Abstract
BRCA1 is a nuclear phosphoprotein expressed in a broad spectrum of tissues during cell division. The inheritance of a mutant BRCA1 allele dramatically increases a woman’s lifetime risk for developing both breast and ovarian cancers. A number of mouse lines carrying mutations in the Brca1 gene have been generated, and mice homozygous for these mutations generally die before day 10 of embryonic development. We report here the survival of a small number of mice homozygous for mutations in both the p53 and Brca1 genes. The survival of these mice is likely due to additional unknown mutations or epigenetic effects. Analysis of the Brca1−/− p53−/− animals indicates that BRCA1 is not required for the development of most organ systems. However, these mice are growth retarded, males are infertile due to meiotic failure, and the mammary gland of the female mouse is underdeveloped. Growth deficiency due to loss of BRCA1 was more thoroughly examined in an analysis of primary fibroblast lines obtained from these animals. Like p53−/− fibroblasts, Brca1−/− p53−/− cells proliferate more rapidly than wild-type cells; however, a high level of cellular death in these cultures results in reduced overall growth rates in comparison to p53−/− fibroblasts. Brca1−/− p53−/− fibroblasts are also defective in transcription-coupled repair and display increased sensitivity to DNA-damaging agents. We show, however, that after continued culture, and perhaps accelerated by the loss of BRCA1 repair functions, populations of Brca1−/− p53−/− fibroblasts with increased growth rates can be isolated. The increased survival of BRCA1-deficient fibroblasts in the absence of p53, and with the subsequent accumulation of additional growth-promoting changes, may mimic the events that occur during malignant transformation of BRCA1-deficient epithelia.
Germ line mutations in BRCA1 account for 45% of all hereditary cases of breast cancer (35). Inheritance of one defective copy of BRCA1 confers an estimated 80 to 90% overall lifetime risk for breast or ovarian cancer (15). Analysis of the primary structure of BRCA1 identified a number of potential functional motifs, including a RING finger domain in the N terminus and two putative nuclear localization sequences (5, 50, 52). In addition, two BRCT domains, characteristic of proteins involved in DNA repair, were identified in the C terminus (3). Expression of BRCA1 is detected early in embryonic development and continues to be found in a broad spectrum of tissues in the adult animal (27, 34). It has now been established that BRCA1 is a nuclear phosphoprotein expressed in dividing cells in a cell-cycle-specific manner, with maximal expression of BRCA1 occurring in the S phase (6, 42). Phosphorylation levels of BRCA1 also change throughout the cell cycle, peaking in late S phase (6, 42).
Despite extensive study, the function(s) of BRCA1 has not yet been clearly defined. The expression and phosphorylation of BRCA1 in a cell-cycle-specific manner suggest that this protein may be involved in the regulation of cell cycle transition. BRCA1 has been shown to induce expression of p21 and, more recently, BRCA1 has been shown to act as a p53 coactivator (38, 48, 54). In addition, a potential role as a transcriptional activator has been shown by the fusion of the C terminus of BRCA1 with a GAL4 DNA-binding domain (4, 36).
The hyperphosphorylation and relocalization of BRCA1 with RAD51 to PCNA-containing foci after exposure to DNA-damaging agents suggest an additional or alternate function for this protein (43). RAD51 has been implicated in DNA repair, as demonstrated by the high sensitivity to DNA-damaging agents of Saccharomyces cerevisiae rad51 mutants (17, 46). This increased sensitivity is thought to be due to a defect in recombinational repair of double-strand breaks. The recent demonstration that embryonic stem (ES) cells deficient in BRCA1 are more sensitive to some DNA-damaging agents provides direct evidence supporting a role for BRCA1 in maintaining genomic integrity (19). These BRCA1-deficient cells are unable to carry out transcription-coupled repair (TCR) after exposure to DNA-damaging agents, demonstrating the participation of this protein in at least one cellular repair pathway. This repair system, which preferentially repairs the transcribed strand of active genes, is important for the removal of lesions for which the global repair process is too slow.
Several mouse lines carrying mutations in the Brca1 gene have been generated (18, 21, 32, 33). Unlike humans, mice carrying a mutant Brca1 allele do not display an increased risk for tumor formation. This observation and the early embryonic lethality of mice homozygous for the mutant allele have to date limited the contribution of these models to understanding the role of BRCA1 in tumorigenesis. However, we have recently reported the generation of five mammary tumors from mice heterozygous for both a mutant Brca1 allele and p53 allele after exposure to ionizing radiation (9). Furthermore, loss of heterozygosity of both Brca1 and p53 could be demonstrated in tumor tissue obtained from three of these tumors. This suggests that exposure of Brca1+/− mice to specific environmental risk factors may be necessary for the development of mammary tumors during the short lifespan of the mouse. It also suggests that mutations in genes, such as p53, which confer a growth advantage and in particular allow the continued growth of cells having incurred DNA damage may be critical to the development of these tumors.
Evidence for a relationship between p53 and BRCA1 is further suggested by the demonstration that survival of BRCA1-deficient embryos is extended in the absence of p53 expression (22, 33). However, the Brca1−/− p53−/− embryos still die early in embryogenesis, precluding examination of the loss of BRCA1 in later stages of development and in tumorigenesis. The early lethality of the Brca1−/− p53−/− embryos did not permit the generation of Brca1−/− p53−/− embryonic fibroblast cell lines.
We report here the survival and characterization of a small number of Brca1−/− p53−/− mice after extensive breeding of Brca1+/− p53−/− animals. Although growth retarded, the development of the somatic tissues of these mice is largely normal. Male mice are infertile due to a defect in spermatogenesis, thus defining a role for BRCA1 in meiosis. Brca1−/− p53−/− animals die of tumors typical of p53−/− mice; however, tumor latency is reduced compared to that observed in the p53−/− population in our colony. Fibroblast lines were generated from the Brca1−/− p53−/− mice and compared to the fibroblast lines from both wild-type and p53−/− animals. While, as expected, p53−/− fibroblasts grow rapidly, the growth of early passage Brca1−/− p53−/− fibroblasts is similar to that of wild-type cells. This decreased growth rate is largely the result of a decrease in the survival of the BRCA1-deficient cells. We also show here that the Brca1−/− p53−/− cells are more sensitive than p53−/− cells to DNA-damaging agents and that the ability of the cells to carry out transcription-coupled repair in response to DNA damage is compromised. These observations support the hypothesis that BRCA1 plays an important role in DNA repair pathways. In summary, our results suggest that loss of heterozygosity of BRCA1 is not an initiating event in tumorigenesis. Cells must instead attain a growth advantage, by mechanisms such as loss of cell cycle checkpoint control or responsiveness to trophic factors, for loss of BRCA1 function to predispose towards tumorigenesis.
MATERIALS AND METHODS
Animal husbandry.
p53+/− mice were obtained from Jackson Labs and bred to Brca1+/− mice generated in our colony (18, 26). Genomic DNA was recovered from tail biopsy, and genotypes were determined by PCR amplification, as described elsewhere (18, 26). All three Brca1−/− p53−/− mice were generated from matings of Brca1+/− p53−/− females with a Brca1+/− p53−/− male. The Brca1 genotype was verified by Southern blotting analysis. Genomic DNA was digested with EcoRV and analyzed by Southern blot with a probe containing a portion of intron 9 and exon 10 of the Brca1 gene. Each mouse was euthanized when moribund and then necropsied. Tissues were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Sections were cut (5 μm) and stained with hematoxylin and eosin. Testes sections were also stained with toluidine blue. The TUNEL (terminal deoxynucleotidytransferase-mediated dUTP-biotin nick end labeling) assay was performed on testes sections to determine the level of apoptosis, as per the manufacturer’s instructions (Trevigen, Gaithersburg, Md.). Additional testes sections were immunostained with a rabbit polyclonal antibody for HSP70-2 (13) and staining was detected by a Peroxidase Elite ABC Kit (Vector Laboratories, Burlingame, Calif.) as per the manufacturer’s instructions. Whole-mount preparations were done as described elsewhere (37a). Briefly, mammary epithelium was fixed in 10% phosphate-buffered neutral formalin and then rehydrated. The mammary glands were stained overnight with carmine alum, dehydrated, cleared in xylene, and mounted in Permount.
Generation of cell lines.
Primary fibroblasts were obtained from the skin and ears of all three Brca1−/− p53−/− mice. These tissues were washed repeatedly with phosphate-buffered saline (PBS), finely minced, and digested overnight with collagenase (Life Technologies, Grand Island, N.Y.). The fibroblasts released were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and supplemented with l-glutamine, penicillin, streptomycin, and gentamicin at 37°C with 5% CO2. Cells were passaged before they reached 100% confluence. Fibroblast lines were also generated from two Brca1+/− p53−/− mice and a Brca1+/+ p53+/+ mouse.
Analysis of cellular growth.
Determination of cellular growth was performed as described elsewhere (12). Briefly, asynchronous fibroblasts from each cell line were plated onto a series of 35-mm dishes. Each of the Brca1−/− p53−/− lines was plated at a density of 4 × 104 cells per well, while 2 × 104 cells of each of the Brca1+/− p53−/− lines per well were plated. These values were determined based on the slower growth of the Brca1−/− p53−/− cells. Cells from two wells per line were counted daily by using trypan blue exclusion. The medium was changed daily for the remaining cells. For cell cycle analysis, asynchronous fibroblasts from each cell line were harvested after a 4-h incubation with 10 μM bromodeoxyuridine (BrdU; Boehringer Mannheim, Indianapolis, Ind.) and then fixed in cold 70% ethanol. Fixed cells were stored at 4°C until analysis. Nuclei were released by treatment with 0.08% pepsin in 0.1 N HCl at 37°C for 20 min. Nuclei were then treated with 2 N HCl at 37°C for 20 min, followed by neutralization with 0.1 M sodium borate. Cells were then incubated with the fluorescein isothiocyanate-conjugated anti-BrdU antibody (Becton Dickinson, San Jose, Calif.) and counterstained with propidium iodide (50 μg/ml) containing RNase (5 μg/ml) overnight at 4°C. Cell cycle analysis was performed on a FACScan cell sorter (San Jose, Calif.) with Cytomation data acquisition software (Fort Collins, Colo.). WinMDI software was used to determine the percentage of cells in each cell cycle phase.
Analysis of cellular death.
To determine the ratio of dead to live cells, asynchronous cells from each line were plated onto six 100-mm dishes and grown to 70 to 80% confluence. The medium was removed, and the cells were incubated with 8 ml of fresh medium for 7 h. For quantitation of live cells, each plate was trypsinized, and cells were counted by trypan blue exclusion with a hemocytometer. For the dead cell count, the medium from each plate was centrifuged in separate tubes. Pelleted cells were resuspended in 10% FBS in PBS and cytospun onto a microscope slide. The dead cells were stained with Diff-Quik stain (Dade Diagnostics of P.R., Inc., Miami, Fla.), and photographs from random areas of each slide were taken to facilitate counting. Only cells containing nuclei were counted.
To confirm the decrease in cell viability, two additional assays were performed. For the first assay, nonadherent cells from two Brca1−/− p53−/− cell lines and one Brca1+/− p53−/− cell line were harvested as described above. The nonadherent cells were counted and plated in 1 ml of medium in 24-well dishes. Adherent cells were then trypsinized and plated at an equivalent density. Live adherent cells (as determined by trypan blue exclusion) were counted from each well 24 h later. The second assay utilized the MTT assay (Roche Molecular Biochemicals, Indianapolis, Ind.) with the following modifications. Nonadherent cells from two Brca1−/− p53−/− lines and one Brca1+/− p53−/− line were harvested as described above, resuspended in <1 ml of medium, and counted. Adherent cells were trypsinized and counted, and 5,000, 10,000, 20,000, and 40,000 cells (nonadherent and adherent) were plated in duplicate. All cells were immediately incubated with the MTT labeling reagent for 4 h and then incubated with the solubilization solution for 24 h. Conversion to formazan dye was detected at 595 nm.
To quantitate apoptosis in this dead cell population, the TUNEL assay was performed on the nonadherent cells immobilized on a microscope slide (as described above) as per the manufacturer’s directions (Trevigen). To determine the percentage of apoptotic cells, photographs from random areas of each slide were taken.
For p21 protein analysis, asynchronous Brca1+/− p53−/−, Brca1−/− p53−/−, and wild-type fibroblasts were washed three times with PBS and mechanically lysed for 15 min at 4°C in 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer (containing 20 mM Tris, 143 mM KCl, 5 mM EDTA, 10 mM dithiothreitol, 20 mM NaCl) supplemented with a Complete Mini Protease Inhibitor Cocktail Tablet (Roche Molecular Biochemicals, Indianapolis, Ind.). Supernatants of the cell lysates were collected following centrifugation (15,000 × g at 4°C for 15 min). Five micrograms of protein was resolved on a sodium dodecyl sulfate–12% polyacrylamide gel and then transferred to nitrocellulose. p21 was detected by a monoclonal antibody (F-5, 1:1,000 dilution; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), followed by incubation with a horseradish peroxidase-conjugated secondary antibody and visualized with chemiluminescence regents (Pierce, Rockford, Ill.).
DNA damage analysis.
The survival of the fibroblasts following treatment with DNA-damaging agents was determined as described elsewhere (40). In brief, to determine survival after ionizing radiation, asynchronous cells were irradiated in suspension at room temperature by using a 137Cs source. The medium was changed immediately after irradiation. Brca1−/− p53−/− cells were then plated at a density of 4 × 104 cells per 35-mm dish and Brca1+/− p53−/− cells were plated at a density of 2 × 104 cells per dish. The surviving cells were counted 4 days later by trypan blue exclusion and expressed as a percentage, by using untreated, nonconfluent cells as the 100% level. To measure survival after hydrogen peroxide treatment, Brca1−/− p53−/− cells were plated at a density of 4 × 104 cells per 35-mm dish, and Brca1+/− p53−/− cells were plated at a density of 2 × 104 cells per dish. When cells were adherent 10 h later, cells were incubated with hydrogen peroxide for 15 min at 37°C. Cells were then washed twice with medium. Quantitation of cells was done 4 days later by trypan blue exclusion, and this is expressed as a percentage, using untreated, nonconfluent cells as the 100% value. To determine survival after UV irradiation, cells were plated as described above for the hydrogen peroxide treatment. The cells were incubated for 10 h to allow adherence. The medium was then aspirated, and cells were irradiated by using a Stratalinker (Stratagene, La Jolla, Calif.). Medium was then immediately added. Quantitation was done 4 days later by trypan blue exclusion and expressed as a percentage, by using untreated, nonconfluent cells as the 100% level. Each of these experiments was conducted in duplicate, and in each replicate, the cell survival was determined four times at each experimental point.
Transcription-coupled repair analysis was performed as previously described (29). For labeling of parental DNA, cells were grown for 3 days in medium containing 1 μCi of [3H]thymidine (TdR; Amersham) per ml in the presence of 5 μg of unlabelled TdR per ml. The medium was removed, and the cells were grown for an additional 2 days in nonradioactive medium. Prior to irradiation, cells were incubated for 1 h in medium containing 10 μM BrdU and 1 μM fluorodeoxyuridine (FdUrd). Cultures were washed with PBS and were irradiated with either a 60Co gamma source at dose rates of 1.3 to 1.6 Gy/min or at 254 nm by using a germicidal lamp at an incident dose rate of 0.63 J/m2/s. After irradiation, the cultures were either harvested immediately or incubated for various lengths of time in medium containing 10 μM BrdU and 1 μM FdUrd. For measurements of thymine glycol production and repair, cell cultures were exposed to 10 mM H2O2 for 15 min at 37°C. The cells were then incubated for various lengths of time in this medium. After incubation, the cultures were washed twice with PBS and harvested by lysis in 10 mM Tris–10 mM EDTA–0.5% sodium dodecyl sulfate.
Repair analysis of UV and ionizing-radiation-induced DNA damage was carried out as previously described (28) by using a monoclonal antibody against BrUra. Briefly, purified DNA, digested with BamHI, was centrifuged to equilibrium in a CsCl gradient to separate parental density DNA (containing BrUra-substituted repair patches) from hybrid density DNA (synthesized by semiconservative replication). Unreplicated, parental-density DNA was then reacted with the monoclonal antibody against BrUra. The DNA bound by the antibody was separated by centrifugation. Aliquots of the supernatant and pellet were assayed for radioactivity by liquid scintillation counting to determine the relative amount of DNA bound by the antibody.
Repair analysis of thymine glycols was carried out as previously described (30) with a monoclonal antibody that recognizes thymine glycols in DNA. Heat-denatured DNA (50 to 100 μg) was incubated with the antibody (1:1,000 dilution) in PBS containing 0.1% bovine serum albumin for 1 h at 37°C, followed by an overnight incubation at 4°C. An equal volume of ice-cold saturated ammonium sulfate in PBS was added, and the mixture was incubated at 4°C for 15 min. The DNA bound by the antibody was collected as a pellet by centrifugation. Aliquots of the supernatant and pellet were assayed for radioactivity by liquid scintillation counting to determine the relative amount of DNA bound by the antibody.
Equal amounts of DNA, based on the 3H prelabel, from the supernatant and pellet were electrophoresed on 0.7% neutral agarose gels. After electrophoresis, the DNA was transferred to a GeneScreen Plus (NEN) membrane. RNA probes were prepared as previously described (29). After hybridization, the membranes were washed and exposed to Kodak XAR-5 X-ray film. The intensity of hybridization to the fragments of interest was measured by using a Bio-Rad phosphorimager. The value for the density of each fragment was multiplied by the amount of DNA in the bound or free fractions to obtain the total amount of each gene in both fractions. The percentage of the dihydrofolate reductase gene (DHFR) gene bound by the antibody was then calculated from the total amount of the gene in the bound fraction divided by the total amount of the gene in the bound plus free fractions. For the studies on the time course of repair of thymine glycols, the percentage of the genes containing thymine glycols immediately after treatment was set at 100%.
RESULTS
Identification of Brca1−/− p53−/− mice.
Mice heterozygous for the Brca1 mutation, Brca1Δ223–763, were intercrossed with mice carrying a mutant p53 allele to obtain animals heterozygous for both mutations (18, 26). While litters obtained from the intercrossing of the derived double-heterozygous animals yielded p53−/− animals at reported frequencies, no Brca1−/− animals were identified among either the p53−/− or p53+/− offspring. Brca1+/− p53−/− mice were further intercrossed and again no double-homozygous animals were obtained on examination of 56 offspring obtained from nine different litters (9). Further expansion of this mouse population, however, led to the identification of three Brca1−/− mice, two males and a single female, all also homozygous for the p53 mutant allele. Southern blot analysis revealed that only the targeted Brca1 allele was present (data not shown). The absence of the normal Brca1 mRNA transcript was further verified by Northern blot analysis (data not shown). Interestingly, two of these animals not only shared one parent but were further related in that the dam of the second Brca1−/− p53−/− mouse was the sibling of the first Brca1−/− p53−/− animal identified.
All three double-homozygous animals were severely growth retarded. The female mouse reached a maximum weight of 14 g at 12 weeks of age compared to the average weight of 21 g seen in the p53-deficient females in our colony. Similarly, the two Brca1−/− p53−/− males weighed only 16.3 and 14.5 g compared to an average weight of 27 g seen in p53−/− males at this age.
Similar to the majority of the p53−/− mice, the Brca1−/− p53−/− mice developed tumors and were killed when moribund. All three mice died of lymphomas, the most common tumor seen in the p53−/− populations (14, 26). Necropsy of one of the male mice also revealed the presence of a hemangiosarcoma. Lymphomas and hemangiosarcomas are seen in 59 and 18%, respectively, of the p53−/− population (23). While the tumor types observed in the Brca1−/− p53−/− mice are consistent with the tumors seen with p53 deficiency, the survival age of the double-homozygous animals was substantially less than the average 19 weeks of survival for p53−/− animals in our colony. All three Brca1−/− p53−/− mice died at between 10 and 12 weeks of age.
Alopecia (generalized loss of fur), as well as an unusually high percentage of white hairs dispersed through their coats, was noted in two of the three double-homozygous animals. On gross observation the skin of all the double homozygous animals appeared thinner and more transparent than that of the p53-deficient mice. Histological analysis revealed atrophy of follicles and adnexa with a reduction in the size of both the sebaceous glands and follicles. Follicles were also reduced in number in comparison to p53−/− mice.
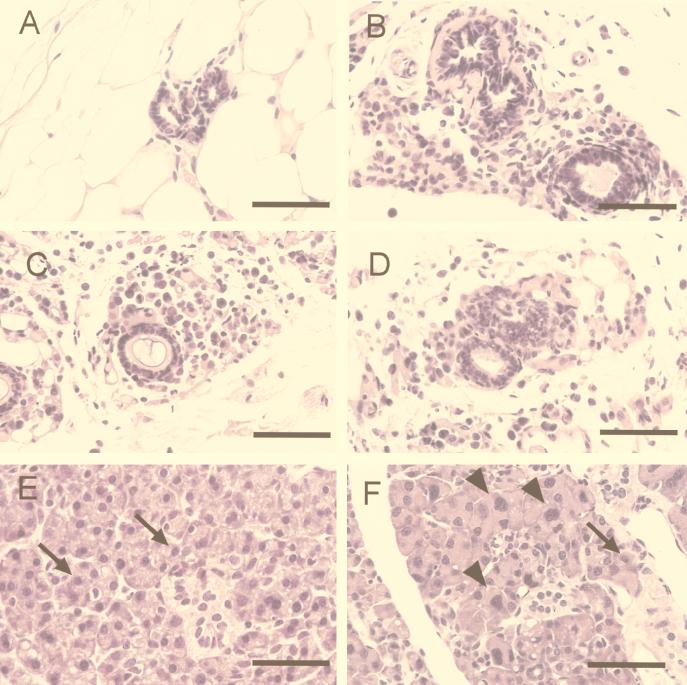
Loss of BRCA1 did not result in morphological or histological changes in the kidney, liver, brain, lungs, pancreas, or gastrointestinal tract, other than those secondary to metastatic hematopoietic neoplasms. Histopathologic lesions were primarily confined to the epithelia of the parotid, mammary, and prostate glands. Parotid salivary glands exhibited diffuse acinar atrophy that is usually observed only in geriatric animals. Mild and multifocal cytomegaly and karyomegaly suggestive of polypoidy were seen in the acinar epithelium (Fig. 2E and F). Parotid ductal cells were normal, as was the histology of the submandibular and sublingual glands. While the coagulated and vesicular glands of the male Brca1−/− p53−/− did not differ from control mice, a rare benign granular cell tumor, usually found in older animals, was observed in the prostate glands. Cytomegaly and karyomegaly were also observed in the prostate glands.
FIG. 2.
Cellular abnormalities within the mammary and salivary glands of the Brca1−/− p53−/− mice. (A) Histological analysis of the mammary gland ducts from a p53−/− mouse shows a duct lined by one to two layers of low cuboidal epithelia. Mammary ducts of the Brca1−/− p53−/− female are dilated and lined by a single layer of flattened-to-low cuboidal cells. (B to D) In some regions, the ducts are surrounded by an array of individual cells. While acinar cells in the p53−/− salivary gland appear to be uniform (E), cytomegaly and karyomegaly (arrows) are seen sporadically throughout the acinar units of the salivary glands of the Brca1−/− p53−/− animals (F). Staining was done with hematoxylin and eosin. Magnification bar, 50 μm.
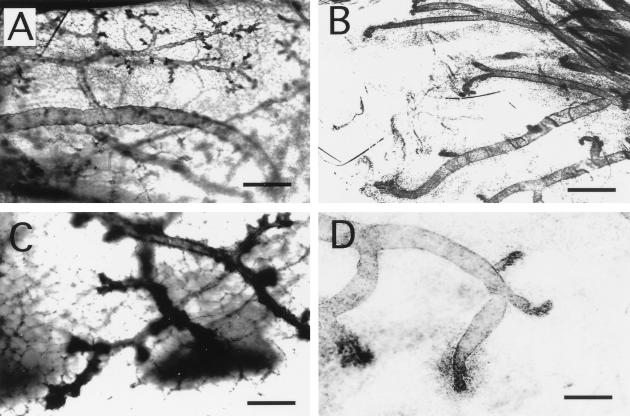
BRCA1 is expressed in the mouse mammary gland during puberty, pregnancy, and regression after discontinuation of lactation (34). The mammary gland in the female Brca1−/− p53−/− mouse was examined by whole mount. While the primary ducts were easily identified, a substantial reduction in the branching is seen in comparison to whole mounts prepared from an age-matched p53−/− virgin mouse (compare Fig. 1A and B). In addition, the end buds of the BRCA1-deficient female appeared underdeveloped (compare Fig. 1C and D), and the fat pad in the region of the end buds contains finely dispersed cellular material. To further examine the structure of the mammary gland, the fat pad was embedded for histological analysis (Fig. 2A through D). Primary ducts in mammary glands from Brca1−/− p53−/− mice were fewer and more dilated than those in the mammary gland of the p53−/− mice. In the Brca1−/− p53−/− mice (Fig. 2B through D), the ducts were surrounded by loose, concentric aggregates of individualized oval cells. These cells had variably sized round nuclei and scant-to-modest eosinophilic cytoplasm. Some of the periductal cells had pyknotic nuclei suggestive of cell death. These cells may be myoepithelial cells or ductal cells that failed to migrate and differentiate normally. We cannot, however, exclude the presence of inflammatory cells in these cellular aggregates on the basis of morphology.
FIG. 1.
Abnormal end bud formation and decreased ductal branching in BRCA1-deficient mice. (A) Mammary epithelium of a 12-week-old p53−/− female mouse displays extensive branching and numerous end buds. (B) In contrast, very few branches from the primary ducts are seen in the mammary gland of the Brca1−/− p53−/− female mouse. At a higher magnification, many end buds can be visualized in the p53−/− female (C), while only a few underdeveloped end buds are seen in the Brca1−/− p53−/− mouse (D). Magnification bars: A and B, 500 μm; C and D, 200 μm.
Spermatogenesis is disrupted in the Brca1−/− p53−/− mice.
Male Brca1−/− p53−/− mice failed to impregnate female mice. This deficiency was not the result of a failure of the mice to copulate, since copulation plugs were observed in females following mating with a Brca1−/− p53−/− male. On gross examination, the testes of both of the double-homozygous males were smaller in size than those of wild-type males of similar weight. We next compared the histological development of the seminiferous tubules of these testes to those from p53−/− control animals of similar age. Sertoli cells, the supporting cells within the tubules, were seen throughout the testes of the Brca1−/− p53−/− males, suggesting that spermatogenesis was not aborted due to loss of this cell population.
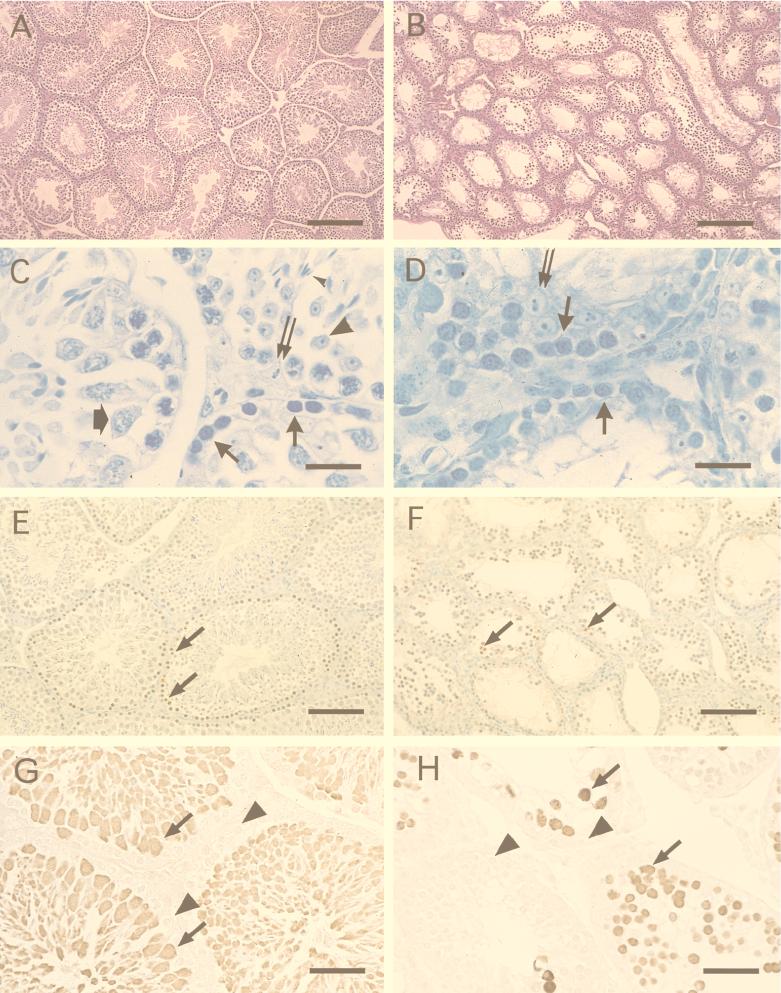
As expected, the seminiferous tubules of 10-week-old p53−/− male mice contain cells in all stages of spermatogenesis, including spermatogonia, spermatocytes and round and fully elongated spermatids (Fig. 3A and C). The spermatogonia, the stem cells of the testes, are located at the periphery of a seminiferous tubule. The developing spermatocytes, located just interior to the spermatogonia, undergo meiosis I and II to form the haploid round spermatids, which are easily identified by their light-staining nuclei. Fully elongated spermatids with tails extending into the lumen form from these round spermatids after spermiogenesis. The seminiferous tubules of the Brca1−/− p53−/− males appeared smaller in diameter in comparison to those of an age-matched p53−/− male and contained fewer cells (Fig. 3B). While the spermatogonia appeared relatively normal in both Brca1−/− p53−/− males, spermatids and spermatozoa were not observed (Fig. 3D). In addition, only two pachytene spermatocytes, identified by their distinct chromatin structure, were observed on survey of the entire section. To further define the defect in spermatogenesis of the double-homozygous animals, sections were stained with an antibody to HSP70-2 (13). This antibody is specific for a heat shock protein whose expression is initiated as cells enter into meiosis. As expected, the peripheral layer of cells, the spermatogonia, of both the p53−/− (Fig. 3G) and Brca1−/− p53−/− (Fig. 3H) mice failed to stain with this antibody. Cells luminal to this single layer of spermatogonia present in some of the seminiferous tubules of the testes of the Brca1−/− p53−/− males stained brightly with this antibody, indicating that these cells had likely entered into meiosis. Together with the absence of pachytene spermatocytes, this suggests that meiotic failure occurs during prophase I of meiosis in the Brca1−/− p53−/−- mice.
FIG. 3.
Absence of spermatids and spermiogenesis in a BRCA1-deficient male. The testes and seminiferous tubules of a Brca1−/− p53−/− male (B) are smaller than those of an age-matched p53−/− control male (A). While the seminiferous tubules of the p53−/− control male mouse (C) contained all of the cells involved in spermatogenesis (spermatogonia, thin arrows; spermatocyte, wide arrow; spermatid, large arrowhead; spermatozoa, small arrowhead), only spermatogonia (solid arrows) can be seen in the testes of the Brca1−/− p53−/− male (D). Sertoli cells (double arrow) were seen in abundance in the seminiferous tubules of the Brca1−/− p53−/− males and the p53−/− males (C and D). (E) Apoptosis as determined by the TUNEL assay (apoptotic cells, arrow) occurs rarely in spermatogenesis in p53−/− males. Apoptotic cells (arrows in panels E and F) appear to be more numerous in the testes of the Brca1−/− p53−/− male. However, to correct for the decreased size of the seminiferous tubules of the Brca1−/− p53−/− male in a given area, we counted the number of apoptotic cells present in 25 randomly chosen seminiferous tubules from the two animals. (F) TUNEL-positive cells were only increased approximately threefold in testes of the Brca1−/− p53−/− male. (G) Spermatocytes (arrows), spermatids, and spermatozoa can be detected in the testes of a p53−/− control male stained with HSP70-2 antibody. Incubation with the HSP70-2 antibody revealed the presence of spermatocytes (arrows) in the testes of a Brca1−/− p53−/− male. Staining: A and B, hematoxylin and eosin; C and D, toluidine blue; E and F, diaminobenzidine and methyl green; G and H, diaminobenzidine. Magnification bars: A and B, 200 μm; C and D, 20 μm; E and F, 100 μm; G and H, 67 μm.
To determine whether meiotic failure observed in the Brca1−/− p53−/− mice was paralleled by increased levels of apoptosis of spermatocytes, tissue sections obtained from the testes of both control p53−/− mice and Brca1−/− p53−/− animals were analyzed by using the TUNEL assay. Only a modest increase in the number of stained cells was observed (Fig. 3E and F). In 25 randomly examined seminiferous tubules, 13 apoptotic cells were seen in the testes of the p53−/− control mice compared to 37 apoptotic cells per 25 tubules in the testes of the Brca1−/− p53−/− mouse. Examination of other tissues from the double-homozygous animals also failed to reveal an increased level of apoptosis.
Growth properties of primary skin fibroblasts.
Skin fibroblasts were prepared from Brca1−/− p53−/− mice, Brca1+/− p53−/− mice, and wild-type mice, and the growth properties of these primary cultures were examined. Brca1+/− p53−/− fibroblasts had growth characteristics similar to Brca1+/+ p53−/− lines. For simplicity we will refer to the Brca1+/− p53−/− lines as p53−/− lines throughout the peper. Before these experiments were begun, the plating efficiency of the fibroblasts of each genotype was established. This was done by plating known numbers of cells, allowing the cells to adhere, and then determining the number of viable surviving cells 7 h later. At this time point, cell numbers reflect the plating efficiency of the cells rather than differences in the growth rates of the various lines. No differences between the plating efficiencies of the Brca1−/− p53−/−, p53−/−, and wild-type lines were observed (data not shown).
Proliferation rates and saturation densities of fibroblast cultures prepared from wild-type mice and from two p53−/− and two Brca1−/− p53−/− mice were determined at both low and high passage points (Fig. 4A and B, respectively). Early-passage p53−/− fibroblasts grew more quickly and reached higher saturation densities than cells obtained from wild-type control animals. Both the growth rate and the saturation density of the early passage Brca1−/− p53−/− fibroblasts resembled more closely those of the wild-type cells than those observed for the p53−/− cells.
FIG. 4.
Growth rate of Brca1−/− p53−/− fibroblasts. (A) At early passages, Brca1−/− p53−/− and p53−/− control lines and a wild-type fibroblast line were plated at a low density and were counted daily. Brca1−/− p53−/− fibroblasts have a growth rate similar to that of the wild-type fibroblasts. In contrast, the Brca1+/− p53−/− control fibroblast lines have a higher growth rate, and contact inhibition occurs at a higher density. (B) However, at later passages, the growth rate of the Brca1−/− p53−/− fibroblasts has increased, and contact inhibition occurs at a higher cell density. (C) Brca1−/− p53−/− fibroblasts were incubated with BrdU and stained with anti-BrdU antibody and propidium iodide. Quantitation of the percentage of cells in each cell cycle stage was performed by flow cytometry. One Brca1+/− p53−/− fibroblast line and one wild-type fibroblast line were used as controls. Cells at passage numbers 3 and 4 were considered early-passage cells, while cells at passage numbers 10 through 14 were considered late-passage cells.
As an increase in the expression of p21 in BRCA1-deficient embryos (embryonic days 6 to 8) was previously reported (21), it was of interest to determine whether a similar increase might underlie the decreased growth rate of the Brca1−/− p53−/− cells. Because these cells are deficient in p53, such an increase if observed would be dependent on stimulation of p21 expression by p53-independent pathways (1, 39, 55). Protein levels of p21 were determined by Western blot analysis in asynchronous, proliferating Brca1−/− p53−/−, p53−/−, and wild-type fibroblasts. While high levels of p21 were observed in the wild-type fibroblasts, a finding consistent with the rapid senescence of these cells, only very low levels of p21 were detected in the Brca1−/− p53−/− and p53−/− fibroblasts. These results fail to support a role of p21 in the decreased growth rate of the Brca1−/− p53−/− cells (Fig. 5A).
FIG. 5.
A higher rate of cellular death occurs in Brca1−/− p53−/− primary fibroblast cultures. (A) p21 protein levels are not elevated in the Brca1−/− p53−/− fibroblasts (lanes a and b) or the Brca1+/− p53−/− fibroblasts (lane c). In contrast, high levels of p21 were detected in wild-type fibroblasts (lanes d and e). (B) Nonadherent Brca1−/− p53−/− fibroblasts were collected and counted. This number was normalized to the number of adherent live cells in each fibroblast population. Nonadherent and adherent counts were obtained from six plates for each of the cell lines. The bars represent the standard errors.
We next wished to determine whether the growth differences between the cell lines would be maintained after extended in vitro culture (passage 15 to 25). Senescence of the wild-type cells by passage 8 precluded their inclusion in this experiment. While a small increase in the growth rate and saturation density of one of the p53−/− lines was observed on comparison of early- and late-passage cell cultures, a dramatic increase in both parameters was seen in both late passage Brca1−/− p53−/− lines (Fig. 4B).
Cell cycle kinetics of Brca1−/− p53−/− fibroblasts.
Previous studies of p53−/− and wild-type embryonic fibroblasts indicated that the increased proliferation rate of the p53−/− cells correlated with an increase in the percentage of cells in S phase and a decrease in the number of cells in G0/G1 (24). These alterations are believed to stem largely from a loss of the G1/S checkpoint in the p53−/− lines. To determine whether further alterations in the cell cycle kinetics underlie the decreased growth rate of the Brca1−/− p53−/− lines, asynchronous early-passage fibroblasts of each of the three genotypes were labeled with BrdU, stained with propidium iodide, and subjected to flow cytometry analysis (Fig. 4C). As reported previously and consistent with observed growth rates, a higher percentage of p53−/− cells are observed in S phase than seen in similarly labeled wild-type cells (24). In addition, loss of the spindle checkpoint due to p53 deficiency results in an increased percentage of tetraploid cells (11). Surprisingly, in light of the growth properties of the Brca1−/− p53−/− cells, the percentage of these cells in S phase was similar to that observed in the p53−/− fibroblast cultures. Thus, while at low passage the growth rate of the Brca1−/− p53−/− lines is similar to wild-type lines, the percentage of cells in S phase is approximately two to three times greater than seen in this control population.
To determine whether the increased proliferation rate seen in the late-passage Brca1−/− p53−/− cells was a result of an increased rate through the cell cycle, BrdU and DNA content analyses were performed on asynchronous cells from a p53−/− control line and two Brca1−/− p53−/− lines. Again, the percentage of cells in S phase was similar in all three lines (p53−/− control, 58.2%; Brca1−/− p53−/− line 1, 64.2%; Brca1−/− p53−/− line 2, 57.6%).
Decreased growth rate of Brca1−/− p53−/− cells is due to an increased rate of cellular death.
In order to resolve the discrepancy between the observed growth rate of the Brca1−/− p53−/− cells and the percentage of the cells in S phase, we next determined the rate of cellular death in the cultures of Brca1−/− p53−/−, p53−/−, and wild-type fibroblasts. Dead cells can be collected and enumerated easily in these cultures because after cellular death, the fibroblasts become nonadherent, lift off from the surface of the culture dishes, and can be harvested by aspiration and centrifugation of the tissue culture medium. Collection of these cells from cultures of all three genotypes and analysis with trypan blue exclusion dye confirmed that virtually all of the cells harvested in this manner were dead and that the collection procedure had not resulted in disturbance of the loosely attached mitotic cells (Fig. 5B).
To confirm that this nonadherent population of cells was in fact dead and did not simply reflect a change in anchorage dependence of the Brca1−/− p53−/− fibroblasts during various stages of the cell cycle, we examined the plating efficiency of these cells. Brca1−/− p53−/− nonadherent cells were collected and plated in 24-well plates at a density similar to that used in passage of adherent cells. After 24 h, the numbers of adherent and nonadherent cells were determined. A decrease in the total number of cells present had occurred, and only 3% of the nonadherent cells plated had attached. Analysis of these cells and the nonadherent population indicated that only the attached cells excluded the dye. The viability of the Brca1−/− p53−/− cells was further examined by using a colorimetric MTT-based assay capable of reliably detecting small numbers of viable cells (37). Between 5 × 103 and 4 × 104 adherent or nonadherent cells were collected, and the conversion of MTT to formazan dye was examined quantitatively. In two Brca1−/− p53−/− fibroblast lines and one p53−/− line, fewer than 15% of the nonadherent cells examined were viable.
To determine whether cellular death was occurring through an apoptotic pathway, the TUNEL assay was performed on the nonadherent cells of two Brca1−/− p53−/− lines and two Brca1+/− p53−/− lines. Although nonadherent cells are not viable as determined by trypan blue exclusion, fewer than 25% stained positively for apoptosis in the four lines examined (data not shown).
Brca1−/− p53−/− fibroblasts show increased sensitivity to DNA-damaging agents.
A number of lines of evidence support a role for BRCA1 in the maintenance of genome integrity (19, 43). If increased cellular death in the Brca1−/− p53−/− lines is directly related to the inability of these cells to repair DNA damage, it would be expected that the growth of these cells would be further compromised by exposure to DNA-damaging agents. We therefore determined the survival rate of the two Brca1−/− p53−/− lines and two p53−/− lines after treatment with ionizing radiation, hydrogen peroxide, and UV light. A dose-dependent decrease in cellular survival was seen in cells of both genotypes after exposure to 2.5, 5.0, and 7.5 Gy of ionizing radiation (Fig. 6A). However, the decrease in survival after treatment was significantly greater in the double-homozygous lines. A similar increase in sensitivity to DNA-damaging agents was observed after exposure of the cells to hydrogen peroxide (Fig. 6B). Again, while hydrogen peroxide treatment decreased cellular survival of both Brca1−/− p53−/− and p53−/− fibroblasts, the impairment of survival of the double-homozygous cells was significantly greater. The difference between the Brca1−/− p53−/− and p53−/− cells was most pronounced after exposure to 500 μM hydrogen peroxide. Although differences in the survival of the Brca1−/− p53−/− and p53−/− cells were not as dramatic after exposure of these lines to ultraviolet light (Fig. 6C), survival impairment of the double-homozygous lines was greater than that observed in the p53−/− cultures. These differences achieved statistical significance in two of the UV radiation doses examined.
FIG. 6.
Brca1−/− p53−/− fibroblasts are sensitive to gamma radiation, hydrogen peroxide, and UV radiation. Fibroblasts from two Brca1−/− p53−/− cell lines and two Brca1+/− p53−/− cell lines were exposed to 0, 2.5, 5.0, or 7.5 Gy of gamma radiation (A); 0, 250, 500, or 750 μM hydrogen peroxide (B); or 0, 5, 10, or 30 J of UV radiation (C). Survival was determined in each cell line by normalizing surviving cells following DNA damage to the number of cells which were untreated. An asterisk indicates statistical significance (P < 0.05).
Transcription-coupled repair in p53−/− and Brca1−/− p53−/− cells.
We have previously demonstrated an increased sensitivity to ionizing radiation and hydrogen peroxide in an ES cell line that is BRCA1 deficient and that this increased sensitivity paralleled a defect in TCR (19). However, as this BRCA1-deficient ES cell line was a single isolate and as no additional lines could be identified in numerous additional experiments, it is possible that this phenotype was not directly related to the loss of BRCA1 but rather to the accumulation of other mutations in the cell line. To determine if TCR is dependent on normal BRCA1 function, a Brca1−/− p53−/− fibroblast line and a Brca1+/− p53−/− control line were exposed to ionizing radiation, UV light, or hydrogen peroxide, and the rate of repair on the DHFR gene was measured. After exposure to 10 Gy of ionizing radiation, a rapid repair of the transcribed strand of the DHFR gene was found in the Brca1+/− p53−/− cells (Fig. 7A). In contrast, the rate of repair on the transcribed strand of the DHFR gene was similar to that of the nontranscribed strand and the genome overall in the Brca1−/− p53−/− cells, similar to what was observed in the BRCA1-deficient ES cells. Similarly, when cells were exposed to 10 mM H2O2 and the TCR of thymine glycols was examined, a deficiency in the rapid removal of this oxidized base from the transcribed strand of the DHFR gene was also observed in Brca1−/− p53−/− cells (Fig. 7B). However, when cells were exposed to 10 J of UV per m2, no deficiency in TCR was observed in the Brca1−/− p53−/− cells compared to the Brca1+/− p53−/− control line (Fig. 7C). These results indicate that TCR of oxidative DNA damage is absent in cells defective in BRCA1.
FIG. 7.
TCR is defective in the DHFR gene of Brca1−/− p53−/− fibroblasts after exposure to ionizing radiation and hydrogen peroxide but not after exposure to UV light. Cells were exposed to 10 Gy of gamma rays (A) or 10 J of UV radiation per m2 (C) and allowed to repair in the presence of 10 μM BrdU. Genomic DNA, digested with BamHI, was reacted with an antibody to BrdU. DNAs from the bound and free fractions were electrophoresed and transferred to a GeneScreen Plus membrane. The percentage of total DNA (▴) bound by the antibody was determined from the 3H prelabel. The percentage of the transcribed strand (■) and nontranscribed strand (●) of the DHFR gene was analyzed with a Bio-Rad phosphorimager. (B) Cells were exposed to 10 mM H2O2 for 15 min at 37°C. Purified DNA was digested with BamHI and incubated with an antibody against thymine glycols, and the extent of removal was determined as described above. The percentage of total DNA (▴) bound by the antibody was determined from the 3H prelabel. The percentage of the transcribed strand (■) and nontranscribed strand (●) of the DHFR gene was analyzed with a Bio-Rad phosphorimager.
DISCUSSION
We report here the survival to adulthood of a small number of mice homozygous for a mutation in the Brca1 gene. BRCA1-deficient males are infertile, and the paucity of spermatocytes is consistent with a role for BRCA1 in meiosis. Surprisingly, the presence of growing follicles in the ovary of the BRCA1-deficient female suggests that BRCA1 is not required for the early meiotic events in female germ line cells. Examination of these BRCA1-deficient mice and primary cells derived from these mice indicates that loss of normal BRCA1 expression results in growth retardation: mice homozygous for the mutation are smaller than littermates, and the growth rate of the Brca1−/− p53−/− cells is reduced in comparison to p53−/− control lines. Furthermore, we show that the mechanism underlying this growth deficit is largely due to an increase in the percentage of cells in these cultures that die and not to a decrease in the transition rate of the cell cycle. This cellular death does not occur by apoptosis. After extensive passage, the growth rate of the Brca1−/− p53−/− cultures increases to approach that of the p53−/− fibroblasts. The rate of transition through the cell cycle increases in the Brca1−/− p53−/− fibroblasts; however, this is also true of the p53−/− fibroblasts maintained in culture over a similar length of time. We believe that it is primarily the decreased rate of cellular death of the Brca1−/− p53−/− fibroblasts that leads to a decrease in the difference in growth rates observed on comparison of the later passage Brca1−/− p53−/− and p53−/− fibroblasts.
Mechanisms for survival of BRCA1-deficient, p53-deficient mice.
We and others have reported that loss of normal BRCA1 expression results in death early in mouse embryogenesis and that BRCA1-deficient embryos are severely growth retarded (21, 32, 33, 47). Heterogeneity has been observed in the embryonic stage at which death occurs, both within a mouse line and between lines carrying different mutations. The embryos homozygous for the Brca1Δ223–763 mutation (the mutation carried by the animals described in this paper) on average survived the longest of all of the Brca1−/− embryos described to date. While in general embryos homozygous for other Brca1 mutations do not survive past embryonic day 8, the majority of the Brca1−/− embryos in our colony survive to embryonic day 9 or 10. There are a number of possible reasons for these findings. The Brca1−/− embryos generated in our mouse colony were maintained on a mixed genetic background consisting of 129, C57BL/6, and DBA/2. In contrast, other BRCA1-deficient embryos examined were on a mixed genetic background consisting of only 129 and C57BL/6. It is possible that the additional vigor of the more heterogeneous background, or a specific modifier gene(s) present in the DBA/2 strain, contributed to this extended survival. Interestingly, mice homozygous for the Brca2 truncation mutation were reported to survive only when on a mixed genetic background consisting of 129, C57BL/6, and DBA/2 (7, 16). No adult animals were obtained on the 129, C57BL/6 background (33, 45, 49).
It is also possible that the difference in the survival of the various BRCA1-deficient lines is related to the mutation carried by these animals. A neomycin resistance gene replaces the 3′ portion of intron 10 and the 5′ portion of exon 11 of the Brca1 gene in our BRCA1-deficient mouse line. Analysis of RNA from cell lines heterozygous and homozygous for the mutant allele shows that a Brca1 mRNA, 4 kb smaller than the wild-type message, is transcribed from the mutant allele. Sequence analysis of this message indicates that it is a splice variant deficient in all exon 11-derived sequences (9). Exon 11 encodes the two nuclear localization motifs and a region of the gene believed to be essential for binding with RAD51 (5, 44, 50). In multiple cases, mutations resulting in deficiencies only in exon 11 are associated with increased risk for mammary tumorigenesis in BRCA1+/− patients (8, 20). In addition, this mutation has recently been shown to represent a null allele for at least some functions of BRCA1, since TCR is undetected in the ES cell line homozygous for this mutation (19). It is possible, however, that this splice variant can produce a protein with some activity and that this activity extends the survival of the embryos.
The survival of BRCA1-deficient embryos on a p53 null background has been examined by introduction of a p53 mutation into two different BRCA1-deficient lines (22, 33). In both cases, a modest extension of the survival of the embryos was reported, although no embryos survived beyond 14 days. Similarly, p53 deficiency has not resulted, in general, in the survival of embryos homozygous for the Brca1Δ223–763 mutation (9). Therefore, while it is likely that lack of p53 expression contributed to the survival to adulthood of the mice reported here, it is also apparent that genetic factors in addition to loss of p53 are required for the survival of mice without normal BRCA1 function. The close relationship of the two initial Brca1−/− animals identified and the derivation of the third Brca1−/− animal from mice related to these two initial survivors suggest that this survival is dependent on additional genetic factors. The increased survival of Brca2−/− mice carrying DBA/2 loci would suggest that alleles specific to the DBA/2 strain may not only be responsible for the extended survival of our Brca1−/− embryos (7, 16) but also play a role in the survival of the three Brca1−/− p53−/− mice described here. However, until larger numbers of surviving mice are identified and the relationship between these animals is examined, it will not be possible to determine whether alleles of a single gene or a number of modifier loci confer this survival advantage. While the number of BRCA1-deficient mice is small, the phenotype observed in these animals and the fibroblasts isolated from these animals was never seen on examination of siblings or related mice in this colony. Therefore, while the phenotype is undoubtedly modified by the p53-null background and the presence of a modifier gene, it is most likely dependent on homozygosity for the mutant Brca1 allele. However, we cannot rule out the possibility that the Brca1−/− p53−/− mice independently acquired spontaneous somatic mutations that enabled embryonic survival and that these mutations contribute in part to the observed phenotype.
Characterization of BRCA1-deficient, p53-deficient mice.
BRCA1 expression can be detected at high levels in the adult testes, and in situ analysis indicates that expression is highest in spermatocytes, in particular those in the late pachytene and diplotene stages of the first meiotic prophase (2, 27, 35, 53). In contrast, Sertoli cells and Leydig interstitial cells do not express BRCA1. Immunohistological studies have further localized BRCA1 within the meiotic cells (44). Extensive morphological changes accompany the pairing or synapsis of homologous chromosomes during the first meiotic prophase. A proteinaceous axis forms between sister chromatids and, as pairing continues, the axes of homologues adhere to each other, thereby initiating the formation of the synaptonemal complex. Lateral elements and additional proteins associate with the paired chromosomal region, completing the formation of a synaptonemal complex. BRCA1 is associated with both unsynapsed axial elements and the axes in the process of synapsing. BRCA1 was also detected on unsynapsed centromeric heterochromatin, on remaining univalent chromosomes, and at pairing forks. The identification of cells which express HSP70-2 (a 70-kDa heat shock protein) indicates that spermatogonia develop into spermatocytes and initiate meiosis in the double-homozygous animals. However, the virtual absence of pachytene spermatocytes in the BRCA1-deficient males suggests that the association of BRCA1 with the prophase chromosomes is essential for normal completion of this process.
The comparable expression pattern of BRCA1 and BRCA2, the association of loss of both genes with mammary tumors, and the related structure of the two genes suggest that these proteins carry out similar functions. Interestingly, however, the effect of the loss of each gene on gamete formation is very different. In contrast to BRCA1-deficient mice, no germ cells were detected in mice homozygous for a Brca2 truncation mutation (7). However, we cannot rule out the possibility that the expressed Brca1 mRNA from the mutant allele can confer functions of BRCA1 in these mitotic cells but cannot carry out the function of BRCA1 during meiosis. Alternatively, it is possible that the absence of p53 contributes to the survival of the mitotic spermatogonia but cannot rescue the meiotic cells deficient in BRCA1.
Although BRCA1 expression has been demonstrated in the rapidly dividing thecal cells of the ovary, expression during the formation of female gametes has not been established (2, 41). Histological examination of the female BRCA1-deficient animal revealed the presence of both primary and growing follicles. In normal mice, growing follicles have completed the pachytene stage of prophase of meiosis I, the stage of highest expression of BRCA1 in spermatogenesis (25, 53). The presence of these follicles in the Brca1−/− p53−/− female suggests that BRCA1 is not required for completion of these stages of meiosis I.
Despite the growth retardation of the Brca1−/− p53−/− mice, with the exception of the testes, mammary gland, and skin, the organ systems of the BRCA1-deficient mice appeared in general to develop normally. In the female mouse, the branching of the mammary epithelium was reduced and the end buds were smaller. Histological analysis showed individual cells arrayed around the duct. One interpretation of these observations is that these cells represent epithelial cells that fail to differentiate into normal gland structures and instead migrate away from the developing end bud into the surrounding fat pad. The observed alterations in the development of the mammary gland are of particular interest given that both in vivo and in vitro loss of BRCA1 function are associated primarily with mammary and ovarian tumors in humans. These observations will require confirmation by analysis of additional animals or by the generation of animals in which the loss of BRCA1 function is limited to mammary epithelium. However, they do suggest that the impact of the loss of BRCA1 on growth and differentiation is more severe than that seen in other cell lineages. While these changes were not observed in the analysis of a number of wild-type and p53-deficient animals, we cannot rule out the possibility that the developmental change in the mammary gland of the Brca1−/− p53−/− female is in whole or in part dependent on the loss of both BRCA1 and p53. Cytomegaly and karyomegaly were noted in the parotid gland of all of the double-homozygous animals and in the prostate glands of the male animals. This is of particular interest in light of evidence suggesting that BRCA1 is associated with centrosomes and may be important in segregation of the chromosome during mitosis (13).
BRCA1 in DNA damage repair.
Consistent with the observed growth retardation of Brca1−/− p53−/− mice and the phenotype of BRCA1-deficient embryos, primary cell cultures obtained from the Brca1−/− p53−/− mice grow more slowly than those obtained from p53−/− animals. The overall growth rate of a cell culture is determined by both the rate of cellular division and the rate of cellular death. Differences in the growth of the p53−/− and Brca1−/− p53−/− cultures appear to stem largely from an increased frequency of cellular death in the absence of normal BRCA1 expression. If, as has been proposed, BRCA1 is required for DNA repair, this increased rate of cellular death may result from the rapid accumulation of chromosomal abnormalities and other mutations incompatible with the survival of normal cells. A similar mechanism may underlie the growth retardation of BRCA1-deficient embryos. Variability in the stage to which BRCA1-deficient embryos develop, even when carrying identical mutations, may reflect the fact that environmental factors influence the rate at which mutations accumulate, with embryos surviving longer if a number of cell divisions occur before accumulation of mutations leads to cell death or senescence.
This interpretation is supported by our observation that the growth of the primary BRCA1-deficient cells is compromised to a greater extent than that of the p53-deficient control cells by exposure to UV light, gamma irradiation, and H2O2. Brca1−/− ES cells have also been reported to display increased sensitivity to gamma irradiation and H2O2 in comparison to the parental Brca1+/+ lines (19). However, unlike the Brca1−/− p53−/− fibroblasts, no difference in the response of the Brca1−/− ES cells to UV radiation was observed. This may reflect differences in the dependence of various cell lineages on a particular DNA repair pathway after exposure to UV light. The loss of one repair pathway due to BRCA1 deficiency may result in cellular death in fibroblasts but may be compensated adequately by another repair pathway in ES cells. It is also possible that the contribution of BRCA1 in the response to UV damage is detected only when p53-dependent repair pathways are disabled.
The Brca1−/− ES cell line was shown to be deficient in TCR. However, this interpretation was dependent on results obtained from a single ES cell line isolate. Attempts to isolate similar lines have failed, raising the possibility that the cell line carries other mutations that have accumulated, allowing it to survive without normal BRCA1 function, and that these alterations, not the loss of BRCA1, lead to the altered TCR. A role for BRCA1 in TCR is confirmed here by the demonstration that Brca1−/− p53−/− primary fibroblasts are also deficient in TCR. Again, this deficiency was observed after exposure to H2O2 and gamma irradiation but not to UV light. In contrast to the Brca1−/− ES cell line, the growth sensitivity of the Brca1−/− p53−/− fibroblasts to DNA-damaging agents did not correlate directly with TCR deficiency. This suggests that defects in TCR repair may not account for all of the growth deficiency observed in BRCA1-deficient cells. The fact that TCR is unlikely to be the only function of BRCA1 is supported by the comparison of the phenotype of mice homozygous for a mutant allele of the CSB gene, which results in a loss of TCR (51). These mice survive, appear normal, and are fertile. Our results, therefore, support the hypothesis that BRCA1 is a multifunctional protein either as a result of the presence of multiple functional domains or due to the ability of BRCA1 to activate a number of different cellular pathways.
As discussed above, loss of p53 extends the survival of the embryos carrying other Brca1 mutations and is likely to play a role together with other as-yet-unidentified genetic factors in the survival to adulthood of the mice described here. p53 is involved in multiple cellular pathways, and we cannot at present determine whether the inactivation of one specific pathway or a number of these pathways contributes to survival of the BRCA1-deficient embryos and cells (31). For example, it is possible that in the absence of p53-mediated apoptosis, BRCA1-deficient cells that have accumulated DNA damage survive. However, our data, as well as the observations made on examination of histological sections from BRCA1-deficient embryos stained for apoptotic cells by TUNEL, do not support this hypothesis. In Brca1−/− embryos, in which the p53-mediated apoptotic pathway is intact, no increase in the number of apoptotic cells was observed (21, 32). Furthermore, the high percentage of dead cells in primary Brca1−/− p53−/− cultures indicates that p53-independent pathways can lead to the death of BRCA1-deficient cells.
p53 has also been implicated in numerous cell cycle checkpoints, and it is possible that it is the disruption of these pathways that contributes to the survival of the Brca1−/− p53−/− animals (31). It is possible that, in normal cells, accumulation of genetic alterations as a result of loss of BRCA1 leads to growth arrest via p53-dependent pathways. Analysis of growth-arrested BRCA1-deficient embryos indicated increased expression of p21 (22). This and the finding that p21 deficiency also extends the survival of the BRCA1-deficient embryos support the hypothesis that the loss of the p53-dependent G1/S checkpoint contributes to the extended survival of the BRCA1-deficient cells (21). p53 has also been implicated in other cell cycle checkpoints, including the spindle checkpoint (11). Deficiency in the spindle checkpoint contributes to the increased chromosomal aberrations in cells lacking normal p53 expression. It is not possible to discern at this time whether deficiencies in this and other checkpoints also contribute to the survival of the Brca1−/− p53−/− mice.
Contribution of mutations in BRCA1 to tumorigenesis.
The studies reported here indicate that loss of p53, in combination with other genetic modifiers, can extend the survival of BRCA1-deficient mice and cells. Even then, BRCA1-deficient cells grow slowly, making it difficult to reconcile the BRCA1-deficient phenotype with the observation of increased tumor risk of individuals heterozygous for mutant BRCA1 alleles. Examination of the growth of the primary BRCA1-deficient cells after extended passage in cell culture provides a possible explanation: once a number of mutational events have occurred that allow survival of BRCA1-deficient cells, the loss of BRCA1 may accelerate the accumulation of additional mutations necessary for malignant transformation. Thus, after extended passage in tissue culture, clones of Brca1−/− p53−/− cells can be identified that have an increased percentage of cells in the S phase. The percentage of dead cells in these cultures also decreases, resulting in a growth rate that equals that of the p53-deficient primary fibroblasts.
Together, our analysis of the Brca1−/− p53−/− mice and fibroblasts supports the following models for the role of BRCA1 in tumorigenesis (Fig. 8). In the first model the loss of the wild-type BRCA1 allele is not the first event in tumor progression. Cells must first accumulate mutations in multiple genes, such as p53, that confer a growth advantage. Only after having acquired an increased growth potential will loss of BRCA1 function be compatible with cell survival. With loss of wild-type BRCA1 function, the resulting cell populations display growth rates that approach those of normal cells. However, if as suggested by our results BRCA1 plays a role in multiple DNA repair pathways, loss of BRCA1 function would be expected to accelerate the accumulation of additional mutations leading to malignant transformation. This model is supported by the recent demonstration that virtually all BRCA1−/− tumors examined to date have lost p53 function (10). In addition, we have identified mammary tumors in Brca1+/− p53+/− mice after exposure to irradiation (9). A number of these tumors have lost the wild-type p53 and Brca1 alleles.
FIG. 8.
Models for the role of mutations in BRCA1 in tumorigenesis. Experiments described in this study suggest that the loss of BRCA1 results in deficits in cellular growth. However, loss of BRCA1 is consistently seen in rapidly proliferating tumors from patients carrying a germ line mutation in BRCA1. Two models attempt to resolve these two conflicting concepts. In model 1, mutations in genes resulting in a growth advantage are necessary for the survival of cells which have lost BRCA1 function. The loss of BRCA1, as well as the growth-promoting mutations, results in an increased mutation rate, thus accelerating tumorigenesis. Therefore, in order for loss of BRCA1 to promote tumorigenesis, other growth promoting mutations must first occur. In model 2, cellular growth or death after the loss of BRCA1 function is dependent on the ability of the cell type to compensate by utilizing trophic factors. This model accounts for the specificity of tumor types resulting from the loss of BRCA1 in humans.
In a second model, the impact of loss of BRCA1 on cellular survival depends on the cell type, the growth state of the cell, and the presence of trophic factors capable of overriding signals that would, under other growth conditions, lead to the death of cells with damaged DNA. For example, it is possible that during the growth of the mammary epithelia, high levels of hormones and growth factors can take the place of growth-enhancing mutations such as loss of p53, allowing BRCA1 heterozygous cells that lose the wild-type allele to survive. This differential sensitivity of cells to loss of BRCA1 would lead to the observed tumor types associated with inheritance of BRCA1 mutations. This model is not supported by our observation that the mammary epithelium of the Brca1−/− p53−/− mice was underdeveloped, suggesting that mammary epithelia may in fact have increased sensitivity to loss of BRCA1 function. It is possible, however, that this dependency on BRCA1 function reflects an underlying difference between mice and humans in the growth and responsiveness of mammary epithelium to hormones and that this characteristic contributes to the differential effect that heterozygosity of BRCA1 has on mammary tumorigenesis in the two species. While further studies will be required to test these and additional models, the results presented here suggest that in a number of normal cell populations, the loss of BRCA1 results in a growth disadvantage. Only in combination with other mutations, under specific growth conditions or in specific populations of cells, can a loss of BRCA1 contribute to tumorigenesis.
ACKNOWLEDGMENTS
We thank R. Bagnell and V. Madden for assistance with microscopy; E. M. Eddy for generously providing the HSP70-2 antibody; D. O’Brien for examination of tissue sections and helpful discussions on spermatogenesis; K. Burns and T. Bartolotta for assistance with histology; L. Arnold and B. Nostrom for assistance with flow cytometry; V. Allen and J. Morris for assistance with animal husbandry; and J. Snouwaert, A. Pace, and M. Hawley for helpful comments on the manuscript.
This work was supported by NIH grants CA70490 and IP50CA58223 (to B.H.K.) and CA40453 (to S.A.L.) and the Department of Defense USAMRMC grant DAMD17-97-1-7102 (to V.L.C.).
REFERENCES
- 1.Akashi M, Hachiya M, Osawa W, Spirin K, Suzuki G, Koeffler H. Irradiation induces WAF1 expression through a p53-independent pathway in KG-1 cells. J Biol Chem. 1995;270:19181–19187. doi: 10.1074/jbc.270.32.19181. [DOI] [PubMed] [Google Scholar]
- 2.Blackshear P E, Goldsworthy S M, Foley J F, McAllister K A, Bennett L M, Collins N K, Bunch D O, Brown P, Wiseman R, Davis B J. Brca1 and Brca2 expression patterns in mitotic and meiotic cells of mice. Oncogene. 1998;16:61–68. doi: 10.1038/sj.onc.1201506. [DOI] [PubMed] [Google Scholar]
- 3.Callebaut I, Mornon J-P. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 4.Chapman M S, Verma I M. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-F, Li S, Chen Y, Chen P-L, Sharp Z D, Lee W-H. The nuclear localization sequences of the BRCA1 protein interact with the importin-α subunit of the nuclear transport signal receptor. Proc Natl Acad Sci USA. 1996;271:32863–32868. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Farmer A A, Chen C-F, Jones S C, Chen P-L, Lee W-H. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- 7.Connor F, Bertwistle D, Mee P J, Ross G M, Swift S, Grigorieva E, Tybulewicz V L J, Ashworth A. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 8.Couch F J, Weber B L. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- 9.Cressman V L, Backlund D C, Hicks E M, Gowen L C, Godfrey V, Koller B H. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 1999;10:1–10. [PubMed] [Google Scholar]
- 10.Crook T, Crossland S, Crompton M R, Osin P, Gusterson B A. p53 mutations in BRCA1-associated familial breast cancer. Lancet. 1997;350:638–639. doi: 10.1016/S0140-6736(05)63327-2. [DOI] [PubMed] [Google Scholar]
- 11.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 12.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 13.Dix D J, Allen J W, Collins B W, Mori C, Nakamura N, Poorman-Allen P, Goulding E H, Eddy E M. Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc Natl Acad Sci USA. 1996;93:3264–3268. doi: 10.1073/pnas.93.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 15.Easton D F, Bishop D T, Ford D, Crockford C P the Breast Cancer Linkage Consortium. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman L S, Thistlethwaite F C, Pael K J, Yu V P C C, Lee H, Venkitarman A R, Abel K J, Carlton M B L, Hunter S M, Colledge W H, Evans M J, Ponder B A J. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 1998;58:1338–1343. [PubMed] [Google Scholar]
- 17.Game J C. Radiation-sensitive mutants and repair in yeast. In: Spencer D H, Smith A R W, editors. Yeast genetics: fundamental and applied aspects. New York, N.Y: Springer-Verlag; 1983. pp. 109–137. [Google Scholar]
- 18.Gowen L, Johnson B L, Latour A M, Sulik K K, Koller B H. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 19.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;2841:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 20.Grade K, Jandrig B, Scherneck S. BRCA1 mutation update and analysis. J Cancer Res Clin Oncol. 1996;122:702–706. doi: 10.1007/BF01209036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakem R, de la Pompa J L, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmar A, Billia F, Firpo E, Hui C C, Roberts J, Rossant J, Mak T W. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 22.Hakem R, Pompa J L, Elia A, Potter J, Mak T W. Partial rescue of Brca15-6 early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 23.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 24.Harvey M, Sands A, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 25.Hogan B, Costantini F, Lacy E. Summary of mouse development. In: Hogan B, Costantini F, Lacy E, editors. Manipulating the mouse embryo. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. pp. 17–79. [Google Scholar]
- 26.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 27.Lane T F, Deng C, Elson A, Lyu M S, Kozak C A, Leer P. Expression of Brca1 is associated with terminal differentiation of ectodermally and mesodermally derived tissues in mice. Genes Dev. 1995;9:2712–2722. doi: 10.1101/gad.9.21.2712. [DOI] [PubMed] [Google Scholar]
- 28.Leadon S A, Cooper P K. Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leadon S A, Lawrence D A. Preferential repair of DNA damage on the transcribed strand of the human metallothionein genes requires RNA polymerase II. Mutat Res. 1991;255:67–78. doi: 10.1016/0921-8777(91)90019-l. [DOI] [PubMed] [Google Scholar]
- 30.Leadon S A, Lawrence D A. Strand-selective repair of DNA damage in the yeast GAL7 gene requires RNA polymerase II. J Biol Chem. 1992;267:23175–23182. [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Liu C-Y, Flesken-Nikitin A, Li S, Yeng Y, Lee W-H. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 34.Marquis S T, Rajan J V, Wynshaw-Boris A, Xu J, Yin G-Y, Abel K J, Weber B L, Chodosh L A. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat Genet. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]
- 35.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennet L M, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow P K, Norris F H, Helvering L, Morrison P, Rosteck P, Lai M, Barrett J C, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick M H. A strong candidate for the breast camcer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 36.Monteiro A N, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 37a.National Institute of Diabetes and Digestive and Kidney Diseases. Biology of the mammary gland [Online.] htt://mammary.nih.gov. [6 August 1999, last date accessed.]
- 38.Ouchi T, Monteiro A N, August A, Aaronson S A, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker S, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olsen E, Harper J W, Elledge S J. p53-independent expression of p21 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 40.Patel K J, Yu V P C C, Lee H, Corcoran A, Evans M J, Colledge W H, Friedman L S, Ponder B A J, Venkitarman A R. Involvement of Brca2 in DNA Repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 41.Phillips K W, Goldsworthy S M, Bennet L M, Brownlee H A, Wiseman R, Davis B J. Brca1 is expressed independently of hormonal stimulation in the mouse ovary. Lab Investig. 1997;76:419–425. [PubMed] [Google Scholar]
- 42.Ruffner H, Verma I M. BRCA1 is a cell cycle-regulated nuclear phosphoprotein. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Dynamic changes of BRCA1 subnuclear localization and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 44.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 45.Sharan S K, Morimatsu M, Albrecht U, Lim D-S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 46.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 47.Snouwaert J N, Gowen L C, Lee V, Koller B H. Characterization of Brca1 deficient mice. Breast Dis. 1998;10:33–44. doi: 10.3233/bd-1998-101-206. [DOI] [PubMed] [Google Scholar]
- 48.Somasundaram K, Zhang H, Zeng Y-X, Houvras Y, Peng Y, Wu G S, Licht J D, Weber B L, El-Deiry W S. Arrest of the cell cycle by the tumor-suppressor BRCA1 requires the CDK-inhibitor p21waf1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, Koo W, Billia P, Ho A, Fukumoto M, Hui C C, Mak T W. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 50.Thakur S, Zhang H B, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid L M, Couch F J, Wilson R B, Weber B L. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol Cell Biol. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Horst G T J, van Steeg H, Berg R J W, van Gool A J, de Wit J, Weeda G, Morreau H, Beems R B, van Kreijl C F, Gruijl F R, Bootsma D, Hoeijmakers J H J. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 52.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M-C W, Hwang L-Y, Bowcock A M, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 53.Zabludoff S, Wright W W, Harshman K W B J. BRCA1 mRNA is expressed highly during meiosis and spermiogenesis but not during mitosis of male germ cells. Oncogene. 1996;13:649–653. [PubMed] [Google Scholar]
- 54.Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber B L, El-Deiry W S. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W, Grasso L, McClain C D, Gambel A M, Cha Y, Travali S, Deisseroth A B, Mercer W E. p53-independent induction of WAF1/CIP1 in human leukemia cells is correlated with growth arrest accompanying monocytes/macrophage differentiation. Cancer Res. 1995;55:668–674. [PubMed] [Google Scholar]