Abstract
Most Acinetobacter baumannii strains are naturally competent. Although some information is available about factors that enhance or reduce the frequency of the transformation of this bacterium, the regulatory elements and mechanisms are barely understood. In this article, we describe studies on the role of the histone-like nucleoid structuring protein, H-NS, in the regulation of the expression of genes related to natural competency and the ability to uptake foreign DNA. The expression levels of the natural transformation-related genes pilA, pilT, pilQ, comEA, comEC, comF, and drpA significantly increased in a Δhns derivative of A. baumannii A118. The complementation of the mutant with a recombinant plasmid harboring hns restored the expression levels of six of these genes (pilT remained expressed at high levels) to those of the wild-type strain. The transformation frequency of the A. baumannii A118 Δhns strain was significantly higher than that of the wild-type. Similar, albeit not identical, there were consequences when hns was deleted from the hypervirulent A. baumannii AB5075 strain. In the AB5075 complemented strain, the reduction in gene expression in a few cases was not so pronounced that it reached wild-type levels, and the expression of comEA was enhanced further. In conclusion, the expression of all seven transformation-related genes was enhanced after deleting hns in A. baumannii A118 and AB5075, and these modifications were accompanied by an increase in the cells’ transformability. The results highlight a role of H-NS in A. baumannii’s natural competence.
Keywords: Acinetobacter baumannii, H-NS, natural transformation, naturally competent, DNA acquisition
1. Introduction
The histone-like nucleoid structuring protein (H-NS) is a global regulator, widely distributed among different genera of bacteria. H-NS functions to directly repress transcription across the genome. H-NS-like proteins are shown to assist horizontal DNA transmission and have important implications for bacterial evolution [1]. In Enterobacteriaceae, H-NS acts as a transcriptional repressor of the type I-E CRISPR-Cas system leading to natural transformation events [2,3]. A correlation between H-NS-mediated regulation and the lack of conservation of the respective potential horizontally acquired gene clusters in different Acinetobacter sp. genomes was observed. This evidence indicated that H-NS acts as a xenogenic repressor in A. baumannii [4]. A. baumannii H-NS disruption is known to regulate genes associated with quorum sensing, type VI secretion system, type I pili, phenylacetic acid degradation, and acetoin metabolism, among other functions [4]. Horizontal gene transfer (HGT) mechanisms play a crucial role in the dissemination of antimicrobial resistance [5,6]. Natural transformation, one of the main HGT mechanisms that promotes the integration of exogenous DNA, has been documented in approximately 80 bacterial species [5,7]. Many of the Acinetobacter sp. are naturally competent, making transformation a critical strategy for evolution and acquiring novel genetic material [8,9,10,11,12,13,14,15,16,17,18,19,20]. A. baumannii’s genomes are highly variable, showing large segments of DNA of different origins, which often code for virulence factors, adaptability systems, and antibiotic resistance [21,22,23,24].
The transformation frequency of A. baumannii increases in the presence of human pleural fluid and human serum albumin [13,20,25]. Furthermore, A. baumannii DNA uptake occurs while moving across wet surfaces [12]. Further studies refined our understanding of natural competency, which correlates with the growth phase-dependent synthesis of a type IV pilus [26]. The studies described above conclusively show that motility and natural competence are intimately associated. As a result, it is possible that other factors affecting motility also impact the capability of A. baumannii to take up DNA. The recent report that the disruption of the A. baumannii ATCC 17978 hns gene by an insertion sequence result in hyper-motility [27], as well as the role of H-NS in genome stability [1,2] raised the question that an H-NS function may also be associated with the natural transformation in A. baumannii. This analysis describes the significant enhancement in expression levels of genes related to natural competence when hns is deleted.
2. Results and Discussion
2.1. H-NS Role in Natural Transformation in the First Naturally Competent A. baumannii Clinical Isolate
To determine the effect of the H-NS global regulator in natural transformation, we deleted the genes in two experimentally validated A. baumannii strains and determined the expression of competence-associated genes. Pilus-related genes and twitching motility are essential for A. baumannii’s transformability [8,26]. Therefore, we compared the expression levels of pilA, pilT, pilQ, comEA, comEC, comF, and drpA in the wild type, the Δhns, and a complemented strain. The latter was constructed by introducing the hns-carrying plasmid pMBLe-hns into the Δhns mutant.
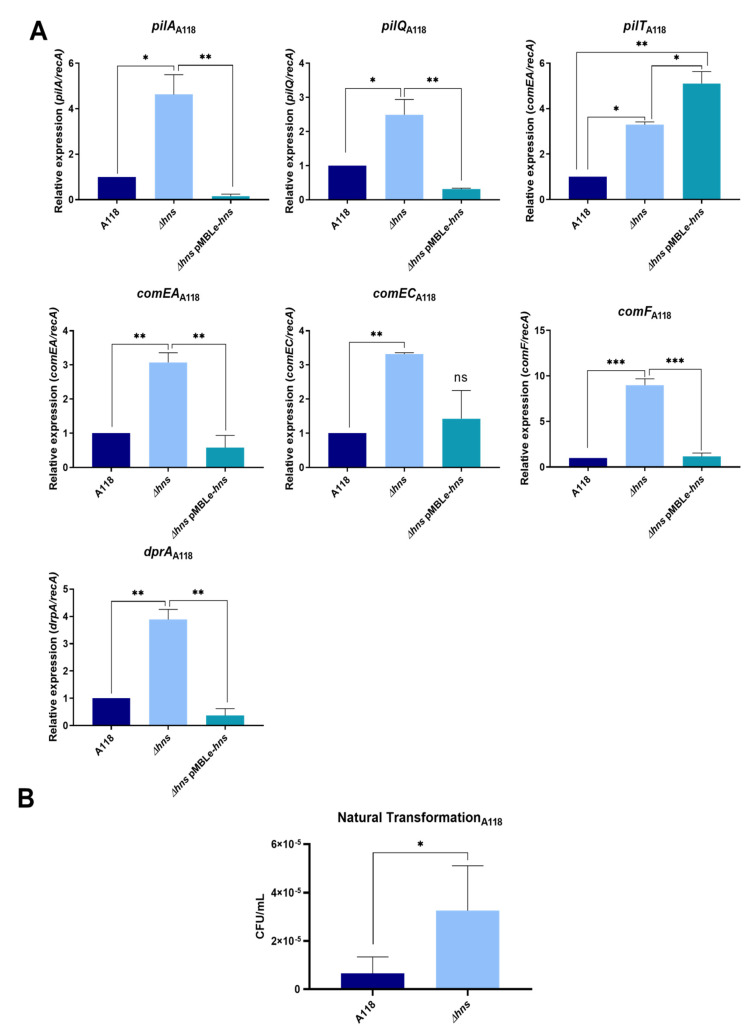
Quantitative RT-PCR (qRT-PCR) assays using total RNA demonstrated that the expression levels of all tested genes were significantly increased (Figure 1A). Exceptionally, significant differences were not observed between Δhns and Δhns pMBLe-hns for the comEC gene in A. baumannii A118. Furthermore, except for pilT, the expression levels of these genes in the complemented mutant were reduced to wild-type levels (Figure 1A). Consistent with these results, the assessment of the wild-type and the Δhns mutant transformation frequencies showed a five-fold increase in A. baumannii A118 Δhns (p < 0.05) (Figure 1B).
Figure 1.
(A) The qRT-PCR of A. baumannii A118, A118 Δhns and A118 Δhns pMBLe-hns genes associated with competence and type IV pilus: pilA, pilQ, pilT, comEA, comEC, comF and dprA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were tested, and three technical replicates were performed from each sample. Statistical significance (* p < 0.05) was determined by ANOVA followed by Tukey’s comparison test; one asterisk: * p < 0.05; two asterisks: ** p < 0.01 and three asterisks: *** p < 0.001. (B) Natural transformation frequencies for A. baumannii A118 and A118 Δhns strains in LB broth. At least three independent replicates were performed and p < 0.05 was considered significant (t test).
2.2. The Expression of Natural Competence Associated Genes Is also under the Control of H-NS in a Hypervirulent and Resistant Model Strains
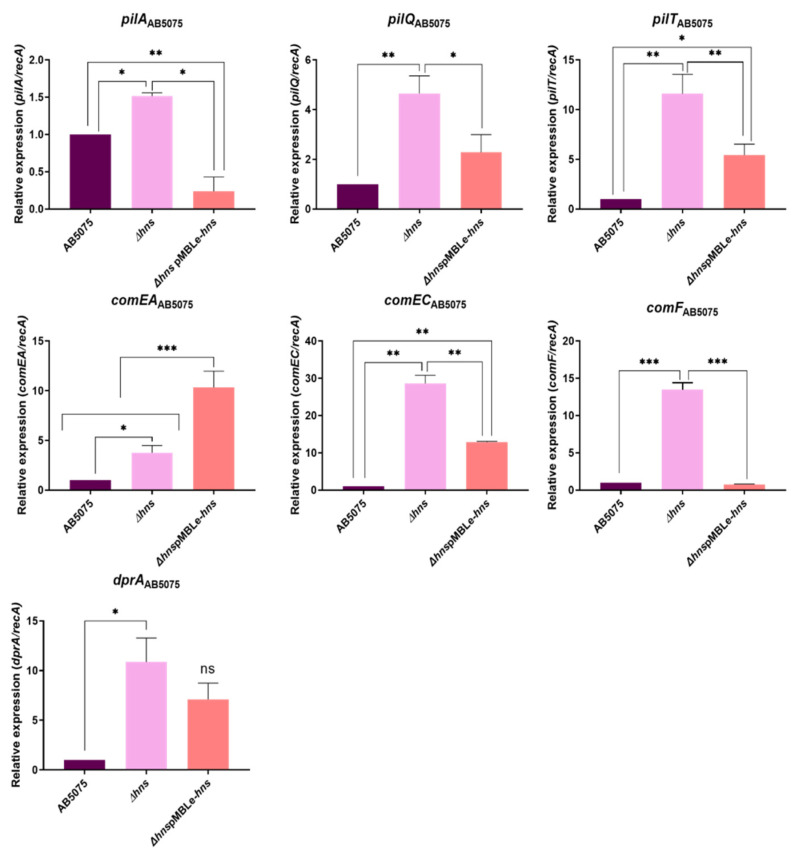
The effect of H-NS in natural competence was also studied in the hypervirulent A. baumannii AB5075, its Δhns derivative, and a complemented strain carrying pMBLe-hns. As was the case for A. baumannii A118 and A118 Δhns, all seven genes, pilA, pilT, pilQ, comEA, comEC, comF, and drpA, were expressed at higher levels in the mutant (Figure 2). The complementation via the introduction of pMBLe-hns caused a reduction in the expression levels in six genes. The expression of comEA was the only exception to this behavior. The levels of expression of the comEC gene in the absence of H-NS were higher with respect to the other genes; this may have been due to the essential comEC function. In natural transformation, the single-stranded DNA translocated across the inner membrane through the comEC channel [26]. Underscoring the importance of comEC in DNA acquisition, other inner-membrane DNA translocation channels have not been described to date. It is common that essential functions are equipped to be expressed at levels much higher than needed, but tight mechanisms negatively regulate them. When H-NS is deleted, the high levels of expression of comEC, are potentially one of these cases. comEC may be a target of interest as we observed a slight reduction in the expression of this gene after complementation in both strains.
Figure 2.
qRT-PCR of A. baumannii AB5075, AB5075 Δhns and AB5075 Δhns pMBLe-hns genes associated with competence and type IV pilus: pilA, pilQ, pilT, comEA, comEC, comF and dprA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used, and three technical replicates were performed from each sample. Statistical significance (* p < 0.05) was determined by ANOVA followed by Tukey’s comparison test; one asterisks: * p < 0.05; two asterisks: ** p < 0.01 and three asterisks: *** p < 0.001.
The levels of expression of comEA gene in AB5075 Δhns pMBLe-hns could play a role in virulence in this strain since this result was not observed in the A118 complemented strain, and the virulence and susceptibility profile were different for both strains. As was the case with pilA in the A. baumannii A118 Δhns (pMBLe-hns), we still do not know the significance and impact of these responses to the production of H-NS from an extrachromosomal element. The natural transformation frequency of A. baumannii AB5075 Δhns was five-fold higher than that of the AB5075 parent strain (Figure 2).
3. Materials and Methods
3.1. Bacterial Strains
The susceptible A. baumannii A118 model strain, the isogenic A118 Δhns variant, and A118 Δhns containing the plasmid pMBLe-hns, which expressed a wild-type copy of hns under the control of own promoter, were used (Rodgers et al., 2021, submitted). In addition, to extend the role of H-NS in the A. baumannii response, the multidrug-resistant and hypervirulent A. baumannii AB5075 strain, AB5075 Δhns [28], and AB5075 Δhns pMBLe-hns, were used in the present study.
3.2. RNA Extraction and qRT-PCR
A. baumannii A118 and AB5075 and their derivate strains were grown in lysogeny broth (LB) and incubated with agitation for 18 h at 37 °C. Then, a 1:10 dilution in fresh LB broth was realized and incubated for 7 h at 37 °C and 200 rpm. The RNA extraction was performed using the Direct-zol RNA Kit (Zymo Research, Irvine, CA, USA) in triplicate. RNA samples were treated with DNAse (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instruction. A PCR amplification of the 16S rDNA gene was performed to verify that DNA contamination was not present. The qRT-PCR was next performed to analyze the expression of natural transformation associated genes. The cDNA was prepared using the iScript™ Reverse Transcription Supermix (BioRad, Hercules, CA, USA) and iQ™SYBR® Green Supermix (BioRad, Hercules, CA, USA) per the manufacturer’s recommendations, respectively. Relative gene expression to recA was calculated using the comparative 2−ΔΔCt method [29]. Each cDNA sample was run in triplicate and repeated in at least three independent sets of samples. The ANOVA test followed by Tukey’s comparison was used to determine statistical significance (p-value < 0.05) using GraphPad Prism (GraphPad software, San Diego, CA, USA).
3.3. Natural Transformation Assays
Natural transformation assays were performed as described [25,26]. Briefly, 20 uL of A. baumannii cells grown overnight in LB medium at 37 °C were mixed with 1 μg of pMBLe-OA-ArK (apramycin resistance) plasmid and the mixture was spotted onto twitching motility plates (10). After 4 h of incubation at 37 °C, the cells were scraped off from the plate and resuspended in a microcentrifuge tube containing 200 μL of LB medium. Apramycin (15 µg/mL) resistant colonies were counted (transformants cells) and scored. In parallel, total colony forming units (CFUs) were performed by plating serial dilutions on LB agar plates. Experiments were conducted in technical and biological triplicates including negative controls. Student’s t test analysis was performed using GraphPad Prism (GraphPad software, San Diego, CA, USA). A p-value < 0.05 was considered significant.
4. Conclusions
The global repressor H-NS modulates the expression of a plethora of A. baumannii genes with functions related to virulence, biosynthetic pathways, cell adhesion, quorum sensing, and autotransporters, among others. Previous analyses suggested that H-NS plays a major role in the acquisition of genes transferred horizontally [1,30]. While the acquisition of DNA is a mechanism to better adapt to the environment, as it is the case with numerous mobile genetic elements, too many of these events could have deleterious effects. The acquisition and insertion of DNA fragments into the chromosome were mechanisms that must be tightly regulated to limit the detrimental consequences. H-NS could act as a negative regulator of DNA acquisition.
The focused analysis of the role of H-NS in the regulation of expression of genes related to the mobility and DNA transformation carried out in this analysis showed that: (a) seven genes that code functions associated with natural competence are overexpressed in the absence of H-NS; and (b) the transformation frequency is higher in the absence of this protein. Consequently, H-NS modulates the DNA uptake by A. baumannii strains, suggesting a participation in gene acquisition and, concomitantly, evolution during infectious processes.
Author Contributions
C.L., C.P., M.R.T., T.S., J.E., B.N., S.A., D.R., R.A.B., R.S., M.E.T. and M.S.R. conceived the study and designed the experiments; C.L., C.P., M.R.T., T.S., J.E., B.N., S.A., D.R., R.S., and M.S.R. performed the experiments and genomics and bioinformatics analyses.; M.R.T., T.S., R.S., R.A.B., M.E.T. and M.S.R. analyzed the data and interpreted the results; R.A.B., M.E.T. and M.S.R. contributed reagents/materials/analysis tools; M.R.T., T.S., R.S., R.A.B., M.E.T. and M.S.R. analyzed the data, wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors’ work was supported by NIH SC3GM125556 to M.S.R, R01AI100560 to R.A.B., R01AI063517, R01AI072219 to R.A.B and 2R15 AI047115 to M.E.T., C.P. and J.E. were supported by grant MHRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health. D.R. has a MARC U*STAR fellowship by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T34GM008612. S.A. was supported by Project RAISE, U.S. Department of Education HSI-STEM award number P031C160152. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. M.R.T. and T.S. are recipients of a postdoctoral fellowship from CONICET. R.S. is a staff member from CONICET.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data pertaining to the study described in the manuscript is de-scribed in the report.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Doyle M., Fookes M., Ivens A., Mangan M., Wain J., Dorman C.J. An H-NS-like Stealth Protein Aids Horizontal DNA Transmission in Bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 2.Sun D., Mao X., Fei M., Chen Z., Zhu T., Qiu J. Histone-like Nucleoid-Structuring Protein (H-NS) Paralogue StpA Activates the Type I-E CRISPR-Cas System against Natural Transformation in Escherichia coli. Appl. Environ. Microbiol. 2020;86:e00731-20. doi: 10.1128/AEM.00731-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin T.-L., Pan Y.-J., Hsieh P.-F., Hsu C.-R., Wu M.-C., Wang J.-T. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci. Rep. 2016;6:31644. doi: 10.1038/srep31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eijkelkamp B., Stroeher U., Hassan K., Elbourne L., Paulsen I., Brown M.H. H-NS Plays a Role in Expression of Acinetobacter baumannii Virulence Features. Infect. Immun. 2013;81:2574–2583. doi: 10.1128/IAI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston C., Martin B., Fichant G., Polard P., Claverys J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Genet. 2014;12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 6.von Wintersdorff C., Penders J., Van Niekerk J.M., Mills N.D., Majumder S., Van Alphen L.B., Savelkoul P.H.M., Wolffs P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubnau D., Blokesch M. Mechanisms of DNA Uptake by Naturally Competent Bacteria. Annu. Rev. Genet. 2019;53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 8.Harding C.M., Tracy E.N., Carruthers M.D., Rather P.N., Actis L.A., Munson R.S. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio. 2013;4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pour N.K., Dusane D., Dhakephalkar P.K., Zamin F.R., Zinjarde S.S., Chopade B.A. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol. Med. Microbiol. 2011;62:328–338. doi: 10.1111/j.1574-695X.2011.00818.x. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez M.S., Don M., Merkier A.K., Bistué A.J.S., Zorreguieta A., Centron D., Tolmasky M.E. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol. 2010;48:1488–1490. doi: 10.1128/JCM.01264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramírez M.S., Merkier A.K., Quiroga M.P., Centrón D. Acinetobacter baumannii is able to gain and maintain a plasmid harbouring In35 found in Enterobacteriaceae isolates from Argentina. Curr. Microbiol. Curr. Microbiol. 2012;64:211–213. doi: 10.1007/s00284-011-0052-9. [DOI] [PubMed] [Google Scholar]
- 12.Wilharm G., Piesker J., Laue M., Skiebe E. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J. Bacteriol. 2013;195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn B., Traglia G.M., Nguyen M., Martinez J., Liu C., Fernandez J.S., Ramirez M.S. Effect of Host Human Products on Natural Transformation in Acinetobacter baumannii. Curr. Microbiol. 2018;76:950–953. doi: 10.1007/s00284-017-1417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn B., Martinez J., Liu C., Nguyen M., Ramirez M.S. The effect of sub-inhibitory concentrations of antibiotics on natural transformation in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2018;51:809–810. doi: 10.1016/j.ijantimicag.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traglia G.M., Quinn B., Schramm S.T.J., Soler-Bistue A., Ramirez M.S. Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016;60:4920–4929. doi: 10.1128/AAC.00529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godeux A.-S., Svedholm E., Lupo A., Haenni M., Venner S., Laaberki M.-H., Charpentier X. Scarless Removal of Large Resistance Island AbaR Results in Antibiotic Susceptibility and Increased Natural Transformability in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2020;64:e00951-20. doi: 10.1128/AAC.00951-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingues S., Rosário N., Cândido Â., Neto D., Nielsen K.M., Da Silva G.J. Competence for Natural Transformation Is Common among Clinical Strains of Resistant Acinetobacter spp. Microorganisms. 2019;7:30. doi: 10.3390/microorganisms7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domingues S., Rosário N., Ben Cheikh H., Da Silva G.J. ISAba1 and Tn6168 acquisition by natural transformation leads to third-generation cephalosporins resistance in Acinetobacter baumannii. Infect. Genet. Evol. 2018;63:13–16. doi: 10.1016/j.meegid.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y., He L., Tao X., Meng F., Zhang J. High DNA Uptake Capacity of International Clone II Acinetobacter baumannii Detected by a Novel Planktonic Natural Transformation Assay. Front. Microbiol. 2019;10:2165. doi: 10.3389/fmicb.2019.02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez J., Liu C., Rodman N., Fernandez J.S., Barberis C., Sieira R., Perez F., Bonomo R.A., Ramirez M.S. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2018;93:183–187. doi: 10.1016/j.diagmicrobio.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier P.-E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith M.G., Gianoulis T.A., Pukatzki S., Mekalanos J.J., Ornston L.N., Gerstein M., Snyder M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touchon M., Cury J., Yoon E.-J., Krizova L., Cerqueira G.C., Murphy C., Feldgarden M., Wortman J., Clermont D., Lambert T., et al. The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol. Evol. 2014;6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traglia G.M., Chua K., Centron D., Tolmasky M., Ramírez M.S. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol. 2014;6:2235–2239. doi: 10.1093/gbe/evu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le C., Pimentel C., Tuttobene M.R., Subils T., Nishimura B., Traglia G.M., Perez F., Papp-Wallace K.M., Bonomo R.A., Tolmasky M.E., et al. Interplay between meropenem and human serum albumin on expression of carbapenem resistance genes and natural competence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021:AAC0101921. doi: 10.1128/aac.01019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vesel N., Blokesch M. Pilus production in Acinetobacter baumannii is growth phase dependent and essential for natural transformation. J. Bacteriol. 2021;203:e00034-21. doi: 10.1128/JB.00034-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J.-H., Hassan K., Begg S.L., Rupasinghe T.W.T., Naidu V., Pederick V.G., Khorvash M., Whittall J.J., Paton J.C., Paulsen I.T., et al. Identification of Novel Acinetobacter baumannii Host Fatty Acid Stress Adaptation Strategies. mBio. 2019;10:e02056-18. doi: 10.1128/mBio.02056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher L.A., Ramage E., Weiss E.J., Radey M., Hayden H.S., Held K.G., Huse H.K., Zurawski D.V., Brittnacher M.J., Manoil C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn B., Rodman N., Jara E., Fernandez J.S., Martinez J., Traglia G.M., Montaña S., Cantera V., Place K., Bonomo R.A., et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018;8:14741. doi: 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hüttener M., Paytubi S., Juárez A. Success in incorporating horizontally transferred genes: The H-NS protein. Trends Microbiol. 2015;23:67–69. doi: 10.1016/j.tim.2014.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data pertaining to the study described in the manuscript is de-scribed in the report.