Figure 4.
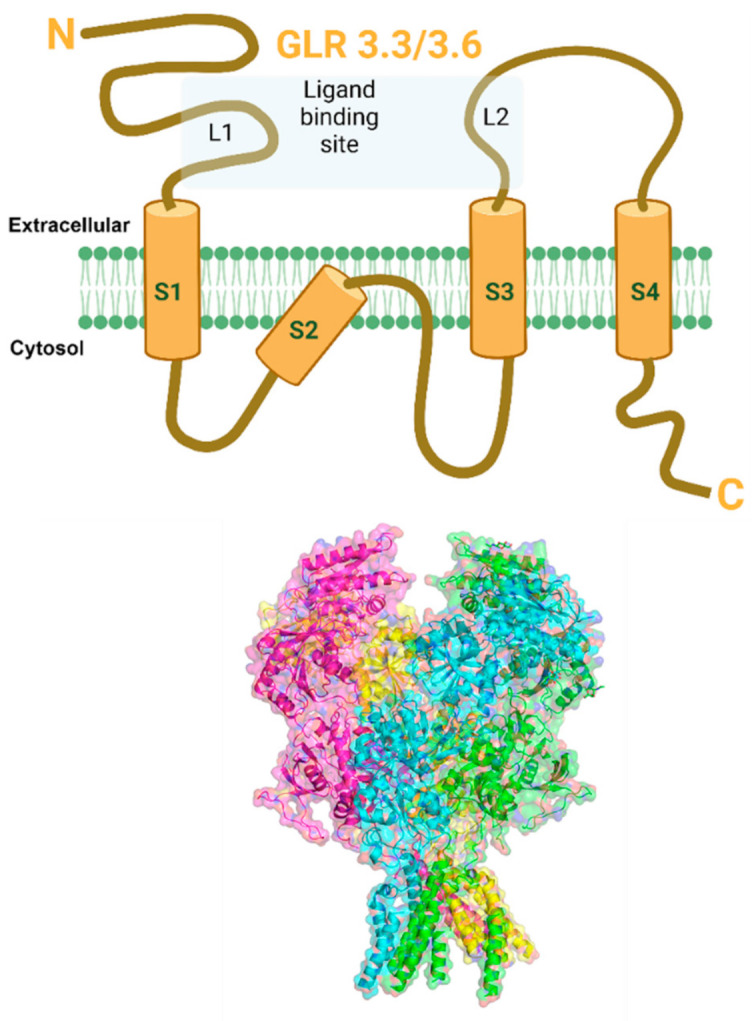
Putative structure of GLR3.3/3.6 channel. (Top) Schematic cartoon representation of GLR3.3/3.6 channel subunit showing extracellular N-terminus, four membrane-spanning regions (S1–S4), 2 extracellular ligand-binding sites (L1, L2), and intracellular C-terminus. (Bottom) The structure of GLR3.3/3.6 has not been solved to date but is likely to show similarities with the animal NMDA receptor family of channels. Therefore, the structure shown in the figure is an approximation based on homology to other channels. The predicted GLR3.3/3.6 secondary 3D structure model showing four subunits in transparent surface view was developed from closest homolog PDB structure 4TLL (Xenopus laevis GluN1/GluN2B NMDA receptor), using PHYRE 2.0 program. The image was prepared using PyMol software (PyMOL Molecular Graphics System, Version 2.4, Schrödinger, LLC, New York, NY, USA). Created with BioRender.com (accessed on 30 August 2021).
