Abstract
Cytochrome P450 (CYP) 2A6 is a monooxygenase involved in the metabolism of various endogenous and exogenous chemicals, such as nicotine and therapeutic drugs. The genetic polymorphisms in CYP2A6 are a cause of individual variation in smoking behavior and drug toxicities. The enzymatic activities of the allelic variants of CYP2A6 were analyzed in previous studies. However, the three-dimensional structures of the mutants were not investigated, and the mechanisms underlying activity reduction remain unknown. In this study, to investigate the structural changes involved in the reduction in enzymatic activities, we performed molecular dynamics simulations for ten allelic mutants of CYP2A6. For the calculated wild type structure, no significant structural changes were observed in comparison with the experimental structure. On the other hand, the mutations affected the interaction with heme, substrates, and the redox partner. In CYP2A6.44, a structural change in the substrate access channel was also observed. Those structural effects could explain the alteration of enzymatic activity caused by the mutations. The results of simulations provide useful information regarding the relationship between genotype and phenotype.
Keywords: cytochrome P450, drug-metabolizing enzyme, molecular dynamics simulation, genetic polymorphism, structural analysis
1. Introduction
Cytochrome P450 (CYP) is a monooxygenase that plays a central role in the synthesis pathways of steroids, fatty acids, lipid-soluble vitamins, and eicosanoids [1]. In addition, CYP is a primary enzyme for drug metabolism [2,3,4]. The active site of CYP includes an heme iron that mediates electron transfer related to oxidation reactions [5,6]. Drug metabolism by CYP proceeds via oxidation, reduction (phase I), hydrolysis, and conjugation (phase II). The phase I oxidation requires electron carriers such as nicotinamide adenine dinucleotide phosphate (NADPH), and electron transfers are mediated by the membrane-bound flavor protein CYP redox partner [7]. The binding of CYP and the redox partner is necessary for electron transfer, and the helices C, K, and L of CYP are predicted to interact with the redox partner [8,9,10,11].
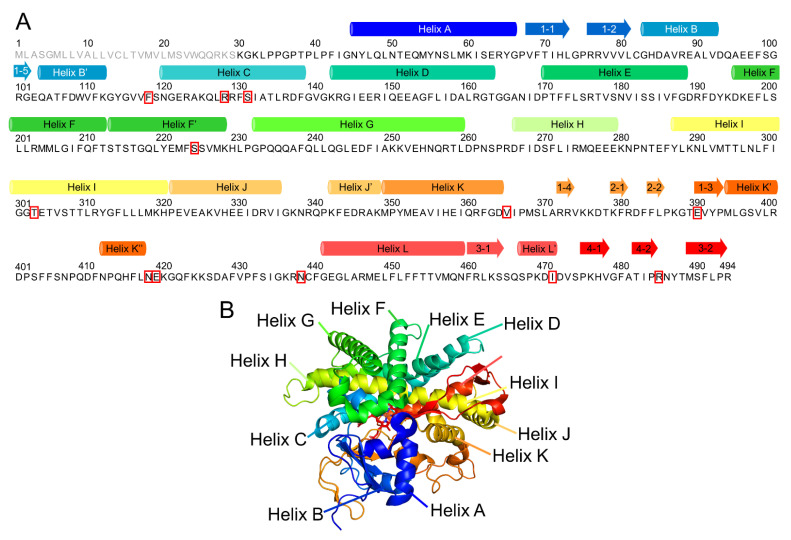
One of the members of CYP, CYP2A6, is involved in the metabolism of nicotine and various therapeutic drugs such as tegafur, valproic acid, pilocarpine, propofol, and cisapride [12,13]. In addition, Jiang et al. reported that CYP2A6 was a protective factor of hepatocellular carcinoma [14]. The cavity in the active site of CYP2A6 is about 25% of the volume of those in CYP2C8, CYP2C9, and CYP3A4 [15]. Therefore, the substrates and inhibitors for CYP2A6 are relatively small molecules [16,17]. The amino acid sequence and whole structure of CYP2A6 is shown in Figure 1. The structure includes the helices A–L and β1–4. CYP2A6 has six substrate-recognizing sites (SRS-1–6) [18]. SRS-1 is composed of B’ and a flanking area (residues 102–112), SRS-2 is composed of the C-terminal end of helix F (residues 197–204), SRS-3 is composed of the N-terminal end of helix G (residues 232–239), SRS-4 is composed of the N-terminal half of helix I (residues 288–296), SRS-5 is composed of the β2 area (residues 358–368), and SRS-6 is composed of the central region of β5 (residue 468–476). The flexibilities of helices F and G, including SRS-2 and SRS-3, respectively, are markedly reduced by substrate binding. Therefore, these helices are considered important for the accommodation of enzymatic activities.
Figure 1.
Numbering of the secondary structures of CYP2A6. (A) Correspondence between the amino acid sequence and secondary structures. The circular column and arrow indicate helix and β-strand formation regions, respectively. The gray letters of the amino acid sequence are the disordered region in the crystal structure. The mutation sites relevant to 10 variants investigated in this study are shown in red squares. (B) Correspondence between the three-dimensional structure and secondary structures.
The CYP2A6 gene is mapped to chromosome 19 and located in a gene cluster composed of 350 kbp, together with the CYP2A7, CYP2A13, and CYP2B subfamilies, and the CYP2F subfamily [13]. CYP2A7 is known as a nonfunctional pseudogene. Phenotyping and in vitro studies indicate the prevalence of CYP2A6 polymorphisms. Although the CYP2A6 poor metabolizers are <1% of the Caucasian population, they comprise 20% of the Orientals population [19,20]. The presence of CYP2A6 polymorphisms is a risk factor for drug toxicity and tobacco-related lung cancer in male Japanese smokers [21]. Several CYP2A6 polymorphisms are characterized. CYP2A6*3, CYP2A6*4, CYP2A6*12, CYP2A6*20, CYP2A6*27, and CYP2A6*34 are inactive because of frame-shift mutations, gene conversion with the pseudogene CYP2A7, or whole gene deletion [22,23]. In CYP2A6*9, a point mutation in the TATA box located in the 5′ flanking region reduces the transcriptional level. CYP2A6*29, CYP2A6*30, CYP2A6*32, and CYP2A6*33 alleles are still being evaluated. Otherwise, 34 CYP2A6 mutants are characterized, and their metabolizing activities are evaluated by the assays for nicotine C-oxidization and coumarin 7-hydroxylation [22,24]. In CYP2A6*14 and CYP2A6*15, the nicotine and coumarin metabolizing activities are higher than those of the wild type. These activities of CYP2A6*28 and CYP2A6*31 are the same levels as those of the wild type. Other mutations reduce metabolizing activities. However, the detailed mechanisms of the metabolizing activity reductions are obscure because experimental structures of CYP2A6 mutants are unsolved.
In this study, to investigate the structures of CYP2A6 mutants, we performed molecular dynamics (MD) simulations for 10 mutants (CYP2A6.6, CYP2A6.11, CYP2A6.17, CYP2A6.25, CYP2A6.26, CYP2A6.35, CYP2A6.36, CYP2A6.37, CYP2A6.43, and CYP2A6.44). The selected mutants and their metabolizing activities are shown in Table 1. To assess the reason for metabolizing activity reduction, we selected variants with lower activities than the wild type. The allele frequencies of CYP2A6.6 and CYP2A6.11 were 0.4 and 0.6%, respectively, in healthy Japanese people [22,25]. CYP2A6.17 was frequently detected in African Americans (9.4%) [22]. Although the frequencies of CYP2A6.43 and CYP2A6.44 were lower (0.2% in African American) [26], the enzymatic activities were drastically lower than that of CYP2A6.17 (Table 1). The frequencies of CYP2A6.25 and CYP2A6.26 were 0.5 and 0.7%, respectively, in African Americans [27]. Those of CYP2A6.35 were 2.8% African Canadian and 0.8% Japanese [28]. CYP2A6.36 and CYP2A6.37 were detected in 0.3% of the Taiwanese population [28]. The MD simulations for CYP2A6.25, CYP2A6.26, CYP2A6.35, CYP2A6.36, and CYP2A6.37 were performed to investigate the effects of mutation combinations on three-dimensional (3D) structures. MD simulations were variable methods to investigate the structural features of mutant proteins including CYP [29,30,31,32,33,34]. For comparison, the simulation of the wild type (CYP2A6.1) was also performed.
Table 1.
Kinetic parameters for the wild type (CYP2A6.1) and mutants used in this study. Kinetic parameters for nicotine and coumarin are shown. The percentages of intrinsic clearances (CLint) are compared with CYP2A6.1 CLint. [22].
| Nicotine | Coumarin | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Amino Acid Changes | Km/μM |
Vmax/pmol min−1 |
CLint/ nL·min−1 |
Km/μM | Vmax/pmol min−1 |
CLint/ μL·min−1 |
| CYP2A6.1 | – | 33.3 ± 5.44 | 5.91 ± 1.26 | 176 ± 11.2 (100%) | 1.94 ± 0.06 | 5.89 ± 0.34 | 3.04 ± 0.08 (100%) |
| CYP2A6.6 | R128Q | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.11 | S224P | 142 ± 31.3 | 4.31 ± 0.34 | 31.6 ± 5.88 * (18%) | 3.30 ± 0.37 | 2.57 ± 0.03 | 0.79 ± 0.009 *** (26%) |
| CYP2A6.17 | V365M | 25.9 ± 6.16 | 1.70 ± 0.21 | 68.3 ± 12.2 * (39%) | 1.72 ± 0.03 | 2.76 ± 0.08 | 1.60 ± 0.002 ** (53%) |
| CYP2A6.25 | F118L | 59.9 ± 5.94 | 1.67 ± 0.09 | 28.0 ± 2.48 * (16%) | N.D. | N.D. | N.D. |
| CYP2A6.26 | F118L R128L S131A |
N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.35 | N438Y | 75.7 ± 14.9 | 3.98 ± 0.51 | 53.5 ± 5.31 * (30%) | 2.50 ± 0.28 | 4.63 ± 0.35 | 1.86 ± 0.08 *** (61%) |
| CYP2A6.36 | N438Y I471T |
N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.37 | N438Y I471T R485L |
N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.43 | T303I | 1049 ± 683 | 4.76 ± 2.00 | 5.33 ± 1.28 * (3%) | 8.55 ± 1.47 | 1.34 ± 0.09 | 0.16 ± 0.02 *** (5%) |
| CYP2A6.44 | E390K N418D E419D |
1097 ± 284 | 2.39 ± 0.43 | 2.22 ± 0.16 * (1%) | N.D. | N.D. | N.D. |
(% of CYP2A6.1), * p < 0.05, ** p < 0.01, and *** p < 0.005 compared with CYP2A6.1. N.D.: Not determined.
2. Results and Discussion
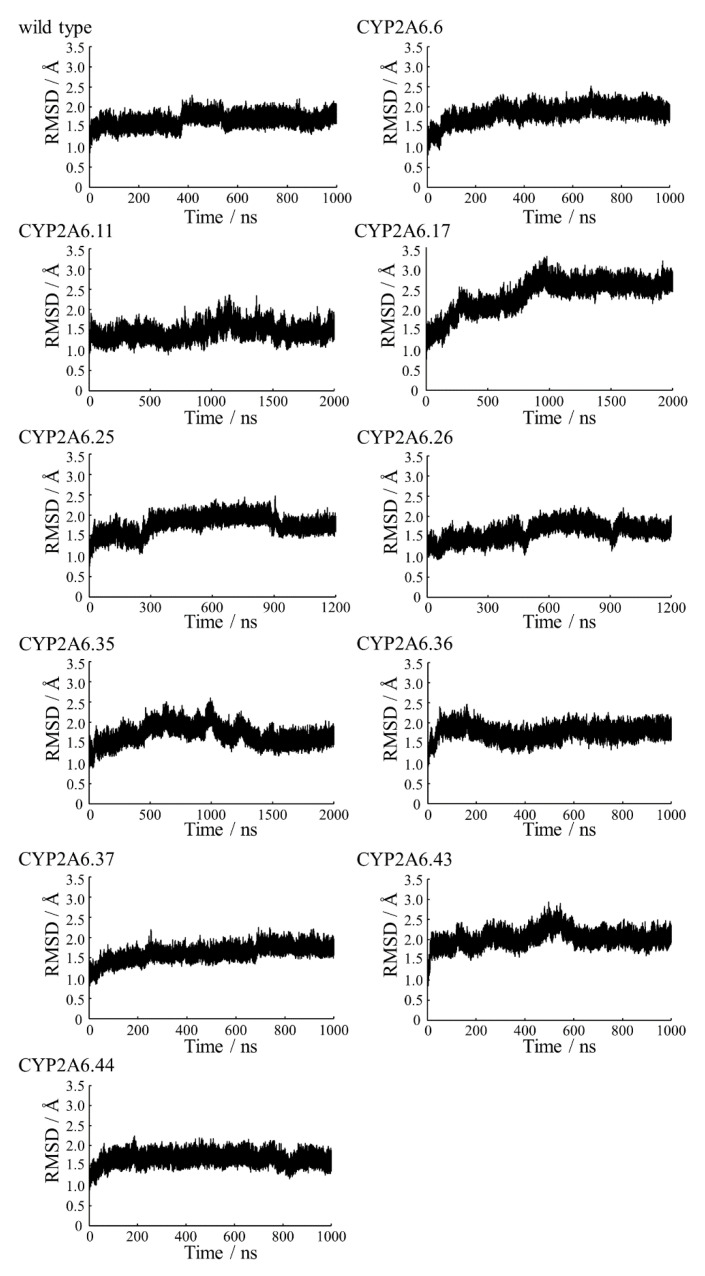
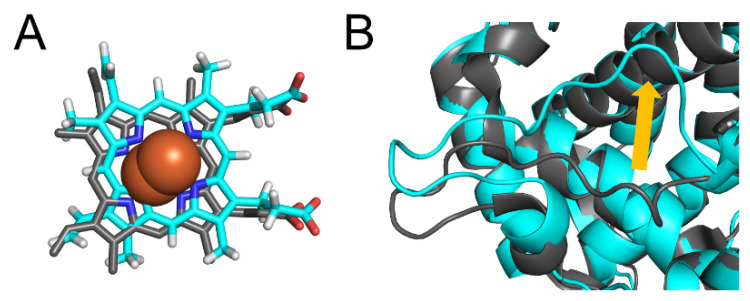
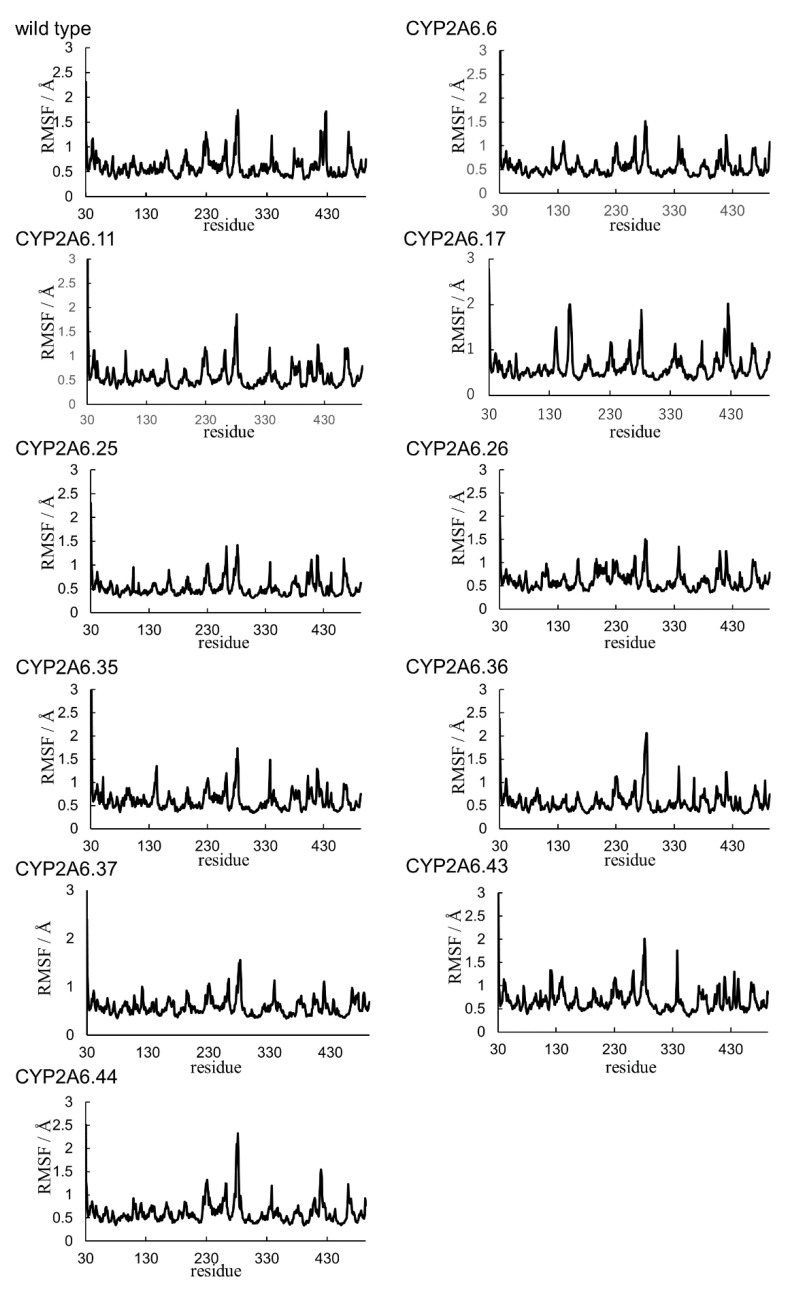
To investigate the structures of CYP2A6 mutants, the MD simulations were performed, and the convergences were evaluated by RMSDs (Figure 2). Each calculation was converged during the simulation times. The final structures obtained by the simulations are shown in Figure S1. From the final structures and RMSDs, large structural collapses were not observed within the simulations for all mutants. In the wild type, the location of heme in the calculated structure was similar to that of the experimental structure (Figure 3A). In previous studies, short MD simulations for the wild type were performed [35,36]. In [36], the simulation converged within 20 ns and continued until 100 ns. The simulations of the wild type in this study seemed to converge within 100 ns; however, the RMSD was largely changed at around 400 ns. Therefore, the simulation of CYP2A6 needed to be over 400 ns. In this calculated structure, there was almost no deviation from the experimental structure, except for the K”L loop (Figure 3B). The root mean square fluctuation (RMSF) values of residues 427 and 428 in the K”L loop were relatively high (≥1.5 Å) (Figure 4). Therefore, the shifts in K”L loops in the final structure were likely due to their flexibility. The RMSF value of Pro282 in the HI-loop was 1.75 Å, but the flexibilities of other regions were low. The alterations of secondary structure formations are shown in Table 2 and Table 3. In CYP2A6.6, CYP2A6.26, CYP2A6.36, CYP2A6.37, and CYP2A6.44, no significant alterations were observed. Helices F’ and L were separated into two helices in CYP2A6.11. Helix I was separated into two helices in CYP2A6.25 and CYP2A6.43. In addition, helix L was separated into three helices in CYP2A6.17. The formation of β4 was impaired in CYP2A6.35.
Figure 2.
Root mean square deviations (RMSD) plots of the main-chain atoms for CYP2A6 wild type and mutants.
Figure 3.
Structural deviation between the experimental and calculated structures. (A) The deviation of heme and (B) the K”L loop. The experimental structure and calculated structures are shown in gray and cyan, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The yellow arrow indicates the deviation of the K”L loop.
Figure 4.
RMSF plots of Cα atoms in the last 10 ns of simulations.
Table 2.
Secondary structure formation in >50% of trajectories for last 100 ns of simulations of the wild type 2A6.6, 2A6.11, 2A6.17, and 2A6.25 a.
| Wild Type | 2A6.6 | 2A6.11 | 2A6.17 | 2A6.25 | |
|---|---|---|---|---|---|
| HA | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 |
| β1-1 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 |
| β1-2 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 |
| HB | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 |
| β1-5 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 |
| HB’ | 105–111 | 105–108 | 105–111 | 105–108 | 105–111 |
| HC | 121–138 | 122–135 | 122–138 | 125–134 | 121–138 |
| HD | 143–163 | 145–162 | 143–162 | 145–160 | 142–162 |
| HE | 171–186 | 170–186 | 170–186 | 170–186 | 170–186 |
| HF | 196–212 | 196–211 | 196–212 | 196–212 | 196–213 |
| HF’ | 215–227 | 215–227 | 215–222, 224–227 |
215–227 | 215–227 |
| HG | 233–256 | 233–257 | 233–257 | 233–256 | 233–256 |
| HH | 267–277 | 267–277 | 267–277 | 267–277 | 267–279 |
| HI | 288–319 | 288–318 | 288–319 | 288–319 | 288–296, 302–319 |
| HJ | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 |
| HJ’ | 343–347 | 343–347 | 343–348 | 343–348 | 343–347 |
| HK | 350–362 | 350–362 | 350–362 | 350–362 | 350–362 |
| β1-4 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 |
| β2-1 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 |
| β2-2 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 |
| β1-3 | 391–393 | 391–393 | 390–393 | 390–393 | 390–393 |
| HK’ | 395–399 | 395–398 | 395–398 | 395–398 | 395–398 |
| HK’’ | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 |
| HL | 447–459 | 442–459 | 441–443, 447–459 |
435–437, 441–443, 448–457 |
442–459 |
| β3-1 | 460–463 | 460–463 | 460–463 | 460–462 | 460–464 |
| HL’ | 468–470 | 468–470 | 468–470 | 468–470 | 468–470 |
| β4-1 | 477–478 | 477–478 | 477–478 | 477–478 | 477–478 |
| β4-2 | 482–483 | 482–483 | 482–483 | 482–483 | 482–484 |
| β3-2 | 489–493 | 489–493 | 489–493 | 491–493 | 489–493 |
a HX shows helix X, and β shows β-strand formation moiety.
Table 3.
Secondary structure formation in >50% of trajectories for last 100 ns of simulations of 2A6.26, 2A6.35, 2A6.36, 2A6.37, 2A6.43, and 2A6.44 a.
| 2A6.26 | 2A6.35 | 2A6.36 | 2A6.37 | 2A6.43 | 2A6.44 | |
|---|---|---|---|---|---|---|
| HA | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 |
| β1-1 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 |
| β1-2 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 |
| HB | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 |
| β1-5 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 |
| HB’ | 105–110 | 108–111 | 105–110 | 105–111 | 105–110 | 107–111 |
| HC | 124–138 | 121–139 | 121–138 | 122–137 | 122–138 | 121–137 |
| HD | 145–162 | 145–162 | 143–161 | 143–161 | 145–161 | 143–161 |
| HE | 171–186 | 171–186 | 171–186 | 171–186 | 171–186 | 171–186 |
| HF | 196–212 | 196–212 | 196–211 | 196–213 | 196–213 | 196–212 |
| HF’ | 215–227 | 215–227 | 215–227 | 215–227 | 215–227 | 215–227 |
| HG | 233–256 | 233–257 | 233–256 | 233–256 | 233–257 | 233–256 |
| HH | 267–278 | 267–277 | 267–278 | 267–278 | 267–280 | 267–277 |
| HI | 288–319 | 288–319 | 288–319 | 288–319 | 288–297, 305–319 |
288–319 |
| HJ | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 |
| HJ’ | 343–347 | 343–348 | 343–347 | 343–347 | 343–346 | 343–348 |
| HK | 350–363 | 350–362 | 350–362 | 350–362 | 350–362 | 350–362 |
| β1-4 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 |
| β2-1 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 |
| β2-2 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 |
| β1-3 | 390–393 | 390–393 | 390–393 | 390–393 | 390–393 | 390–393 |
| HK’ | 395–398 | 395–398 | 395–399 | 395–398 | 395–398 | 395–398 |
| HK’’ | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 |
| HL | 442–459 | 442–459 | 446–457 | 447–459 | 449–457 | 442–459 |
| β3-1 | 460–463 | 460–464 | 460–464 | 460–463 | 460–463 | 460–463 |
| HL’ | 468–470 | 468–470 | 468–470 | 470–472 | 468–470 | 468–470 |
| β4-1 | 477–478 | – | 477–478 | 477–478 | 477–478 | 477–478 |
| β4-2 | 482–483 | – | 482–483 | 482–483 | 482–483 | 482–483 |
| β3-2 | 489–493 | 489–493 | 489–493 | 489–493 | 489–493 | 489–493 |
a HX shows helix X, and β shows β-strand formation moiety.
2.1. CYP2A6.6 Affected the Interaction with the Redox Partner
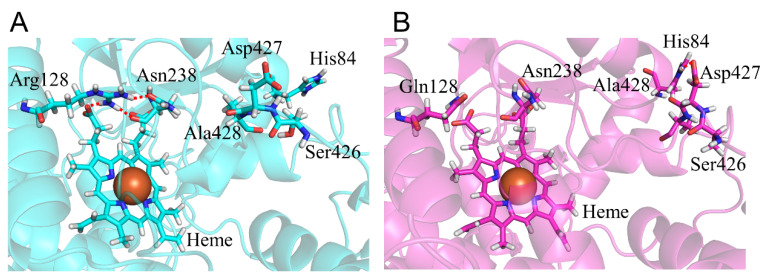
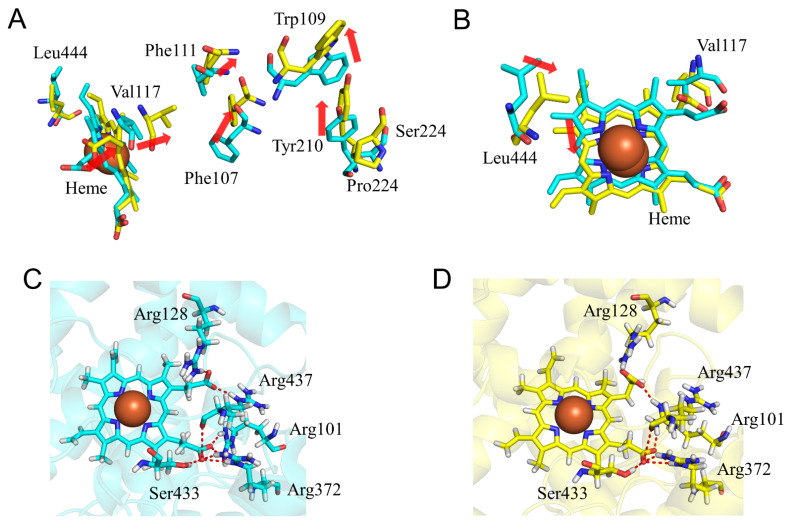
To evaluate the structure of CYP2A6.6, we compared the calculated results of the R128Q mutant with those of the wild type. The catalytic activity of CYP2A6.6 for nicotine and coumarin was reportedly undetectable [22]. Although, for Arg128 locates on helix C, the helix structure was not collapsed by the R128Q mutation (Table 2). However, the length change in the side chain and the loss of the positive charge of residue 128 affected hydrogen bond formation and interaction with the K”L loop. In the wild type, Arg128 formed an hydrogen bond with heme (Table 4 and Figure 5). On the other hand, Gln128 of the R128Q mutant formed no hydrogen bond with heme. The R128Q mutation was considered to decrease heme-binding affinity. Arg128 formed hydrogen bonds with Asn438 located on the K”L loop in the wild type, but these were lost in R128Q mutants. The loss of the hydrogen bond formation affected the structure and flexibility of K”L loop. In CYP2A6.6, Ser426–Ala428 were close to helix B, and Asp427 formed the ionic bonds with His84. This ionic bond was not observed in the wild type. In addition, the RMSF values of Ser426-Ala428 were smaller than the wild type (Figure 4B). Therefore, R128Q mutation shifted the K”L loop to helix B and decreased the flexibility of the K”L loop. Those alterations in the K”L loop structure could affect the interaction with the CYP redox partner. For this interaction with redox partner, R128Q mutation on helix C could have a direct effect because arginine residues on helix C were reported to be important for the interaction with the redox partner [10]. Therefore, the hindrance of the complex formation with the redox partner was suggested to be one of the reasons for loss of function in CYP2A6.6.
Table 4.
Hydrogen bond formation involved in the structural change in CYP2A6.6.
| Donor | Donor H | Acceptor | Wild Type | 2A6.6 |
|---|---|---|---|---|
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Asn438 O | 99.92 | 0 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 Oδ | 88.72 | 0 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 O | 33.52 | 0 |
| Phe429 N | Phe429 H | Ser425 O | 0 | 77.88 |
| Ala428 N | Ala428 H | Ser425 Oγ | 42.66 | 61.68 |
| His84 Nε2 | His84 Hε2 | Asp427 Oδ2 | 0 | 46.76 |
| His84 Nε2 | His84 Hε2 | Asp427 Oδ1 | 0 | 38.64 |
| Val430 N | Val430 H | Ala428 O | 0 | 52.86 |
Figure 5.
Structural change in CYP2A6.6. The wild type (A) and CYP2A6.6 (B) are shown in cyan and magenta, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as the van der Waals radius. The red dotted lines indicate the hydrogen bonds.
2.2. CYP2A6.11 Affected Secondary Structure Formation and Interaction with Heme
To evaluate the structural change in CYP2A6.11, the calculated results of the S224P mutant were compared with those of the wild type. The intrinsic clearances (CLint) for nicotine and coumarin in CYP2A6.11 were 18% and 26%, respectively, of those in the wild type [22]. Ser224 was included in helix F’, and the S224P mutation interrupted helix formation. Namely, helix F’ of CYP2A6.11 was separated into two helices composed of Ser215–Met222 and Pro224–Met227 (Table 2). The proline residing in an α-helix was known to cause a kink and the destabilization of the helical structure [37,38]. Therefore, the S224P mutation caused the separation of helix F’ by destabilizing the helical structure. In addition, helix L without the mutation was also separated into two helices composed of Gly441–Gly443 and Met447–Asn459. This alteration of helical structure was also caused by the effect of the S224P mutation. The structural change of helix F’ by S224P mutation altered the location of Tyr210 on helix F’ (Figure 6A). The shifted side chain of Tyr210 displaced Trp109 and helix B’, including this tryptophan residue. Val117 on the B’C loop moved into the space generated by the migration of Phe107 and Phe111 on helix B’. These phenylalanine residues were associated with the interaction with coumarin in the complex structure [15,39]. In addition, Val117 shifted the location of heme to the same direction as the shift of this valine residue. According to the deviation of the heme location, Leu444 on helix L was moved, and the helix L was separated (Figure 6B). The structural change in helix L could affect the interaction with the redox partner. The deviation of heme altered the hydrogen bond formation between heme and surrounding residues (Table 5). Although the wild type Arg101 formed an hydrogen bond only with OA, in the S224P mutant Arg101 formed hydrogen bonds with the oxygen of carbonic acid moiety, binding the pyrrole ring A (OA) and D (OD) of heme. Hydrogen bond formation between Arg372 and OA was attenuated in the S224P mutant. We concluded that the catalytic activity of CYP2A6.11 decreased due to the deviation of heme caused by the changes in the helix B’ structure and the decrease in the binding affinity of the redox partner.
Figure 6.
Structural change in CYP2A6.11 (A,B). The wild type (C) and CYP2A6.11 (D) are shown in cyan and yellow, respectively. In (A,B), all hydrogen atoms are omitted. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red dotted lines indicate the hydrogen bonds. The red arrows indicate the shift in CYP2A6.11.
Table 5.
Hydrogen bond formation with heme involved in the structural change in CYP2A6.11.
| Donor | Donor H | Acceptor | Wild Type | 2A6.11 |
|---|---|---|---|---|
| Arg101 Nη1 | Arg101 Hη1 | Heme O1A | 100 | 0 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O1A | 99.98 | 0.06 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O2A | 0 | 98.88 |
| Arg101 Nη1 | Arg101 Hη1 | Heme O2D | 0 | 97.48 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O2D | 0 | 88.66 |
| Arg101 Nε | Arg101 Hε | Heme O2A | 0 | 65.4 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 81.86 |
| Arg372 Nη2 | Arg372 Hη2 | Heme O2A | 99.1 | 48.12 |
| Arg372 Nε | Arg372 Hε | Heme O1A | 98.88 | 74.96 |
| Arg372 Nη2 | Arg372 Hη2 | Heme O1A | 57.9 | 0 |
| Arg372 Nε | Arg372 Hε | Heme O2A | 51.34 | 94.66 |
| Ser433 Nγ | Ser433 Hγ | Heme O2A | 100 | 99.9 |
| Arg437 Nε | Arg437 Hε | Heme O2D | 99.32 | 0.1 |
| Arg437 Nη1 | Arg437 Hη1 | Heme O2D | 65.76 | 0.08 |
| Arg437 Nε | Arg437 Hε | Heme O1D | 4.5 | 99.78 |
2.3. CYP2A6.17 Affected Secondary Structure Formation and Interaction with Heme and Redox Partoner
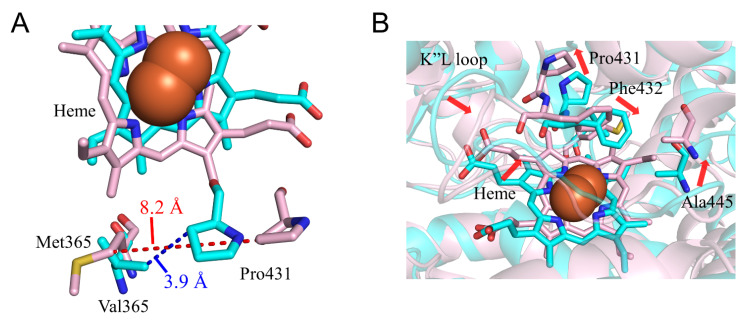
To evaluate the structural change in CYP2A6.17, the calculated results for the V365M mutant were compared with the wild type. The CLint for nicotine and coumarin in CYP2A6.17 were 39% and 53% of those in the wild type, respectively. Although Val365 was included in SRS-5, the range of Michaelis constant (Km) value did not increase in comparison with the wild type [22]. Thus, the V365M mutation was not expected to affect substrate binding in the nicotine–coumarin binding site. Helix C was composed of residues 121–138 in the wild type; however, it was formed by residues 125–134 in CYP2A6.17. In addition, helix L was separated into residues 441–443 and 448–457, and residues 435–437 formed a new helix despite no direct contact with the mutated residue (Table 2). Val365 located on the loop between the helix K and β1-4 interacted with the K”L loop in the wild type, and the distance between the nearest carbon atoms of Val365 and Pro431 was 3.9 Å in the final structure (Figure 7A). On the other hand, the distance between the nearest carbon atoms of Met365 and Pro431 was 8.2 Å in the final structure of CYP2A6.17. In addition, the RMSFs of Pro431–Phe432 were 1.5-fold higher than those of the wild type (Figure 4D). Therefore, the interaction between the K”L loop and the residue 365 decreased and the flexibility of the K”L loop increased in CYP2A6.17. Although the RMSF for other regions of the K”L loop did not increase, a structural change occurred. Residues 434–438 in the K”L loop shifted toward the heme and their percentage of helix formation increased. These shifted residues pushed the heme as shown in Figure 7B, and caused the shift of Ala445, resulting in the separation of helix L. These structural changes around heme and helix L were the cause for the decrease in enzymatic activity of CYP2A6.17. On the other hand, the nicotine and coumarin binding site was on the opposite side of the structurally changed region to the heme. This may be the reason why the V365M mutation had no effect on nicotine and coumarin binding affinity.
Figure 7.
Structural change in CYP2A6.17 (A,B). The wild type and CYP2A6.17 are shown in cyan and pink, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red and blue dotted lines indicate the distances between Val/Met365 and Pro431. The red arrows indicate the shift in CYP2A6.17. All hydrogen atoms are omitted.
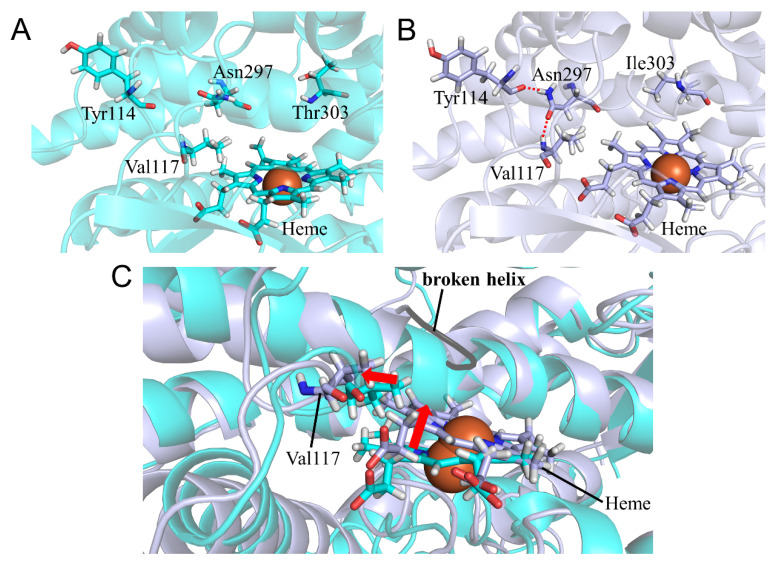
2.4. CYP2A6.25 and CYP2A6.26 Affected Secondary Structure Formation and Interaction with Heme and Substrates
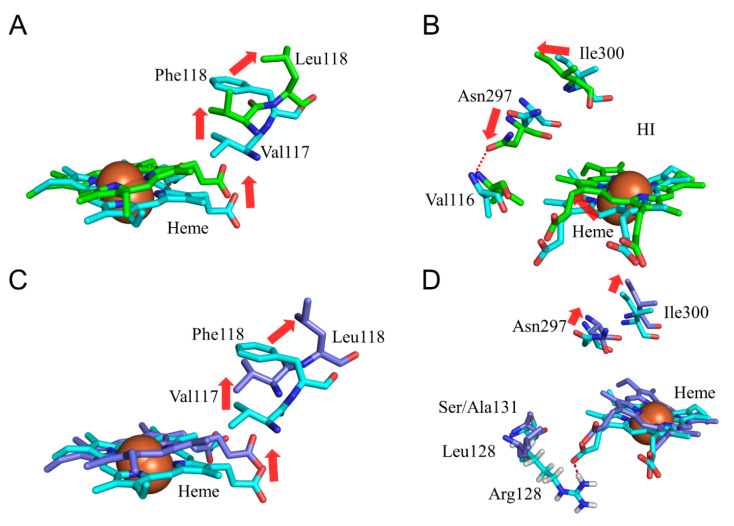
To evaluate the structural changes in CYP2A6.25 and CYP2A6.26, the calculated results of F118L and F118L/R128L/S131A mutants were compared with those of the wild type. In CYP2A6.25, CLint for nicotine was 16% of that of the wild type, and the kinetic parameter for coumarin was not detectable [22]. In CYP2A6.26, the kinetic parameters for both compounds were not detectable [22]. Phe118 was one of the interacting residues with coumarin in the complex structure [15,39], and the adjacent Val117 interacted with nicotine [40]. The location of the side chain of residue 118 moved away from heme in the F118L mutant, and Val117 similarly shifted in CYP2A6.25 (Figure 8A). These alterations were significant in decreasing the binding affinity of nicotine and coumarin. In addition, the location change of heme was caused by the shift of Val117, and the crash of heme with helix I separated the helix into two helices composed of residues 288–296 and 302–319 (Table 2). Due to these alterations, Asn297 and Ile300 moved away from heme, and Asn297 formed an hydrogen bond with Val116 (Figure 8B and Table 6). Asn297 and Ile300 were reported to interact with nicotine and coumarin in the crystal structures of these complexes [15,39]. Therefore, location changes for Val117, Leu118, Asn297, and Ile300 decreased the substrate binding affinity in CYP2A6.25. The deviation of heme might reduce the enzymatic activity of CYP2A6.25. In CYP2A6.26, the shifts of heme and Val117 caused by F118L mutation were also observed (Figure 8C). However, the locations of Asn297 and Ile300 were similar to those of the wild type, and helix I was not separated (Figure 8D). The crash of heme to helix I was weak and had no effect on helix I formation in CYP2A6.26. The loss of the hydrogen bond between Arg128 and heme was caused by R128L mutation and could decrease the binding stability of heme. The crash of heme with helix I could have insufficient influence to separate the helix structure of helix I due to the weakness of the binding stability of heme in CYP2A6.26. In addition, the change in Asn297 location was not observed in this mutant. On the other hand, the structural effects for the S131A mutation were not observed. Therefore, the main factors for the reduction in the activity of CYP2A6.26 could be the decrease in the interaction with heme due to R128L mutation and a decrease in substrate binding affinity by the F118L mutation.
Figure 8.
Structural change in CYP2A6.25 and CYP2A6.26 (A–D). The wild types, CYP2A6.25, and CYP2A6.26 are shown in cyan, green, and marine, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red arrows indicate the shift in CYP2A6.25 and CYP2A6.26. The hydrogen atoms are omitted except for Those of Arg128.
Table 6.
Hydrogen bond formation involved in the structural change in CYP2A6.25 and CYP2A6.26.
| Donor | Donor H | Acceptor | Wild Type | 2A6.25 | 2A6.26 |
|---|---|---|---|---|---|
| Val116 N | Val116 H | Asn297 Oδ1 | 0.02 | 96.38 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O2D | 99.94 | 99.84 | 0 |
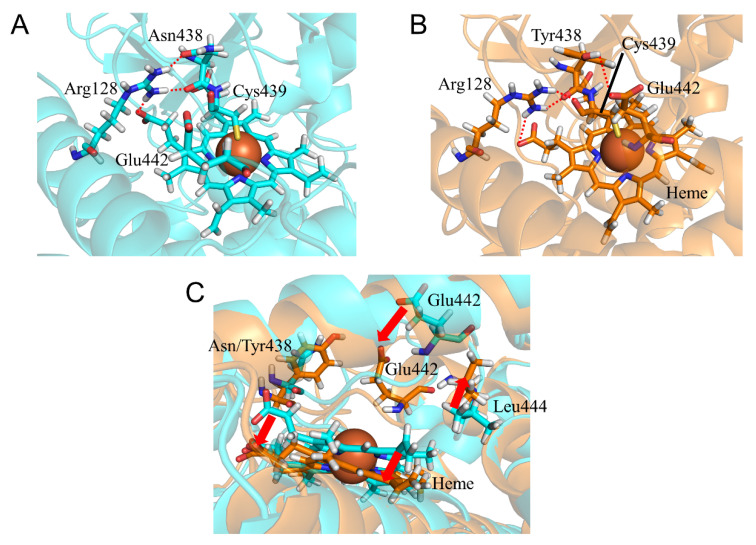
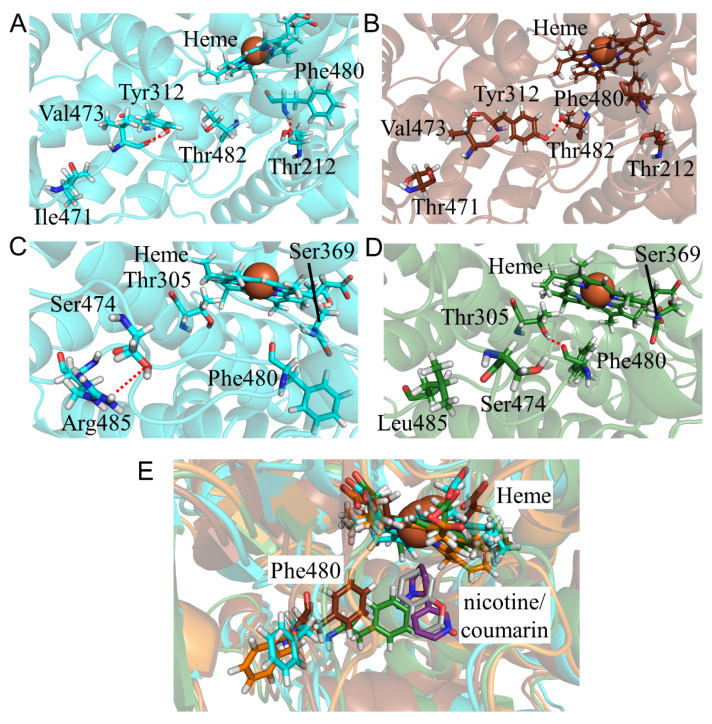
2.5. CYP2A6.35, CYP2A6.36, and CYP2A6.37 Affected Interaction with Heme and Substrates
To evaluate the structural changes in CYP2A6.35, CYP2A6.36, and CYP2A6.37, the calculated results for N438Y, N438Y/I471T, and N438Y/I471T/R485L mutants were compared with those of the wild type. The CYP2A6.35 had higher Km and lower Vmax values for nicotine and coumarin than those of the wild type, and the CLint parameters for nicotine and coumarin were 30% and 61% of those in the wild type, respectively [22]. In addition, the kinetic parameters for the CYP2A6.36 and CYP2A6.37 mutants were not detectable. Asn438 is a residue adjacent to Cys439 which is an axial ligand of heme iron. The N438Y mutation affected the hydrogen bond formation around heme. In the wild type, the side- and main-chain oxygen atoms of Asn438 formed hydrogen bonds with Arg128 (Figure 9A and Table 7). In CYP2A6.35, Tyr438 formed an hydrogen bond with Glu442 but the hydrogen bond between the side chain of Tyr438 and Arg128 was not observed (Figure 9B and Table 7). The loss of this hydrogen bond caused interaction changes between the side chain of Arg128 and heme, resulting in the rotation of the carboxylate moiety of heme. In addition, the hydrogen bond formation between Tyr438 and Glu442 caused conformational changes to the K”L loop (Figure 9C). Leu444 was shifted away from heme, and the interaction between Leu444 and heme was attenuated. Those changes in hydrogen bond formation and the K”L loop conformation caused the shift of heme. Therefore, the structural changes around heme were considered the reason for a lower enzymatic activity in CYP2A6.35. Similar structural changes were observed in CYP2A6.36 and CYP2A6.37. In addition, the I471T and R485L mutations affected the structure near the nicotine and coumarin binding sites. In CYP2A6.36, the structural change of the C-terminal region near the mutated residue caused the loss of the hydrogen bond between Tyr312 and Val473, and Tyr312 formed a new hydrogen bond with Thr482 (Figure 10A,B and Table 7). This alteration in hydrogen bond formation shifted the location of Phe480. In CYP2A6.37, the loss of the hydrogen bond formation between Ser474 and Arg485 caused a shift in the location of Phe480 close to heme, and Phe480 formed an hydrogen bond with Thr305 (Figure 10C,D and Table 7). The shifted Phe480 in CYP2A6.36 and CYP2A6.37 invaded the nicotine and coumarin binding sites. Therefore, these structural changes involved in Phe480 were the cause for the decrease in the enzymatic activities of CYP2A6.36 and CYP2A6.37.
Figure 9.
Structural change in CYP2A6.35 (A–C). The wild type and CYP2A6.35 are shown in cyan and orange, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red dotted lines indicate the hydrogen bonds. The red arrows indicate the shift in CYP2A6.35.
Table 7.
Hydrogen bond formation involved in the structural change in CYP2A6.35, CYP2A6.36, and CYP2A6.37.
| Donor H | Donor | Acceptor | Wild Type | 2A6.35 | 2A6.36 | 2A6.37 |
|---|---|---|---|---|---|---|
| Arg128 Nη1 | Arg128 Hη1 | Asn/Tyr438 O | 99.92 | 99.98 | 83.84 | 99.78 |
| Arg128 Nη2 | Arg128 Hη2 | Asn/Tyr438 O | 88.28 | 77.68 | 0.20 | 97.34 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 Oδ/Tyr438 Oη | 33.52 | 0 | 7.22 | 0.02 |
| Arg128 Nη2 | Arg128 Hη2 | Cys439 O | 13.86 | 71.68 | 99.44 | 37.70 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 0 | 0 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O2D | 0 | 76.96 | 100 | 83.90 |
| Tyr312 Nη | Tyr312 Hη | Val473 O | 58.62 | 0 | 0 | 0 |
| Tyr312 Nη | Tyr312 Hη | Thr482 Oγ | 12.90 | 0 | 54.78 | 0 |
| Phe480 N | Phe480 H | Thr212 O | 73.92 | 0 | 5.10 | 17.24 |
| Ser369 N | Ser369 H | Phe480 O | 39.4 | 0 | 28.08 | 0 |
| Arg485 Nη2 | Arg485 Hη2 | Ser474 Oγ | 69.98 | 74.78 | 62.9 | – |
| Arg485 Nε | Arg485 Hε | Ser474 Oγ | 47.80 | 20.36 | 45.84 | – |
| Arg485 Nε | Arg485 Hε | Ser474 O | 32.84 | 83.22 | 44.02 | – |
| Thr305 Oγ | Thr305 Hγ | Phe480 O | 0 | 0 | 0 | 82.26 |
Figure 10.
Structural change in CYP2A6.36 and CYP2A6.37 (A–E). The wild type, CYP2A6.36, and CYP2A6.37 are shown in cyan, deep green and brawn, respectively. Nicotine and coumarin are illustrated in gray and purple, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red dotted lines indicate the hydrogen bonds.
2.6. CYP2A6.43 Affected the Interaction with Substrates
The structural change in CYP2A6.43 was investigated by the comparison between the calculated structures of the wild type and the T303I mutant. In this mutant, the nicotine and coumarin binding affinity for CYP2A6 were much lower than those for wild type [22]. The CLint for nicotine and coumarin in CYP2A6.43 were 1% and 5% of those in the wild type, respectively. In the calculated results, helix I including the mutated residue was separated into residues 288–297 and 305–319 (Table 3). The helix-broken moiety contained Ile300, which was an important residue for the interaction with nicotine and coumarin [15,39]. In addition, Asn297 shifted away from heme and formed hydrogen bonds with Tyr114 and Val117 (Figure 11A,B and Table 8). These structural changes to helix I were expected to reduce the nicotine and coumarin binding affinity for CYP2A6.43. A shift of heme was also observed and thought to reduce the enzymatic activity. This shift was caused by the structural change of helix I and the shift of Val117. The results indicated that the break of helix I was the main reason for the reduction in the enzymatic activity of CYP2A6.43.
Figure 11.
Structural change in CYP2A6.43 (A–C). The wild type and CYP2A6.43 are shown in cyan and light blue, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red dotted lines indicate the hydrogen bonds. The red arrows indicate the shift in CYP2A6.43.
Table 8.
Hydrogen bond formation involved in the structural change in CYP2A6.43.
| Donor | Donor H | Acceptor | Wild Type | 2A6.43 |
|---|---|---|---|---|
| Val117 N | Val117 H | Asn297 Oδ1 | 0 | 98.28 |
| Asn297 Nδ2 | Asn297 Hδ2 | Tyr114 O | 0 | 91.92 |
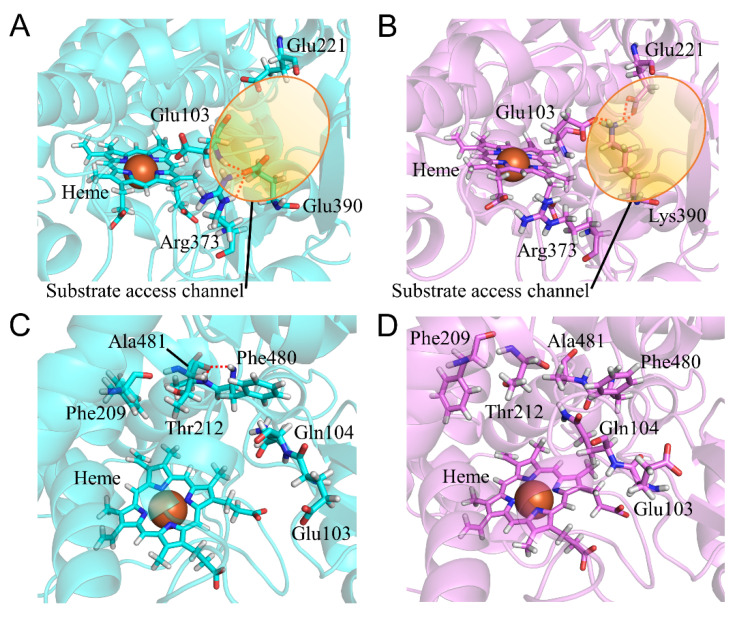
2.7. CYP2A6.44 Changed Substrate Access Channel
To evaluate the structure of CYP2A6.44, we compared the calculated results of the E390K/N418D/E419D mutant with those of the wild type. The CLint for nicotine was 1% of that in the wild type, and the kinetic parameter for coumarin was not detectable [22]. This mutant had the highest Km value for nicotine in all of the CYP2A6 mutants. However, the kinetic parameters (Km, Vmax, and CLint) of the N418D/E419D mutant (CYP2A6.28) were at the same level as that of the wild type. Thus, E390K mutation was assumed to strongly affect substrate binding. Glu390 was located on β1-3 between helices K and K’ and next to the BC loop. β1 and BC loops reportedly composed the substrate access channel [1,41,42,43]. In the wild type, Glu390 formed hydrogen bonds with the main chain of Glu103 and the side chain of Arg373 (Figure 12A and Table 9). In CYP2A6.44, Lys390 formed hydrogen bonds with the main-chain oxygen of Glu103, the side-chain oxygen of Glu103, and the side-chain oxygen of Glu221. The hydrogen bond formation with Glu221 could impair the opening of the substrate access channel. In addition, Glu103 was a residue which formed the substrate access channel and the side-chain atoms of Glu103 in CYP2A6.44, which were in the opposite direction to that in the wild type, located inside of the substrate access channel. The substrate access channel was not seen clearly both in the experimental and calculated structures. Although the difference in the state of substrate access channel was not clearly seen, the E390K mutation was considered to impair access of the substrate to the substrate binding site because of the alteration of the interaction around the substrate access channel. The changes in hydrogen bond formation affected the structures of the substrate binding site. The conformational change of the main chain of Glu103 by the hydrogen bond formation with Lys390 altered the location of Gln104, and the side chain of Gln104 entered the substrate binding site (Figure 12C,D). Displacement of Gln104 triggered a shift in helix F. The hydrogen bonds between Phe480 and Ala481 observed in the wild type were lost in CYP2A6.44 (Table 9). Phe209 moved away from heme. Phe209 interacted with nicotine and coumarin in those complex structures; therefore, the shift of Phe209 reduced the substrate binding affinity for CYP2A6.44 [15,40]. The structural change in the substrate access channel and the substrate binding site could be the key effects of E390K/N418D/E419D mutations on low enzymatic activity.
Figure 12.
Structural change in CYP2A6.44 (A–D). The wild type and CYP2A6.44 are shown in cyan and purple, respectively. Nitrogen, oxygen, and hydrogen are displayed in blue, red, and white, respectively, in the stick model. Iron is shown as an orange sphere by a model written as van der Waals radius. The red dotted lines indicate the hydrogen bonds.
Table 9.
Hydrogen bond formation involved in the structural change in CYP2A6.44.
| Donor | Donor H | Acceptor | Wild Type | 2A6.44 |
|---|---|---|---|---|
| Glu103 N | Glu103 H | Glu390 Oε1 | 96.67 | – |
| Arg373 Nε | Arg373 Hε | Glu390 Oε1 | 99.40 | – |
| Arg373 Nη2 | Arg373 Hη2 | Glu390 Oε1 | 95.10 | – |
| Arg373 Nη2 | Arg373 Hη2 | Glu390 Oε2 | 73.22 | – |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 Oε1 | – | 14.18 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 Oε1 | – | 19.30 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 Oε1 | – | 35.42 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 Oε2 | – | 17.50 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 Oε2 | – | 8.72 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 Oε2 | – | 6.76 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 O | – | 19.96 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 O | – | 32.06 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 O | – | 22.28 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu221 Oε1 | – | 38.20 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu221 Oε1 | – | 28.04 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu221 Oε1 | – | 25.60 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu221 Oε2 | – | 36.88 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu221 Oε2 | – | 27.58 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu221 Oε2 | – | 25.52 |
| Phe480 N | Phe480 H | Thr212 O | 73.92 | 0 |
| Ala481 N | Ala481 H | Thr212 O | 78.72 | 0 |
3. Computational Methods
To construct the initial structures, the experimental structure was retrieved from the protein data bank (PDB). The complex structures of CYP2A6 with nicotine or coumarin were registered (PDB ID: 4EJJ and 1Z10, respectively) [15,40]. Each crystal structure formed tetramers and was devoid of residues 1–29 by the replacement of the N-terminal region for crystallization. Since the resolution and each R-value of 1Z10 was better than those of 4EJJ, we used 1Z10 to construct the initial structure. Chain B–C, water molecules, and coumarin were deleted from the experimental structure, and the bond between heme iron and the ligand Cys439 was constructed using AmberTool16. The system was a solvated TIP3P model [44] and was neutralized by adding a counter ion. Structural minimizations of water and counter ion were performed for 1000 steps. Minimizations of the whole system were performed at 2500 steps. Temperature-increasing MD simulations of 20 ps were carried out with the temperature raised from 0 to 300 K, and then, equilibrating MD simulations were performed under constant temperature and pressure. The time step for the MD simulations was 2 fs. The simulation times for the wild type, CYP2A6.6, CYP2A6.36, CYP2A6.37, and CYP2A6.44 were 1000 ns. Other simulation times were extended due to slow convergence. The simulation times for the CYP2A6.25 and CYP2A6.26 were 1200 ns. Those of CYP2A6.11, CYP2A6.17, and CYP2A6.35 were 2000 ns. The cutoff distance for the calculations of the nonbonding interactions was set at 10 Å. The particle mesh Ewald method was used to calculate electrostatic interactions in the periodic boundary condition [45]. The SHAKE algorithm was applied to constrain the lengths of covalent bonds containing hydrogen atoms [46]. All calculations were performed using AMBER16 [47]. Root mean square deviations (RMSDs), RMSFs, and hydrogen bond formations were calculated using the cpptraj module of AmberTools16. The trajectories for the last 10 ns were used in the analysis of RMSF and hydrogen bond formations. AMBER ff14SB force field was used for the amino acid parameters [48]. For heme and the axial ligand Cys439, the parameters determined in a previous study were used [49]. The initial structures of all variants were constructed based on the calculated structure of the wild type after MD simulations.
4. Conclusions
We performed the MD simulation for the wild type and 10 mutants and investigated the structural changes related to the reduction in enzymatic activity for the mutants of CYP2A6. In the wild type, the large structural differences in the experimental structure was not observed. The calculated results including heme were available to understand the mechanism for the reduction in the enzymatic activity for the mutants. The effects of the mutations on 3D structures involved in the enzymatic activity reduction are shown in Table 10. The effects of multi mutations were suggested to be additive. The shifts of phenylalanine residues (Phe107, Phe111, Phe118, and Phe480) involved in the interaction with the substrates were crucial for substrate binding. In addition to drug metabolism, CYP2A6 was reported to be involved in hepatocellular carcinoma by contributing to immune regulation [14]. The results of MD simulations could contribute to the prediction of structural changes in CYP2A6 mutants and provide useful information for the relationship between genotype and phenotype. The clarification of the protein–protein interactions between CYP and redox partner is expected to reveal more detail effects of mutations in CYP2A6 variants.
Table 10.
Structural effects observed in the calculated structures of CYP2A6 mutants.
| Protein | Amino Acid Changes | Predicted Structural Effects Involved in the Enzymatic Activity |
|---|---|---|
| CYP2A6.6 | R128Q | conformational change involved in interaction with the redox partner |
| CYP2A6.11 | S224P | heme deviation, change of secondary structures |
| CYP2A6.17 | V365M | heme deviation, change of secondary structures, conformational change involved in interaction with the redox partner |
| CYP2A6.25 | F118L | structural change of substrate-interacting residues, heme deviation, change of secondary structures |
| CYP2A6.26 | F118L R128L S131A |
structural change of substrate-interacting residues, heme deviation, change of secondary structures |
| CYP2A6.35 | N438Y | heme deviation, Cys439 conformational change |
| CYP2A6.36 | N438Y I471T |
structural change of substrate-interacting residues, heme deviation, Cys439 conformational change |
| CYP2A6.37 | N438Y I471T R485L |
structural change of substrate-interacting residues, heme deviation, Cys439 conformational change |
| CYP2A6.43 | T303I | structural change of substrate-interacting residues |
| CYP2A6.44 | E390K N418D E419D |
structural change of substrate access channel, structural change of substrate-interacting residues |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms221810119/s1.
Author Contributions
Conceptualization, K.K. and A.O.; methodology, A.O.; software, A.O.; validation, H.H., Y.I., E.K. and M.H.; formal analysis, K.K.; investigation, T.N. and R.N.; resources, A.O.; data curation, K.K.; writing—original draft preparation, K.K.; writing—review and editing, T.N. and A.O.; visualization, K.K.; supervision, A.O.; project administration, K.K., T.N., and A.O.; funding acquisition, A.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research (17K08257, 19J23595, and 21K15244) from the Japan Society for the Promotion of Science. We are grateful to the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan for a Grant-in-Aid for Scientific Research on Transformative Research Area (A) “Hyper-Ordered Structures Science” (20H05883).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guengerich F.P. Human Cytochrome P450 Enzymes. In: Ortiz de Montellano P.R., editor. Cytochrome P450 Structure, Mechanism, and Biochemistry. 4th ed. Springer; Cham, Switzerland: 2015. pp. 523–786. [Google Scholar]
- 2.Rendic S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002;34:83–448. doi: 10.1081/DMR-120001392. [DOI] [PubMed] [Google Scholar]
- 3.Williams J.A., Hyland R., Jones B.C., Smith D.A., Hurst S., Goosen T.C., Peterkin V., Koup J.R., Ball S.E. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 4.Wienkers L.C., Heath T.G. Predicting in vivo drug interactions from in vitro drug discovery data. Nat. Rev. Drug Discov. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 5.Schlichting I., Berendzen J., Chu K., Stock A.M., Maves S.A., Benson D.E., Sweet R.M., Ringe D., Petsko G.A., Sligar S.G. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 6.Krest C.M., Onderko E.L., Yosca T.H., Calixto J.C., Karp R.F., Livada J., Rittle J., Green M.T. Reactive intermediates in cytochrome p450 catalysis. J. Biol. Chem. 2013;288:17074–17081. doi: 10.1074/jbc.R113.473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guengerich F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu T., Tateishi T., Hatano M., Fujii-Kuriyama Y. Probing the role of lysines and arginines in the catalytic function of cytochrome P450d by site-directed mutagenesis. Interaction with NADPH-cytochrome P450 reductase. J. Biol. Chem. 1991;266:3372–3375. doi: 10.1016/S0021-9258(19)67801-4. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y.T., Stiffelman O.B., Vakser I.A., Loew G.H., Bridges A., Waskell L. Construction of a 3D model of cytochrome P450 2B4. Protein Eng. 1997;10:119–129. doi: 10.1093/protein/10.2.119. [DOI] [PubMed] [Google Scholar]
- 10.Bridges A., Gruenke L., Chang Y.T., Vakser I.A., Loew G., Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 11.Hamdane D., Xia C., Im S.C., Zhang H., Kim J.J., Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelkonen O., Rautio A., Raunio H., Pasanen M. CYP2A6: A human coumarin 7-hydroxylase. Toxicology. 2000;144:139–147. doi: 10.1016/S0300-483X(99)00200-0. [DOI] [PubMed] [Google Scholar]
- 13.Di Y.M., Chow V.D., Yang L.P., Zhou S.F. Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr. Drug Metab. 2009;10:754–780. doi: 10.2174/138920009789895507. [DOI] [PubMed] [Google Scholar]
- 14.Jiang T., Zhu A.S., Yang C.Q., Xu C.Y., Yang D.Q., Lou Z.H., Zhang G.J. Cytochrome P450 2A6 is associated with macrophage polarization and is a potential biomarker for hepatocellular carcinoma. FEBS Open Bio. 2021;11:670–683. doi: 10.1002/2211-5463.13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano J.K., Hsu M.H., Griffin K.J., Stout C.D., Johnson E.F. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat. Struct. Mol. Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 16.Raunio H., Rautio A., Gullstén H., Pelkonen O. Polymorphisms of CYP2A6 and its practical consequences. Br. J. Clin. Pharmacol. 2001;52:357–363. doi: 10.1046/j.0306-5251.2001.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrychová T., Anzenbacherová E., Hudeček J., Skopalík J., Lange R., Hildebrandt P., Otyepka M., Anzenbacher P. Flexibility of human cytochrome P450 enzymes: Molecular dynamics and spectroscopy reveal important function-related variations. Biochim. Biophys. Acta. 2011;1814:58–68. doi: 10.1016/j.bbapap.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992;267:83–90. doi: 10.1016/S0021-9258(18)48462-1. [DOI] [PubMed] [Google Scholar]
- 19.Shimada T., Yamazaki H., Guengerich F.P. Ethnic-related differences in coumarin 7-hydroxylation activities catalyzed by cytochrome P4502A6 in liver microsomes of Japanese and Caucasian populations. Xenobiotica. 1996;26:395–403. doi: 10.3109/00498259609046718. [DOI] [PubMed] [Google Scholar]
- 20.Hassanzadeh Khayyat M., Vahdati Mashhadian N., Eghbal S., Jalali N. Inter-individual variability of coumarin 7-hydroxylation (CYP2A6 activity) in an Iranian population. Iran. J. Basic Med. Sci. 2013;16:610–614. [PMC free article] [PubMed] [Google Scholar]
- 21.Fujieda M., Yamazaki H., Saito T., Kiyotani K., Gyamfi M.A., Sakurai M., Dosaka-Akita H., Sawamura Y., Yokota J., Kunitoh H., et al. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 2004;25:2451–2458. doi: 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- 22.Hosono H., Kumondai M., Maekawa M., Yamaguchi H., Mano N., Oda A., Hirasawa N., Hiratsuka M. Functional characterization of 34 CYP2A6 allelic variants by assessment of nicotine C-Oxidation and coumarin 7-hydroxylation activities. Drug Metab. Dispos. 2017;45:279–285. doi: 10.1124/dmd.116.073494. [DOI] [PubMed] [Google Scholar]
- 23.López-Flores L.A., Pérez-Rubio G., Falfán-Valencia R. Distribution of polymorphic variants of CYP2A6 and their involvement in nicotine addiction. EXCLI J. 2017;16:174–196. doi: 10.17179/excli2016-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miles J.S., McLaren A.W., Forrester L.M., Glancey M.J., Lang M.A., Wolf C.R. Identification of the human liver cytochrome P-450 responsible for coumarin 7-hydroxylase activity. Biochem. J. 1990;267:365–371. doi: 10.1042/bj2670365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daigo S., Takahashi Y., Fujieda M., Ariyoshi N., Yamazaki H., Koizumi W., Tanabe S., Saigenji K., Nagayama S., Ikeda K., et al. A novel mutant allele of the CYP2A6 gene (CYP2A6*11) found in a cancer patient who showed poor metabolic phenotype towards tegafur. Pharm. Genom. 2002;12:299–306. doi: 10.1097/00008571-200206000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Mwenifumbo J.C., Al Koudsi N., Ho M.K., Zhou Q., Hoffmann E.B., Sellers E.M., Tyndale R.F. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum. Mutat. 2008;29:679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 27.Al Koudsi N., Ahluwalia J.S., Lin S.K., Sellers E.M., Tyndale R.F. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharm. J. 2009;9:274–282. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piliguian M., Zhu A.Z., Zhou Q., Benowitz N.L., Ahluwalia J.S., Sanderson Cox L., Tyndale R.F. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharm. Genom. 2014;24:118–128. doi: 10.1097/FPC.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouard C., Terreux R., Tissier A., Jacqueroud L., Vigneron A., Ansieau S., Puisieux A., Payen L. Destabilization of the TWIST1/E12 complex dimerization following the R154P point-mutation of TWIST1: An in silico approach. BMC Struct. Biol. 2017;17:6. doi: 10.1186/s12900-017-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narang S.S., Shuaib S., Goyal D., Goyal B. Assessing the effect of D59P mutation in the DE loop region in amyloid aggregation propensity of β2-microglobulin: A molecular dynamics simulation study. J. Cell. Biochem. 2018;119:782–792. doi: 10.1002/jcb.26241. [DOI] [PubMed] [Google Scholar]
- 31.Kato K., Nakayoshi T., Sato M., Kurimoto E., Oda A. Molecular dynamics simulations for three-dimensional structures of orotate phosphoribosyltransferases constructed from a simplified amino acid set. ACS Omega. 2020;5:13069–13076. doi: 10.1021/acsomega.0c01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi K., Takahashi O., Hiratsuka M., Yamaotsu N., Hirono S., Watanabe Y., Oda A. Evaluation of influence of single nucleotide polymorphisms in cytochrome P450 2B6 on substrate recognition using computational docking and molecular dynamics simulation. PLoS ONE. 2014;9:e96789. doi: 10.1371/journal.pone.0096789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuyoshi S., Kometani M., Watanabe Y., Hiratsuka M., Yamaotsu N., Hirono S., Manabe N., Takahashi O., Oda A. Molecular dynamics simulations to investigate the influences of amino acid mutations on protein three-dimensional structures of cytochrome P450 2D6.1, 2, 10, 14A, 51, and 62. PLoS ONE. 2016;11:e0152946. doi: 10.1371/journal.pone.0152946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y., Fukuyoshi S., Hiratsuka M., Yamaotsu N., Hirono S., Takahashi O., Oda A. Prediction of three-dimensional structures and structural flexibilities of wild-type and mutant cytochrome P450 1A2 using molecular dynamics simulations. J. Mol. Graph. Model. 2016;68:48–56. doi: 10.1016/j.jmgm.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Skopalík J., Anzenbacher P., Otyepka M. Flexibility of human cytochromes P450: Molecular dynamics reveals differences between CYPs 3A4, 2C9, and 2A6, which correlate with their substrate preferences. J. Phys. Chem. B. 2008;112:8165–8173. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 36.Hendrychova T., Berka K., Navratilova V., Anzenbacher P., Otyepka M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr. Drug. Metab. 2012;13:177–189. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- 37.Vanhoof G., Goossens F., de Meester I., Hendriks D., Scharpe S. Proline motif in peptides and their biological processing. FASEB J. 1995;9:9736–9744. doi: 10.1096/fasebj.9.9.7601338. [DOI] [PubMed] [Google Scholar]
- 38.Kato K., Furuhashi T., Kato K., Oda A., Kurimoto E. The assembly mechanism of coiled-coil domains of the yeast cargo receptors Emp46p/47p and the mutational alteration of pH-dependency of complex formation. J. Biochem. 2018;163:441–446. doi: 10.1093/jb/mvy011. [DOI] [PubMed] [Google Scholar]
- 39.Qi X., Dou T., Wang Z., Wu J., Yang L., Zeng S., Deng M., Lü M., Liang S. Inhibition of human cytochrome P450 2A6 by 7-hydroxycoumarin analogues: Analysis of the structure-activity relationship and isoform selectivity. Eur. J. Pharm. Sci. 2019;136:104944. doi: 10.1016/j.ejps.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 40.DeVore N.M., Scott E.E. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes. J. Biol. Chem. 2012;287:26576–26585. doi: 10.1074/jbc.M112.372813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cojocaru V., Winn P.J., Wade R.C. The ins and outs of cytochrome P450s. Biochim. Biophys. Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Schoch G.A., Yano J.K., Sansen S., Dansette P.M., Stout C.D., Johnson E.F. Determinants of cytochrome P450 2C8 substrate binding: Structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J. Biol. Chem. 2008;283:17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishelovitch D., Shaik S., Wolfson H.J., Nussinov R. Theoretical characterization of substrate access/exit channels in the human cytochrome P450 3A4 enzyme: Involvement of phenylalanine residues in the gating mechanism. J. Phys. Chem. B. 2009;113:13018–13025. doi: 10.1021/jp810386z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joung I.S., Cheatham T.E., III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darden T., York D., Pedersen L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 46.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 47.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., et al. AMBER16. University of California; San Francisco, CA, USA: 2012. [Google Scholar]
- 48.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oda A., Yamaotsu N., Hirono S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J. Comput. Chem. 2005;26:818–826. doi: 10.1002/jcc.20221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.