Abstract
One of the most common chromosomal abnormalities in acute leukemia is a reciprocal translocation involving the HRX gene (also called MLL, ALL-1, or HTRX) at chromosomal locus 11q23, resulting in the formation of HRX fusion proteins. Using the yeast two-hybrid system and human cell culture coimmunoprecipitation experiments, we show here that HRX proteins interact directly with the GADD34 protein. We have found that transfected cells overexpressing GADD34 display a significant increase in apoptosis after treatment with ionizing radiation, indicating that GADD34 expression not only correlates with apoptosis but also can enhance apoptosis. The amino-terminal third of the GADD34 protein was necessary for this observed increase in apoptosis. Furthermore, coexpression of three different HRX fusion proteins (HRX-ENL, HRX-AF9, and HRX-ELL) had an anti-apoptotic effect, abrogating GADD34-induced apoptosis. In contrast, expression of wild-type HRX gave rise to an increase in apoptosis. The difference observed here between wild-type HRX and the leukemic HRX fusion proteins suggests that inhibition of GADD34-mediated apoptosis may be important to leukemogenesis. We also show here that GADD34 binds the human SNF5/INI1 protein, a member of the SNF/SWI complex that can remodel chromatin and activate transcription. These studies demonstrate, for the first time, a gain of function for leukemic HRX fusion proteins compared to wild-type protein. We propose that the role of HRX fusion proteins as negative regulators of post-DNA-damage-induced apoptosis is important to leukemia progression.
The disruption of the human homologue of the Drosophila Trithorax (trx) gene, HRX, by chromosomal translocations resulting in the juxtaposition of genetic elements and formation of HRX fusion genes is one of the most common genetic alterations in human acute leukemia (52). These translocations occur in approximately 10% of acute lymphoid leukemias (ALLs), 5% of acute myeloid leukemias (AMLs), and 85% of topoisomerase II inhibitor-related secondary leukemias in adults. Furthermore, these translocations are present in half of all the de novo leukemias in children younger than 1 year (26).
HRX, also referred to as ALL-1, MLL-1, or HTRX (13, 18, 52, 60), is a ubiquitously expressed 3,969-amino-acid nuclear protein (28) with unknown biologic function. HRX shares at least two regions of strong homology with the similarly sized Drosophila Trx: a series of centrally located zinc finger-like domains and a carboxy-terminal stretch of 210 amino acids. In Drosophila, Trx controls body segment patterning as a positive transcriptional regulator of the homeotic selector genes of the Antennapedia and bithorax complexes (7). Studies with transgenic mice have shown that the function of Hrx in mice has features in common with that of Trx in Drosophila. Hrx has been demonstrated to be required for proper segment identity and to function positively as a regulator of Hox gene expression in Hrx heterozygous and homozygous deficient mice (58).
To date, more than 26 different human leukemic HRX fusion proteins, resulting from reciprocal translocations between the HRX gene at chromosome 11q23 and partner genes at other loci, have been predicted to exist by cytogenetic studies (4, 20, 42). At least 15 of the partner genes have been cloned and characterized (5, 24, 27, 34, 35, 47, 48, 50, 57). The derivative 11 fusion products, usually referred to as HRX fusion proteins, are composed of common amino-terminal HRX sequences fused to carboxy-terminal residues donated from one of the partner proteins. Cytogenetic and Northern blot analyses on patient specimens and leukemia cell lines consistently show the presence of the derivative 11 fusion products, suggesting that this product is the critical leukemogenic factor (30). This hypothesis is further supported by the occurrence of leukemia in transgenic mice expressing a derivative 11-fusion product (11).
How HRX fusion proteins cause leukemia is unknown. Although most of the fusion partners are structurally and functionally unrelated, eight of them are involved in transcriptional regulation. ENL, AF9, and AF4 activate transcription from synthetic reporter genes in vivo (36, 39, 43). AFX and AF6q21 are forkhead proteins known to possess DNA-binding and transcriptional regulation properties (5, 24). CBP and p300 are transcriptional coactivators with histone acetylation activity (27, 48, 50). Yet another HRX fusion partner, ELL, is an RNA polymerase II elongation factor (45, 51). Since neither truncated versions of HRX nor HRX fusion proteins with a partner fused out of frame have been found in leukemias, it is likely that both N-terminal HRX and partner sequences are critical to the leukemogenic effect of the HRX fusion proteins. This contention is further supported by a study in which the expression of the HRX-ENL fusion protein resulted in immortalization of hematopoietic stem cells ex vivo, but neither the expression of full-length ENL nor a deletion mutant of HRX-ENL lacking ENL had this effect (32).
Of the HRX motifs present in all the fusion proteins, the AT hook region, containing three closely spaced AT hooks composed of conserved basic amino acids, is the best characterized. A similar triplet of conserved AT hooks exists in the architectural transcription factors HMG-C and HMG-I(Y). HMG-I(Y) bends or distorts promoter/enhancer DNA sequences, possibly making these sequences accessible and thereby overcoming the repressive effects of nucleosomes upon transcription (15). Recently, a binding site for the HRX AT hooks was found 1.7 kb upstream of the ARP1 gene that is down-regulated in HRX double-knockout mouse embryonic stem cells (3). The AT hook region of HRX is necessary for immortalization of murine hematopoietic progenitors in vitro (46). Furthermore, homozygous Hrx-deleted embryonic stem cells are blocked in hematopoietic differentiation in vitro (16). In vitro differentiation assays of HRX +/+, +/−, and −/− yolk sac progenitor cells show that HRX is required for myeloid and macrophage differentiation of early hematopoietic progenitors (23). We have previously shown that the AT hook region of HRX contains binding sites for the leukemia-associated SET protein (1). Taken together, the AT hook region is important to the role of HRX in targeting and regulating transcriptional units important for normal hematopoietic growth and differentiation.
Genetic studies of Drosophila Trx suggest that the Trx protein is involved in the stable propagation of gene activity. Trx does not initiate transcription itself but maintains a positive transcriptional state in sets of specific genes initiated by other factors (6). Trx genetically opposes the action of the Polycomb group proteins that are thought to inactivate transcription through a mechanism involving modification of chromatin structure analogous to heterochromatin formation (12). It has been suggested, therefore, that Trx maintains target genes in a transcriptionally active state through subsequent cell divisions by an epigenetic mechanism that probably involves chromatin remodeling. Although HRX has not been shown to remodel chromatin, the carboxy-terminal SET domain of HRX interacts with hSNF5/INI1, a component of the SNF/SWI complex, a chromatin-remodeling system (41). Drosophila Trx was shown to interact analogously with Snr1, the fly homologue of hSNF5/INI1, by the same investigators, suggesting that wild-type Trx and HRX associate with the SNF/SWI apparatus to remodel chromatin and maintain active transcription. It is important to note that the SET domain is lost when amino-terminal HRX and carboxy-terminal partner residues fuse to form HRX fusion proteins during 11q23 translocations in leukemia. The loss of the SET domain and associated SNF/SWI function may explain the down-regulation of the ARP1 gene in HRX double-knockout mouse embryonic stem cells and six leukemic cell lines expressing HRX fusion proteins (3).
In this study we report that HRX proteins interact with GADD34, a DNA damage-inducible factor. We show here that GADD34 expression leads to apoptosis following gamma irradiation and that leukemic HRX fusion proteins, in contrast to wild-type HRX, inhibit this GADD34-induced apoptosis. We propose that this gain of function by HRX fusion proteins is important in disrupting normal cellular growth arrest following DNA damage and thereby provides a proliferative advantage to cells harboring 11q23 chromosomal translocations.
MATERIALS AND METHODS
Cell lines.
Human 293T cells and human SW480 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (14, 33).
Construction of expression vectors.
Portions of HRX were cloned into the expression vector pCS2+MT, which allows for in-frame fusions with six copies of the myc epitope tag under the control of a simian cytomegalovirus promoter; these constructs have been described previously (1). pCS884 encodes the portion of HRX used as bait in the yeast two-hybrid screen and was made by first cloning the SmaI-SspI fragment of HRX (amino acids 110 to 405) into the SmaI site of pBSSK+. Then an XbaI-HincII fragment was cloned into the XbaI and SnaBI sites of pCS2+MT. pCS1385 begins at the NotI site of HRX (amino acid 79) and continues to the XbaI site (amino acid 710). pCSARQ2 contains the entire HRX-ENL coding sequence beginning at the AvrII site at amino acid 27, blunted with Klenow fragment, and cloned into the StuI and EcoRI sites of pCS2+MT by a multi-step cloning procedure. A series of pCSARQ2 deletion constructs were made ending at nucleotides 4332, 4602, and 4913; these constructs were designated pCSAR4332, pCSAR4602, and pCSARBst, respectively, and result in the expression of proteins that begin at amino acid 27 and end at amino acids 1444, 1534, and 1637, respectively. The construct pCSARBst was made by digesting the clone with BstEII, treating it with Klenow fragment to blunt the construct, and finally digesting it with SnaBI. The resulting clones were then religated. The constructs pCSAR4332 and pCSAR4602 were made by a PCR approach involving the PflMI site in HRX-ENL and a reverse primer with a 5′ extension incorporating a SnaBI restriction enzyme site. PCSHRX contains the entire wild-type HRX coding sequence beginning at the AvrII site at amino acid 27, blunted with Klenow fragment, and cloned into the StuI and EcoRI sites of pCS2+MT by a multistep cloning procedure.
The pCSHHE1 vector expresses a protein that contains HRX sequences (amino acids 181 to 405) fused to ENL sequences (amino acids 1445 to 1534 of HRX-ENL). The pCSHHE1 vector was made with the XhoI-SnaBI PCR fragment of ENL sequences cloned in frame into the XhoI and SnaBI sites of an EcoRI-XhoI PCR fragment of HRX sequences amino acids 181 to 405. The pCSHHE1CENL vector is similar to the pCSHHE1 vector, except that it codes for a protein that, in addition to the HRX and ENL sequences included in pCSHHE1, includes all the ENL sequences as they appear in the HRX-ENL fusion protein as described by Adler et al. (1). The pCSHHE1CENL vector was made with the MluI-NotI fragment of pCSARQ2 cloned into the MluI and NotI sites of pCSHHE1.
The pCSGADD34 vector, containing the partial GADD34 cDNA recovered from the yeast two-hybrid screen, was made with the EcoRI-XbaI fragment of pBSGADD34 cloned into the EcoRI and XbaI sites of pCS2+MT. The pCSGADD34FL vector, encoding full-length GADD34, was made by ligating a PCR product encoding amino acids 1 to 180 amplified from GADD34 pCMV (25) to the pCSGADD34 clone. pCSGADDA vector, containing GADD34 amino acids 180 to 483, was made by religating pCSGADD34 following double digestion with EcoRV and SnaBI.
The pSGADD34 vector, containing the partial GADD34 cDNA, was made with the EcoRI (filled in)-BglII fragment of p1(1T-)1 (the original yeast two-hybrid vector pSE1107 containing the partial GADD34 clone) cloned into the SmaI and BglII sites of pSGFL2. pSGFL2 is the expression vector pSG5 (Stratagene) that has been modified for in-frame fusions with two tandem copies of the 8-amino-acid IBI FLAG marker peptide (Kodak). The pSGADD34FL vector, containing a full-length GADD34 cDNA, was constructed in plasmid pSGADD34 by using the partial GADD34 cDNA as the starting point and using the same PCR product described above to insert amino acids 1 to 180 of a cDNA from the vector human GADD34 pCMV (25). Two pSGADD34 deletion constructs, ending at nucleotides 1458 and 1830, were made. These constructs were designated pSGADDA and pSGADDC, respectively, and result in the expression of proteins that begin at amino acid 180 and end at amino acids 483 and 610, respectively. The pSGADDA construct was made by digesting pSGADD34 with BglII, treating it with Klenow fragment to blunt the construct, digesting it with EcoRV, and then religating to create a blunt-blunt deletion construct. SGADDC was made by digesting a PCR product amplified with a forward primer incorporating the internal GADD34 EcoRV site and a reverse primer designed to produce a BglII site ending at GADD34 nucleotide 1808 and cloned into EcoRV- and BglII-digested pSGADD34. pSGADD484 was designed to encode a FLAG-tagged GADD34 deletion protein including amino acids 338 to 555 and was made by digesting a PCR product amplified with a forward primer starting at bp 1014 incorporating an EcoRI site and a reverse primer starting at bp 1665 incorporating a BglII site and cloned in frame into EcoRI- and BglII-digested pSGFL2. The pSGSNF5 vector, containing a nearly complete hSNF5/INI1 cDNA, was constructed with a PCR fragment containing amino acids 27 to 385 of the hSNF5/INI1 protein. The pSGSNF5 vector was made with the EagI (filled in)-PstI fragment of pCR3Ini1 cloned into the SmaI and PstI sites of pSGFL2. The pCSHRXAF9 vector contains the entire HRX-AF9 coding sequence beginning at the AvrII site at amino acid 27. The pCSHRXAF9 construct was made by replacing a XbaI fragment from pCSARQ2 with one from pCMVHRXAF9. The pCSHRXELL vector contains the entire HRX-ELL coding sequence beginning at the AvrX-AF9 coding sequence at the AvrII site at amino acid 27. The pCSHRXAF9 construct was made by replacing a XbaI fragment from pCSARQ2 with one from pCMVHRXAF9. The pCSHRXELL vector contains the entire HRX-ELL coding sequence beginning at the AvrII site at amino acid 27. The pCSHRXELL construct was made with the BglII-BsaBI fragment of pMLLELL cloned into the BglII and SnaBI sites of pCSARQ2. The pCSmycTEL vector contains the coding sequence for the TEL protein and is described by Kwiatkowski et al. (31).
Coimmunoprecipitation and Western blotting.
Human 293T cells were transiently transfected with plasmids by the calcium phosphate transfection method, grown for 2 days, and lysed in a Nonidet P-40 (NP-40) lysis buffer (150 mM NaCl, 0.5% NP-40, 50 mM Tris HCl [pH 8.0], 5 mM EDTA, 4 mg of leupeptin per ml, 4 mg of phenylmethylsulfonyl fluoride per ml, 2 mg of pepstatin per ml) and sonicated. After centrifugation, the lysates were immunoprecipitated with anti-myc mouse monoclonal antibody 9E10 (a gift from Jim Roberts, Fred Hutchinson Cancer Research Center, Seattle, Wash.) followed by protein A-Sepharose CL-4B (Pharmacia Biotech). To allow binding of HRX-associated proteins, the pellet (after centrifugation) was resuspended in 250 μl of lysis buffer and diluted with 900 μl of binding buffer (20 mM HEPES [pH 7.5], 10% glycerol, 12.5 mM MgCl2, 0.1 mM EDTA, 50 mM NaCl), and gently rocked for 30 min before being given three final washes with binding buffer. Anti-GADD34 coimmunoprecipitates were done as described above, except that rabbit anti-GADD34 (Santa Cruz Antibodies Inc.) was used at a dilution of 1:100 instead of anti-myc.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes as described previously (1). However, hSNF5/INI1 proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes at 12 V for 30 min in a Trans-Blot (Bio-Rad Laboratories) semidry electrophoretic transfer cell as specified by the manufacturer. Western blotting was performed with the anti-myc mouse monoclonal antibody 9E10 described previously (1) or with anti-FLAG monoclonal antibody (Kodak). To detect primary antibodies, horseradish peroxidase-conjugated goat antibody to either rabbit immunoglobulin or murine (Sigma) immunoglobulin G was used at a dilution of 1:5000. Bands were visualized with enhanced chemiluminescence reagents (Bio-Rad Laboratories).
Yeast two-hybrid library screening and protein-protein interaction assay.
The yeast two-hybrid library screening was performed as described by Wu et al. (55) and was described in detail previously (1). A clone, designated p1(1T-)1, containing a partial GADD34 cDNA in the vector pSE1107 was recovered and was found to code for amino acids 180 to 674 of the published human cDNA sequence (25). The GADD34 sequences in p1(1T-)1 were removed from the original pSE1107 vector, subcloned into pBSK(+), and renamed pBSKGADD34.
Apoptosis assays.
Human SW480 colon carcinoma cells were transiently transfected with HRX and GADD34 constructs, and the experiments were carried out in triplicate. A total of 6 × 104 cells were plated in each well of a 24-well cluster and were transfected for 5 h with a total of 6 μg of plasmid DNA by using Cellfectin (Gibco BRL). At 24 h posttransfection, the cells were treated with 10 Gy of ionizing radiation (IR) from a 137Cs irradiator (J. L. Shepherd and Associates). At 48 h posttransfection, the number of apoptotic cells was determined by light microscopy with the ApopTag Plus in situ apoptosis detection kit (Oncor, Gaithersburg, Md.) as described by the manufacturer. Nuclei were counterstained with Meyer’s hematoxylin. In each experiment, approximately 2,500 cells were evaluated.
In vitro GADD34-hSNF5/INI1 affinity-binding assays.
In vitro transcription and translation of GADD34 protein from pSGADD34 was carried out with the TNT kit (Promega) with T7 RNA polymerase and labeled [35S]methionine and [35S]cysteine (Translabel; ICN), as specified by the manufacturer. Glutathione S-transferase (GST) and GST-hSNF5/INI1 fusion proteins were expressed from plasmids pGEX4Tk (Pharmacia) and pGEX-Ini1 in Escherichia coli as previously described (56). Cells were harvested 3 h following induction. After sonication and centrifugation, the supernatant was incubated with glutathione-linked agarose beads (Sigma) overnight at 4°C. The beads were collected by centrifugation and washed extensively with phosphate-buffered saline supplemented with 0.25% NP-40. In vitro affinity binding assays were carried out by incubating in vitro-transcribed-translated [35S]methionine-labeled GADD34 with 1 to 2 μg of GST-hSNF5/INI1 or GST bound to glutathione-agarose beads in IPB (20 mM HEPES [pH 6.9], 100 mM NaCl, 0.1 mM EDTA, 12.5 mM MgCl2, 0.1 mM dithiothreitol, 10% glycerol) at 4°C for 1 h. The agarose beads were precipitated and washed three times with IPB plus 0.2 to 0.5% NP-40. The bound proteins were eluted from the beads with SDS sample buffer (62.5 mM Tris [pH 6.8], 10% glycerol, 2.2% SDS, 1% β-mercaptoethanol, 0.0005% bromophenol blue) at 98°C for 5 min, separated by SDS-PAGE (10% polyacrylamide), and dried for autoradiography.
RESULTS
Yeast two-hybrid screen reveals that HRX binds GADD34.
To identify proteins that interact with HRX and therefore may participate in HRX fusion protein-mediated leukemogenesis, we performed a yeast two-hybrid screen with HRX884, an amino-terminal HRX clone that includes the three AT hooks (Fig. 1A). The HRX884 bait was selected because of its similarity to the small architectural transcription factor HMG-I(Y), a protein of only 100 amino acids that also contains three AT hooks. HMG-I(Y) has protein-protein-binding sites near its AT hooks that are essential to the role of this factor in facilitating transcription factor binding to DNA (29). The HRX884 bait fragment, as depicted in Fig. 1A, encodes a 295-amino-acid region including the three AT-hooks. To identify HRX884-interacting proteins, we used a LexA-based yeast two-hybrid screen as described previously (1). One of the candidate clones was sequenced and found to encode an amino-terminally truncated GADD34 protein (residues 180 to 674). GADD34 was originally isolated as a transcript induced by growth arrest or DNA damage following genotoxic stress (59).
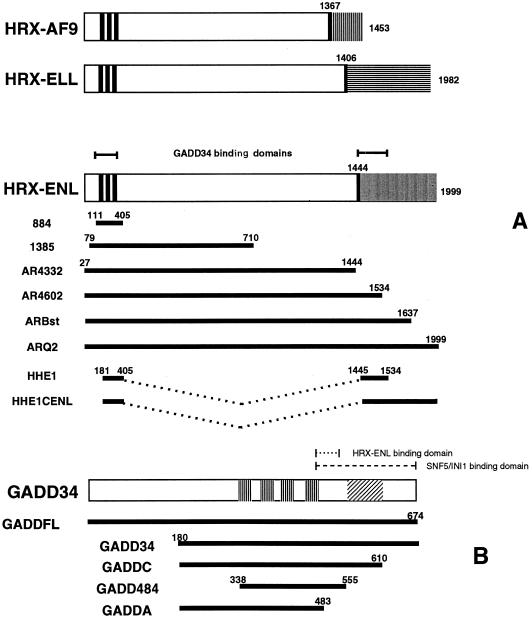
FIG. 1.
HRX fusion proteins and deletion mutants of HRX-ENL and GADD34. (A) Schematic representation of full-length human HRX-AF9, HRX-ELL, and HRX-ENL fusion proteins (top) and various HRX-ENL deletion constructs (bottom). Numbers represent amino acid residues. The three thick vertical lines designate the AT hooks. Shaded areas represent C-terminal residues donated by each of the respective fusion partners. Fusion sites are delineated by single thick vertical lines. (B) Schematic representation of the GADD34 protein (top) and GADD34 deletion constructs (bottom). The vertical stripes represent the four repeated 20- to 23-amino-acid motifs, FXXXWXYRPGXXTEEEEXXX, in GADD34. The area designated by diagonal lines represents the conserved 63-amino-acid region present in the carboxy terminus of GADD34, MyD116, and HSV ICP34.5.
Wild-type HRX and HRX fusion proteins bind GADD34 in vivo.
To confirm the yeast two-hybrid results and to delimit the region of HRX and HRX-ENL interacting with GADD34, a coimmunoprecipitation procedure was used as reported previously (1). Wild-type HRX and fragments of the HRX-ENL fusion protein, tagged with the myc epitope, were coexpressed with FLAG-tagged GADD34 protein (amino acids 180 to 674) in human kidney 293T cells. Cells were lysed, and the lysates were immunoprecipitated with anti-myc epitope antibodies. Coimmunoprecipitated GADD34 proteins were identified by Western analysis with anti-FLAG antibody (Fig. 2A, C, and D). pCSHRX884, containing the original yeast two-hybrid bait fragment, and pCS1385 are shown here to bind the 69-kDa GADD34, albeit weakly (Fig. 2A, C, and D). TEL, an unrelated protein, and HMG-C, another member of the AT hook family of proteins, expressed as a negative controls, failed to bind GADD34 (Fig. 2A and C, respectively). As shown in Fig. 2D, full-length HRX bound GADD34 weakly whereas HRX-ENL showed strong binding. Full-length HRX-ENL (pCSARQ2), as well as various carboxy-terminal deletion mutants of HRX-ENL, pCSAR4602, and pCSARBst, strongly bound GADD34 (Fig. 2A, lanes 4 and 5). Schematic representations of the HRX-ENL deletion constructs used in this assay are depicted in Fig. 1A. Since the carboxy-terminal deletion mutant of HRX-ENL lacking all ENL residues expressed from pCSAR4332 consistently bound less GADD34 than did pCSAR4602, pCSARBst, and pCSAR-Q2 (Fig. 2A, lane 3), we tested whether ENL residues actually contribute to GADD34 binding. pCSHHE1 and pCSHHE1CENL were constructed as described and illustrated above (see Materials and Methods and Fig. 1A), and both strongly bound overexpressed FLAG-tagged GADD34 (data not shown). We also tested other HRX fusion proteins, namely, HRX-AF9 and HRX-ELL. Both of these HRX fusion proteins bind GADD34 (Fig. 2A, lanes 7 and 8). An anti-myc immunoblot of the cellular lysates, demonstrating that the interactions observed do not correlate with expression of the myc-tagged proteins, is shown in Fig. 2B. Of note, the full-length myc-tagged HRX-ENL protein (pCSARQ2) was not seen on the blot at the exposures shown (Fig. 2B, lane 6); however, longer exposures revealed a protein of the expected size and anti-myc blots demonstrated abundant protein in the immunoprecipitated fractions (data not shown). The same results were obtained when the same coimmunoprecipitation experiments were repeated with the same myc-tagged HRX proteins and full-length FLAG-tagged GADD34 (data not shown). Additionally, in reciprocal coimmunoprecipitation experiments with cotransfected FLAG-tagged HHE1CENL and myc-tagged GADD34 constructs, we were able to reproduce the results described above using both anti-FLAG and anti-myc coimmunoprecipitations (data not shown). In these experiments, we have defined the amino-terminal 1444 amino acids of HRX as sufficient for GADD34 interaction. In addition to the AT hook region, other sequences located between amino acids 794 and 1444 of HRX either bind directly to or facilitate the binding of the AT hook region to GADD34. In addition, these results suggest that the amino-terminal third of ENL (amino acids 1445 to 1534 or HRX-ENL) facilitates GADD34 binding. Wild-type HRX binds GADD34 much less avidly than the HRX fusion proteins do.
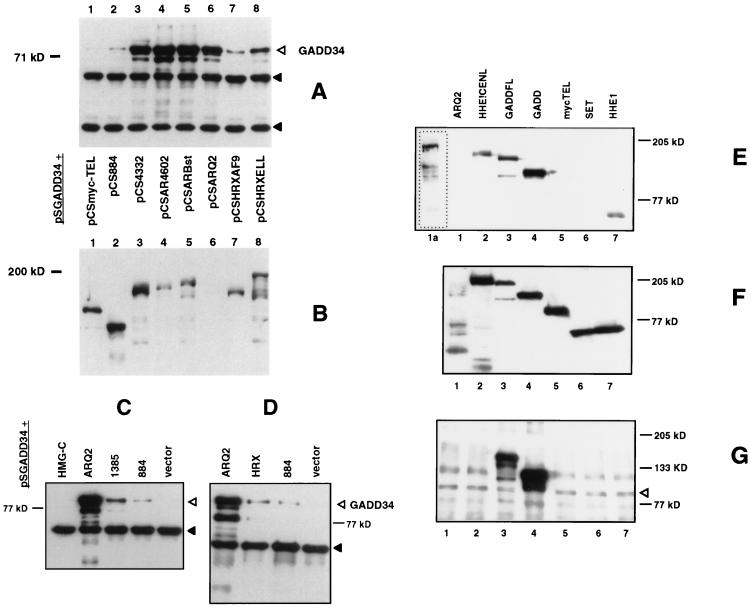
FIG. 2.
HRX proteins interact with GADD34 in vivo. (A) An anti-FLAG Western blot of anti-myc immunoprecipitates showing the amount of coimmunoprecipitated FLAG-tagged amino-truncated GADD34 protein, pSGADD34 (open triangle), bound to myc-tagged HRX fusion proteins and HRX-ENL deletion proteins expressed in the transfections described below. Solid triangles show cross-reacting murine immunoglobin bands in coimmunoprecipitates. (B) An anti-myc Western blot of cell lysates, using an anti-myc antibody, showing expression levels of the myc-tagged HRX fusion proteins and HRX-ENL deletion proteins expressed in the transfections described below. Longer exposure of lane 6 showed expression of the appropriately sized HRX-ENL protein (not shown). pSGADD34 (amino-truncated FLAG-tagged GADD34) was cotransfected with the following myc-tagged constructs: pCSmyc-TEL, pCS884, pCSAR4332, pCSAR4602, pCSARBst, pCSARQ2, pCSHRXAF9, and pCSHRXELL. (C and D) Anti-FLAG Western blot of anti-myc immunoprecipitates showing the amount of coimmunoprecipitated FLAG-tagged amino-truncated GADD34 protein, pSGADD34 (open triangle), bound to myc-tagged HRX-ENL, HRX N-terminal deletion proteins, and wild-type HRX expressed in the transfections described below. Solid triangles show cross-reacting murine immunoglobin bands in coimmunoprecipitates. The results shown here in are from two separate sets of transfections. pSGADD34 (amino-truncated FLAG-tagged GADD34) was cotransfected with the following myc-tagged constructs: pCSHMG-C, pCSARQ2, pCS1385, pCS884, and pCS2+MT vector only. (E to G) HRX fusion proteins bind endogenous GADD34 in vivo. (E) An anti-myc Western blot of anti-GADD34 immunoprecipitates showing the amount of coimmunoprecipitated myc-tagged proteins expressed in the transfections described below. Lane 1a is a longer exposure of lane 1. (F) An anti-myc Western blot of lysates from the transfections described below. (G) An anti-GADD34 Western blot of anti-GADD34 immunoprecipitates showing the amount of coimmunoprecipitated proteins from the transfections with the following constructs: pCSARQ2 (lane 1), pCSHHE1CENL (lane 2), pCSGADDFL (lane 3), pCSGADD34 (lane 4), myc-TEL (lane 5), pCSSET (lane 6), and pCSHHE1 (lane 7). The open arrow delineates endogenous GADD34 protein, shown here as a 90-kDa doublet.
Deletion analyses define a region of GADD34 that binds HRX fusion proteins.
To determine the region of GADD34 responsible for binding to HRX-ENL, we generated various FLAG-tagged GADD34 deletion constructs (as depicted in Fig. 1B). In this experiment, the GADD34 deletion constructs were coexpressed with full-length myc-tagged HRX-ENL constructs in 293T cells. Coimmunoprecipitation and Western analysis were carried out as described for the preceding experiment. As shown in Fig. 3A, the GADD34 proteins expressed from constructs pSGADD34, pSGADDC, and pSGADD484 all bound strongly to the myc-HRX-ENL fusion protein. However, the GADD34 deletion protein expressed from the pSGADDA construct did not bind to the myc-HRX-ENL fusion protein, thus defining the HRX-ENL binding region of GADD34 as residues 483 through 555. These 72 amino acids include part of the last GADD34 repeat element and the intervening sequences up to but not including the conserved 63-amino-acid region present in the carboxy terminus of the GADD34, MyD116, and herpes simplex virus (HSV) ICP34.5 proteins.
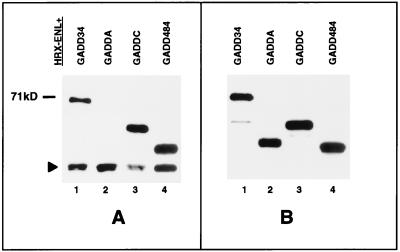
FIG. 3.
GADD34 deletion proteins interact with HRX-ENL in vivo. (A) Anti-FLAG Western blot of anti-myc immunoprecipitates, showing the amount of coimmunoprecipitated FLAG-tagged GADD34 deletion proteins bound to the myc-tagged HRX-ENL fusion protein (pCSARQ2) expressed in the transfections described below. A solid triangle shows a cross-reacting murine immunoglobin band in the coimmunoprecipitates. (B) Anti-FLAG Western blot of cell lysates with an anti-FLAG antibody, showing expression levels of the FLAG-tagged GADD34 deletion proteins expressed in the transfections described below. pCSARQ2 (myc-tagged HRX-ENL) was cotransfected with the following FLAG-tagged constructs: pSGADD34, pSGADDA, pSGADDC, and pSGADD484.
HRX fusion proteins bind endogenous GADD34.
To date, it has not been possible to detect endogenous GADD34 protein by Western analysis with available antisera, nor has it been possible to reliably distinguish wild-type from leukemic fusion HRX proteins on Western blots. We found that we could detect endogenous GADD34 as a prominent 90-kDa band on Western blots from coimmunoprecipitations with the polyclonal rabbit anti-GADD34 antibody (Santa Cruz Biotechnology Inc.) (see Materials and Methods). Using this approach, we were able to specifically pull down transfected HRX fusion constructs, pCSARQ2 (HRX-ENL), pCSHHE1CENL, and pCSHHE1 bound to endogenous GADD34, thus further confirming a physiologic interaction between HRX and GADD34 (Fig. 2E). As can be seen in Fig. 2E and F, in addition to the GADD34-binding HRX proteins, the anti-GADD34 antibody appropriately pulled down the two myc-tagged GADD34 proteins expressed from pCSGADD34 and pCSGADD34FL, (lanes 3 and 4) but failed to bring down the negative control proteins TEL and SET (lanes 5 and 6). Figure 2G is an anti-GADD34 Western blot of the anti-GADD34 immunoprecipitates showing endogenous GADD34 as a 90-kDa doublet.
GADD34 induces apoptosis following irradiation.
It has been demonstrated that GADD34 expression is upregulated, independent of p53 status, in several cell lines following exposure to ionizing irradiation (25). In the SW480 colorectal cell line, ionizing irradiation upregulates GADD34 expression by 3.8-fold and induces apoptosis. We expressed GADD34 in SW480 cells, treated the cells with 10 Gy of ionizing radiation, and measured apoptosis by DNA fragmentation detection and light microscopy with the ApoTag Plus in situ apoptosis detection kit. The results of two independent experiments, each done in triplicate, are depicted in Fig. 4A and B. As baseline, between 35 and 55% of the cells transfected with empty vector underwent apoptosis following transfection and irradiation (vector-only experiments in Fig. 4A and B). The number of cells undergoing apoptosis was not influenced by transfection of vectors expressing HRX-ENL (pCSARQ2-Expt. 4A), HRX-AF9 (pCSHRXAF9-Expt. 4A), HRX884 (pCS884 [Fig. 4B]), HRX-ELL (pCSHRXELL [Fig. 4B]), and Tel (pCS2mycTEL [Fig. 4B]). The expression of full-length GADD34 induced an additional 25 to 30% of the cells to undergo apoptosis (64% verses 35% in Fig. 4A, and 87% versus 55% in Fig. 4B). The truncated GADD34 protein, missing the first 179 amino acids, failed to induce a statistically significant apoptotic effect (GADD [Fig. 4A]). Since GADD34 alone without ionizing radiation induces only a minor apoptotic affect (10% cell apoptosis [data not shown]), the present result suggests that GADD34 facilitates cellular apoptosis associated with ionizing radiation. The amino-terminal 179 amino acids of GADD34 are essential for this effect.
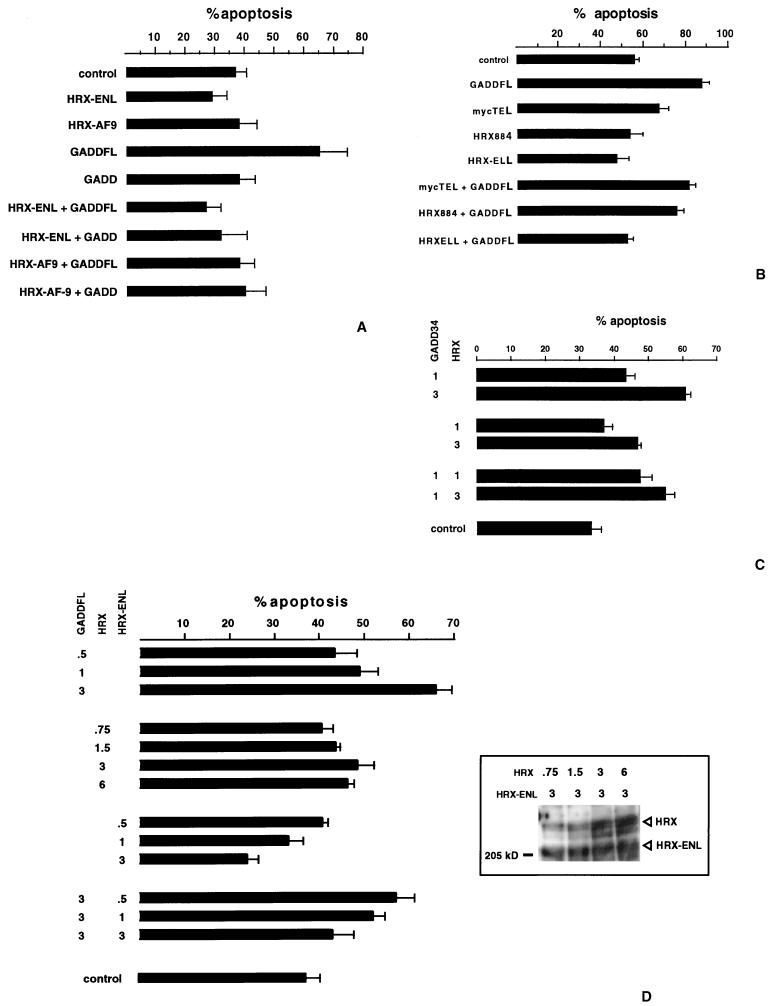
FIG. 4.
GADD34 induces apoptosis in SW480 cells. HRX fusion proteins abrogate GADD34-induced apoptosis and wild-type HRX induces apoptosis of SW480 cells after transfection and treatment with ionizing radiation. The mean percent apoptosis is shown for the transfected constructs below. Each column represents three separate experiments. (A) GADD34 induces apoptosis in SW480 cells, and the amino terminus is necessary for this effect. HRX fusion proteins, HRX-ENL and HRX-AF9, abrogate GADD34-induced apoptosis. The percent apoptosis of SW480 cells without transfection and without IR is approximately 8%, and after transfection of pSGADD34FL without IR it is approximately 17% (results of one trial only). GADD34FL encodes the full-length cDNA, and GADD34 encodes an amino-terminal deletion GADD34 protein (the clone retrieved from the yeast two-hybrid screen). (B) The HRX fusion protein HRX-ELL abrogates GADD34- induced apoptosis. (C) Wild-type HRX induces apoptosis in a dose-dependent fashion and does not inhibit GADD34-induced apoptosis. To the left is designated, in micrograms, the amount of wild-type HRX and GADD34 plasmid transfected. (D) GADD34 induces apoptosis in a dose-dependent fashion. HRX-ENL inhibits GADD34-induced apoptosis in a dose-dependent fashion. The inset shows an anti-myc Western blot from a SDS-PAGE gel (6% polyacrylamide) in a series of cotransfection experiments with increasing amounts of transfected pCSHRX plasmid (encoding myc-tagged wild-type HRX protein) with constant amounts of transfected pCSARQ2 plasmid (encoding myc-tagged HRX-ENL). The amounts of transfected plasmids are shown above in micrograms.
HRX fusion proteins inhibit GADD34-induced apoptosis.
We next addressed the question whether HRX fusion proteins affect GADD34-induced apoptosis. Various constructs encoding HRX884, HRX-ENL, HRX-AF9, and HRX-ELL were coexpressed with full-length GADD34 (GADD34FL) in SW480 cells. The cells were then similarly irradiated and quantitated for apoptosis. As shown in Fig. 4A and B, all three HRX fusion proteins, HRX-ENL (Fig. 4A), HRX-AF9 (Fig. 4A), and HRX-ELL (Fig. 4B), abrogated the apoptotic effect of full-length GADD34, since the number of cells undergoing apoptosis returned to the baseline level. HRX884, encoding the amino-terminal HRX fragment originally used as the yeast two-hybrid bait, failed to influence the level of apoptosis, and the unrelated TEL construct also had no effect (Fig. 4B). Western analysis of transfected cells revealed equivalent expression of the GADD34 protein in these cotransfection experiments, thus eliminating reduction of cellular GADD34 protein level as an explanation of the observed HRX fusion protein effect (data not shown). We therefore conclude that HRX fusion proteins can specifically inhibit GADD34-induced apoptosis. Of note, there was an observed decrease in apoptosis, albeit minor and statistically insignificant, when HRX fusion proteins were transfected compared to vector alone. Although we do not have proof, it is interesting to postulate that this minor effect may be attributed to the antiapoptotic effect of HRX fusion proteins upon endogenous GADD34.
Wild-type HRX induces apoptosis.
Experiments were carried out to measure the effect of wild-type HRX expression upon apoptosis following irradiation in SW480 cells. In contrast to HRX fusion proteins, full-length wild-type HRX exhibited a statistically significant dose-related mild proapoptotic effect (1 μg of HRX = 37% and 3 μg of HRX = 47% compared to 33% for control) and an additive effect when coexpressed with GADD34 (1 μg of GADD34 alone = 43%, 1 μg of GADD34 + 1 μg of HRX = 47%, and 1 μg of GADD34 plus 3 μg of HRX = 55%) (Fig. 4C). Western blots from transfected cells confirmed a linear correlation between the amounts of HRX plasmid transfected and the amounts of proteins expressed (Western blot inset, Fig. 4D). We proceeded to carry out a series of cotransfection experiments with SW480 cells involving increasing amounts of expressed proteins (the total amount of transfected plasmids was constant) to determine if the proapoptotic effect of GADD34 and the antiapoptotic effects of HRX-ENL were dose related. As can be seen in Fig. 4D, increasing the amount of GADD34 did significantly increase apoptosis (0.5, 1, and 3 μg resulting in 43, 49, and 66% apoptotic cells, respectively). Cotransfection experiments with constant amounts of GADD34 expressed (3 μg of GADD34 transfected) and increasing amounts of HRX-ENL revealed a dose-related antiapoptotic effect of HRX-ENL upon GADD34-induced cell death (0.5, 1, and 3 μg of HRX-ENL corresponding to 57, 52, and 43% apoptosis, respectively).
GADD34 binds hSNF5/INI1.
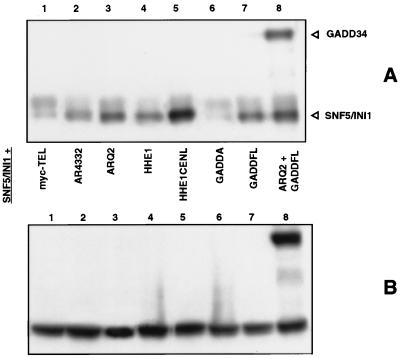
It has been previously reported that yeast Snf5 interacts with the yeast homologue of ENL (9). We were interested in determining whether HRX-ENL bound hSNF5/INI1 and what effect GADD34 might have on this interaction. With this in mind, we performed coimmunoprecipitation assays on cell lysates from 293T cells overexpressing a combination of GADD34, HRX, ENL, and hSNF5/INI1 proteins (Fig. 5). As shown here in anti-myc coimmunoprecipitation assays on cotransfected 293T cells, the two myc constructs that include carboxy-terminal ENL residues, pCSAR- Q2 and pCSHHE1CENL, strongly pulled down FLAG-tagged hSNF5/INI1 (Fig. 5A, lanes 3 and 5). FLAG-tagged hSNF5/INI1 did not coimmunoprecipitate with the negative control, pCSmyc-TEL. Since a construct lacking ENL residues altogether (pCSAR4332 [lane 2]) and one containing only the amino-terminal third of ENL (pCSHHE1 [lane 4]) both pulled down significantly less hSNF5/INI1 protein, a domain responsible for the interaction resides in the carboxy-terminal two-thirds of ENL. Furthermore, when a third protein, full-length GADD34 (pSGADD34FL), was expressed and coimmunoprecipitated with HRX-ENL (pCSAR-Q2), more hSNF5/INI1 appeared to be brought down. To confirm this unexpected result, we then cotransfected GADD34 and hSNF5/INI1 together and showed that these two proteins associated strongly on their own (lane 8). A GADD34 deletion mutant, pSGADDA, encoding residues 180 through 483, failed to bind, thus defining an hSNF5/INI1-binding domain between residues 483 to 610. It may be that the small amount of hSNF5/INI1 observed with the two constructs lacking the carboxy-terminal ENL interaction domain in lanes 2 and 4 is due to small amounts of bound endogenous GADD34. Because a direct interaction between GADD34 and hSNF5/INI1 would greatly enhance our understanding of both GADD34 and HRX leukemic fusion protein function, we tested for such an interaction by using in vitro binding assays.
FIG. 5.
HRX-ENL and GADD34 interact with hSNF5/INI1 in vivo. (A) Anti-FLAG Western blot of anti-myc immunoprecipitates showing the amount of coimmunoprecipitated FLAG-tagged hSNF5/INI1 (lanes 1 to 7) and FLAG-tagged hSNF5/INI1 and GADD34 (lane 8) proteins bound to the myc-tagged proteins expressed in the transfections described below. (B) Anti-FLAG Western blot of cell lysates with an anti-FLAG antibody, showing expression levels of full-length FLAG-tagged GADD34 and FLAG-tagged hSNF5/INI1 proteins expressed in the transfections described below. pSGSnf5 (full length FLAG-tagged hSNF5/INI1) was cotransfected with the following constructs: myc-TEL (negative control) (lane 1), pCSAR4332 (lane 2), pCSARQ2 (HRX-ENL) (lane 3), pCSHHE1 (lane 4), pCSHHE1CENL (lane 5), pCSGADDA (lane 6), pCSGADDFL (lane 7), and pCSARQ2 plus pSGADD34FL (lane 8).
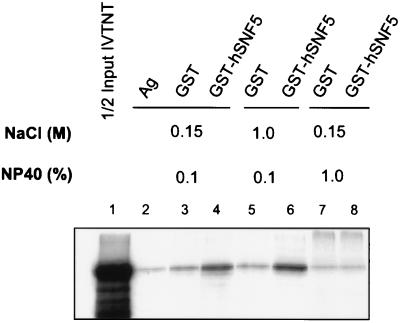
To confirm that GADD34 can directly bind hSNF5/INI1, we performed an in vitro binding assay with in vitro-transcribed/translated 35S-labeled GADD34 and GST-hSNF5/INI1 fusion protein bound to glutathione agarose beads (Fig. 6). GADD34 is shown here to bind specifically to hSNF5/INI1, as reflected by increased retention of GADD34 by GST-hSNF5/INI1 (lanes 4 and 6) compared with that by GST alone (lanes 3 and 5). This protein-protein interaction appears to be hydrophobic, since the association is stable over a wide NaCl concentration range of 0.1 to 1.0 M, but is readily disrupted at 1% NP-40 (lane 8). In summary, the coimmunoprecipitation and in vitro binding results show that HRX-ENL, GADD34, and hSNF5/INI1 associate in a trimeric protein complex in vivo. It is possible that the human SNF/SWI complex, of which hSNF5/INI1 is a member, is important to the function of GADD34.
FIG. 6.
In vitro binding of GADD34 to GST-hSNF5/INI1. 35S-labeled GADD34 was transcribed and translated in vitro from pSGFL2GADD34FL (lane 1) and used directly to bind agarose (lane 2), GST-agarose (lanes 3, 5, and 7), and GST-hSNF5-agarose (lanes 4, 6, and 8) in binding buffers with the NaCl and NP-40 concentrations shown. After three washes with binding buffer, the bound 35S-labeled GADD34 was eluted, subjected to SDS-PAGE, and visualized by autoradiography.
DISCUSSION
We report here the finding of a novel HRX-interacting protein partner, GADD34. The interaction was discovered in a yeast two-hybrid screen, using the HRX AT hook region as bait, and confirmed by in vivo coimmunoprecipitation analysis with transfected cells. GADD34 was originally discovered as a UV-inducible transcript in Chinese hamster ovary cells (17). A later study correlated the onset of apoptosis with GADD34 expression in selected cell lines following ionizing irradiation or treatment with the alkylating agent methyl methanesulfate (25). We show here that, in combination with irradiation, GADD34 overexpression enhances apoptosis (Fig. 4). We have also shown that three different leukemic HRX fusion proteins are negative regulators of GADD34-induced apoptosis whereas wild-type HRX has a proapoptotic effect. Taken together, these data show for the first time that there are functional differences between the HRX fusion proteins and wild-type HRX and suggest that abrogation of GADD34-mediated post-DNA damage apoptosis is an important function for these leukemic fusion proteins.
How does GADD34 lead to apoptosis? Insight into GADD34 function may be elucidated from examining the functional domains of MyD116, a protein that shows high degrees of sequence homology to GADD34. MyD116 is expressed in the absence of protein synthesis in M1 myeloblastic leukemia cells induced to terminal differentiation by interleukin-6 (59). GADD34 and MyD116, originally believed to be homologues, have now been shown to be distinct proteins in human, mouse, and hamster cells (49). Both proteins have virtually identical amino-terminal regions, the portion of GADD34 shown in this study to be essential for mediating apoptosis. In addition, GADD34 and MyD116 share a conserved carboxy-terminal 63 amino acid domain, also present in the HSV virulence factor ICP34.5A. This domain is necessary for suppression of apoptosis mediated by the HSV ICP34.5 protein in virally infected cells, suggesting this region interacts with the cellular apoptotic machinery (10). Moreover, this domain is functionally interchangeable, since a chimeric HSV ICP34.5 protein containing the MyD116 domain can also suppress apoptosis (21). Additionally, the carboxy-terminal regions of HSV ICP34.5 and MyD116 have been shown separately to bind protein phosphatase 1α and proliferating-cell nuclear antigen (PCNA) (8, 22). Upon binding to the HSV ICP34.5 protein, protein phosphatase 1α is redirected to dephosphorylate eIF-2A, prohibiting infected neuroblastoma cells from triggering the total shutoff of protein synthesis that is characteristic of apoptosis in neuronal cells. PCNA, a replication factor involved in regulating cell cycle progression, is a binding target for multiple proteins, including GADD45, a p53-induced growth arrest and DNA damage-associated protein (19). GADD45 binding to PCNA results in a block in replication of viral DNA in infected cells, whereas HSV ICP34.5 binding to PCNA has a permissive effect upon replication. Although the role of GADD34 in replication is unknown and it has yet to determined whether GADD34 associates with PCNA, the C-terminal domain that GADD34 shares with MyD116 and HSV IVP34.5A is probably active in the control of apoptosis. GADD34 therefore has at least two domains implicated in control of apoptosis, an amino-terminal domain (present in the first third of the protein) and a 63-amino-acid carboxy-terminal domain.
Our finding that HRX fusion proteins bind GADD34 and abrogate GADD34-induced apoptosis supports a hypothesis whereby HRX fusion proteins interfere with programmed cell death following DNA damage. In our proposed model, the leukemic HRX chimeric protein, expressed from fusion genes arising from the juxtaposition of genetic elements during chromosomal translocation, in a gain of function over wild-type HRX, acts to bypass the GADD34-mediated cellular program for regulating growth of cells following DNA damage. This hypothesis suggests that DNA damage is the important initial event in HRX leukemias and that HRX fusion proteins provide a proliferative advantage to cells following DNA damage by inhibiting cellular responses aimed at either repairing or eliminating damaged genomes. An attractive hypothesis is that proliferation and apoptosis may exist in a near equilibrium in cells that are not fully neoplastic. A perturbation of this equilibrium would constitute a step toward a more aggressive cancer, termed progression. Interestingly, Shibata et al. have studied the transition from preneoplasia to carcinoma during mammary tumor progression in C3(1)/SV40 large T-antigen transgenic mice (44). From studies with p53−/− mice, they found that preneoplastic cells have both increased proliferation and increased apoptosis over control cells and that eventually these cells develop into carcinoma cells, in which p53-independent apoptosis is suppressed. A similar result was found in the transition to carcinoma for T-antigen-induced pancreatic β-cell tumors (38). Our contention that suppression of apoptosis as an important step in the progression of 11q23 leukemias is supported by a recent study in which cells expressing HRX fusion transcripts underwent apoptosis following the addition of antisense oligonucleotides directed against the fusion transcripts (2). These data suggest that the inhibition of GADD34 p53-independent apoptosis by HRX fusion proteins may play a role in the progression of 11q23 leukemias.
Interestingly, PEG-3, another protein with approximately 69% homology to GADD34, has been implicated in cancer progression. PEG-3 was initially discovered by subtraction hybridization analysis from virus- and oncogene-transformed rat embryo cells. PEG-3 has been implicated in cancer progression because overexpression in cells results in a transformed phenotype, as shown by increased anchorage-independent growth and tumorigenic potential (49). One interpretation of these data is that GADD34 and PEG-3 share motifs critical to cancer progression and that when complexed with HRX fusion proteins, GADD34 has the potential to affect cancer progression akin to PEG-3. An alternative hypothesis could invoke opposing roles on the p53-independent apoptotic pathway by PEG-3 and GADD34, with PEG-3 potentially interfering with the proapoptotic function of GADD34. In this regard, it is interesting that the PEG-3 protein conspicuously lacks the 63-amino-acid carboxy-terminal domain implicated in promoting apoptosis that is found in the GADD34, MyD116, and ICP34.5 proteins.
Consistent with the previously reported interaction between SNF5 and the ENL homologue in yeast (9), we show here that HRX-ENL forms a complex with the hSNF5/INI1 protein (Fig. 5). We also show by in vivo coimmunoprecipitation studies and an in vitro affinity binding assay that GADD34 associates with hSNF5/INI1 (Fig. 5 and 6). Our results suggest the presence of a protein complex that consists of at least three members, the HRX fusion protein, GADD34, and hSNF5/INI1. hSNF5/INI1 is a component of the multiunit SNF/SWI protein complex (54). Current evidence suggests that the SNF/SWI complex can affect nucleosome positioning at target genes and thus overcome the repressive effect of nucleosomes upon transcription (40). Since hSNF5/INI1 has not been reported to function independently of SNF/SWI, it is plausible that GADD34 is involved in recruiting the SNF/SWI complex to HRX fusion proteins, an event that may be necessary for the transcriptional transactivation essential to the immortalization of cells by these leukemic fusion proteins. It has previously been shown that HRX-ENL deletion mutants lacking the transcriptional transactivation domain of ENL are unable to immortalize murine myeloid cells (46). The human SNF/SWI complex may indeed play a role in regulating cell growth, since BRG1, one of two known homologues of an essential SNF/SWI complex member, SNF2, is required for RAS-mediated transformation of SW13 cells (37). Additionally, mutations resulting in the truncation of hSNF5/INI1 have recently been described in childhood cancers (53). The GADD34-hSNF5/INI1 interaction described here may therefore play a role in neoplastic transformation, and further work is required to determine whether this interaction is important in 11q23 leukemias.
In summary, we report here a general biological function for leukemic HRX-fusion proteins distinct from wild-type HRX. We have shown that HRX proteins associate with and affect the function of another protein, GADD34, that is involved in cell growth regulation. Our results suggest that HRX fusion proteins probably disregulate hematopoietic cell growth and differentiation through multiple pathways that include abrogation of apoptosis.
ACKNOWLEDGMENTS
We thank Hye Son Yi and Thanh Nyugen for providing valuable technical support, Boguslaw Kwiatkowski for providing the pCSmycTEL DNA, Nancy Zeleznik-Le for providing an HRX-AF9 cDNA, Mike Thirman for providing an HRX-ELL cDNA, and Michael Cleary for a full-length HRH cDNA.
Funding from the T. J. Martell Foundation and a National Cancer Institute Cancer Center Support Grant (CA 68485) to R.C. supported this work. A National Institutes of Health grant (CA73969) and a Department of Veterans Affairs Merit Review grant to D.C.T. also supported this work.
REFERENCES
- 1.Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. doi: 10.1074/jbc.272.45.28407. [DOI] [PubMed] [Google Scholar]
- 2.Akao Y, Mizoguchi H, Misiura K, Stec W J, Seto M, Ohishi N, Yagi K. Antisense oligodeoxyribonucleotide against the MLL-LTG19 chimeric transcript inhibits cell growth and induces apoptosis in cells of an infantile leukemia cell line carrying the t(11;19) chromosomal translocation. Cancer Res. 1998;58:3773–3776. [PubMed] [Google Scholar]
- 3.Arakawa H, Nakamura T, Zhadanov A B, Fidanza V, Yano T, Bullrich F, Shimizu M, Blechman J, Mazo A, Canaani E, Croce C M. Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc Natl Acad Sci USA. 1998;95:4573–4578. doi: 10.1073/pnas.95.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard O A, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 5.Borkhardt A, Repp R, Haas O A, Leis T, Harbott J, Kreuder J, Hammermann J, Henn T, Lampert F. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 6.Breen T R, Chinwalla V, Harte P J. Trithorax is required to maintain engrailed expression in a subset of engrailed-expressing cells. Mech Dev. 1995;52:89–98. doi: 10.1016/0925-4773(95)00393-f. [DOI] [PubMed] [Google Scholar]
- 7.Breen T R, Harte P J. Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- 8.Brown S M, MacLean A R, McKie E A, Harland J. The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J Virol. 1997;71:9442–9449. doi: 10.1128/jvi.71.12.9442-9449.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns B R, Henry N L, Kornberg R D. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N, King G, Rabbitts T H. An M11-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. . (Erratum, 4:431, 1993.) [DOI] [PubMed] [Google Scholar]
- 14.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 16.Fidanza V, Melotti P, Yano T, Nakamura T, Bradley A, Canaani E, Calabretta B, Croce C M. Double knockout of the ALL-1 gene blocks hematopoietic differentiation in vitro. Cancer Res. 1996;56:1179–1183. [PubMed] [Google Scholar]
- 17.Fornace A J, Alamo I J, Hollander M C. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 19.Hall P A, Kearsey J M, Coates P J, Norman D G, Warbrick E, Cox L S. Characterisation of the interaction between PCNA and Gadd45. Oncogene. 1995;10:2427–2433. [PubMed] [Google Scholar]
- 20.Harrison C J, Cuneo A, Clark R, Johansson B, LafagePochitaloff M, Mugneret F, Moorman A V, SeckerWalker L M. Ten novel 11q23 chromosomal partner sites. Leukemia. 1998;12:811–822. doi: 10.1038/sj.leu.2401017. [DOI] [PubMed] [Google Scholar]
- 21.He B, Chou J, Liebermann D A, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the gamma (1)34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B, Gross M, Roizman B. The gamma (1)34.5 protein of herpes simplex virus I complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess J L, Yu B D, Li B, Hanson R, Korsmeyer S J. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- 24.Hillion J, LeConiat M, Jonveaux P, Berger R, Bernard O A. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- 25.Hollander M C, Zhan Q M, Bae I, Fornace A J. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J Biol Chem. 1997;272:13731–13737. doi: 10.1074/jbc.272.21.13731. [DOI] [PubMed] [Google Scholar]
- 26.Hunger S P, Tkachuk D C, Amylon M D, Link M P, Carroll A J, Welborn J L, Willman C L, Cleary M L. HRX involvement in de novo and secondary leukemias with diverse chromosome 11q23 abnormalities. Blood. 1993;81:3197–3203. [PubMed] [Google Scholar]
- 27.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 28.Joh T, Kagami Y, Yamamoto K, Segawa T, Takizawa J, Takahashi T, Ueda R, Seto M. Identification of MLL and chimeric MLL gene products involved in 11q23 translocation and possible mechanisms of leukemogenesis by MLL truncation. Oncogene. 1996;13:1945–1953. [PubMed] [Google Scholar]
- 29.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi H, Espinosa R, Thirman M J, Davis E M, Diaz M O, Le B M M, Rowley J D. Variability of 11q23 rearrangements in hematopoietic cell lines identified with fluorescence in situ hybridization. Blood. 1993;81:3027–3033. [PubMed] [Google Scholar]
- 31.Kwiatkowski B A, Bastian L S, Bauer T R J, Tsai S, Zielinska K A G, Hickstein D D. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J Biol Chem. 1998;273:17525–17530. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 32.Lavau C, Szilvassy S J, Slany R, Cleary M L. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leibovitz A, Stinson J C, McCombs W B, McCoy C E, Mazur K C, Mabry N D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 34.Majidi M, Hubbs A E, Lichy J H. Activation of extracellular signal-regulated kinase 2 by a novel Abl-binding protein, ST5. J Biol Chem. 1998;273:16608–16614. doi: 10.1074/jbc.273.26.16608. [DOI] [PubMed] [Google Scholar]
- 35.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. t(11;22)(q23;q11.2) in acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrissey J J, Raney S, Cleary M L. The FEL (AF-4) protein donates transcriptional activation sequences to HRX-FEL fusion proteins in leukemias containing t(4;11)(q21;q23) chromosomal translocations. Leuk Res. 1997;21:911–917. doi: 10.1016/s0145-2126(97)00012-x. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt C, Bourachot B, Reyes J C, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naik P, Karrim J, Hanahan D. The rise and fall of apoptosis during multistage tumorigenesis: down-modulation contributes to tumor progression from angiogenic progenitors. Genes Dev. 1996;10:2105–2116. doi: 10.1101/gad.10.17.2105. [DOI] [PubMed] [Google Scholar]
- 39.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 41.Rozenblattt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubnitz J E, Behm F G, Downing J R. 11q23 rearrangements in acute leukemia. Leukemia. 1996;10:74–82. [PubMed] [Google Scholar]
- 43.Rubnitz J E, Morrissey J, Savage P A, Cleary M L. ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood. 1994;84:1747–1752. [PubMed] [Google Scholar]
- 44.Shibata M A, Maroulakou I G, Jorcyk C L, Gold L G, Ward J M, Green J E. p53-independent apoptosis during mammary tumor progression in C3(1)/SV40 large T antigen transgenic mice: suppression of apoptosis during the transition from preneoplasia to carcinoma. Cancer Res. 1996;56:2998–3003. [PubMed] [Google Scholar]
- 45.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 46.Slany R K, Lavau C, Cleary M L. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So C W, Caldas C, Liu M M, Chen S J, Huang Q H, Gu L J, Sham M H, Wiedemann L M, Chan L C. EEN encodes for a member of a new family of proteins containing an Src homology 3 domain and is the third gene located on chromosome 19p13 that fuses to MLL in human leukemia. Proc Natl Acad Sci USA. 1997;94:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznikle N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su Z Z, Shi Y J, Fisher P B. Subtraction hybridization identifies a transformation progression-associated gene PEG-3 with sequence homology to a growth arrest and DNA damage-inducible gene. Proc Natl Acad Sci USA. 1997;94:9125–9130. doi: 10.1073/pnas.94.17.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 51.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 53.Versteege I, S’evenet N, Lange J, Rousseau M M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 54.Wang W, Cot’e J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young B D, Saha V. Chromosome abnormalities in leukaemia:The 11q23 paradigm. Cancer Surv. 1996;28:225–245. [PubMed] [Google Scholar]
- 58.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 59.Zhan Q, Lord K A, Alamo I J, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa III R, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]