. eCIM can only be performed for Enterobacterales isolates that give a positive mCIM test result. This test was performed and interpreted as described in CLSI guidelines 2020 [25].
2.4. Phenotypic Analysis Using Heatmap Signature
The antimicrobial resistance profiles and carbapenemase production results were used to generate a dendrogram showing heatmap signatures of the isolates to determine their phenotypic relatedness. It was generated by Morpheus online software (https://software.broadinstitute.org/morpheus/, accessed on 15 April 2021) using Euclidean distances.
2.5. Molecular Detection of Plasmid-Borne Carbapenemase Genes
2.5.1. Extraction of Plasmid DNA from Carbapenem-Resistant Isolates
Overnight cultures of the CR-GNB isolates were grown in Luria Bertani broth containing 8 μg/mL meropenem as a selective pressure to enhance plasmid recovery. The extraction of plasmid DNA was carried out using GeneJet Plasmid Miniprep Kit Catalog number: K0502 for extraction or large-sized plasmids (Thermo Fisher Scientific, Waltham, MA, USA; https://www.thermofisher.com/order/catalog/product/K0502#/K0502, accessed on 15 April 2021). The extracted plasmids were analyzed via agarose gel electrophoresis (Sub-Cell® GT Agarose Gel Electrophoresis Systems, Bio-Rad Laboratories, Hercules, CA, USA) and visualized by UV transilluminator (Benchtop 3UV transilluminator, UVP, LLC, Upland, CA, USA) [27].
2.5.2. Amplification of Some Plasmid-Encoded Carbapenemase Genes
The extracted plasmids were used as templates for PCR using the appropriate primers synthesized by invitrogen® (Thermo Fisher Scientific, Waltham, MA, USA), and Dream Taq™ Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The annealing temperatures (Ta) and primers for blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP genes are shown in Table 1, and the PCR conditions are listed in Table 2. The amplified PCR products were analyzed using agarose gel electrophoresis, and the sizes of the DNA fragments were determined by comparison to a 100 bp DNA ladder (GeneRuler 100 bp DNA ladder, Thermo Fisher Scientific, Waltham, MA, USA).
Table 1.
Primers used in this study, expected PCR product sizes, and annealing temperatures (Ta).
| PCR Reaction | Gene | Primer | Primer Sequence (5′ → 3′) | Expected PCR Product Size (bp) | Ta (°C) | References |
|---|---|---|---|---|---|---|
| Multiplex | bla KPC | Pf | TGTCACTGTATCGCCGTC | 1011 | 50 | [28] |
| Pr | CTCAGTGCTCTACAGAAAACC | |||||
| bla NDM | Pf | GGTTTGGCGATCTGGTTTTC | 621 | [29,30] | ||
| Pr | CGGAATGGCTCATCACGAT | |||||
| Multiplex | bla VIM | Pf | TCTACATGACCGCGTCTGTC | 748 | 50 | [31] |
| Pr | TGTGCTTTGACAACGTTCGC | |||||
| bla OXA-48 | Pf | GCGTGGTTAAGGATGAACAC | 438 | [28,30] | ||
| Pr | CATCAAGTTCAACCCAACCG | |||||
| Monoplex | bla IMP | Pf | CTACCGCAGCAGAGTCTTTG | 587 | 50 | [32] |
| Pr | AACCAGTTTTGCCTTACCAT | |||||
| Multiplex | aac(6′)-Ib | Pf | TTGCGATGCTCTATGAGTGG | 358 | 49 | [33] |
| Pr | CGTTTGGATCTTGGTGACCT | |||||
| bla SHV | Pf | GGTTATGCGTTATATTCGCC | 867 | [34] | ||
| Pr | TTAGCGTTGCCAGTGCTC | |||||
| Multiplex | bla CTX-M | Pf | CGCTTTGCGATGTGCAG | 550 | 51 | [35] |
| Pr | ACCGCGATATCGTTGGT | |||||
| bla TEM | Pf | ATGAGTATTCAACATTTCCG | 867 | [34] | ||
| Pr | CTGACAGTTACCAATGCTTA | |||||
| ERIC-PCR | Pf | AAGTAAGTGACTGGGGTGAGCG | Variable | 45 | [36] | |
| Pr | ATGTAAGCTCCTGGGGATTCAC | |||||
Notes: blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP genes code for KPC, NDM, VIM, OXA-48-like, and IMP carbapenemases, respectively, aac(6ʹ)-Ib gene codes for aminoglycoside 6ʹ-N-acetyltransferase type Ib, blaCTX-M, blaSHV, and blaTEM genes code for ESBLs. Abbreviations: Pf, forward primer; Pr, reverse primer; Ta, annealing temperature.
Table 2.
Conditions of PCR cycles (thermocycler input data).
| PCR Reaction | Primary Denaturation | Secondary Denaturation | Annealing | Extension | No. of Cycles | Final Extension |
|---|---|---|---|---|---|---|
| Multiplex blaKPC/blaNDM |
95 °C 4 min |
95 °C 30 s |
50 °C 45 s |
72 °C 1 min |
30 | 72 °C 10 min |
| Multiplex blaVIM/blaOXA-48 |
95 °C 4 min |
95 °C 30 s |
50 °C 45 s |
72 °C 1 min |
30 | 72 °C 10 min |
| Monoplex blaIMP |
95 °C 4 min |
95 °C 30 s |
50 °C 45 s |
72 °C 1 min |
30 | 72 °C 10 min |
| Multiplex aac(6′)-Ib/blaSHV |
95 °C 4 min |
95 °C 30 s |
49 °C 45 s |
72 °C 1 min |
30 | 72 °C 10 min |
| Multiplex blaCTX-M/blaTEM |
95 °C 4 min |
95 °C 30 s |
51 °C 45 s |
72 °C 1 min |
30 | 72 °C 10 min |
| ERIC-PCR | 95 °C 15 min |
94 °C 30 s |
45 °C 45 s |
72 °C 7 min |
45 | 72 °C 10 min |
Notes: blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP genes code for KPC, NDM, VIM, OXA-48-like, and IMP carbapenemases, respectively, aac(6ʹ)-Ib gene codes for aminoglycoside 6ʹ-N-acetyltransferase type Ib, blaCTX-M, blaSHV, and blaTEM genes code for ESBLs.
2.5.3. Sequencing of PCR Products
Some PCR products of the amplified genes were sent for sequencing at Macrogen Inc. (Seoul, Korea) using an Applied Biosystems 3730XL sequencer. The assembly of the obtained forward and reverse sequence files into the final consensus sequence was completed using BioEdit v7.2.5 software [37]. The open reading frames (ORFs) of the final contigs were detected using FramePlot 4.0beta (http://nocardia.nih.go.jp/fp4/, accessed on 23 April 2021) [38]. The sequencing data were analyzed using the basic local alignment search tool BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 23 April 2021). The blaOXA-48, blaNDM, blaKPC, and blaVIM nucleotide sequences were annotated and submitted into the NCBI GenBank database under the accession codes MW562895, MZ092838, MZ092839, and MZ092840, respectively.
2.5.4. Transformation
E. coli DH5α is a standard strain that does not harbor any resistance genes. Competent E. coli DH5α cells were prepared according to the modified Hanahan method [39]. The extracted plasmids were transformed into competent E. coli DH5α to compare the phenotypic properties of the transformants with those of the parent clinical isolates. Chemical transformation was carried out as described in Sambrook and Russell [27]. Transformants were cultured on LB/ampicillin and LB/meropenem agar plates at concentrations of 100 µg/mL and 8 µg/mL, respectively. Untransformed E. coli DH5α was used as a negative control. Plasmid DNA was extracted from transformants that showed growth on LB/ampicillin and/or LB/meropenem plates, and was used as templates for PCR to confirm the presence of carbapenemase genes on the transformed MGEs, as well as to detect the presence of other antimicrobial resistance genes including ESBL genes (blaSHV, blaCTX-M, and blaTEM), and the aac(6′)-Ib gene which confers resistance to aminoglycosides. The annealing temperatures (Ta) and primers for blaSHV, blaCTX-M, blaTEM, and aac(6′)-Ib genes are shown in Table 1, and the PCR conditions are in Table 2. Phenotypic tests including antimicrobial susceptibility testing, Blue-Carba, and mCIM, were also carried out for the transformants as previously described in this study, to detect the acquired phenotypic properties of the transformants.
2.6. Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR) for Isolates Containing Plasmids
ERIC-PCR is a sequence analysis tool used for epidemiological analysis of bacterial isolates to determine their genetic relatedness. DNA template preparation of the isolates was performed as described by Doyle et al. [28] and the templates were used for PCR. The primers (Table 1) and conditions used for ERIC-PCR (Table 2) were described by Codjoe et al. [36]. ERIC fingerprinting data were transformed into a binary code depending on the presence (denoted 1) or absence (denoted 0) of each band. A dendrogram was generated by the unweighted pair group method with arithmetic average (UPGMA) and Ward’s hierarchical clustering routine using Statistical Package for the Social Sciences software IBM® SPSS® version 23 (IBM Corp., Armonk, NY, USA) [40,41], and a proximity matrix by Jaccard measure was generated to calculate the similarity index of the isolates.
2.7. Statistical Analysis
Frequency tables, crosstabs, dendrogram construction, similarity index calculation, and chi-square analysis were carried out using Statistical Package for the Social Sciences software IBM® SPSS® version 23 (IBM Corp., Armonk, NY, USA) [41].
3. Results
3.1. Collection of Clinical Isolates
Based on the hospitals’ records, a total of 256 non-duplicate GNB clinical isolates were collected from patients with cUTIs (patients having colony count of uropathogen >105 cfu/mL in the urine sample and one or more risk factors of cUTIs). Among the patient population, 111 (43.4%) were females, while 145 (56.6%) were males. Based on the hospitals’ records, the chronological age of the patients ranged from 3 months to 82 years, 40 of which (15.6%) were below the age of 18 years, 96 (37.5%) were in the age range of 18–40 years, 56 (21.9%) were in the age range of 41–60 years, and 64 (25%) were above the age of 60 years.
3.2. Identification of the Collected GNB Clinical Isolates
Out of the collected 256 GNB clinical isolates, 210 (82%) were identified as Enterobacterales, including 96 (37.5%) E. coli, 87 (34%) Klebsiella spp., 18 (7%) Proteus spp., and 9 (3.5%) Enterobacter spp. The remaining GNB isolates included 31 (12%) Pseudomonas spp., and 15 (6%) Acinetobacter spp.
3.3. Antimicrobial Susceptibility of the Collected Isolates
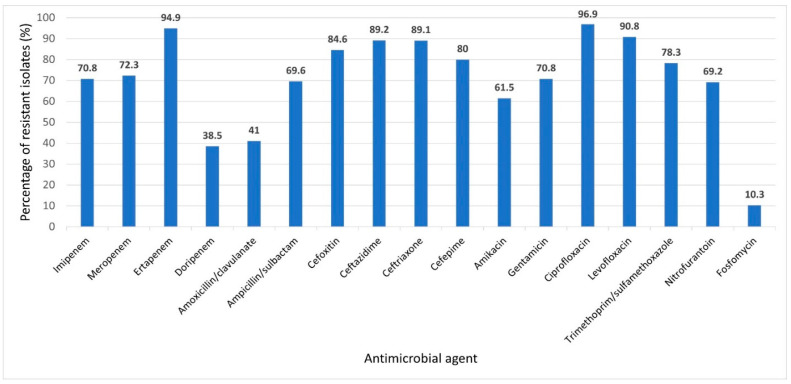
Among the 256 GNB isolates, 65 (25.4%) showed resistance to one or more of the tested carbapenems; they were categorized as carbapenem-resistant Gram-negative bacterial (CR-GNB) isolates and were selected for further studying. The CR-GNB isolates were Klebsiella spp. (28; 43.1%), including 22 K. pneumoniae and 6 K. terrigena, P. aeruginosa (19; 29.2%), Acinetobacter baumannii (8; 12.3%), E. coli (6; 9.2%), Proteus mirabilis (2; 3.1%), and Enterobacter cloacae (2; 3.1%). The results of antimicrobial susceptibility testing showed that 64 out of 65 CR-GNB isolates were MDR, showing the highest resistance rates to ciprofloxacin (96.9%), ertapenem (94.7%), levofloxacin (90.8%), ceftazidime (89.2%), and ceftriaxone (89.1%). The lowest rates of resistance were to fosfomycin (10.5%), followed by doripenem (38.5%). The percentage of antimicrobial resistance of the 65 CR-GNB isolates are demonstrated in Figure 1, and their resistance patterns relative to identity are shown in Table 3. The results of antibiogram analysis of the CR-GNB isolates in this study, as well as the identity of the isolates, are included in the supplementary material (Supplementary Tables S1–S3).
Figure 1.
Prevalence of antimicrobial resistance of 65 carbapenem-resistant Gram-negative bacterial uropathogens to various tested antimicrobial agents. Prevalence was expressed as percent of resistant isolates relative to the total tested bacterial species for each antimicrobial agent.
Table 3.
Resistance patterns to various antimicrobial agents among different tested carbapenem-resistant Gram-negative bacterial isolates (n = 65).
| Antimicrobial Class | Antimicrobial Agent | Percentage of Resistance (%) | |||||
|---|---|---|---|---|---|---|---|
|
Klebsiella spp. (n = 28) |
P. aeruginosa (n = 19) |
A. baumannii (n = 8) |
E. coli (n = 6) |
Proteus mirabilis (n = 2) |
Enterobacter cloacae (n = 2) |
||
| Carbapenems | Imipenem (10 µg) | 64.3 | 89.5 | 100 | 33.3 | 50 | 0 |
| Meropenem (10 µg) | 71.4 | 94.7 | 87.5 | 16.7 | 50 | 0 | |
| Ertapenem (10 µg) | 92.9 | ND | ND | 100 | 100 | 100 | |
| Doripenem (10 µg) | 46.4 | 42.1 | 25 | 16.7 | 50 | 0 | |
| β-lactam combination agents | Amoxicillin/clavulanic acid (30 µg) | 46.4 | ND | ND | 16.7 | 50 | 0 |
| Ampicillin/sulbactam (20 µg) | 89.3 | ND | 25 | 50 | 50 | 50 | |
| Cephalosporins | Cefoxitin (30 µg) | 89.3 | ND | ND | 83.3 | 50 | 50 |
| Ceftazidime (30 µg) | 85.7 | 94.7 | 87.5 | 100 | 50 | 100 | |
| Ceftriaxone (30 µg) | 89.3 | ND | 87.5 | 100 | 50 | 100 | |
| Cefepime (30 µg) | 78.6 | 78.9 | 87.5 | 100 | 50 | 50 | |
| Aminoglycosides | Amikacin (30 µg) | 78.6 | 47.4 | 100 | 0 | 50 | 0 |
| Gentamicin (10 µg) | 64.3 | 89.5 | 100 | 0 | 50 | 100 | |
| Fluoroquinolones | Ciprofloxacin (5 µg) | 92.9 | 100 | 100 | 100 | 100 | 100 |
| Levofloxacin (5 µg) | 92.9 | 100 | 62.5 | 100 | 50 | 100 | |
| Folate pathway inhibitors | Trimethoprim/sulfamethoxazole (25 µg) | 78.6 | ND | 50 | 100 | 100 | 100 |
| Nitrofurans | Nitrofurantoin (300 µg) | 92.9 | ND | ND | 0 | 0 | 0 |
| Fosfomycins | Fosfomycin (200 µg) | 10.7 | ND | ND | 0 | 0 | 50 |
Abbreviations: ND, not determined.
3.4. Phenotypic Detection of CPs
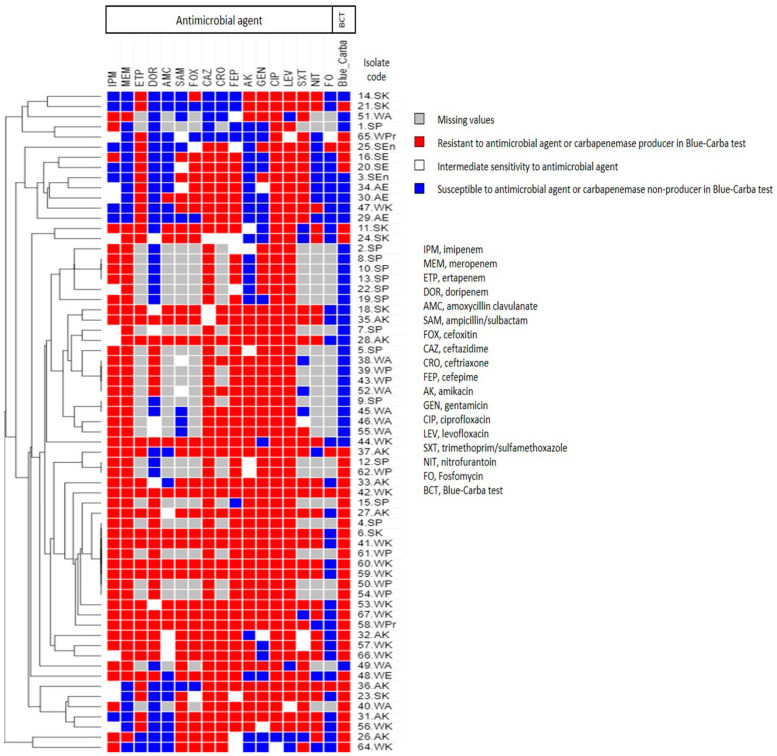
Carbapenemase enzyme production by the CR-GNB isolates (n = 65) was determined phenotypically using the Blue-Carba and mCIM tests. The production of MBLs was detected using the eCIM test, the results of which are shown in Table 4. Based on the results of Blue-Carba test, 36 out of 65 (55.4%) CR-GNB isolates were CPs, 29 were carbapenemase-producing Enterobacteriaceae (CPE) as confirmed by mCIM test. The results of antimicrobial resistance and carbapenemase production were used to generate a dendrogram showing heatmap signature of the isolates (Figure 2). This was in hopes of providing more insight on the phenotypic relatedness of the isolates. The heatmap signature of five pairs of P. aeruginosa isolates and one pair of A. baumannii isolates were found to be similar based on their phenotypic properties.
Table 4.
Phenotypic detection of carbapenemase production in carbapenem-resistant Gram-negative bacterial isolates.
| Tested Isolates | Blue-Carba Test (n = 65) [26] |
Modified Carbapenem Inactivation Method (mCIM) (n = 57) [25] |
EDTA-Modified Carbapenem Inactivation Method (eCIM) (n = 29) [25] |
|||
|---|---|---|---|---|---|---|
| No. of CPs/Total No. of Tested Isolates | % | No. of CPs/Total No. of Tested Isolates | % | No. of MBL Producers/Total No. of Tested Isolates | % | |
| Klebsiella spp. | 22/28 | 78.6 | 23/28 | 82.1 | 12/23 | 52.2 |
| P. aeruginosa | 7/19 | 36.8 | 2/19 | 10.5 | ND | ND |
| A. baumannii | 1/8 | 12.5 | ND | ND | ND | ND |
| E. coli | 3/6 | 50 | 4/6 | 66.7 | 3/4 | 75 |
| Proteus mirabilis | 2/2 | 100 | 1/2 | 50 | 0/1 | 0 |
| Enterobacter cloacae | 1/2 | 50 | 1/2 | 50 | 1/1 | 100 |
Notes: Blue-Carba test is a phenotypic test for the detection of carbapenemase production in Enterobacterales, Pseudomonas spp., and Acinetobacter spp. Modified carbapenem inactivation method (mCIM) test is a phenotypic test for the detection of carbapenemase production in Enterobacterales and Pseudomonas spp., but not Acinetobacter spp. EDTA-modified carbapenem inactivation method (eCIM) is carried out only for Enterobacterales that give a positive result in mCIM. CPs, carbapenemase-producers; MBL, metallo-β-lactamase; ND, not determined.
Figure 2.
Heatmap of carbapenem-resistant Gram-negative bacterial isolates (n = 65) in this study based on their antimicrobial resistance patterns and carbapenemase enzyme production (Blue-Carba test results). This heatmap was generated by using Morpheus online software using Euclidean distances (https://software.broadinstitute.org/morpheus/, accessed on 15 April 2021).
3.5. Extraction of Plasmid DNA and Amplification of Carbapenemase Genes
Out of the 65 CR-GNB isolates, 31 (47.7%) harbored plasmids. Agarose gel electrophoresis of some of the extracted plasmids is shown in Supplementary Figure S1. The plasmid extracts were used as templates for PCR amplification of the carbapenemase genes blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP. The PCR results of multiplex blaVIM/blaOXA-48 revealed that blaOXA-48 gene was detected in the plasmids of 24 (77.4%) of the tested isolates, while blaVIM gene was detected in 8 (25.8%) of the isolates. The PCR results of multiplex blaKPC/blaNDM showed that both blaKPC and blaNDM genes were co-present in one (3.2%) isolate. The blaIMP gene was not detected in any of the plasmid extracts. The results of agarose gel electrophoresis of multiplexes blaKPC/blaNDM and blaVIM/blaOXA-48 of some tested CR-GNB isolates are demonstrated in Supplementary Figures S2 and S3, respectively.
3.6. Transformation of Plasmids into Competent E. coli DH5α
Plasmid extracts that harbored any of the tested carbapenemase genes were transformed into competent E. coli DH5α to compare the phenotypic properties of the transformants to those of the parent clinical isolates. Transformants were cultured on LB/ampicillin and LB/meropenem agar plates at concentrations of 100 µg/mL and 8 µg/mL, respectively. Untransformed E. coli DH5α was used as a negative control.
The successfully transformed MGEs in the current study were all extracted from K. pneumoniae clinical isolates, transformation from other clinical isolates was not successful. One transformant (code: TS24.SK) showed growth on both LB/ampicillin and LB/meropenem plates, indicating successful transformation of a MGE carrying at least one carbapenem resistance gene. A single colony of this transformant was picked and subcultured on an LB/meropenem plate. Visible growth was observed after overnight incubation. Antimicrobial susceptibility testing revealed the newly acquired properties of the transformant, where it acquired resistance to meropenem, ertapenem, ampicillin/sulbactam, cefoxitin, ceftazidime, ciprofloxacin, and levofloxacin, and intermediate sensitivity to imipenem, ceftriaxone, and cefepime.
Three other transformants (codes: TS37.AK, TS59.WK, and TS66.WK) showed growth on LB/ampicillin agar plates, indicating successful transformation of an MGE carrying at least one antimicrobial resistance gene. A single colony of each transformant was picked and subcultured on LB/ampicillin plates to increase the plasmid copy number by selective pressure, then further subcultured on LB/meropenem plates. Weak but visible growth was observed on the LB/meropenem plates after overnight incubation. Antimicrobial susceptibility testing revealed that these transformants acquired resistance to ampicillin/sulbactam, cefoxitin, and ceftazidime, and intermediate sensitivity to ceftriaxone. Both TS37.AK and TS59.WK acquired intermediate sensitivity to imipenem, while TS66.WK became resistant to it.
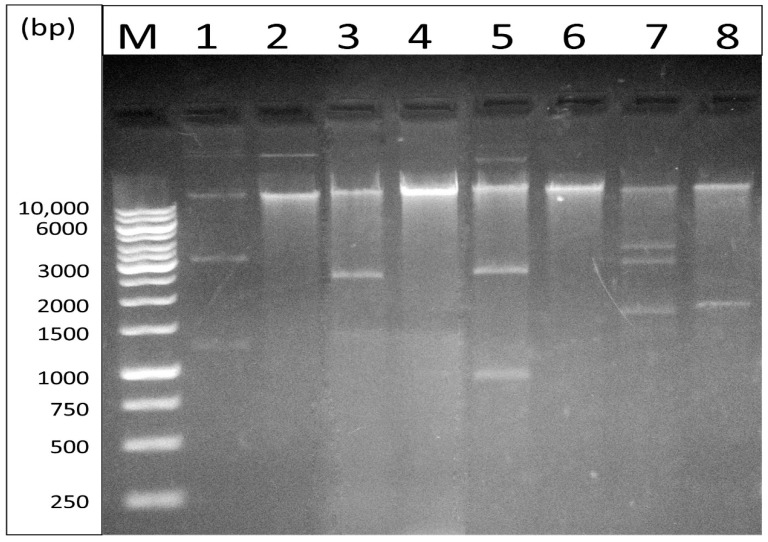
Plasmid DNA was extracted from the transformants (n = 4). Agarose gel electrophoresis of plasmid DNA extracted from CR-GNB isolates and their corresponding transformants are shown in Figure 3. Plasmid DNA from the transformants was used as templates for PCR to confirm the presence of carbapenemase genes in the transformants, as well as to detect the presence of other antimicrobial resistance genes including ESBL genes (blaSHV, blaCTX-M, and blaTEM), and the aac(6′)-Ib gene which confers resistance to aminoglycosides. Untransformed E. coli DH5α was used as a negative control.
Figure 3.
Agarose gel electrophoresis of plasmid DNA extracted from some carbapenem-resistant Gram-negative bacterial isolates and their corresponding transformants; lane M, a Gene Ruler 1 kb ladder; lane 1, clinical isolate 37.AK; lane 2, transformant TS37.AK; lane 3, clinical isolate 59.WK; lane 4, transformant TS59.WK; lane 5, clinical isolate 66.WK; lane 6, transformant TS66.WK; lane 7, clinical isolate 24.SK; lane 8, transformant TS24.SK.
The results of PCR and gel electrophoresis confirmed that all the transformants were carrying blaOXA-48 gene, two of which carried the blaCTX-M gene, one carried the blaTEM gene, and one carried the aac(6′)-Ib gene. All transformants also gave positive results with Blue-Carba and mCIM tests. Phenotypic properties of the transformants were compared to those of untransformed E. coli DH5α, and the parent clinical isolates (24.SK, 37.AK, 59.WK, 66.WK, which were identified as K. pneumoniae) from which the plasmids were extracted and transformed, as shown in Table 5. Phenotypic and genotypic characteristics of the transformed plasmids are summarized in Table 6.
Table 5.
Phenotypic properties of transformants, their corresponding K. pneumoniae parent clinical isolates, and untransformed E. coli DH5α.
| TS24.SK | 24.SK | TS37.AK | 37.AK | TS59.WK | 59.WK | TS66.WK | 66.WK | E. coli DH5α | |
|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial susceptibility testing | |||||||||
| Imipenem (10 µg) | I | I | I | R | I | R | I | I | S |
| Meropenem (10 µg) | R | R | S | R | S | R | S | R | S |
| Ertapenem (10 µg) | R | R | S | R | S | R | S | R | S |
| Doripenem (10 µg) | S | I | S | S | S | R | S | R | S |
| Amoxicillin/clavulanic acid (30 µg) | S | R | S | S | S | R | S | I | S |
| Ampicillin/sulbactam (20 µg) | R | R | R | R | R | R | R | R | S |
| Cefoxitin (30 µg) | R | R | R | R | R | R | R | R | S |
| Ceftazidime (30 µg) | R | R | R | R | R | R | R | R | S |
| Ceftriaxone (30 µg) | I | I | I | R | I | R | R | R | S |
| Cefepime (30 µg) | SDD | SDD | S | R | S | R | SDD | R | S |
| Amikacin (30 µg) | S | S | S | R | S | R | S | R | S |
| Gentamicin (10 µg) | S | S | S | R | S | R | S | S | S |
| Ciprofloxacin (5 µg) | R | R | S | R | S | R | S | R | S |
| Levofloxacin (5 µg) | R | R | S | R | S | R | S | R | S |
| Trimethoprim/sulfamethoxazole (25 µg) | S | S | S | R | S | R | S | R | S |
| Nitrofurantoin (300 µg) | S | R | S | S | S | R | S | R | S |
| Fosfomycin (200 µg) | S | S | S | R | S | S | S | S | S |
| Blue-Carba test | |||||||||
| + | + | + | + | + | + | + | + | - | |
| Modified carbapenem inactivation method (mCIM) | |||||||||
| + | + | + | + | + | + | + | + | - | |
Notes: TS24.SK, TS37.AK, TS59.WK, and TS66.WK are transformants of K. pneumoniae clinical isolates 24.SK, 37.AK, 59.WK, and 66.WK, respectively. Untransformed E. coli DH5α was used as negative control. Abbreviations: S, sensitive; R; resistant; I, intermediate sensitivity; SDD, susceptible dose-dependent (a breakpoint category for which the susceptibility of an isolate depends on the dosing regimen used).
Table 6.
Phenotypic and genotypic characteristics of the transformed plasmids.
| Parental Strain | Transformant Code | Plasmid Code | Acquired Transformant Phenotype | Plasmid Genotype | Plasmid Size in Gel | |
|---|---|---|---|---|---|---|
| 24.SK | TS24.SK | pKPT24 | Imipenem (10 µg) | I | blaOXA-48/blaCTX-M | 2 plasmid bands: Large plasmid: >10 kb Small plasmid: ≈1700 bp |
| Meropenem (10 µg) | R | |||||
| Ertapenem (10 µg) | R | |||||
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| Cefepime (30 µg) | SDD | |||||
| Ciprofloxacin (5 µg) | R | |||||
| Levofloxacin (5 µg) | R | |||||
| 37.AK | TS37.AK | pKPT37 | Imipenem (10 µg) | I | bla OXA-48 | 2 plasmid bands: Large plasmid: >10 kb Small plasmid: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| 59.WK | TS59.WK | pKPT59 | Imipenem (10 µg) | I | blaOXA-48/blaTEM/blaaac(6′)-Ib | 1 plasmid band: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| 66.WK | TS66.WK | pKPT66 | Imipenem (10 µg) | I | blaOXA-48/blaCTX-M | 1 plasmid band: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | R | |||||
| Cefepime (30 µg) | SDD | |||||
Notes: blaOXA-48 gene codes for OXA-48-like carbapenemase; blaCTX-M and blaTEM genes code for ESBLs; aac(6ʹ)-Ib gene codes for aminoglycoside 6ʹ-N-acetyltransferase type Ib.
3.7. Genotyping of CR-GNB Containing Plasmids
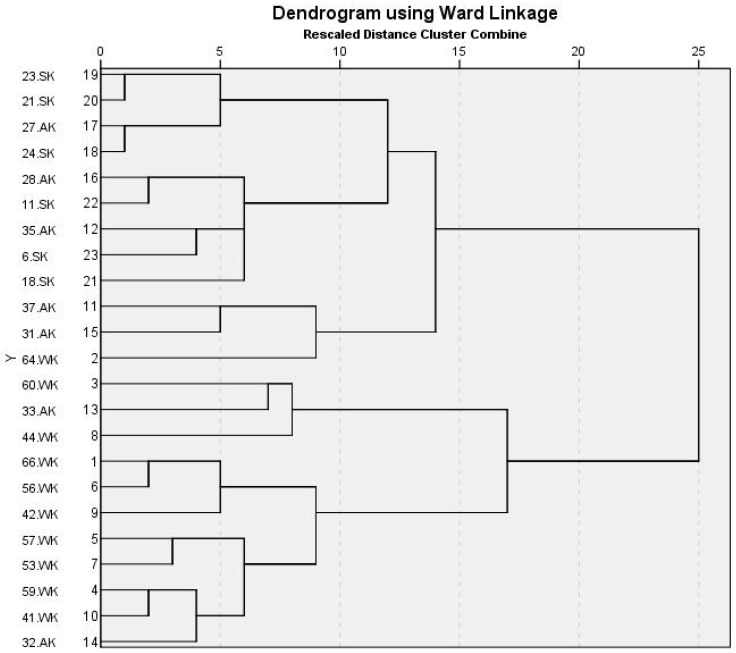
ERIC-PCR was performed for the 31 CR-GNB isolates harboring plasmids to determine their genetic relatedness. The tested isolates included 23 Klebsiella spp., 3 E. coli, 3 P. aeruginosa, and 2 A. baumannii. The ERIC-PCR gel analysis exhibited a range from 3 to 13 bands between the sizes of 111 to 1778 bp (Supplementary Figure S4).
As shown in Figure 4, the dendrogram generated from ERIC-PCR genomic DNA products of Klebsiella spp. isolates reveals that the tested isolates were not clonal, which ensures the genetic diversity of the isolates. However, based on the calculated Jaccard similarity index, isolates 21.SK and 23.SK were found to be 100% similar, as well as isolates 24.SK and 27.AK, although they were all collected from different patients. Aside from these four isolates, all the other plasmid-bearing isolates in this study were genetically dissimilar, as confirmed by the calculated similarity index of the isolates.
Figure 4.
Dendrogram generated from ERIC-PCR genomic DNA products of 23 carbapenem-resistant Klebsiella spp. isolates harboring plasmids.
3.8. Statistical Analysis
Categorical variables were analyzed using chi-square analysis to detect statistically significant associations between phenotypic and genotypic properties of the tested isolates. Significance was two-sided, and a value of p < 0.05 was considered statistically significant.
The results have revealed significant associations between the phenotypic resistance of the isolates to amoxicillin/clavulanic acid and the presence of blaVIM and blaOXA genes on their plasmids, with Pearson chi-square values of 0.048 and 0.01, respectively. There was also a significant association between the phenotypic resistance of the isolates to ampicillin/sulbactam and the presence of the blaVIM gene on their plasmids, with a Pearson chi-square value of 0.004.
4. Discussion
GNB are common causes of nosocomial and community-acquired UTls [42]. Owing to limited therapeutic options, UTls caused by antibiotic-resistant GNB are an increasing concern [43]. Such pathogens are prone to acquiring genes encoding multiple mechanisms of antibiotic resistance, including ESBLs and carbapenemases [44]. CR occurs mainly among GNB, such as Enterobacterales, P. aeruginosa, and A. baumannii, and may either be intrinsic or conferred by transferable carbapenemase-encoding genes. Thus, our aim was to detect the carbapenemase-producing ability of GNB isolated from urine specimens of cUTI patients admitted to two of the most important tertiary care hospitals in Cairo, Egypt, to determine the prevalence of carbapenemase genes among plasmid-bearing isolates, and to explore the possible transferability of such resistance to other bacterial host(s).
Out of 210 Enterobacterales in the current study, 38 (18.1%) were carbapenem-resistant Enterobacteriaceae (CRE). Contrary to our results, a study on the epidemiology of CRE in Egyptian intensive care units reported that 117/236 (49.6%) Enterobacterales isolates from urine specimens were CRE, suggesting the presence of healthcare transmission [45]. Antimicrobial susceptibility testing showed that 64 out of 65 CR-GNB isolates were MDR, which was in accordance to previously published studies that underscore the considerable resistance of many CPs [46,47,48].
Among the collected uropathogens in this study, 29 out of 210 (13.8%) Enterobacterales were CPE. A study by Woodford et al. reported that the rate of confirmed CPE among urinary GNB isolates from five UK laboratories participating in the study was 0.13% [49]. The proportion of CPE was 2.3% in another study by Giani et al. on urinary GNB isolates from five Italian hospitals [50]. At a referral hospital in northwest Ethiopia, 5/183 (2.73%) Enterobacterales isolates from UTI patients were CPE [51]. These rates are substantially lower than the one observed in the current study (13.8%), indicating weak infection control strategies. This might be attributed to the overuse of broad-spectrum antimicrobials as empiric treatments for all gravely ill patients, out of fear of missing an individual with a highly resistant pathogen.
In the present study, PCR amplification was used to detect some plasmid-borne carbapenemase genes including blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP. The most prevalent of which was blaOXA-48 gene (77.4%), followed by blaVIM gene (25.8%). Both blaKPC and blaNDM genes were co-present in one isolate. The blaIMP gene was not carried in the plasmids of any of the tested isolates, which agreed with the results of other studies from Egypt reporting the absence of blaIMP gene from the tested CR-GNB isolates [48,52]. However, this does not negate the presence of IMP carbapenemase in Egypt, as it was reported to be harbored by P. aeruginosa isolates in Egypt in 2017 [53].
Plasmid DNA extracted from CR-GNB isolates was transformed into competent E. coli DH5α. Four transformants were able to grow on LB/ampicillin agar plates two successive generations, then on LB/meropenem plates, indicating successful transformation of one or more MGEs carrying resistance genes. The transformed MGEs were extracted using the GeneJet Plasmid Miniprep Kit, and were found to carry the blaOXA-48 gene by PCR amplification, which in turn confirmed the possibility of horizontal transfer of carbapenemase-encoding genes. In accordance with our findings, a study performed by Göttig et al. established the in vivo transfer of a plasmid carrying a blaOXA-48 gene from K. pneumoniae to E. coli in the gut of an infected patient [54].
Antibiogram analysis was carried out for the transformants. The results of this revealed the newly acquired properties of the transformants, where one transformant acquired resistance to meropenem, ertapenem, ampicillin/sulbactam, cefoxitin, ceftazidime, ciprofloxacin, and levofloxacin, and intermediate sensitivity to imipenem, ceftriaxone, and cefepime. The remaining three transformants showed resistance to ampicillin/sulbactam, cefoxitin, and ceftazidime, and intermediate sensitivity to ceftriaxone, two of which acquired intermediate sensitivity to imipenem, while one acquired imipenem resistance. It is well known that the OXA-48 enzyme has a high capacity for the hydrolysis of penicillins, a low capacity for the hydrolysis of carbapenems, and does not hydrolyze broad-spectrum cephalosporins [21]; thus, the acquired resistance of the transformants to cephalosporins and carbapenems may be attributed to other resistance determinants carried on the same OXA-encoding plasmid, that was transferred to the previously susceptible E. coli DH5α.
All transformants gave a positive result with Blue-Carba and mCIM tests, indicating the production of carbapenemase enzymes. It is worth noting that three of the transformants initially failed to grow on LB/meropenem plates, and only grew on LB/ampicillin. The copy number of the plasmids might have increased on LB/ampicillin plates, allowing the sufficient replication of the CR genes, and enabling the transformants to grow on LB/meropenem plates upon subculturing from LB/ampicillin plates.
Our future perspectives include whole plasmid sequencing of the successfully transformed plasmids to detect all the other resistance determinants carried on such plasmids, in addition to fully characterizing these transferable plasmids.
5. Conclusions
This study shows that resistance to carbapenems, one of the last resort classes of antibiotics, became prevalent and distributed among Gram-negative bacterial uropathogens in two major tertiary care hospitals in Cairo, Egypt. This type of resistance can be transferred horizontally to other bacterial hosts causing limitations and challenges in treatment options of bacterial infections. Antibiotic stewardship programs must be implemented to reduce the emergence and spread of CR and improve the outcomes of infectious diseases treatment programs.
Acknowledgments
The authors would like to acknowledge the Microbiology and Immunology Department, Faculty of Pharmacy, Ain Shams University for providing the laboratory facilities for this study. The authors are also grateful to the staff at the microbiology laboratories of El-Demerdash and Kasr Al-Ainy Hospitals, Cairo, Egypt, for providing the clinical specimens and data records. The authors thank kKing Khalid University for the administrative support and collaboration.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10090889/s1, Figure S1: Agarose gel electrophoresis of plasmid DNA extracted from some carbapenem-resistant Gram-negative bacterial isolates; lane M, a Gene Ruler 1 kb ladder; lanes 1 through 8 are positive for plasmid bands; Figure S2: Agarose gel electrophoresis of multiplex-PCR amplification of blaKPC/blaNDM genes in some carbapenem-resistant Gram-negative isolates, lane M, a Gene Ruler 100 bp ladder; lane 1 is positive for blaKPC and blaNDM with expected sizes of 1011 and 621 bp, respectively; lanes, 2,3, and 4 were negative; lane 5 is negative control. Arrows indicate positive bands; Figure S3: Agarose gel electrophoresis of multiplex-PCR amplification of blaVIM/blaOXA-48 genes in some carbapenem-resistant Gram-negative isolates, lane M, a Gene Ruler 100 bp ladder; lanes 2, 5, 7 are positive for blaVIM with expected size of 748 bp; lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 are positive for blaOXA-48 with expected size of 438 bp; lane 10 is negative control; Figure S4. Agarose gel electrophoresis of ERIC fingerprints of 31 carbapenem-resistant Gram-negative bacterial isolates harboring plasmids on agarose gel electrophoresis; lane L, DNA size marker; lanes 1 through 31, numbered fingerprints. By visual inspection, samples 8 and 9 show close relatedness on the gel; Table S1: Antibiogram analysis of carbapenem-resistant Enterobacteriaceae isolates (n = 38); Table S2: Antibiogram analysis of carbapenem-resistant Pseudomonas aeruginosa isolates (n = 19); Table S3: Antibiogram analysis of carbapenem-resistant Acinetobacter baumannii isolates (n = 8)).
Author Contributions
Conceptualization, A.A.E., S.E.S., M.Y.A., K.M.A., M.M.A. and N.A.H.; methodology, A.A.E., S.E.S., M.M.A. and K.M.A.; funding acquisition, M.Y.A. and K.M.A.; writing—original draft preparation, A.A.E. and S.E.S.; writing—review and editing, M.Y.A., K.M.A., M.M.A. and N.A.H.; supervision, K.M.A., M.M.A. and N.A.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Faculty of Pharmacy Ain Shams University Ethics Committee (ENREC-ASU-2019-98).
Informed Consent Statement
Informed consents were obtained from the patients or their legal guardians after explaining the purpose of the study.
Data Availability Statement
All the data supporting the findings are included in the manuscript and supplementary material.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McLellan L., Hunstad D. Urinary tract infection: Pathogenesis and complications. Trends Mol. Med. 2016;22:946–957. doi: 10.1016/j.molmed.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doesschate T.T., Van Der Vaart T., Damen J., Bonten M., Van Werkhoven C. Carbapenem-alternative strategies for complicated urinary tract infections: A systematic review of randomized controlled trials. J. Infect. 2020;81:499–509. doi: 10.1016/j.jinf.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Barber A.E., Norton J.P., Spivak A.M., Mulvey M.A. Urinary tract infections: Current and emerging management strategies. Clin. Infect. Dis. 2013;57:719–724. doi: 10.1093/cid/cit284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenberger P., Hooton T.M. Complicated urinary tract infections. Curr. Infect. Dis. Rep. 2008;10:499–504. doi: 10.1007/s11908-008-0081-0. [DOI] [PubMed] [Google Scholar]
- 5.Neal D.E. Complicated Urinary Tract Infections. Urol. Clin. N. Am. 2008;35:13–22. doi: 10.1016/j.ucl.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Pallett A., Hand K. Complicated urinary tract infections: Practical solutions for the treatment of multiresistant Gram-negative bacteria. J. Antimicrob. Chemother. 2010;65:iii25–iii33. doi: 10.1093/jac/dkq298. [DOI] [PubMed] [Google Scholar]
- 7.Pitout J.D., Laupland K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 8.Elshamy A.A., Aboshanab K.M., Yassien M.A., Hassouna N.A. Prevalence of plasmid-mediated resistance genes among multidrug-resistant uropathogens in Egypt. Afr. Health. Sci. 2020;20:190–198. doi: 10.4314/ahs.v20i1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meletis G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2015;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diene S.M., Rolain J.-M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014;20:831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 12.Bush K., Bradford P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris P.N.A., Tambyah P.A., Paterson D.L. β-lactam and β-lactamase inhibitor combinations in the treatment of extended-spectrum β-lactamase producing Enterobacteriaceae: Time for a reappraisal in the era of few antibiotic options? Lancet Infect. Dis. 2015;15:475–485. doi: 10.1016/S1473-3099(14)70950-8. [DOI] [PubMed] [Google Scholar]
- 14.Codjoe F.S., Donkor E.S. Carbapenem Resistance: A Review. Med. Sci. 2017;6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshamy A.A., Aboshanab K.M. A review on bacterial resistance to carbapenems: Epidemiology, detection and treatment options. Futur. Sci. OA. 2020;6:FSO438. doi: 10.2144/fsoa-2019-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby G.A., Munoz-Price L.S. Mechanisms of disease: The New β-Lactamases. N. Engl. J. Med. 2005;352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 18.Gotte M., Berghuis A., Matlashewski G., Wainberg M.A., Sheppard D. In: Handbook of Antimicrobial Resistance. Berghuis A., Matlashewski G., Wainberg M.A., Sheppard D., editors. Springer; New York, NY, USA: 2017. [Google Scholar]
- 19.Cantón R., Ruiz-Garbajosa P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011;11:477–485. doi: 10.1016/j.coph.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Naas T., Oueslati S., Bonnin R.A., Dabos M.L., Zavala A., Dortet L., Retailleau P., Iorga B.I. Beta-lactamase database (BLDB)–structure and function. J. Enzym. Inhib. Med. Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel L., Potron A., Nordmann P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 22.Findlay J., Hopkins K., Meunier D., Woodford N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J. Antimicrob. Chemother. 2015;70:1338–1342. doi: 10.1093/jac/dku571. [DOI] [PubMed] [Google Scholar]
- 23.Bergey D.H., Holt J.G. Bergey’s Manual of Determinative Bacteriology. 9th ed. Williams & Wilkins; Baltimore, MD, USA: 1994. [Google Scholar]
- 24.CLSI . Performance Standards for Antimicrobial Disk Susceptibility Tests. CLSI Standard M02. 13th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 25.CLSI Performance Standards for Antimicrobial Susceptibility Testing . Informational Supplement. CLSI Document M100. 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. [Google Scholar]
- 26.Pires J., Novais A., Peixe L. Blue-Carba, an Easy Biochemical Test for Detection of Diverse Carbapenemase Producers Directly from Bacterial Cultures. J. Clin. Microbiol. 2013;51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J.J., Russell D.D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Volume 3. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 28.Doyle D., Peirano G., Lascols C., Lloyd T., Church D.L., Pitout J.D.D. Laboratory Detection of Enterobacteriaceae That Produce Carbapenemases. J. Clin. Microbiol. 2012;50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordmann P., Poirel L., Carrër A., Toleman M., Walsh T.R. How To Detect NDM-1 Producers. J. Clin. Microbiol. 2011;49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordmann P., Naas T., Poirel L. Global Spread of Carbapenemase-producingEnterobacteriaceae. Emerg. Infect. Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L., Naas T., Nicolas D., Collet L., Bellais S., Cavallo J.-D., Nordmann P. Characterization of VIM-2, a Carbapenem-Hydrolyzing Metallo-β-Lactamase and Its Plasmid- and Integron-Borne Gene from a Pseudomonas aeruginosa Clinical Isolate in France. Antimicrob. Agents Chemother. 2000;44:891–897. doi: 10.1128/AAC.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodford N., Tierno P.M., Young K., Tysall L., Palepou M.-F.I., Ward E., Painter R.E., Suber D.F., Shungu D., Silver L.L., et al. Outbreak of Klebsiella pneumoniae Producing a New Carbapenem-Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004;48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamed S., Aboshanab K.M.A., Elkhatib W.F., Ashour M.S. Aminoglycoside Resistance Patterns of Certain Gram Negative Uropathogens Recovered from Hospitalized Egyptian Patients. Br. Microbiol. Res. J. 2013;3:678–691. doi: 10.9734/BMRJ/2013/5167. [DOI] [Google Scholar]
- 34.Rasheed J.K., Jay C., Metchock B., Berkowitz F., Weigel L., Crellin J., Steward C., Hill B., Medeiros A., Tenover F.C. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 1997;41:647–653. doi: 10.1128/AAC.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnet R., Dutour C., Sampaio J.L.M., Chanal C., Sirot D., Labia R., De Champs C., Sirot J. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240 Gly. Antimicrob. Agents Chemother. 2001;45:2269–2275. doi: 10.1128/AAC.45.8.2269-2275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Codjoe F.S., Brown C.A., Smith T.J., Miller K., Donkor E.S. Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS ONE. 2019;14:e0222168. doi: 10.1371/journal.pone.0222168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 38.Ishikawa J. FramePlot: A new implementation of the Frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 40.Hunter P.R. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkpatrick L.A., Feeney B.C. A Simple Guide to IBM SPSS Statistics for Version 20.0. 12th ed. Wadsworth Cengage Learning; Belmont, CA, USA: 2013. [Google Scholar]
- 42.Tandogdu Z., Wagenlehner F.M. Global epidemiology of urinary tract infections. Curr. Opin. Infect. Dis. 2016;29:73–79. doi: 10.1097/QCO.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch E.B., Zucchi P.C., Chen A., Raux B.R., Kirby J.E., McCoy C., Eliopoulos G.M. Susceptibility of Multidrug-Resistant Gram-Negative Urine Isolates to Oral Antibiotics. Antimicrob. Agents Chemother. 2016;60:3138–3140. doi: 10.1128/AAC.02961-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bader M.S., Loeb M., Brooks A. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad. Med. 2016;129:242–258. doi: 10.1080/00325481.2017.1246055. [DOI] [PubMed] [Google Scholar]
- 45.Kotb S., Lyman M., Ismail G., El Fattah M.A., Girgis S.A., Etman A., Hafez S., El-Kholy J., Zaki M.E.S., Rashed H.-A.G., et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare–associated Infections Surveillance Data, 2011–2017. Antimicrob. Resist. Infect. Control. 2020;9:1–9. doi: 10.1186/s13756-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livermore D.M., Mushtaq S., Warner M., Zhang J.C., Maharjan S., Doumith M., Woodford N. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J. Antimicrob. Chemother. 2011;66:48–53. doi: 10.1093/jac/dkq408. [DOI] [PubMed] [Google Scholar]
- 47.Livermore D.M., Warner M., Mushtaq S., Doumith M., Zhang J., Woodford N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents. 2011;37:415–419. doi: 10.1016/j.ijantimicag.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Mabrouk S.S., Abdellatif G.R., El-Ansary M.R., Aboshanab K.M., Ragab Y.M. Carbapenemase Producers Among Extensive Drug-Resistant Gram-Negative Pathogens Recovered from Febrile Neutrophilic Patients in Egypt. Infect. Drug Resist. 2020;13:3113–3124. doi: 10.2147/IDR.S269971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodford N., Xu-McCrae L., Mushtaq S., Wu H.H.T., Ellington M.J., Lancaster O., Davies F., Donaldson H., Rao G.G., Verma A., et al. Prevalence of carbapenem resistance and carbapenemase production among Enterobacteriaceae isolated from urine in the UK: Results of the UK infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-UK) J. Antimicrob. Chemother. 2018;73:698–702. doi: 10.1093/jac/dkx471. [DOI] [PubMed] [Google Scholar]
- 50.Giani T., Antonelli A., Sennati S., Di Pilato V., Chiarelli A., Cannatelli A., Gatsch C., Luzzaro F., Spanu T., Stefani S., et al. Results of the Italian infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-IT): Activity of ceftazidime/avibactam against Enterobacterales isolated from urine. J. Antimicrob. Chemother. 2020;75:979–983. doi: 10.1093/jac/dkz547. [DOI] [PubMed] [Google Scholar]
- 51.Eshetie S., Unakal C., Gelaw A., Ayelign B., Endris M., Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob. Resist. Infect. Control. 2015;4:12. doi: 10.1186/s13756-015-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamel N.A., El-Tayeb W.N., El-Ansary M., Mansour M.T., Aboshanab K.M. Phenotypic screening and molecular characterization of carbapenemase-producing Gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PLoS ONE. 2018;13:e0202119. doi: 10.1371/journal.pone.0202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Domany R.A., Emara M., El-Magd M.A., Moustafa W.H., Abdeltwab N.M. Emergence of Imipenem-ResistantPseudomonas aeruginosaClinical Isolates from Egypt CoharboringVIMandIMPCarbapenemases. Microb. Drug Resist. 2017;23:682–686. doi: 10.1089/mdr.2016.0234. [DOI] [PubMed] [Google Scholar]
- 54.Göttig S., Gruber T.M., Stecher B., Wichelhaus T.A., Kempf V.A.J. In Vivo Horizontal Gene Transfer of the Carbapenemase OXA-48 During a Nosocomial Outbreak. Clin. Infect. Dis. 2015;60:1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings are included in the manuscript and supplementary material.