Abstract
Using whole-genome sequencing and cloning of the target gene, we identified blaOXA-900 carbapenemase, a novel blaOXA belonging to a distant and distinct sub-family of blaOXA-48-like. The plasmid-mediated gene was identified in a C. freundii isolate with elevated carbapenem MICs that evaded detection by commercial DNA-based methods. The novel gene, an OXA-48 family carbapenem-hydrolyzing class D β-lactamase, OXA-900, likely originates from marine environmental Shewanella. Since this plasmid-mediated gene has entered a member of the Enterobacterales and evades detection by commonly used tests, it may gain wide dissemination among Enterobacterales.
Keywords: carbapenemase, β-lactamase, bla OXA-48-like , Enterobacterales
1. Introduction
Detection of organisms carrying carbapenemases is important for infection control and clinical decision making. Commercial molecular tests target specific, known carbapenemases. Thus, novel carbapenemases may evade detection. OXA-48 family carbapenem-hydrolyzing [Ambler] class D β-lactamases (CHDL) in Enterobacterales are a common enzyme group that was first reported in 2004 in Klebsiella pneumoniae isolated from a patient in Turkey [1]. blaOXA-48-like genes are found mainly in K. pneumoniae but also in other pathogenic Enterobacterales [2]. The hydrolytic activity of OXA-48-like CHDLs against carbapenems is relatively low and only moderately increases MIC values when it is the sole resistance mechanism [2]. However, high MICs for carbapenems are reported in Enterobacterales when OXA-48-like enzymes are produced in strains lacking OmpF and/or OmpC type porins [3]. The latest version (Nov. 2019) of the NCBI Bacterial Antimicrobial Resistance Reference Gene Database held about 40 variants of blaOXA-48-like genes. As blaOXA-48-like genes are genetically conserved, DNA-based tests for their detection are reliable. In clinical settings, most tests for the detection of blaOXA-48-like/OXA-48-like are DNA based, focusing mainly on the blaOXA-48 family variants including blaOXA-181 sub-type but not blaOXA-54. Here, we describe a novel blaOXA belonging to a distant and distinct sub-family of blaOXA-48-like genes identified in a C. freundii isolate with elevated carbapenem MICs, which evaded detection by DNA-based methods.
2. Materials and Methods
2.1. Description of the Isolate
The specimen was a rectal swab obtained for routine surveillance of carbapenemase-producing Enterobacterales (CPEs) in an Israeli long-term care facility in 2019. A suspected CRE colony was identified by visual inspection on selective chromogenic media (CHROMagarTM mSuperCarbaTM, HyLabs, Rehovot, Israel). The isolate was identified by Vitek 2™ (bioMérieux, Marcy-l′Étoile, France) as C. freundii with intermediate resistance to ertapenem (1 µg/mL). Tests for the presence of common carbapenemase genes using Xpert® Carba-R assay on a GeneXpert® system (Cepheid, Sunnyvale, CA, USA) were negative. The specimen was named isolate ISCF142 and sent to the National Laboratory for Antibiotic Resistance for further evaluation.
2.2. Antibiotic Susceptibly Testing
Antibiotic susceptibility testing was performed using broth microdilution (Sensititre™ GN6F plate, ThermoFisher Scientific, Oakwood Village, OH, USA) according to CLSI M07 guidelines and manufacturer’s instructions. For isolates with carbapenem MIC values below the detection level of the Sensititre™ GN6F plate, homemade assays containing ertapenem, imipenem, and meropenem were used. These were produced according to CLSI guidelines using recommended control strains [4]. Susceptibility was determined using CLSI 2019 breakpoints.
2.3. Detection of Carbapenemases
Carbapenemase activity was determined by the β-CARBA™ assay (Bio-Rad Laboratories, Hercules, CA, USA) as well as by a modified carbapenem inactivation method [5].
Three commercial tests and a homemade method were used to detect carbapenemase genes: (1) Xpert® Carba-R assay, (2) Pneumonia plus Panel assay on a BIOFIRE® FILMARRAY® system (bioMérieux), and (3) CarbaR+ panel on a Novodiag® device (Mobidiag, Keilaranta, Finland). These tests detect blaKPC, blaNDM, blaVIM, blaIMP-1 and blaOXA-48/181. We performed a homemade PCR that detects these carbapenemases as described previously [6] and also blaIMI using the IMI-F (5′-GCCATATCACCTAATGACATTCC-3′) and IMI-R (5′-GCAAATGAACGATTTCCATTATGTA-3′) primers.
We used the NG-test CARBA 5 (NG Biotech, Guipry, France) immunochromatographic assay, a rapid multiplex lateral flow assay, for phenotypic detection and differentiation of KPC, OXA-48-like, VIM, IMP, and NDM carbapenemases according to the manufacturer’s instructions.
2.4. WGS Analysis
DNA was extracted using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and sequenced at the Sequencing Core at the University of Illinois at Chicago according to their standard protocol for NextSeq500 (Illumina Inc., San Diego, CA, USA). Long-read sequencing was carried out using the SQK-RBK004 kit (Oxford Nanopore Technologies (ONT), Oxford, UK) on the MinION (ONT) with Guppy software (ONT) according to the manufacturer’s instructions. Illumina reads were quality screened using Fastp [7], assembled with long reads using Unicycler [8], and annotated using Prokka [9]. Strain type was predicted using pubMLST’s C. freundii scheme (https://pubmlst.org/cfreundii/, accessed on 1 August 2020). ORFs were searched against the CARD and NCBI databases using DIAMOND BLAST [10]. Whole-genome sequencing (WGS) using both short-read and long-read technologies was used as previously described [11].
2.5. Insertion Sequences (IS)
IS were identified by Prokka annotation, and the inverted repeat sequences, left and right (IRL and IRR, respectively), were identified using the ISfinder database [12]. Their location was plotted manually on a genomic illustration produced by EasyFig [13].
2.6. Carbapenemase Gene Cloning and Functional Confirmation
Cloning was performed by amplifying the suspected carbapenemase gene using a primer set targeting the first and last 20 bases of the gene with added restriction sites XbaI and EcoRI. The resulting fragment was cleaned using the commercial kit Nucleospin gel and PCR cleanup (MACHEREY-NAGEL GmbH & Co, Düren, Germany). The clean fragment was subjected to cutting by XbaI and EcoRI restriction enzyme (New England Biolabs, Ipswich, MA, USA) and cleanup, as stated above. The vector pHSG396 (Takara Bio, Saint-Germain-en-Laye, France) was subjected to the same restriction enzymes and separated by agarose gel electrophoresis, followed by Nucleospin gel and PCR cleanup kit. The vector and insert were ligated by T4 DNA Ligase (New England Biolabs). The circular plasmid with the gene was inserted into MAX Efficiency™ DH10β Competent Cells (Invitrogen, Waltham, MA, USA). The resulting plasmids were extracted using NucleoSpin® Plasmid EasyPure (MACHEREY-NAGEL). The success of cloning was evaluated by positive PCR for the target gene. The modified carbapenem inactivation method was used on the C. freundii isolate, the transformant E. coli DH10β with the suspected carbapenemase ORF, and E. coli DH10β with the pHSG396 plasmid but no insert (as a negative control).
2.7. Phylogenetic Analysis of Selected blaOXA Genes
The suspected blaOXA gene was compared to the known alleles found at the NCBI Bacterial Antimicrobial Resistance Reference Gene Database (PRJNA313047). All sequences were aligned using MAFFT software [14]. The resulting alignment (only of genes diversified from blaOXA-55) was interpreted by constructing a maximum likelihood phylogenetic tree with 100 permutations using RAxML [15]. The resulting analysis was visualized using Dendroscope software [16].
2.8. Conjugation Efficiency
E. coli DH10β with and without a chloramphenicol resistance gene was used as the recipient for mating experiments; the donor strain was C. freundii ISF142. These experiments included solid mating (0.45 µm pore-size nitrocellulose MF membrane filters, Merck Millipore Ltd., Cork, Ireland) placed on 5% blood agar plates (Hylabs)) and liquid mating (mixed culture left overnight at 35 °C) with both selective media (ertapenem 0.5 µg/mL and 25 µg/mL chloramphenicol) and selective chromogenic media (CHROMagarTM mSuperCarbaTM, HyLabs, Rehovot, Israel) for resistant recipient screening.
2.9. Data Availability
Data were submitted under NCBI BioSample Accession Number SAMN13412315 and Assembly Numbers CP046502-CP046506. The new blaOXA was named blaOXA-900 and can be found under the accession number MN936180.
3. Results
The three commonly used commercial molecular tests targeting blaKPC, blaNDM, blaVIM, blaIMP-1, and blaOXA-48/181 failed to detect a carbapenemase, as did an in-house PCR method. However, two carbapenem hydrolysis tests, the β-CARBA™ assay and the modified carbapenem inactivation method, confirmed carbapenemase activity. The lateral flow assay confirmed the presence of an OXA-48 type carbapenemase.
WGS enabled the assembly of a circular chromosome and four plasmids (NCBI BioSample Accession Number SAMN13412315 and Assembly Numbers CP046502-CP046506). The isolate was identified as C. freundii, ST111. ISCF142 carried two chromosomal antibiotic resistance genes (ARGs), qnrB38 and ampC (blaCMY-65), and several plasmid-coded ARGs (Table A1). Among these ARGs were the β-lactamase genes blaCTX-M-3, blaCTX-M-39, and blaPER-2, which may contribute to C. freundii ISCF142’s resistance to third-generation cephalosporins and aztreonam (Table 1). A fifth gene was also found, encoding a novel putative β-lactamase first annotated as a blaOXA-54 distant variant: the gene blaOXA-900.
Table 1.
Antibiotic susceptibility profile by broth microdilution of C. freundii ISCF142, E. coli DH10β-harboring recombinant β-lactamase blaOXA-900, and E. coli DH10β without the β-lactamase gene insert (negative control).
| MIC in µg/mL (CLSI Breakpoints) | ||||
|---|---|---|---|---|
| Antibiotic Class | Antibiotic | C. freundii Isolate ISCF142 | Transformant DH10β (OXA-900) |
DH10β (Negative Control) |
| Carbapenems | Meropenem | 1 (S) | 1 (S) | <0.12 (S) |
| Imipenem | 1 (S) | 2 (S) | <0.5 (S) | |
| Ertapenem | 1 (I) | 1 (I) | <0.12 (S) | |
| Cephalosporins | Cefotaxime | 8 (R) | <0.5 (S) | <0.5 (S) |
| Ceftazidime | >16 (R) | <0.5 (S) | <0.5 (S) | |
| Cephalosporin combinations | Ceftazidime/ avibactam a |
<0.5 (S) | <0.5 (S) | <0.5 (S) |
| Ceftolozane/ tazobactam a |
>32 (R) | 8 (S) | <0.5 (S) | |
| Monobactams | Aztreonam | >32 (R) | <0.5 (S) | <0.5 (S) |
| Penicillin combinations | Piperacillin/ tazobactam a |
>32 (R) | >32 (R) | 2 (S) |
| Amoxicillin/clavulanic acid b | >64 (R) | >64 (R) | <4 (S) | |
| Fluoroquinolones | Ciprofloxacin | 0.25 (S) | 0.12 (S) | 0.12 (S) |
| Polymixins | Colistin | 1 | 0.25 | 0.5 |
| Aminoglycosides | Tobramycin | 1 (S) | <1 (S) | <1 (S) |
| Tetracyclines | Tigecycline | 0.25 (S) | <0.25 (S) | <0.25 (S) |
| Sulfonamide combinations | Trimethoprim/ sulfamethoxazole c |
1 (S) | <1 (S) | <1 (S) |
a Tazobactam and avibactam at a fixed concentration of 4 µg/mL. b Clavulanic acid at a fixed concentration of 2 µg/mL. c Sulfamethoxazole at a fixed concentration of 19 µg/mL.
We cloned blaOXA-900 into an E. coli DH10β in order to evaluate its activity toward several antibiotics by broth microdilution. Results of antibiotic susceptibility testing are shown in Table 1. The resulting transformant was susceptible to commonly used cephalosporins, intermediate to ertapenem (1 µg/mL), and had increased MICs to meropenem (1 µg/mL) and imipenem (2 µg/mL), compared to the susceptible E. coli DH10β.
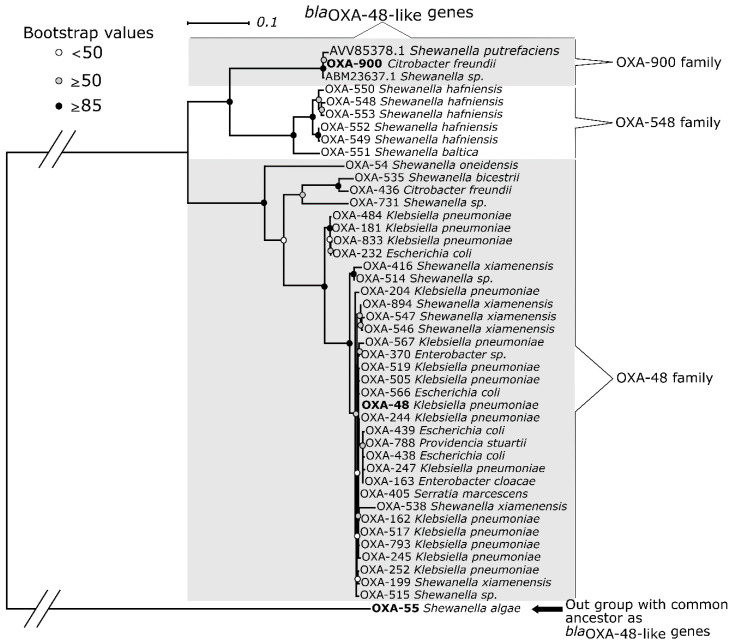
Comparing blaOXA-900 to other blaOXA-48-like genes in the NCBI Bacterial ARG database, the two best hits were from environmentally isolated organisms (Figure 1). blaOXA-900 was most similar (99% and 98% similarity) to two genes found previously in Shewanella putrefaciens, a saprophytic warm-climate marine organism, genomes that were not formally reported or characterized. blaOXA-900 also had high deduced amino acid similarity to blaOXA-548 (83.5%), blaOXA-48 (81.1%), and blaOXA-181 (80.7%).
Figure 1.
Maximum Likelihood phylogenetic tree analysis of all blaOXA that diversify from blaOXA-55 found at the NCBI’s Bacterial Antimicrobial Resistance Reference Gene Database. Two sequences from environmentally isolated Shewanella were also included, the first from S. putrefaciens isolated from Litopenaeus vannamei (whiteleg shrimp) in China (AVV85378.1) and the second (ABM23637.1) from Shewanella sp. from the marine environment.
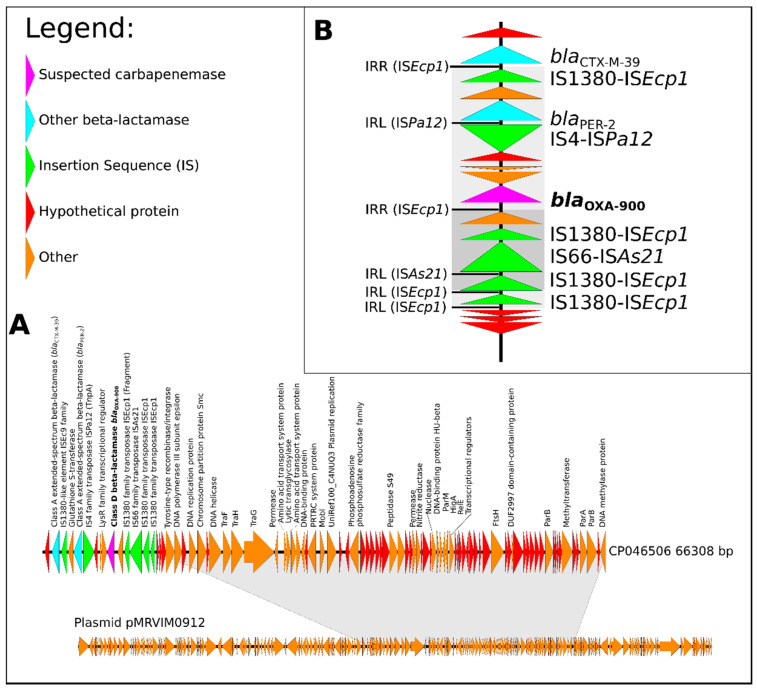
The blaOXA-900 gene was located on an antibiotic resistance island (ARI) on an IncC plasmid (CP046506). Plasmid CP046506 lacks substantial parts compared to a previously reported C. freundii IncC plasmid—namely, pMRVIM0912 [17] (Figure 2a). CP046506 includes replication, conjugation, and addiction elements characteristic of IncC. Mating experiments did not yield any resistant recipient cells, suggesting low conjugative potential with E. coli. Further analysis showed that this plasmid contains six genes previously shown to play a role in conjugation [18]: traFHG, mobI, and the regulatory gene acaCD. Notably, the gene traI was not found in CP046506, nor were other tra genes that were predicted to play a part in conjugation in silico [18], indicating a generally reduced potential for conjugation.
Figure 2.
Genetic analysis of C. freundii plasmid CP046506 and its antibiotic resistance islands (ARI): (A) plasmid CP046506, a version of pMRVIM0912 with an ARI on which the new blaOXA-900 was found (regions of similarity are marked in grey); (B) ARI with two ESBL genes and a novel carbapenemase. All IS inverted repeats on the right (IRR) and on the left (IRL) are marked and the region suggested to be inserted by the ISEcp1is marked in grey shades. Triangles represent ORFs: hypothetical proteins are marked in red, annotated genes in orange, IS fragments in green, the carbapenemase is marked in purple and other β-lactamases in blue.
We analyzed the mechanisms by which OXA-900 entered Enterobacerales. The ARI and the IS involved are grouped by their location on the plasmid backbone [17]. In this study, the CP046506 plasmid includes an ARI with an ISEcp1 insertion sequence carrying a blaCTX-M-39 that was previously reported on type 2 IncC plasmids in a location different from IncC ARI types A and B—namely, RI-5 [17]. The resulting plasmid with its uncommon resistance genes likely reflects recombination next to a RecHS recombination hotspot [17]. The location of the inverted repeats suggests that the fragment carrying the blaOXA-900 gene and several other genes (including blaPER-2) was inserted by the IS1380 family transposase ISEcp1, as it was the sole IS found enclosing the fragment by inverted repeat sequences on the right (IRR) and on the left (IRL) (Figure 2b). This IS, similar to other known mobile elements, has an important role in the evolution of multi-resistant plasmids [19,20].
4. Discussion
Here, we report on a novel OXA-48-like carbapenemase named OXA-900. This enzyme likely originated from marine environmental Shewanella. It avoids detection by commercially available DNA-based tests but is detectable by a lateral flow protein-based assay. The OXA-900 has carbapenemase activity, leading in transformants to increased carbapenem MICs in the range reported for other CHDLs [2]. OXA-900 belongs to a distant family related to OXA-48 that until now has not been described in human pathogens.
Antibiotic resistance genes that originate from environmental organisms are an important source of resistance in human pathogens. For example, blaCTX-m genes likely emerged from Kluyvera [21], blaNDM has origins in Erythrobacter litoralis [22], and qnrA is found in Shewanella algae [23]. Moreover, the blaOXA-48-like genes that gained wide distribution in human pathogens originate from Shewanella spp. [24,25]. Therefore, OXA-900’s introduction from an environmental organism into Enterobacterales poses the imminent threat that it may further spread and establish a reservoir in human pathogens. Since it eludes detection by the common commercial diagnostic methods, it may escape infection control efforts to limit the spread of carbapenemases.
blaOXA-900 represents a distinct family of OXA enzymes. In their overview of Shewanella blaOXA-48-like genes, Tacão et al. [25] presented this genus’s diversity of blaOXA-48-like genes in the environment. One can group these enzymes into three major clusters: the blaOXA-48 family, the blaOXA-548 family, and a third cluster that includes relatives of the blaOXA-900 described here (named the blaOXA-900 family).
We conclude that OXA-900 has the potential to become a widespread resistance determinant in Enterobacterales. Diagnostic tests should be adapted to detect the blaOXA-900 family.
Acknowledgments
We would like to thank Elizabeth Temkin for her helpful comments on this manuscript.
Appendix A
Table A1.
Resistance genes found on genomic fragments assembled from C. freundii ISCF142.
| Resistance Mechanism | Antibiotic Resistance Gene | Best Hit Identity (%) | Query/Template Length | Genetic Fragment |
|---|---|---|---|---|
| Quinolone resistance protein Class C β-lactamases | qnrB38-like | 99.53 | 645/645 | Chromosome (CP046502) |
| bla CMY-65-like | 99.91 | 1146/1146 | ||
| Efflux pumps | tet(D) | 100 | 1185/1185 | Plasmid (CP046504) |
| Class A β-lactamases | bla CTX-M-3 | 100 | 876/876 | |
| Dihydropteroate synthase | sul2 | 100 | 816/816 | |
| Class A β-lactamases | bla CTX-M-39 | 100 | 876/876 | Plasmid (CP046506) |
| Class A β-lactamases | blaPER-2 | 100 | 927/927 | |
| Class D β-lactamases | blaOXA-900 | 81.5 * | 810/795 |
* with blaOXA-54.
Author Contributions
Conceptualization, S.F., N.R., J.L. and Y.C.; methodology, S.F., N.R., J.L., D.S., E.P. and M.N.L.-W.; data collection, H.K., R.R., S.A. and E.P.; formal analysis, S.F. and M.N.L.-W.; writing—original draft preparation, S.F., N.R., J.L., M.N.L.-W. and Y.C.; writing—review and editing, S.F., N.R. and Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute for Antibiotic Resistance and Infection Control, Ministry of Health, Israel as part of the unit’s routine work.
Institutional Review Board Statement
Ethical review and approval were waived for this study because the study involved only microbiological analysis of a de-identified clinical isolate.
Data Availability Statement
Data were submitted under the NCBI BioSample accession number SAMN13412315 and assembly numbers CP046502-CP046506. The new blaOXA was named blaOXA-900 and can be found under the accession number MN936180.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Poirel L., Héritier C., Tolün V., Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes N.T., Lamoureaux T.L., Toth M., Stewart N.K., Frase H., Vakulenko S.B. Class D β-Lactamases: Are they all carbapenemases? Antimicrob. Agents Chemother. 2014;58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans B.A., Amyes S.G.B. OXA β-lactamases. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute (CLSI) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 11th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. CLSI Standard M07. [Google Scholar]
- 5.Pierce V.M., Simner P.J., Lonsway D.R., Roe-Carpenter D.E., Johnson J.K., Brasso W.B., Bobenchik A.M., Lockett Z.C., Charnot-Katsikas A., Ferraro M.J., et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J. Clin. Microbiol. 2017;55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerner A., Solter E., Rachi E., Adler A., Rechnitzer H., Miron D., Krupnick L., Sela S., Aga E., Ziv Y., et al. Detection and characterization of carbapenemase-producing Enterobacteriaceae in wounded Syrian patients admitted to hospitals in northern Israel. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:149–154. doi: 10.1007/s10096-015-2520-9. [DOI] [PubMed] [Google Scholar]
- 7.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 10.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 11.Frenk S., Rakovitsky N., Temkin E., Schechner V., Cohen R., Kloyzner S., Schwaber M.J., Solter E., Cohen S., Stepansky S., et al. Investigation of outbreaks of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in three neonatal intensive care units using whole genome sequencing. Antibiotics. 2020;9:705. doi: 10.3390/antibiotics9100705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huson D.H., Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 17.Ambrose S.J., Harmer C.J., Hall R.M. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid. 2018;99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Hancock S.J., Phan M.D., Luo Z., Lo A.W., Peters K.M., Nhu N.T.K., Forde B.M., Whitfield J., Yang J., Strugnell R.A., et al. Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB. Nat. Microbiol. 2020;5:1340–1348. doi: 10.1038/s41564-020-0775-0. [DOI] [PubMed] [Google Scholar]
- 19.Smet A., Van Nieuwerburgh F., Vandekerckhove T.T.M., Martel A., Deforce D., Butaye P., Haesebrouck F. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: Insertional events of transposons and insertion sequences. PLoS ONE. 2010;5:e11202. doi: 10.1371/journal.pone.0011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts M.C. Antibiotic-resistant environmental bacteria and their role as reservoirs in disease. In: Hurst C.J., editor. Modeling the Transmission and Prevention of Infectious Disease. Springer International Publishing; Berlin/Heidelberg, Germany: 2017. pp. 187–212. [Google Scholar]
- 22.Zheng B., Tan S., Gao J., Han H., Liu J., Lu G., Liu D., Yi Y., Zhu B., Gao G.F. An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell. 2011;2:250–258. doi: 10.1007/s13238-011-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L., Rodriguez-Martinez J.-M., Mammeri H., Liard A., Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuelsen Ø., Hansen F., Aasnæs B., Hasman H., Lund B.A., Leiros H.-K.S., Lilje B., Janice J., Jakobsen L., Littauer P., et al. Dissemination and characteristics of a novel plasmid-encoded carbapenem-hydrolyzing Class D β-Lactamase, OXA-436, found in isolates from four patients at six different hospitals in Denmark. Antimicrob. Agents Chemother. 2017;62:e01260-17. doi: 10.1128/AAC.01260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tacão M., Araújo S., Vendas M., Alves A., Henriques I. Shewanella species as the origin of blaOXA-48 genes: Insights into gene diversity, associated phenotypes and possible transfer mechanisms. Int. J. Antimicrob. Agents. 2018;51:340–348. doi: 10.1016/j.ijantimicag.2017.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data were submitted under the NCBI BioSample accession number SAMN13412315 and assembly numbers CP046502-CP046506. The new blaOXA was named blaOXA-900 and can be found under the accession number MN936180.