Abstract
In Saccharomyces cerevisiae, PHO85 encodes a cyclin-dependent protein kinase (Cdk) catalytic subunit with multiple regulatory roles thought to be specified by association with different cyclin partners (Pcls). Pcl10p is one of four Pcls with little sequence similarity to cyclins involved in cell cycle control. It has been implicated in specifying the phosphorylation of glycogen synthase (Gsy2p). We report that recombinant Pho85p and Pcl10p produced in Escherichia coli reconstitute an active Gsy2p kinase in vitro. Gsy2p phosphorylation required Pcl10p, occurred at physiologically relevant sites, and resulted in inactivation of Gsy2p. The activity of the reconstituted enzyme was even greater than Pho85p-Pcl10p isolated from yeast, and we conclude that, unlike many Cdks, Pho85p does not require phosphorylation for activity. Pcl10p formed complexes with Gsy2p, as judged by (i) gel filtration of recombinant Pcl10p and Gsy2p, (ii) coimmunoprecipitation from yeast cell lysates, and (iii) enzyme kinetic behavior consistent with Pcl10p binding the substrate. Synthetic peptides modeled on the sequences of known Pho85p sites were poor substrates with high Km values, and we propose that Pcl10p-Gsy2p interaction is important for substrate selection. Gel filtration of yeast cell lysates demonstrated that most Pho85p was present as a monomer, although a portion coeluted in high-molecular-weight fractions with Pcl10p and Gsy2p. Overexpression of Pcl10p sequestered most of the Pho85p into association with Pcl10p. We suggest a model for Pho85p function in the cell whereby cyclins like Pcl10p recruit Pho85p from a pool of monomers, both activating the kinase and targeting it to substrate.
The budding yeast, Saccharomyces cerevisiae, contains a small family of some five cyclin-dependent protein kinases (Cdks) (27). Though studied mainly from the perspective of cell cycle progression, yeast Cdks are also involved in other cellular processes, including the regulation of transcription (for reviews, see references 10, 46, and 55) and the direct control of metabolic enzymes (23, 24, 70). However, most knowledge about Cdk regulation and function comes from study of classic cell cycle Cdks, Cdc28p in yeast and its mammalian counterparts Cdc2, Cdk2, Cdk4, and Cdk6 (reviewed in reference 43). These Cdks are activated by high-affinity association with cyclins, which were first identified and named as proteins whose levels oscillated during the cell cycle (26). Since the levels of some cyclins are now known to be invariant, the definition must be revised to include structurally related proteins that bind to and activate Cdks (44). However, this commonality of function does not correspond to a particularly high degree of sequence similarity. All cyclins share a cyclin box (26) which forms a set of five α-helices involved in the interaction with the Cdk (2, 28). The three-dimensional structures of cyclin A and cyclin H revealed a COOH-terminal structural repeat of this fold that was not predicted from inspection of the sequence. In addition to cyclin binding, activation of Cdks requires phosphorylation of a specific threonine residue located in the activation loop of the Cdk by a distinct upstream Cdk-activating kinase (Cak) (reviewed in reference 44). For example, yeast Cdc28p is phosphorylated on Thr169 in a reaction likely catalyzed by Cak1p (15, 31, 69). This mechanism is conserved in mammalian Cdks (45, 46).
Most Cdks interact with multiple cyclin subunits (1, 43). Structural studies suggest that only one cyclin can associate with a Cdk molecule at any given time and so formation of different Cdk-cyclin complexes is thought to allow the catalytic subunit to carry out different functions, presumably by dictating the substrate specificity of the kinase complex. However, direct biochemical evidence has proved difficult to obtain, in part because so few genuine Cdk substrates are known. Recently, we showed that the substrate specificity of one of the yeast Cdks, Pho85p, is indeed determined by association with particular cyclins (24). PHO85 was first identified by virtue of its involvement in the transcriptional repression of nonspecific acid phosphatases, such as Pho5p (37, 71, 73). In this instance, Pho85p acts together with the cyclin Pho80p to phosphorylate and prevent the nuclear localization of a transcription factor, Pho4p, required for PHO5 expression (29, 47). Pho85p is now thought to be involved in a variety of cellular processes (16, 41, 67) and can interact with 10 different cyclin partners, or Pcls (Pho80p, Pcl1p and Pcl2p, Clg1p, and Pcl5p through Pcl10p) (42). Four of these cyclins, Pcl6p, Pcl7p, Pcl8p, and Pcl10p, have the cyclin box located close to the COOH terminus and thus lack the second repeat of the five-helix fold found in the classic cyclins.
We found that Pcl8p and Pcl10p were involved in glycogen metabolism and, more specifically, that Pho85p-Pcl10p directly phosphorylated the predominant form of yeast glycogen synthase, Gsy2p (24), whose phosphorylation regulates glycogen storage (20, 21, 58). In the same study, we showed that association of Pho85p with Pcl10p generated a specific Gsy2p kinase that only poorly phosphorylated Pho4p in vitro. Conversely, Pho85p-Pho80p formed a selective Pho4p kinase that had low activity towards Gsy2p. This cyclin specificity in vitro was mirrored in vivo: disruption of PCL8 and PCL10 affected only glycogen accumulation, whereas deletion of PHO80 was specific for acid phosphatase expression.
The Pho85p system provides a good example of individual cyclins determining the specificity of the Cdk towards known, physiological substrates of the kinase. In the present investigation, we have extended this work to study the role of the cyclin Pcl10p in determining substrate selection by Pho85p. Pcl10p is absolutely required for phosphorylation of the substrate Gsy2p by Pho85p. However, Pcl10p alone can also form complexes with Gsy2p, and we propose that this interaction contributes to the process of substrate recognition and effective phosphorylation. Since most Pho85p in cell extracts is monomeric, it is likely that Pcls are limiting and serve to recruit Pho85p from a cellular pool as needed for targeted protein phosphorylation.
MATERIALS AND METHODS
Strains and media.
Standard bacterial and yeast culture conditions and techniques for manipulation were used throughout (19, 59). The yeast strain WW10 (MATα ura3-52 leu2 trp1 pcl8::TRP1 pcl10::LEU2) was derived from DH96-52 (MATα ura3-52 leu2 trp1 pcl8::TRP1) (24) by disruption of the PCL10 gene with LEU2, using a PCR strategy (3). A similar PCR strategy was used to generate strain WW11 (MATα ura3-52 leu2 trp1 pho85::TRP1) from EG328-1A (MATα ura3-52 leu2 trp1). Strain CC9 (MATα ura3-52 leu2 trp1 glg1-2::LEU2 glg2::URA3) is described elsewhere (7). The strains DH96-52 and CC9 are derived from EG328-1A. Strain BY391a (MATa trp1Δ63 GAL2+ ura3-52 lys2-801am ade2-107o his3 Δ200 leu2-Δ1) is described elsewhere (41).
Plasmids.
Plasmids were constructed and maintained in Escherichia coli DH5α. All DNA segments generated by PCR were sequenced prior to use. The plasmid pET-HisPHO85 was constructed as follows. PCR was employed to amplify the PHO85 open reading frame from a genomic clone of PHO85 kindly provided by Brenda Andrews, University of Toronto, Toronto, Ontario, Canada. The primers used introduced an NdeI site at the initiation codon immediately followed by a tag of six histidine residues and a BamHI site 3 bp beyond the stop codon. For expression in E. coli, this DNA fragment was digested with NdeI and BamHI and ligated into pET-3c (Novagen).
Plasmid pET-HAPCL10 was constructed by first amplifying the PCL10 open reading frame from a bacteriophage λ MG3 clone of yeast chromosomal DNA (49) obtained from the American Type Tissue Collection (ATCC 70674). Amplification was conducted in two stages. First, a portion of PCL10 from the 5′ end to an internal HindIII site was produced. The sense primer used introduced an NdeI site followed by a copy of the hemagglutinin (HA) epitope (MYPYDVPDYA). The remainder of the PCL10 open reading frame, from the internal HindIII site to the 3′ end, was then amplified. The antisense primer used introduced a BamHI site 3 bp beyond the stop codon. The 5′ and 3′ ends of PCL10 were joined by using the HindIII site, and the resulting full-length sequence was ligated into pET-3c as an NdeI/BamHI fragment, generating pET-HAPCL10. To allow for coexpression studies in E. coli, a ClaI/SphI piece containing the T7 promoter and the HAPCL10 coding region was excised from pET-HAPCL10 and used to replace a ClaI/SphI segment of pACYC184 (5), which carries the origin of replication from plasmid p15A, creating pACYC-HAPCL10.
An alternative PCL10 construct was also created. A portion of PCL10 from the 5′ end to the internal HindIII site was again amplified, but in this case, the sense primer introduced an NdeI site followed by a tag of six consecutive histidine residues. This segment of PCL10 was joined to the 3′ end generated above with the HindIII site. The full-length PCL10 thus created was ligated into pET-3c as an NdeI/BamHI fragment, generating pET-HisPCL10.
Plasmids pMET-PHO85 and pMET-PHO85 S166A/S167A were used to express either wild-type Pho85p or Pho85p in which two adjacent serine residues in the activation loop had been changed to alanine in yeast strain BY391a. These vectors were gifts from Dongqing Huang and Brenda Andrews, University of Toronto, and contain the promoter from MET3 and the PHO85 open reading frame in a backbone derived from pRS313 (65).
Synthetic peptides.
Synthetic peptides were synthesized by Macromolecular Resources, Colorado State University. Three peptides were made, based upon the sequences surrounding the sites phosphorylated by Pho85p in Gsy2p (23, 24) and Pho4p (47). The peptide sequences used were as follows: Gsy2-654, KKLSVPGSPRDLRS; Gsy2-667, KKSTVYMTPGDLGT; and Pho4-114, KKPRLLYSPLIHTQ, with the phosphorylated residue underlined in each case.
Protein expression and purification.
HisPho85p and HisPcl10p were isolated after transformation of E. coli BL21(DE3) with the appropriate pET plasmids described above. Cells were grown at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) until an optical density at 600 nm of 0.6 was reached. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.4 mM, and the cultures were transferred to 30°C. After 4 h, cells were collected by centrifugation (5,000 × g, 10 min, 4°C), washed, and then resuspended with homogenization buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 10 mM imidazole, 0.1% [vol/vol] Triton X-100, 0.5 μg of leupeptin per ml, 0.7 μg of pepstatin per ml, 1 μg of aprotinin per ml, 10 μg of trypsin inhibitor per ml, 2 mM benzamidine, 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK], 1 mM phenylmethylsulfonyl fluoride [PMSF]). All subsequent steps were performed at 4°C. Cells were broken by two passes through a French pressure cell, and cell debris was removed by centrifugation (20,000 × g, 15 min). The supernatant was mixed with 1.5 ml of Ni2+-NTA-agarose (Qiagen), equilibrated in homogenization buffer for 1 h, and then packed into a 5-ml chromatography column. The column was washed with 15 ml of homogenization buffer, followed by 15 ml of buffer with the imidazole concentration raised to 50 mM. Bound protein was eluted by washing the column with 7.5 ml of homogenization buffer containing 200 mM imidazole. The eluted material was dialyzed against a storage buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol (DTT), 1 mM benzamidine, and 0.1 mM PMSF and then concentrated with Amicon Centricon-10 (Pho85p) or Centricon-30 (Pcl10p) centrifugal concentration devices. Glycerol was added to 20% (vol/vol), and the material was stored frozen at −80°C where it was stable for at least 4 months, through multiple cycles of freezing and thawing. Typical yields were 1 to 3 mg of purified protein per liter of bacterial culture.
For coexpression studies, E. coli BL21(DE3) was simultaneously transformed with vectors pET-HisPHO85 and pACYC-HAPCL10. Transformants were cultured in LB medium (59) supplemented with both ampicillin (100 μg/ml) and chloramphenicol (40 μg/ml). Expression of recombinant proteins was induced as described above. After induction, samples of culture (2 ml) were collected by centrifugation (10,000 × g, 30 s, 4°C). Cell pellets were resuspended to 200 μl with homogenization buffer, frozen on dry ice, and stored at −80°C prior to use. Cell lysates were prepared by sonication (four 15-s pulses with 2 min of cooling on ice between each pulse) and used directly for phosphorylation of Gsy2p.
To produce Pcl10p in yeast, strain WW10 was transformed with pYES2/GS (Invitrogen) containing the PCL10 open reading frame. Pcl10p production was induced by first growing cultures in supplemented minimal medium lacking uracil (0.67% [wt/vol] yeast nitrogen base without amino acids [Difco] containing complete supplement mixture without uracil [Bio 101 Inc.]) and with 1% raffinose as a carbon source. Cells were then transferred to a medium containing 1% raffinose plus 2% galactose and allowed to grow for 24 h. The six-histidine tag encoded by the pYES2/GS vector was used to facilitate isolation of Pcl10p. Cells were collected by centrifugation (5,000 × g, 10 min, 4°C), washed and resuspended at 1 g (wet weight) of cells/ml in a homogenization buffer comprising 20 mM Tris-HCl (pH 7.9), 450 mM NaCl, 50 mM NaF, 5 mM imidazole, 0.1% (vol/vol) Triton X-100, and protease inhibitors as described above. All subsequent steps were performed at 4°C. Cells were broken by vigorous mixing in the presence of glass beads (0.4-mm diameter). Cell debris was removed by centrifugation (20,000 × g, 30 min). The supernatant was mixed with 0.5 ml of Ni2+-NTA–agarose for 1 h, and the resin was packed into a 2-ml chromatography column. The column was washed first with 5 ml of homogenization buffer and then sequentially with 5 ml (each) of homogenization buffer with the imidazole concentration raised to 20 mM and 40 mM. The material eluted with 40 mM imidazole was dialyzed against a solution containing 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 1 mM benzamidine, and 0.1 mM PMSF and then concentrated as described above. Gsy2p was expressed in E. coli BL21(DE3) and purified as described previously (23). Construction of the phosphorylation site mutants of Gsy2p is described elsewhere (24).
Assay of Gsy2p kinase activity. (i) Indirect measurement of phosphorylation.
Phosphorylation converts Gsy2p to a less active form that requires the presence of the allosteric activator glucose-6-phosphate to elicit full activity. Thus, the ratio of activity without and with glucose-6-phosphate (−/+ glucose-6-P activity ratio) can be used as an index of the phosphorylation state of the enzyme. Purified Gsy2p (5 to 10 μg) was combined with either 3 to 5 μg of total cell lysate from E. coli BL21(DE3) expressing HisPho85p and/or HAPcl10p or purified HisPho85p and HisPcl10p in a final volume of 80 μl of kinase buffer (50 mM Tris-HCl [pH 7.4], 1 mM DTT, 0.1% Triton X-100, and protease inhibitors as described above). The reaction was initiated by addition of 20 μl of a mixture of 25 mM MgCl2 and 1 mM ATP, and the samples were incubated at 30°C. At intervals, aliquots (10 μl) were removed and diluted to 200 μl with a solution containing 50 mM Tris-HCl (pH 7.8), 25 mM KF, and 20 mM EDTA to terminate phosphorylation. Aliquots (30 μl) were then assayed for glycogen synthase activity as described previously (21, 68).
(ii) Direct measurement of phosphorylation.
HisPho85p and HisPcl10p were combined with Gsy2p and kinase buffer (as described above) in a final volume of 20 μl. This mixture was preincubated for 10 min at 30°C, and the reaction was initiated by the addition of 5 μl of a mixture comprising 250 μM [γ-32P]ATP (specific activity, 2,000 to 4,000 dpm/pmol) and 6.25 mM MgCl2. Each set of assays containd control reaction mixtures lacking substrate. The reaction was terminated after 10 to 15 min by withdrawing 15 μl of solution and spotting it onto squares of 31-ET chromatography paper (Whatman) saturated with a mixture of 20% (wt/vol) trichloroacetic acid, 1 mM ATP, and 5 mM Na4P2O7. The squares were washed in a solution containing 10% (wt/vol) trichloroacetic acid, 0.25 mM ATP, and 5 mM Na4P2O7. After the squares were washed for 15 min, the solution was decanted and replaced with 5% (wt/vol) trichloroacetic acid and washing continued for a further 15 min. Two more washes with 5% (wt/vol) trichloroacetic acid were performed, followed by a brief rinse with acetone. The paper squares were dried under a heating lamp, and the incorporation of radioactivity was determined by liquid scintillation counting. When E. coli lysates were used as a source of kinase, 3 to 5 μg of E. coli lysate was used in place of HisPcl10p and HisPho85p, the reaction was terminated by addition of 1 ml of ice-cold buffer (50 mM Tris-HCl [pH 7.9], 500 mM NaCl, 50 mM NaF, 50 mM imidazole, 0.1% [vol/vol] Triton X-100, and protease inhibitors as described above). The Gsy2p was then reextracted from the incubation mixture and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as described previously (24).
Phosphorylation of synthetic peptides.
HisPho85p and HisPcl10p were combined with kinase buffer (as above) and substrate peptide in a final volume of 20 μl. The reaction was initiated by addition of 5 μl of a mixture comprising 250 μM [γ-32P]ATP (specific activity, 2,000 to 4,000 dpm/pmol) and 6.25 mM MgCl2. After 30-min incubation at 30°C, 15 μl of the reaction mixture was spotted onto P-81 paper (Whatman) and dropped into 1% (vol/vol) phosphoric acid. The paper squares were washed three times for 10 min each time with 1% (vol/vol) phosphoric acid, rinsed briefly with acetone, and dried. Incorporated radioactivity was determined by liquid scintillation counting.
Mass spectrometry and analysis of protein digests.
HisPho85p was digested in the gel with sequencing grade trypsin (Boehringer Mannheim) at a 10:1 substrate/protease ratio overnight. Protein digests were chromatographed by using a fused silica column (inner diameter, 500 μm; length, 20 cm) packed with Vydac (218TP5x) C18 material by previously published methods (11, 12). Gradients were controlled with an Applied Biosystems/PE 178 high-performance liquid chromatography system employing standard 0.05% trifluoroacetic acid–water– CH3CN solvents at 10 μl min−1. Column effluents were infused directly into the electrospray ion source of a Finnigan LCQ ion-trap mass spectrometer which was operating under software control to determine base peaks in full-scan mode (400 to 2,000 m/z), perform high-resolution scans on the base peak in a second scan event, and fragmentation analysis of the base peak according to the mass and charge state determined in the second scan event. Data were analyzed both manually and by Seaquest software provided by Finnigan.
Gel filtration. (i) Recombinant Gsy2p and Pcl10p.
Gel filtration was performed with a Superose 6 HR 10/30 column attached to a fast protein liquid chromatography system (Pharmacia). The column was equilibrated with buffer (25 mM Tris-HCl [pH 7.5], 200 mM NaCl) at a flow rate of 0.3 ml min−1. The column was calibrated using a series of protein molecular weight standards (Sigma and Pharmacia). Either purified Gsy2p, purified Pcl10p, or a mixture of Gsy2p and Pcl10p was injected onto the column. Fractions (1 ml for Gsy2p and Gsy2p-Pcl10p mixture; 0.3 ml for Pcl10p) were collected, and the elution positions of the various proteins were determined by activity measurements and examination of silver-stained SDS-polyacrylamide gels.
(ii) Yeast cell lysates.
Saturated cultures were grown in 10 ml of YPD medium (19) (EG328-1A and CC9) or in 20 ml of synthetic medium (19) with 1% (wt/vol) raffinose and 2% (wt/vol) galactose (WW10 transformed with pYES2/GS containing the PCL10 open reading frame). The extracts were clarified by centrifugation (17,000 × g, 15 min, 4°C), and the protein content in the supernatant was determined. Approximately 2 mg of protein was applied to a Superose 6 column under the conditions described above, except that the buffer contained 1 mM DTT and the protease inhibitors benzamidine (2 mM), PMSF (0.1 mM), and TLCK (0.1 mM). Fractions (0.3 ml) were collected, and 15-μl aliquots were analyzed by SDS-PAGE and immunoblotting.
Immunoblotting.
Pho85p was detected by using anti-Pho85p serum kindly provided by Brenda Andrews, University of Toronto. Yeast Pcl10p was detected by using an antibody raised against the V5 epitope (Invitrogen) encoded by the GeneStorm vector sequence, and Gsy2p was detected by using antiserum raised against a Gsy2p peptide (21). Enhanced chemiluminescence (Amersham) and horseradish peroxidase-conjugated secondary antibodies (Sigma) were used for detection.
Immunoprecipitation.
Strain WW10 transformed with pYES2/GS containing the PCL10 open reading frame was grown and PCL10 expression was induced as described above. Cell pellets were collected by centrifugation, washed once, and resuspended with immunoprecipitation buffer (100 mM NaCl, 0.02% [vol/vol] Triton X-100, 50 mM Tris-HCl [pH 7.5], plus protease inhibitors as described above). Cells were broken by using glass beads, and the extract was clarified by centrifugation (17,000 × g, 15 min, 4°C). The clarified extract was transferred to a fresh tube, and centrifugation was repeated. Aliquots (200 μl, corresponding to approximately 3 mg of total protein) of this second supernatant were used for immunoprecipitation. The cell extract was precleared with protein A agarose beads (20 μl; Gibco-BRL), and 2.5 μl of anti-V5 antibody (Invitrogen) was added. After incubation for 2 h at 4°C with rocking, 20 μl of protein A agarose beads were added and the incubation continued for a further 2 h. The beads were collected by low-speed centrifugation and washed with 1 ml of immunoprecipitation buffer. This washing was repeated four more times. The pellets were resuspended with SDS-PAGE sample buffer, boiled, and analyzed by SDS-PAGE and immunoblotting with antibody to Gsy2p.
Data analysis.
To analyze enzyme kinetic data that followed hyperbolic kinetics, the data were fitted to a rectangular hyperbola by nonlinear regression analysis, using the KaleidaGraph software package for Macintosh (Synergy Software).
Other methods.
Protein concentrations were measured by the method of Bradford (4), with bovine serum albumin as a standard. Glycogen concentrations in cell extracts were determined by the method of Hardy and Roach (21), and in each case, the values reported are means of determinations from two separate transformants measured in duplicate.
RESULTS
Expression of a functional Pho85p-Pcl10p complex in E. coli.
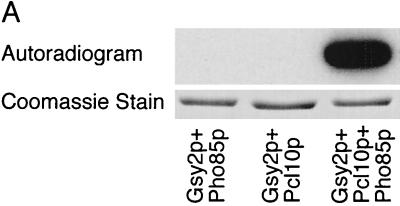
To produce a functional Gsy2p kinase in E. coli, we expressed tagged forms of Pho85p and Pcl10p by using the T7 promoter system. Pho85p had an NH2-terminal six-histidine tag, and Pcl10p had an NH2-terminal HA epitope tag. These constructs were coexpressed in the same cells using vectors with different replication origins (see Materials and Methods). HisPho85p was readily detected in a crude E. coli lysate after Coomassie blue staining of an SDS-polyacrylamide gel (see Fig. 2). No Coomassie blue-stained band corresponding to HAPcl10p was visible, although it was detected by immunoblotting with antibody to the HA epitope (not shown). Lysates of E. coli coexpressing Pcl10p and Pho85p catalyzed the incorporation of 32P from [γ-32P]ATP into Gsy2p, but there was no detectable incorporation of phosphate with extracts from cells expressing either Pho85p or Pcl10p alone (Fig. 1A).
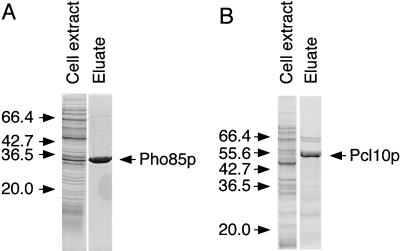
FIG. 2.
Purification of Pho85p and Pcl10p expressed in E. coli. Soluble lysates were prepared from E. coli carrying either pET-HisPHO85 (A) or pET-HisPCL10 (B) as described in Materials and Methods. A Coomassie blue-stained SDS-polyacrylamide gel of the soluble fraction of the cell lysate and the soluble material after purification (∼10 μg) using Ni2+-NTA–agarose chromatography is shown. The positions of molecular size standards (in kilodaltons) are shown to the left of the gels.
FIG. 1.
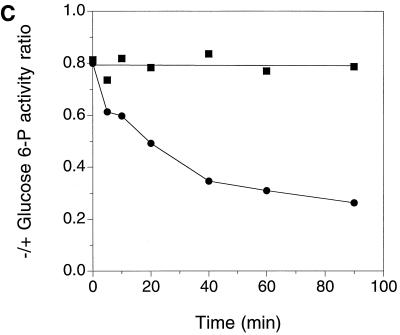
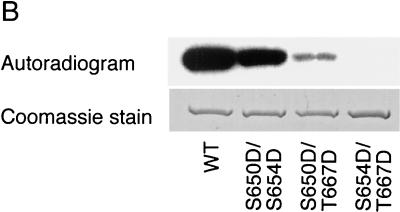
Expression of a functional Pho85p-Pcl10p complex in E. coli. (A) Phosphorylation of Gsy2p by lysates of E. coli coexpressing Pho85p and Pcl10p. Cell lysates were prepared from E. coli carrying either pET-HisPHO85, pACYC-HAPCL10, or both. Purified, recombinant Gsy2p was incubated with these extracts in the presence of [γ-32P]ATP and MgCl2. The Gsy2p was then repurified from the incubation mixture with Ni2+-NTA–agarose and analyzed by SDS-PAGE and autoradiography. (B) Phosphorylation of mutant Gsy2p. Lysates from E. coli expressing both Pho85p and Pcl10p were used to phosphorylate either wild-type (WT) Gsy2p or Gsy2p where combinations of the known phosphorylation sites (Ser650, Ser654, and Thr667) were mutated to aspartic acid. The phosphorylated Gsy2p was recovered and analyzed as described above. (C) Phosphorylation and inactivation of Gsy2p. Purified recombinant Gsy2p was incubated with lysates prepared from E. coli coexpressing Pho85p and Pcl10p either in the presence of MgCl2 and ATP (●) or MgCl2 alone (■). At the indicated times, aliquots were removed and the −/+ glucose-6-P activity of glycogen was determined.
Gsy2p is thought to contain three phosphorylation sites, Ser650, Ser654, and Thr667 (21), and Pho85p-Pcl10p is responsible for the phosphorylation of Ser654 and Thr667, both in vivo and in vitro, with Thr667 being the preferred site (23, 24). To determine whether the recombinant Pho85p-Pcl10p kinase phosphorylated the relevant sites in Gsy2p, two approaches were used. First, we employed a series of mutant Gsy2p species in which two of the three phosphorylation sites were mutated to Asp. The mutant substrates S650D S654D and S650D T667D were phosphorylated, with S650D S654D being the preferred substrate. By comparison, there was no detectable phosphorylation of the mutant S654D T667D which lacks both Pho85p-Pcl10p phosphorylation sites (Fig. 1B). Phosphoamino acid analysis confirmed that the S650D S654D mutant contained only phosphothreonine, while the S650D T667D mutant contained only phosphoserine (not shown). Second, lysates of cells coexpressing Pho85p and Pcl10p catalyzed a time-dependent decrease in the −/+ glucose-6-P activity ratio (Fig. 1C). This ratio is a kinetic index of the extent of phosphorylation of glycogen synthase, with lower values corresponding with increased phosphorylation. Therefore, the Pho85p-Pcl10p kinase phosphorylates the physiologically important sites in Gsy2p and inactivates it.
Although an active glycogen synthase kinase was produced by coexpression of Pho85p and Pcl10p in E. coli, we could not control the relative levels of expression to produce a defined kinase complex. Therefore, Pho85p and Pcl10p were expressed and purified independently. We made a Pcl10p construct with a tag of six histidine residues at the NH2 terminus. Both HisPho85p and HisPcl10p were soluble, stable, and amenable to purification (Fig. 2), routinely yielding 1 to 3 mg of Pho85p or Pcl10p per liter of culture. Densitometric scanning of Coomassie blue-stained gels indicated that the Pho85p was >90% pure, while the Pcl10p was around 80% pure.
Reconstitution of active Gsy2p kinase from recombinant Pcl10p and Pho85p.
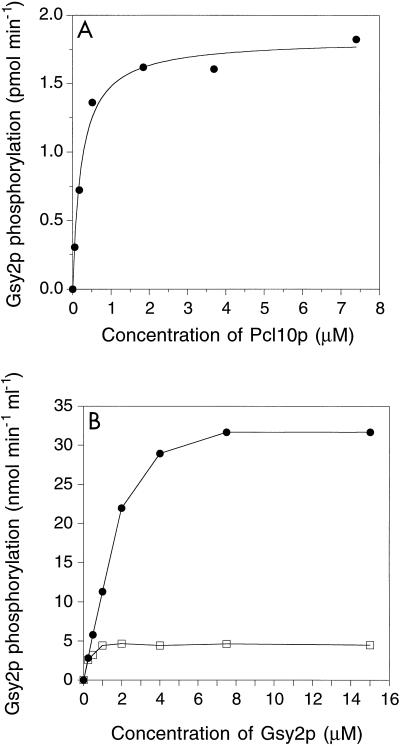
Pho85p alone displayed no detectable activity towards Gsy2p, but addition of Pcl10p generated significant Gsy2p kinase activity (Fig. 3A). The activity increased with Pcl10p concentration and saturated at high levels. The curve was hyperbolic, and the concentration of Pcl10p required for half-maximal activation was approximately 0.2 μM. When the substrate concentration was varied at fixed Pcl10p levels, the reaction rate also reached a saturable, maximum level (Fig. 3B). The curve was not hyperbolic, and at lower Gsy2p concentrations, there was a linear dependence of rate on substrate concentration. This linearity is most clearly seen with the higher Pcl10p concentration in the experiment shown. When the substrate saturation curves at different Pcl10p levels are compared, the Vmax achieved was dependent on the Pcl10p concentration. Thus, with 50 nM Pcl10p, an amount equimolar with the Pho85p catalytic subunit, the rate soon became independent of the Gsy2p concentration even though the same amount of Pho85p could sustain a higher Vmax when Pcl10p was at a higher level (compare the two curves in Fig. 3B). This type of enzyme kinetic behavior is exactly what is predicted for a system in which an obligate activator binds to the substrate prior to the formation of an active enzyme-substrate complex (64). In this case, Pcl10p is the activator and Gsy2p is the substrate.
FIG. 3.
Characterization of Pho85p-Pcl10p kinase reconstituted from polypeptides produced in E. coli. (A) Phosphorylation of Gsy2p by Pho85p requires Pcl10p. Purified Pho85p (80 nM) was incubated together with Gsy2p (3 μM) and the indicated concentrations of Pcl10p. After 10 min at 30°C, the phosphorylation reaction was initiated by the addition of [γ-32P]ATP and MgCl2. After a further 10-min incubation, the reaction was terminated and the incorporation of 32P into Gsy2p was determined by liquid scintillation counting. The data were analyzed as described in Materials and Methods. The concentration of Pcl10p required to give half the maximal rate was calculated to be ∼0.2 μM. (B) Effect of Gsy2p concentration on the rate of phosphorylation. Pho85p (50 nM) and either 50 nM (□) or 2 μM (●) Pcl10p were incubated with the indicated concentrations of Gsy2p, and the initial rate of Gsy2p phosphorylation was determined.
Analysis of recombinant Pho85p by mass spectroscopy.
Many Cdks require phosphorylation of their activation loop, and for Pho85p, Santos et al. (60) had reported that phosphorylation of Ser166 was required for kinase function. Therefore, we attempted to determine whether the recombinant Pho85p isolated from E. coli contained phosphate at this position. Purified Pho85p was digested with trypsin, concentrated, and analyzed by electrospray ionization mass spectrometry. Automated analysis of the fragmentation data led to the identification of peptides representing ∼42% of the Pho85p sequence. Manual examination of data allowed additional identification of peptides comprising 95% of the protein sequence. The peptide containing Ser166 (AFGIPVNTFSSEVVTLWYR) was clearly observed as a major species. The calculated m/z for the doubly charged ion is 1,093.568, and that of the observed ion is 1,093.6. No peptides corresponding to phosphorylated species (+80 Da) or diphosphorylated species (+160 Da) were observed. Given the very high levels of material introduced into the electrospray, we are confident that we should have detected phosphopeptides present at significant proportions, and we conclude that recombinant Pho85p does not require phosphorylation of activation loop residues for kinase activity. This result is consistent with recent results from Huang and Andrews (22) who have found that a mutant form of PHO85 in which both Ser166 and Ser167 are changed to alanine was capable of regulating acid phosphatase expression. We measured the level of glycogen accumulated by a pho85 mutant strain (BY391a) transformed with either empty vector, wild-type PHO85, or the above Ser166Ala Ser167Ala double mutant. The level of glycogen when empty vector was present was 5.6 mg of glycogen/mg of protein. When wild-type PHO85 was expressed, the level was 1.1 mg of glycogen/mg of protein. When the S166A S167A mutant was expressed, the level was 1.8 mg of glycogen/mg of protein. Thus, if phosphorylation of Ser166 and/or Ser167 does occur in vivo, such phosphorylation is not required for any Pho85p function studied so far.
Phosphorylation of peptide substrates by Pcl10p-Pho85p.
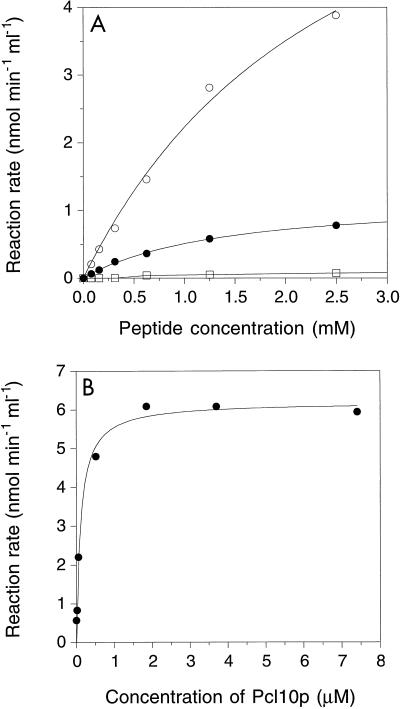
Besides by direct interaction with substrate, cyclins like Pcl10p could also affect Cdk specificity by altering the local sequences preferred by the kinase active site. In fact, if the known Pho85p sites in Gsy2p and Pho4p are compared, there is a subtle difference. Four of the five Pho4p sites have the sequence -S-P-X-I/L- (47), whereas the two Gsy2p sites have the sequence -V-X-X-S/T-P-X-D-L (23). Three peptides were synthesized based upon the sequences surrounding Ser654 and Thr667 in Gsy2p and Ser114 in Pho4p (referred to as Gsy2-654, Gsy2-667, and Pho4-114, respectively). Phosphorylation of both Gsy2-654 and Pho4-114 was readily measured, and there was a hyperbolic dependence of reaction rate on peptide concentration (Fig. 4A). Gsy2-667 was not detectably phosphorylated at the sensitivity of this assay. Although the Vmax values for Pho4-114 and Gsy2-654 were, respectively, 4 and 25% of that for Gsy2p, the Km values were much greater (Table 1). In terms of the pseudo-first-order rate constants (Vmax/Km), the peptides were some 4 orders of magnitude less effective as substrates than was Gsy2p. There was a low but detectable activity towards the synthetic peptides in the absence of added Pcl10p which was not observed when Gsy2p was the substrate (Fig. 4B). As was true for Gsy2p, peptide phosphorylation depended on the Pcl10p concentration, half-maximal activation corresponding to ∼0.2 μM Pcl10p, similar to the value observed with Gsy2p as the substrate. We conclude first that the process of activation, involving interaction of Pho85p and Pcl10p, is similar for both peptide substrates and Gsy2p. Second, the peptides were poor substrates compared to Gsy2p, indicating that local sequence is not the dominant determinant for recognition. Rather, effective phosphorylation of the physiological protein substrates may depend more on targeting interactions between Pcl10p and the substrate, as discussed below.
FIG. 4.
Phosphorylation of synthetic peptide substrates by Pho85p-Pcl10p. (A) Kinetics of peptide phosphorylation. The indicated concentrations of synthetic peptides Gsy2-654 (○), Gsy2-667 (□), and Pho4-114 (●) were incubated with 50 nM Pho85p and 2 μM Pcl10p, and the initial rate of peptide phosphorylation was determined. Data were analyzed as described in Materials and Methods. (B) Phosphorylation of synthetic peptides by Pho85p shows a dependence upon Pcl10p. Synthetic peptide Gsy2-654 (2.5 mM) was incubated with 50 nM Pho85p and the indicated concentrations of Pcl10p. The data were analyzed as described in Materials and Methods. The concentration of Pcl10p required to give half-maximal peptide phosphorylation was calculated to be ∼0.2 μM.
TABLE 1.
Phosphorylation of synthetic peptide substrates and Gsy2pa
| Substrate | Vmax (nmol min−1 ml−1) | Km (mM) | Vmax/Km (min−1) |
|---|---|---|---|
| Gsy2-654 | 8.2 | 2.7 | 3.0 × 10−3 |
| Gsy2-667 | 0b | NDc | ND |
| Pho4-114 | 1.2 | 1.4 | 8.6 × 10−4 |
| Gsy2p | 32d | 1.5 × 10−3e | 11f |
The initial rate of phosphorylation of either synthetic peptides or Gsy2p was determined. The kinetic parameters for the synthetic peptides were obtained by fitting the data in Fig. 4A to the Michaelis-Menten equation by using nonlinear regression analysis. The parameters for Gsy2p phosphorylation were estimated from Fig. 3B, since Gsy2p phosphorylation did not follow hyperbolic kinetics.
Within the limits of the assay, no significant phosphorylation was detected.
ND, not determined.
Measured with 2 μM Pcl10p.
The concentration of Gsy2p required to achieve half the maximum rate. Estimated from Fig. 3B.
Estimated from the initial slope of the graph in Fig. 3B.
Isolation of a Pho85p-Pcl10p complex from yeast.
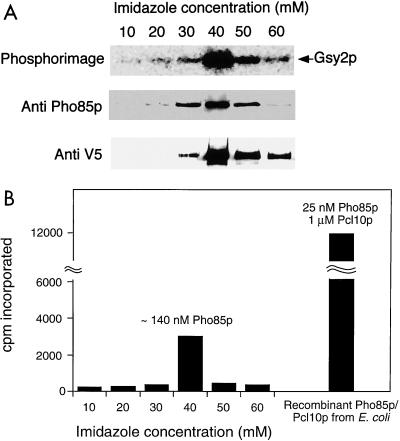
Although the Pho85p-Pcl10p proteins expressed in E. coli were able to reconstitute a functional Gsy2p kinase activity, it was important to determine precisely how active this material was compared to enzyme purified from yeast. It was possible, for example, that the activity detected was low and that in yeast other mechanisms are needed for full activity. We therefore transformed strain WW10 (pcl8 pcl10) with a multicopy vector containing the PCL10 open reading frame. This vector encodes a form of Pcl10p containing a COOH-terminal tag of six histidine residues and the V5 epitope (Invitrogen), under the control of the inducible GAL1 promoter. The vector complemented the glycogen hyperaccumulation defect of the pcl8 pcl10 mutant strain (not shown). Pcl10p was partially purified from yeast lysates by chromatography on Ni2+-NTA–agarose. Material eluted from the Ni2+-NTA–agarose column contained both Pcl10p and Pho85p, as determined by immunoblotting, and was capable of phosphorylating Gsy2p (Fig. 5A). The peaks of Pho85p, Pcl10p, and Gsy2p kinase activity were coincident (Fig. 5A). By analyzing known amounts of recombinant Pho85p in the immunoblotting analysis under the same conditions, we estimated the absolute amount of Pho85p present in the peak of kinase activity isolated from yeast (Fig. 5B). In this way, the specific activity of the Pcl10p-Pho85p kinase from yeast could be compared with that of the kinase reconstituted from proteins expressed in E. coli (Fig. 5B). The Pho85p produced in E. coli, assayed with a saturating concentration of Pcl10p, was 10- to 15-fold more active than Pho85p-Pcl10p isolated from yeast when the rate of Gsy2p phosphorylation was normalized to the amount of Pho85p. However, although the kinase was purified from yeast with a tagged form of Pcl10p, we cannot formally exclude the possibility that Pcl10p was limiting. We conclude that the recombinant kinase has activity at least as high as that purified from yeast.
FIG. 5.
Isolation of Pho85p-Pcl10p from yeast. Cell lysates were prepared from strain WW10 (pcl8 pcl10) expressing PCL10 from the vector pYES2/GS. Pcl10p was partially purified from the lysate by chromatography on Ni2+-NTA–agarose. (A) Column fractions eluting with the indicated concentrations of imidazole analyzed for the presence of Gsy2p kinase activity, Pho85p and Pcl10p. (B) Quantification of the Gsy2p phosphorylation and the degree of phosphorylation achieved with a defined amount of recombinant Pho85p-Pcl10p under the same conditions.
Formation of complexes between Pcl10p and Gsy2p.
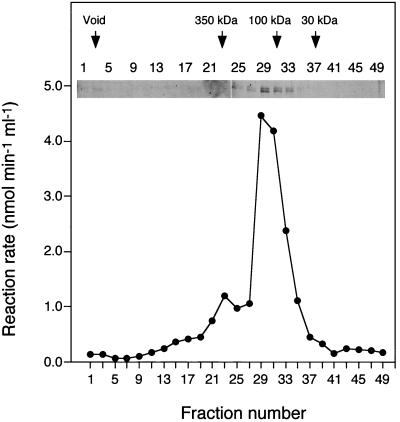
The enzyme kinetic data are consistent with there being interactions between Gsy2p and Pcl10p. Also, we had previously observed a weak interaction between Gsy2p and Pho85p in a two-hybrid assay (24). The availability of purified, recombinant Pcl10p and Gsy2p allowed us to address this interaction in more detail. Pcl10p, Gsy2p, or a mixture of the two proteins was analyzed by gel filtration. Pcl10p eluted predominantly as a peak with an apparent molecular mass of ∼110 kDa, which would be consistent with a dimer of the 48.6-kDa Pcl10p monomer (Fig. 6). A portion of the Pcl10p eluted earlier in the profile, suggesting the possibility of forming even larger aggregates. In order to determine whether the oligomeric forms of Pcl10p were active, fractions from the gel filtration column were assayed for their ability to activate Pho85p, measured using Gsy2p as a substrate. The peak of Pho85p-activating activity coincided with the presence of Pcl10p protein, indicating that all species of Pcl10p detected were capable of activating Pho85p (Fig. 6). After filtration, the Pcl10p ran as a doublet on SDS-PAGE. The reason is not known but it could represent some breakdown of the Pcl10p during the separation.
FIG. 6.
Size distribution of recombinant Pcl10p. Pcl10p was analyzed by gel filtration using a Superose 6 column. Aliquots (15 μl) of each fraction were analyzed by SDS-PAGE and silver staining. To determine whether the eluted Pcl10p was functional, aliquots were combined with Pho85p (65 nM), Gsy2p (3 μM), MgCl2, and [γ-32P]ATP. Phosphorylation of Gsy2p was then measured using the filter paper assay described in Materials and Methods.
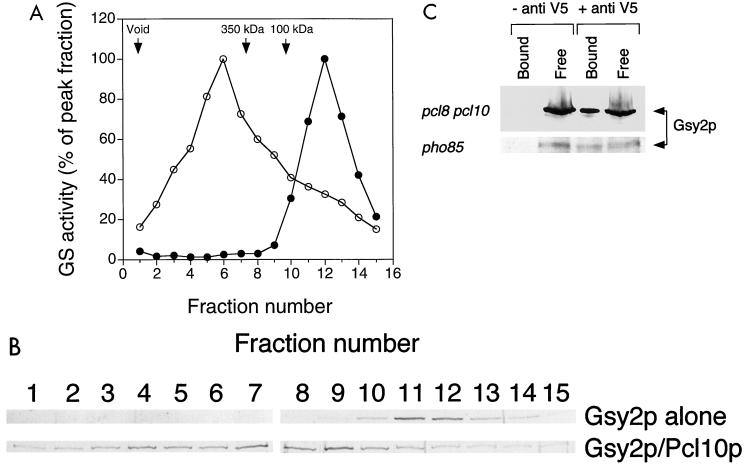
From gel filtration, the estimated molecular mass of Gsy2p was ∼75 kDa, close to the calculated subunit molecular mass of 80 kDa (Fig. 7A). When mixtures of Gsy2p and Pcl10p were analyzed, the profile of Gsy2p activity was shifted to correspond to a substantial increase in size, which we infer to result from interaction with Pcl10p. The peak fraction had an estimated molecular mass on the order of 500 kDa, and the peak was broader than when Gsy2p was analyzed alone (Fig. 7A). Evidently, multiple Gsy2p-Pcl10p complexes can be formed. Note, however, that not all Gsy2p was converted to the larger size, since there was still Gsy2p activity at lower molecular weights. Silver-stained SDS-polyacrylamide gels of the fractions showed that the presence of Gsy2p protein correlated with the activity measurements (Fig. 7B). This result provides strong evidence that Pcl10p and Gsy2p interact in solution.
FIG. 7.
Interaction between Pcl10p and Gsy2p in vitro and in vivo. (A) Gsy2p alone (●) or a mixture of Gsy2p and Pcl10p (in a 1:1.5 molar ratio) (○) was analyzed by gel filtration using a Superose 6 column. Fractions were collected, and the glycogen synthase (GS) activity was measured. (B) Aliquots (15 μl) of each fraction were analyzed by SDS-PAGE and silver staining. (C) Cell lysates were prepared from strain WW10 (pcl8 pcl10) and from strain WW11 (pho85) each expressing PCL10 from the vector pYES2/GS. Immunoprecipitation was carried out as described in Materials and Methods using antibody to the V5 epitope tag encoded by pYES2/GS (+ anti V5) or an equivalent volume of phosphate-buffered saline (− anti V5). The immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with antibody to Gsy2p (Bound). An aliquot (20%) of the supernatant after addition of protein A agarose beads was also analyzed (Free).
To determine whether Gsy2p and Pcl10p might also be associated in vivo, anti-V5 antibody was used to immunoprecipitate Pcl10p from extracts of strains WW10 (pcl8 pcl10) and WW11 (pho85) expressing the V5-tagged PCL10 construct described above. The resulting precipitates were analyzed by SDS-PAGE and immunoblotting with antibody raised against Gsy2p. The anti-V5 antibody coprecipitated Gsy2p from the soluble fraction of a yeast cell lysate, suggesting that Pcl10p and Gsy2p interact in vivo (Fig. 7C). Similar results were obtained, regardless of whether Pho85p was present (WW10) or not (WW11), indicating that the interaction between Gsy2p and Pcl10p is independent of Pho85p in vivo. The amount of Gsy2p precipitated was approximately 20% of the total present in the extract. We conclude that Pcl10p is able to form complexes with Gsy2p that survive gel filtration and immunoprecipitation protocols.
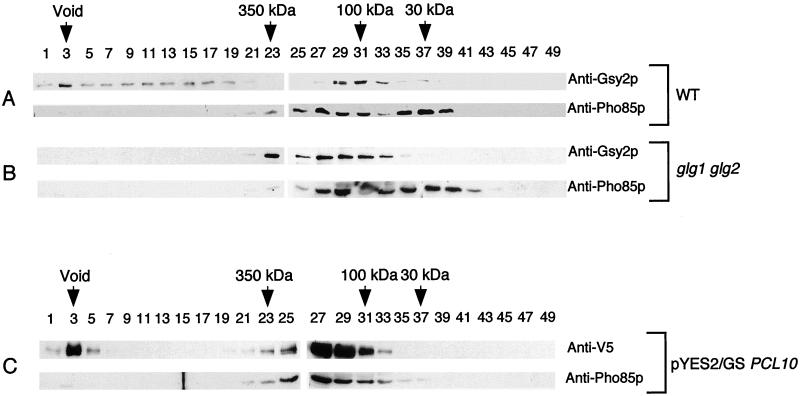
Size distribution of Gsy2p, Pho85p, and Pcl10p in yeast cell lysates.
Since the gel filtration and immunoprecipitation studies indicated interaction between Pcl10p and Gsy2p, the distribution of Gsy2p, Pcl10p, and Pho85p between the free state and higher-order complexes was examined in extracts of yeast. With lysates of wild-type yeast (strain EG328-1A), two distinct pools of Gsy2p were detected (Fig. 8A), one at very high molecular mass, ranging into the void volume, and another at 70 to 100 kDa. This latter pool is around the predicted size of the Gsy2p monomer (80 kDa). Pho85p was also found in two peaks. One peak corresponded to a size of ∼30 kDa, that of free, monomeric Pho85p. The other peak was somewhat broader, extending in size up to ∼170 kDa, and presumably represents Pho85p associated with Pcls or other proteins.
FIG. 8.
Size distribution of Gsy2p, Pho85p, and Pcl10p in yeast cell lysates. Approximately 2 mg of soluble protein from either wild-type (WT) yeast (EG328-1A) (A), a glycogen-deficient strain (CC9 [glg1 glg2]) (B), or a strain expressing V5 epitope-tagged Pcl10p (WW10 [pcl8 pcl10] carrying pYES2/GS containing PCL10) (C) was analyzed by gel filtration using a Superose 6 column. Fractions (300 μl) were collected from the void volume to the total volume, and aliquots (15 μl) were analyzed by SDS-PAGE and immunoblotting.
Since the Gsy2p running at very high molecular mass might be associated with glycogen particles, cell extracts were also prepared from a glg1 glg2 strain that does not make glycogenin, the initiator protein of glycogen biosynthesis. Cells of this strain contain the wild-type level of functional glycogen synthase but are unable to produce glycogen (7). Gel filtration of lysates from glg1 glg2 double mutants revealed the elimination of Gsy2p from the void volume, although some Gsy2p was still detectable in high-molecular-mass fractions (Fig. 8B). There was no major change in the distribution of Pho85p (Fig. 8B).
In order to investigate the size distribution of Pcl10p, we expressed the V5 epitope-tagged Pcl10p construct in pcl10 cells from a high-copy-number plasmid, since we currently do not have anti-Pcl10p antibodies. After gel filtration, Pcl10p was detected with anti-V5 antibody. Two clearly defined pools of Pcl10p were detected, one at very high molecular mass in the column void volume and a second centered around 170 kDa (Fig. 8C). The Pho85p profile was significantly changed compared to that of the wild type, with the majority of the Pho85p now found at higher molecular weight and corresponding to the peak of Pcl10p (Fig. 8C). This result is consistent with overexpression of Pcl10p driving the formation of Pho85p-Pcl10p complexes. We examined these overexpressing cells for any abnormalities associated with a lack of Pho85p function, reasoning that Pho85p complexed with Pcl10p might be unavailable for other functions. However, neither cell morphology nor the ability to regulate acid phosphatase expression differed from that of the wild type (not shown). Therefore, despite being largely sequestered by Pcl10p, sufficient Pho85p was accessible to fulfill these other cellular functions.
DISCUSSION
We have shown that a functional kinase consisting of the Cdk catalytic subunit, Pho85p, and the cyclin, Pcl10p, can be reconstituted from recombinant components expressed in E. coli. The kinase phosphorylates a known in vivo substrate, Gsy2p, at physiologically relevant sites and to an extent that causes inactivation of Gsy2p. The Pho85p-Pcl10p kinase has several properties that differ from the more extensively studied cell cycle-regulated Cdks. First, the kinase catalytic subunit does not appear to require phosphorylation, since the enzyme formed from recombinant components is at least as active as Pho85p-Pcl10p kinase prepared from yeast. Also, the recombinant Pho85p does not appear to contain phosphate at Ser166 due to phosphorylation during expression in E. coli. Second, Pcl10p activation of Pho85p requires substantially more than a one-to-one ratio of cyclin to kinase, with half-maximal activation at ∼0.2 μM Pcl10p. If one infers a binding constant from this value, we are dealing with quite high affinity, enough to explain the clear evidence for interaction between Pho85p and Pcl10p, but not such a high affinity as would imply an irreversible formation of a cyclin-kinase complex. This behavior differs from that of classic cell cycle Cdks. For example, Desai et al. (14) found that Cdc2 and Cdk2 activation by cyclin A and B1 corresponded to a one-to-one titration of Cdc2 or Cdk2 activity by addition of cyclin, indicative of very high affinity and resulting in a complex that was stable to dilution. In this regard, it is important to recognize that Pcl10p belongs to a different family of cyclins. This subset of yeast Pho85p cyclins, Pcl6p, Pcl7p, Pcl8p, and Pcl10p, lacks the COOH-terminal structural repeat of the cyclin box present in cell cycle-regulated cyclins and is also characterized by NH2-terminal extensions of unknown function and no sequence similarity to other cyclins. It is not unreasonable, then, that this class of cyclins differs in how it interacts with the Cdk catalytic subunit.
Our suggestion that Pho85p does not require phosphorylation at Ser166 in the activation loop for activity is in contrast to the conclusions of Santos et al. (60) who reported that mutation of Ser166 to Ala yielded a nonfunctional Pho85p molecule as judged by its inability to complement defects in pho85 cells. In that study, mutation to Glu resulted in a Pho85p molecule which functioned comparably to the wild type. However, others have obtained conflicting results, finding that mutation of Ser166 to Ala has no effect on the ability of Pho85p to regulate acid phosphatase expression (22). Together with the results presented in this study, we conclude that Ser166 phosphorylation is not essential for activity. Indeed, there is a precedent for a Cdk not requiring prior phosphorylation for activity, namely, mammalian Cdk5. This Cdk is thought to be involved in neuronal and muscle development (36, 39, 54, 66). Cdk5 is also expressed in terminally differentiated tissues where it is not believed to have a role in cell cycle controls. It is activated by association with p35, a protein only remotely related in sequence to cyclins (38, 56). Like Pho85p, Cdk5 can be produced in E. coli, and the addition of a truncated form of the p35 activator also produced in E. coli can reconstitute an active kinase complex (56). It is intriguing that Cdk5 is the mammalian Cdk with the amino acid sequence most similar to that of Pho85p, and we have shown that p35 can modestly activate Pho85p (75). However, as noted above, p35 bears almost no resemblance to Pcl10p or other Pcls, and it remains to be established whether it functions mechanistically in a way similar to that of Pcl10p.
In the present study, we provide evidence that Pcl10p forms complexes with Gsy2p. First, in gel filtration studies of recombinant proteins, the Gsy2p elution profile was markedly altered by the presence of Pcl10p. Second, Gsy2p was present in immunoprecipitates from yeast cell extracts using antibodies directed at an epitope tag on Pcl10p. Third, additional evidence for the interaction came from the substrate kinetics of the reconstituted Pho85p-Pcl10p kinase in which the level of Pcl10p determined the Vmax with respect to Gsy2p. This behavior is consistent with an obligate activator interacting with the substrate so that the effective substrate for the enzyme becomes the activator-substrate complex (64), Pcl10p-Gsy2p in this case. Originally, such kinetics were observed with enzymes that utilize ATP, where the MgATP complex is the true substrate (see reference 40 for a comprehensive discussion). The results are also consistent with our observation that Pcl10p gave a weak positive result with Gsy2p in a two-hybrid yeast assay (24). Synthetic peptides based on known Pho85p phosphorylation sites were extremely poor substrates for Pho85p-Pcl10p, with Vmax/Km ratios some 4 orders of magnitude lower than that obtained with Gsy2p. The large difference in Vmax/Km is due mostly to the high Km values for the peptide substrates. We conclude not only that Pcl10p directly interacts with Gsy2p but also that this interaction plays an important role in substrate recognition.
Ideas about the mechanisms of protein kinase substrate specificity have evolved significantly over the last decade or so. Initial efforts to define recognition determinants centered on local sequence features surrounding the modified residue (35, 52). For some kinases, certain critical residues that define consensus sequence motifs can be identified and short synthetic peptides of corresponding sequence serve as effective substrates. For example, cyclic AMP-dependent protein kinase phosphorylates the short peptide known as Kemptide virtually as well as it does liver pyruvate kinase, the protein from which Kemptide is derived (34). However, there are many examples of kinases that, like Pho85p, phosphorylate short synthetic peptides poorly. Such behavior has been attributed to a requirement for additional contacts to be made at points removed from the site of phosphorylation in the substrate (35) as is, we postulate, the case for Pho85p-Pcl10p. Such contacts can be with regions of the kinase catalytic subunit outside of the catalytic site, as in the case of JNK phosphorylation of c-Jun (32) and Elk-1 (76). Alternatively, a separate protein subunit may be involved (17, 51). Examples include scaffolding proteins, like Ste5p, that allow for specific phosphorylations within the MAP kinase cascade by sequestering protein kinases and their kinase substrates (61, 74). Another example is provided by growth factor signaling pathways in which many Tyr phosphorylations are mediated by noncatalytic protein-protein interactions (9, 50). A third example is of proteins that might best be defined as targeting subunits, akin to the multiple targeting subunits that interact with the type 1 protein phosphatase catalytic subunit (13, 25). Different targeting subunits direct the same catalytic subunit to distinct functions. Such is likely to be true for Cdks in general (for example, see references 53 and 72) and for Pho85p in particular. In fact, Pho85p provides one of the better examples where there is evidence that distinct targeting subunits, such as Pcl10p and Pho80p, direct the kinase towards separate substrates both in vitro and in vivo (24). Other recent work has suggested a targeting role also for mammalian cyclins in retinoblastoma protein (Rb) phosphorylation. A region of cyclin E has been identified as directing the phosphorylation of Rb by Cdk2 (33). This region is implicated in direct binding of cyclin E to Rb, and its removal impedes phosphorylation of Rb by Cdk2-cyclin E but not of the generic Cdk substrate histone H1 (33). Cyclin A-mediated phosphorylation involves a hydrophobic patch in cyclin A which is involved in targeting Cdk2-cyclin A to specific substrates (63). Mutation of the hydrophobic patch reduces the ability of Cdk2-cyclin A to phosphorylate Rb.
Mechanistically, targeting subunits could function in two different ways. One mechanism would involve the formation of a complex in which the targeting subunit contributes to the binding interaction with the substrate during catalysis. Alternatively, the targeting subunit could serve to increase the local concentration of substrate at the active site of the kinase rather than to present the substrate to the kinase in a particular orientation. In the example of Rb phosphorylation noted above, addition of a heterologous Rb-binding sequence to the mutant cyclin A that could not bind Rb restored Rb phosphorylation (63). This result is consistent with increased local concentration of substrate contributing significantly to Rb recognition. At present, we favor a similar model for Pho85p-Pcl10p based on the enzyme kinetic data. Half-maximal activation of Pho85p by Pcl10p occurred at ∼0.2 μM whether an efficient substrate, such as Gsy2p, or a poor substrate, such as Gsy2-654, was used. If Pcl10p interacted simultaneously with Gsy2p and Pho85p, one would have expected activation of Gsy2p phosphorylation to occur at lower Pcl10p concentrations than those for peptide phosphorylation. Thus, the results suggest that the enhanced kinetic performance with Gsy2p as a substrate is due to a local concentration effect. However, more work is needed to substantiate this hypothesis.
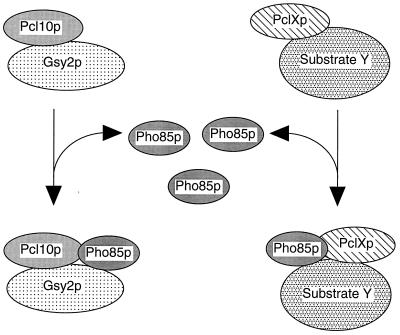
A substantial proportion of the Pho85p in yeast cell extracts exists as a monomer, but it is also detected over a wide range of molecular weights, as was true also for Gsy2p and Pcl10p. There was a clear overlap of the elution of Gsy2p, Pho85p, and Pcl10p, consistent with the proteins being present in the same complex. The Pho85p catalytic subunit is known to interact with 10 different cyclin molecules (16, 29, 41, 42) and the Pho81p inhibitor (48, 62), so a wide size distribution is not surprising. Additionally, if the cyclins mediate other protein-protein interactions, as we propose for Pcl10p, even greater heterogeneity can be expected. Pcl10p itself had an apparent molecular mass of ∼110 kDa or greater which would be consistent with formation of dimers or larger complexes. We have yet to prove formally the presence of Pcl10p dimers, but it is interesting that Desai et al. (14) reported that cyclins A and B1 produced in insect cells had molecular masses of 110 and 160 kDa, respectively, as judged by gel filtration. Other relevant protein-protein interactions include glycogen synthase binding to the glycogenins Glg1p and Glg2p (7), the Gac1p targeting subunit of Glc7p protein phosphatase (8), possibly other phosphatase targeting subunits (6), and glycogen particles (reviewed in reference 57). Thus, there is the potential to form a large variety of different assemblies. From this perspective, it is not surprising that we were able to immunoprecipitate only some 20% of the total Gsy2p from cell lysates using antibody directed to Pcl10p. When Pcl10p was expressed from a high-copy-number plasmid, there was sufficient overexpression to drive the majority of the Pho85p into coelution with Pcl10p. Overexpression of Pcl10p in wild-type yeast cells caused reduced glycogen accumulation, presumably by recruiting more Pho85p to the control of this specific function (18). We envisage a model for Pho85p function in which the Pcls are limiting and Pho85p is in excess, with a significant pool of free monomer (Fig. 9). Pho85p from this free pool could be directed to specific substrates via interactions with substrate-associated Pcls. This sequestration of Pho85p may also involve trafficking between cellular compartments. A large proportion of Pho85p is nuclear (30), whereas glycogen synthesis is thought to be cytosolic. In this regard, it will be of interest to analyze the subcellular localization of Pcl10p. As is true for the control of the classic cell cycle Cdks, the assembly and disassociation of Pho85p-Pcl complexes would provide mechanisms for regulating Pho85p function. To date, little is known of this process. In the case of the classic Cdks, the high-affinity association of the cyclin with the Cdk necessitates regulated proteolytic degradation to deconstruct the complex. Perhaps nonclassic cyclins like Pcl10p, which has lower affinity for its Cdk, are subject to a different type of regulation. The mechanisms for the specific control of Pho85p function through individual Pcls will be an important topic of future investigation.
FIG. 9.
Model for substrate recognition by Pho85p-Pcl10p. In the model, Pho85p is in excess of the Pcls. Free Pho85p is then recruited to phosphorylate particular substrates via interactions between the substrate and the appropriate Pcl. How this recruitment is regulated is not known. Thus, in addition to activating Pho85p, the Pcls also target the kinase to specific substrates.
ACKNOWLEDGMENTS
This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases grants DK27221 and DK42576 and the Indiana University Diabetes Research and Training Center grant DK20542.
We thank Anna DePaoli-Roach, Thomas Hurley, Mark Goebl, and Ron Wek for many helpful discussions during the course of this work and critical comments regarding the manuscript. We thank Brenda Andrews and Dongqing Huang for sharing reagents and results prior to publication.
REFERENCES
- 1.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 2.Bazan J F. Helical fold prediction for the cyclin box. Proteins. 1996;24:1–17. doi: 10.1002/(SICI)1097-0134(199601)24:1<1::AID-PROT1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C, Huang D Q, Roach P J. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast. 1997;13:1–8. doi: 10.1002/(SICI)1097-0061(199701)13:1<1::AID-YEA49>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 7.Cheng C, Mu J, Farkas I, Huang D Q, Goebl M G, Roach P J. Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6632–6640. doi: 10.1128/mcb.15.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, C., and P. J. Roach. Unpublished observations.
- 9.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 10.Cross F. Transcriptional regulation by a cyclin-cdk. Trends Genet. 1995;11:209–211. doi: 10.1016/s0168-9525(00)89047-2. [DOI] [PubMed] [Google Scholar]
- 11.Davis, M. T. 1997. Mass spectrometry methods, ABRF ’97 Tutorial 2. Presented at the 1997 Meeting of the Association of Biomolecular Resource Facilities, Baltimore, Md., 9 February 1997.
- 12.Davis M T, Stahl D C, Swiderek K M, Lee T D. Capillary liquid chromatography/mass spectrometry for peptide and protein characterization. Methods Companion Methods Enzymol. 1994;6:304–314. [Google Scholar]
- 13.Depaoli-Roach A A, Park I K, Cerovsky V, Csortos C, Durbin S D, Kuntz M J, Sitikov A, Tang P M, Verin A, Zolnierowicz S. Serine/threonine protein phosphatases in the control of cell function. Adv Enzyme Regul. 1994;34:199–224. doi: 10.1016/0065-2571(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 14.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 16.Espinoza F H, Ogas J, Herskowitz I, Morgan D O. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 17.Faux M C, Scott J D. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 18.Fujino, M. A., and P. J. Roach. Unpublished observations.
- 19.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 20.Hardy T A, Huang D Q, Roach P J. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 21.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 22.Huang, D. Q., and B. Andrews. Personal communication.
- 23.Huang D Q, Farkas I, Roach P J. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D Q, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard M J, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 26.Hunt T. Cyclins and their partners: from a simple idea to complicated reality. Semin Cell Biol. 1991;2:213–222. [PubMed] [Google Scholar]
- 27.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 28.Jeffrey P D, Ruso A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 29.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 30.Kaffman A, Rank N M, O’Neill E M, Huang L S, O’Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 31.Kaldis P, Sutton A, Solomon M J. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 32.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 33.Kelly B L, Wolfe K G, Roberts J M. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemp B E, Graves D J, Benjamini E, Krebs E G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252:4888–4894. [PubMed] [Google Scholar]
- 35.Kemp B E, Pearson R B. Design and use of peptide substrates for protein kinases. Methods Enzymol. 1991;200:121–134. doi: 10.1016/0076-6879(91)00134-i. [DOI] [PubMed] [Google Scholar]
- 36.Lazaro J B, Kitzmann M, Poul M A, Vandromme M, Lamb N J, Fernandez A. Cyclin dependent kinase 5, cdk5, is a positive regulator of myogenesis in mouse C2 cells. J Cell Sci. 1997;110:1251–1260. doi: 10.1242/jcs.110.10.1251. [DOI] [PubMed] [Google Scholar]
- 37.Lenburg M E, Oshea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 38.Lew J, Huang Q Q, Qi Z, Winkfein R J, Aebersold R, Hunt T, Wang J H. A brain-specific activator of cyclin-dependent kinase 5. Nature. 1994;371:423–426. doi: 10.1038/371423a0. [DOI] [PubMed] [Google Scholar]
- 39.Lew J, Wang J H. Neuronal cdc2-like kinase. Trends Biochem Sci. 1995;20:33–37. doi: 10.1016/s0968-0004(00)88948-3. [DOI] [PubMed] [Google Scholar]
- 40.London W P, Steck T L. Kinetics of enzyme reactions with interaction between a substrate and a (metal) modifier. Biochemistry. 1969;8:1767–1779. doi: 10.1021/bi00832a061. [DOI] [PubMed] [Google Scholar]
- 41.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 42.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 44.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 45.Morgan D O, Fisher R P, Espinoza F H, Farrell A, Nourse J, Chamberlin H, Jin P. Control of eukaryotic cell cycle progression by phosphorylation of cyclin-dependent kinases. Cancer J Sci Am. 1998;4:S77–S83. [PubMed] [Google Scholar]
- 46.Nigg E A. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- 47.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa N, Noguchi K-I, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson M V, Dutchik J E, Graham M Y, Brodeur G M, Helms C, Frank M, MacCollin M, Scheinman R, Frank T. Random-clone strategy for genomic restriction mapping in yeast. Proc Natl Acad Sci USA. 1986;83:7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 51.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 52.Pearson R B, Mitchelhill K I, Kemp B E. Studies of protein kinase/phosphatase specificity using synthetic peptides. In: Hardie D G, editor. Protein phosphorylation: a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 265–291. [Google Scholar]
- 53.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philpott A, Porro E B, Kirschner M W, Tsai L H. The role of cyclin-dependent kinase 5 and a novel regulatory subunit in regulating muscle differentiation and patterning. Genes Dev. 1997;11:1409–1421. doi: 10.1101/gad.11.11.1409. [DOI] [PubMed] [Google Scholar]
- 55.Poon R Y, Hunter T. Cell regulation. Innocent bystanders or chosen collaborators? Curr Biol. 1995;5:1243–1247. doi: 10.1016/s0960-9822(95)00248-x. [DOI] [PubMed] [Google Scholar]
- 56.Qi Z, Huang Q Q, Lee K Y, Lew J, Wang J H. Reconstitution of neuronal Cdc2-like kinase from bacteria-expressed Cdk5 and an active fragment of the brain-specific activator. Kinase activation in the absence of Cdk5 phosphorylation. J Biol Chem. 1995;270:10847–10854. doi: 10.1074/jbc.270.18.10847. [DOI] [PubMed] [Google Scholar]
- 57.Roach P J, Cheng C, Huang D, Lin A, Mu J, Skurat A V, Wilson W A, Zhai L. Novel aspects of the regulation of glycogen storage. J Basic Clin Physiol Pharmacol. 1999;9:139–151. doi: 10.1515/jbcpp.1998.9.2-4.139. [DOI] [PubMed] [Google Scholar]
- 58.Rothman-Denes L B, Cabib E. Two forms of yeast glycogen synthetase and their role in glycogen accumulation. Proc Natl Acad Sci USA. 1970;66:967–974. doi: 10.1073/pnas.66.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Santos R C, Waters N C, Creasy C L, Bergman L W. Structure-function relationships of the yeast cyclin-dependent kinase Pho85. Mol Cell Biol. 1995;15:5482–5491. doi: 10.1128/mcb.15.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaeffer H J, Weber M J. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider K R, Smith R L, O’Shea E K. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 63.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Segel I H. Enzyme kinetics. New York, N.Y: John Wiley and Sons; 1975. pp. 242–250. [Google Scholar]
- 65.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang D, Lee K Y, Qi Z, Matsuura I, Wang J H. Neuronal Cdc2-like kinase: from cell cycle to neuronal function. Biochem Cell Biol. 1996;74:419–429. doi: 10.1139/o96-046. [DOI] [PubMed] [Google Scholar]
- 67.Tennyson C N, Lee J, Andrews B J. A role for the Pcl9-Pho85 cyclin-cdk complex at the M/G(1) boundary in Saccharomyces cerevisiae. Mol Microbiol. 1998;28:69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- 68.Thomas J A, Schlender K K, Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosylltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 69.Thuret J Y, Valay J G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 70.Timblin B K, Tatchell K, Bergman L W. Deletion of the gene encoding the cyclin-dependent protein kinase Pho85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143:57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toh-e A, Tanaka K, Uesono Y, Wickner R B. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 72.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uesono Y, Tokai M, Tanaka K, Tohe A. Negative regulators of the PHO system of Saccharomyces cerevisiae: characterization of PHO80 and PHO85. Mol Gen Genet. 1992;231:426–432. doi: 10.1007/BF00292712. [DOI] [PubMed] [Google Scholar]
- 74.Whitmarsh A J, Davis R J. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, W. A., A. Lin, and P. J. Roach. Unpublished observations.
- 76.Yang S-H, Whitmarsh A J, Davis R J, Sharrocks A D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]