Abstract
Purpose
Despite decreased prevalence of tuberculosis, the incidence of the diseases associated with nontuberculous mycobacteria (NTM) has been increasing in South Korea and around the world. The present retrospective study was conducted to determine longitudinal changes in the epidemiology and distribution of NTM over 13 years at a tertiary care hospital in Korea.
Materials and Methods
We retrospectively analyzed data on Mycobacterium species over 13 years (January 2007 to December 2019) by utilizing the laboratory information system. Mycobacterium species were identified using biochemical tests and PCR-restriction fragment length polymorphism and Mycobacteria GenoBlot assays.
Results
After excluding duplicates from the initial pool of 17996 mycobacterial isolates, 7674 strains were analyzed and 2984 (38.9%) NTM were isolated. The proportion of NTM continuously increased over the 13-year period, from 17.0% in 2007 to 57.5% in 2019. Among the NTM isolates, the most common species were Mycobacterium intracellulare (50.6%), M. avium (18.3%), M. fortuitumcomplex (4.9%), M. abscessus (4.5%), M. gordonae (3.3%), M. kansasii (1.1%), M. chelonae (1.0%), and M. massiliense (0.9%). In patients over the age of 70 years, the proportion of NTM among the isolates increased from 26.6% in 2007 to 62.0% in 2019, and that of M. intracellulare isolates among the NTM increased from 13.9% (11/79) in 2007 to 37.4% (175/468) in 2019.
Conclusion
The number of NTM isolates continuously increased over the study period, and the increase in the proportion of M.intracellulare in patients aged over 70 years was notable.
Keywords: Nontuberculous mycobacteria, Mycobacterium intracellulare, Mycobacterium avium, epidemiology, retrospective studies
INTRODUCTION
Although the prevalence of tuberculosis (TB) has been decreasing,1 the incidence of nontuberculous mycobacteria (NTM) diseases is increasing both worldwide and in South Korea.2,3 As NTM can be found in soil, dust, and water, including natural water sources such as lakes, rivers, and streams, as well as in municipal water sources,4 isolated NTM were initially considered to be merely contaminants and non-pathogenic. However, the incidence of diseases caused by NTM has increased, along with an increase in the number of case reports and case series from diverse countries and regions, showing pronounced differences in the distribution of NTM clinical isolates in different regions and countries.5
The exact cause of the observed increase in NTM diseases is not well-understood; however, increased elderly population, decreased immune function, and exposure to environmental mycobacteria have been reported to be possible causative factors.3,6 Since immune function becomes compromised with age, the incidence of NTM infections is higher in elderly people than in younger populations.7 Immunocompromised individuals are also at high risk for NTM infections. The vast majority (95%) of Acquired Immune Deficiency Syndrome (AIDS)-related Mycobacterium avium complex (MAC) infections are due to M. avium, not M. intracellulare, and M. avium infections typically occur when the CD4 cell count is below 0.05×109/L.8,9,10 Therefore, compromised cellular immunity is likely to be a risk factor for infections due to M. avium but not due to M. intracellulare.10 Although both M. avium and M. intracellulare are MAC species and aquatic organisms, their niches are different: M. avium tends to grow in aqueous suspensions, whereas M. intracellulare forms biofilms.11,12 Han, et al.10 showed that in non-AIDS patients, M. intracellulare is more pathogenic, and M. intracellulare infections in women tends to increase with age beyond menopause (>50 years), regardless of any underlying disease. In addition, their study revealed a high prevalence of M. intracellulare in men with lung cancer, implying that prior lung injury caused by cancer risk factors, such as smoking, predisposes lung tissues to attachment by the microbe, subsequently leading to colonization and infection.
With this background in mind, our retrospective study was performed to determine longitudinal changes in the epidemiology and distribution of NTM species over 13 years at a tertiary care hospital located in the southwestern region of Gangwon-do in South Korea.
MATERIALS AND METHODS
Data collection
From January 2007 to December 2019, relevant data were collected through the laboratory information system at Wonju Severance Christian Hospital, and all data were decoded automatically. Collected specimens were classified as pulmonary or extrapulmonary. If the same mycobacterial species was identified in several lower-respiratory specimens [i.e., sputum and bronchoalveolar lavage (BAL)] from the same patient during follow-up, only one of the initially isolated mycobacterial strains was included in the analyzed set of isolates.
Acid-fast bacillus staining and culturing for Mycobacterium identification
Specimens, with the exception of body fluids, were decontaminated using the N-acetyl-L-cysteine (NALC)-2% sodium hydroxide (NaOH) method. Decontaminated specimens were stained with auramine-rhodamine, and the results were confirmed by Ziehl-Neelsen staining. The presence of acid-fast bacilli (AFB) in a specimen was defined as follows: trace, 1–2 AFB per 300× field; 1+, 1–9 AFB per 100× field; 2+, 1–9 AFB per 10× field; 3+, 1–9 AFB per 1× field; and 4+, more than 9 AFB per 1× field. The NALC-NaOH-pretreated specimens were then inoculated onto Ogawa solid medium (Shinyang Chemical, Seoul, South Korea). Beginning on September 1, 2008, liquid medium was also used [mycobacteria growth indicator tubes (MGITs); Becton Dickinson Diagnostic Systems, Sparks, MD, USA]. After June 2012, Ogawa medium (Union Lab, Seoul, South Korea), sourced from the Korean Institute of Tuberculosis (Osong, South Korea), was used. Inoculum maintained in Ogawa medium was incubated at 37℃ and 5% CO2 for 8 weeks, and inoculum maintained in MGITs was incubated in the BACTEC MGIT 960 system (BD Diagnostic Systems, Sparks, MD, USA) for 6 weeks. A 1 mL aliquot from each MGIT culture was tested, and those that tested positive were subjected to AFB staining for verification. Contaminated cultures were excluded.
Molecular identification of Mycobacterium
DNA purification protocols were medium-type specific. Samples from cultures maintained in Ogawa medium were obtained using an inoculation loop and suspended in 200 µL sterilized distilled water in a 1.5 mL microtube. This mixture was centrifuged at 13000×g for 3 min, and the supernatant was removed. In contrast, for cultures maintained in MGIT tubes, 1 mL sample was removed, placed in a 1.5 mL microtube, and centrifuged at 13000×g for 3 min. The supernatant was removed, and a 1 mL aliquot of distilled water was added to the pellet, vortexed thoroughly, and centrifuged again at 13000×g for 3 min. For both sample types, after the supernatant was removed, 100 µL extraction buffer was added, and the sample was heated at 100℃ for 20 min and centrifuged at 13000×g for 3 min.
For mycobacterial identification, extracted DNA was analyzed using Myco-ID (M&D Inc., Wonju, South Korea), which is based on restriction fragment length polymorphism.13 However, for samples collected after December 2, 2014, the AdvanSure Mycobacteria GenoBlot Assay (LG Chem, Cheongju, South Korea) was used for mycobacterial identification. This assay can classify and identify Mycobacterium tuberculosis complex (MTBC) species, as well as 20 different NTM species. The reverse line blot hybridization assay was processed using an AdvanSure hybridizer (LG Chem), and patterns in the line strips were subjected to automated analysis using AdvanSure GenoLine Scan (LG Chem).14 To distinguish M. abscessus and M. massiliense, the presence of the erythromycin ribosome methyltransferase (erm) gene was confirmed using the ERM-plus real-time PCR kit (LG Chem; not a commercial product). MAC was defined as a group of mycobacteria comprising M. avium, M. intracellulare, and M. avium-intracellulare.
Statistical analysis
The statistical significance of the recovery rates of MTBC and NTM according to sex and age was analyzed using chi-square and Fisher's exact tests. All probability values were two-sided, and p values less than 0.05 were considered statistically significant. Statistical analyses were performed using IBM SPSS Statistics v. 25.0 (IBM Corp., Armonk, NY, USA).
Ethical approval
In accordance with government regulations, the need for ethical review by the Institutional Review Board (IRB) was waived, and the exemption was approved by the IRB of Wonju Severance Christian Hospital (approval no. CR320013).
RESULTS
After excluding duplicates, a final set of 7674 mycobacterial isolates (from a total of 17996) was identified. The most common mycobacterial species were MTBC (61.1%), M. intracellulare (19.7%), M. avium (7.1%), M. fortuitum complex (1.9%), M. abscessus (1.8%), M. gordonae (1.3%), M. kansasii (0.4%), M. chelonae (0.4%), and M. massiliense (0.3%). MAC accounted for 69.3% of the NTM isolates (Table 1).
Table 1. Number and Proportion (%) of Mycobacterial Isolates.
| Mycobacterium species | Isolates (n=7674) | Isolates/total | Isolates among NTM | |
|---|---|---|---|---|
| Mycobacterium tuberculosis complex | 4690 | 61.1 | ||
| Mycobacterium avium complex* | 2067 | 26.9 | 69.3 | |
| Mycobacterium avium * | 546 | 7.1 | 18.3 | |
| Mycobacterium intracellulare * | 1509 | 19.7 | 50.6 | |
| Mycobacterium avium-intracellulare | 12 | 0.2 | 0.4 | |
| Mycobacterium fortuitum complex* | 145 | 1.9 | 4.9 | |
| Mycobacterium abscessus * | 135 | 1.8 | 4.5 | |
| Mycobacterium gordonae * | 99 | 1.3 | 3.3 | |
| Mycobacterium kansasii * | 33 | 0.4 | 1.1 | |
| Mycobacterium chelonae * | 29 | 0.4 | 1.0 | |
| Mycobacterium massiliense | 26 | 0.3 | 0.9 | |
| Mycobacterium terrae complex* | 24 | 0.3 | 0.8 | |
| Mycobacterium lentiflavum/M. genavense* | 18 | 0.2 | 0.6 | |
| Mycobacterium szulgai * | 5 | 0.1 | 0.2 | |
| Mycobacterium flavescens * | 4 | 0.1 | 0.1 | |
| Mycobacterium celatum * | 4 | 0.1 | 0.1 | |
| Mycobacterium mucogenicum | 4 | 0.1 | 0.1 | |
| Mycobacterium scrofulaceum * | 4 | 0.1 | 0.1 | |
| Mycobacterium nonchromogenicum | 4 | 0.1 | 0.1 | |
| Mycobacterium aubagnense | 3 | 0.04 | 0.1 | |
| Mycobacterium phocaicum | 3 | 0.04 | 0.1 | |
| Mycobacterium mucilaginosus | 2 | 0.03 | 0.1 | |
| Mycobacterium malmoense * | 1 | 0.01 | 0.03 | |
| Mycobacterium austroafricanum | 1 | 0.01 | 0.03 | |
| Mycobacterium gastri * | 1 | 0.01 | 0.03 | |
| Mycobacterium phlei | 1 | 0.01 | 0.03 | |
| Mycobacterium vaccae * | 1 | 0.01 | 0.03 | |
| Mycobacterium species | 370 | 4.8 | 12.4 | |
NTM, nontuberculous mycobacteria.
Data are presented as n and %.
*Species that could be identified using the AdvanSure Mycobacteria GenoBlot Assay (LG Chem.).
The frequency of isolation of Mycobacterium species according to specimen type was analyzed. M. gordonae (98.0%), M. massiliense (96.2%), and M. kansasii (93.9%) were mostly found in the sputum/BAL. MTBC (3.9%) and M. chelonae (3.4%) were commonly detected in the pleural fluid. M. chelonae (6.9%), M. fortuitum complex (6.2%), M. abscessus (3.7%), and MTBC (2.4%) were frequently observed in wounds. The isolation frequency of each species according to specimen type is shown in Table 2.
Table 2. Number and Proportion (%) of Mycobacterium Isolates according to Specimen Type.
| Mycobacterium species | Pulmonary system | Extrapulmonary | Other† n=89 (1.2) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sputum/BAL n=6873 (89.6) |
Pleural F n=240 (3.1) |
Wound* n=186 (2.4) |
Catheter n=118 (1.5) |
Urine n=54 (0.7) |
Peritoneal F n=44 (0.6) |
CSF n=37 (0.5) |
Synovial F n=18 (0.2) |
Pericardial F n=15 (0.2) |
|||
| M. tuberculosis complex | 4124 (87.9) | 182 (3.9) | 111 (2.4) | 86 (1.8) | 43 (0.9) | 22 (0.5) | 31 (0.7) | 8 (0.2) | 13 (0.3) | 70 (1.5) | |
| M. avium complex | 1920 (92.9) | 34 (1.6) | 41 (2.0) | 26 (1.3) | 9 (0.4) | 14 (0.7) | 5 (0.2) | 6 (0.3) | 1 (0) | 11 (0.5) | |
| M. avium | 537 (98.4) | 2 (0.4) | 4 (0.7) | 1 (0.2) | 0 (0) | 1 (0.2) | 0 (0) | 1 (0.2) | 0 (0) | 0 (0) | |
| M. intracellulare | 1372 (90.9) | 31 (2.1) | 37 (2.5) | 25 (1.7) | 9 (0.6) | 13 (0.9) | 5 (0.3) | 5 (0.3) | 1 (0.1) | 11 (0.7) | |
| M. avium-intracellulare | 11 (91.7) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. fortuitum complex | 131 (90.3) | 0 (0) | 9 (6.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.7) | 0 (0) | 4 (2.8) | |
| M. abscessus | 126 (93.3) | 2 (1.5) | 5 (3.7) | 0 (0) | 0 (0) | 1 (0.7) | 0 (0) | 1 (0.7) | 0 (0) | 0 (0) | |
| M. gordonae | 97 (98.0) | 0 (0) | 1 (1.0) | 1 (1.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. kansasii | 31 (93.9) | 0 (0) | 1 (3.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3.0) | 0 (0) | 0 (0) | |
| M. chelonae | 26 (89.7) | 1 (3.4) | 2 (6.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. massiliense | 25 (96.2) | 0 (0) | 1 (3.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. terrae complex | 21 (87.5) | 0 (0) | 3 (12.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. lentiflavum/M. genavense | 18 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. szulgai | 5 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. flavescens | 0 (0) | 1 (25.0) | 0 (0) | 2 (50.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (25.0) | |
| M. celatum | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. mucogenicum | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. scrofulaceum | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. nonchromogenicum | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. aubagnense | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. phocaicum | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. mucilaginosus | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. malmoense | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | |
| M. austroafricanum | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. gastri | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. phlei | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. vaccae | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| M. species | 322 (87.0) | 20 (5.4) | 11 (3.0) | 3 (0.8) | 2 (0.5) | 7 (1.9) | 1 (0.3) | 1 (0.3) | 1 (0.3) | 2 (0.5) | |
BAL, bronchoalveolar lavage; F, fluid; CSF, cerebrospinal fluid.
Data are presented as n (%).
*This included abscess and deep wound aspirations, which are regarded as dirty wounds, †This included organ biopsy, gastrointestinal fluid, drain fluid, other fluid, blood, surgical specimens, and stool samples.
The proportion of male patients in this study was 57.7%. In the group aged 40–59 years, the proportion of NTM in samples from female patients was significantly greater than that in samples from male patients (p<0.001). However, in the group aged 70–89 years, the recovery rate of NTM in samples from male patients was significantly greater than that in samples from female patients (p<0.001) (Table 3).
Table 3. Number and Proportion (%) of MTBC and NTM Isolates according to Age and Sex of Patients.
| Age (yr) | MTBC isolates | NTM isolates | Total | p value* | ||||
|---|---|---|---|---|---|---|---|---|
| Female | Male | Subtotal | Female | Male | Subtotal | |||
| 0–9 | 4 (23.5) | 13 (76.5) | 17 (81.0) | 0 (0) | 4 (100) | 4 (19.0) | 21 (0.3) | 0.546 |
| 10–19 | 31 (36.5) | 54 (63.5) | 85 (89.5) | 2 (20.0) | 8 (80.0) | 10 (10.5) | 95 (1.2) | 0.486 |
| 20–29 | 109 (46.4) | 126 (53.6) | 235 (93.3) | 6 (35.3) | 11 (64.7) | 17 (6.7) | 252 (3.3) | 0.375 |
| 30–39 | 149 (44.9) | 183 (55.1) | 332 (85.1) | 32 (55.2) | 26 (44.8) | 58 (14.9) | 390 (5.1) | 0.147 |
| 40–49 | 148 (31.8) | 317 (68.2) | 465 (74.6) | 92 (58.2) | 66 (41.8) | 158 (25.4) | 623 (8.1) | <0.001 |
| 50–59 | 158 (22.9) | 533 (77.1) | 691 (61.2) | 225 (51.4) | 213 (48.6) | 438 (38.8) | 1129 (14.7) | <0.001 |
| 60–69 | 252 (34.9) | 470 (65.1) | 722 (54.7) | 233 (39.0) | 364 (61.0) | 597 (45.3) | 1319 (17.2) | 0.122 |
| 70–79 | 625 (47.9) | 680 (52.1) | 1305 (54.9) | 410 (38.2) | 664 (61.8) | 1074 (45.1) | 2379 (31.0) | <0.001 |
| 80–89 | 444 (59.4) | 304 (40.6) | 748 (56.5) | 241 (41.8) | 336 (58.2) | 577 (43.5) | 1325 (17.3) | <0.001 |
| ≥90 | 55 (61.1) | 35 (38.9) | 90 (63.8) | 29 (56.9) | 22 (43.1) | 51 (36.2) | 141 (1.8) | 0.621 |
| Total | 1975 (42.1) | 2715 (57.9) | 4690 (61.1) | 1270 (42.6) | 1714 (57.4) | 2984 (38.9) | 7674 (100) | |
MTBC, Mycobacterium tuberculosis complex; NTM, nontuberculous mycobacteria.
Data are presented as n (%).
*p value indicates significant difference between the number of female and male patients in each age group.
Of the four NTM species with more than 100 isolates, M. intracellulare, M. avium, and M. fortuitum complex were more frequently isolated from males, while M. abscessus was more frequently isolated from females. For M. intracellulare, the number of isolates increased after 50 years of age: the proportion of total NTM was 46.3% (203/438) for patients in their 50–59 years, 45.6% (272/597) for patients aged 60–69 years, 53.4% (573/1074) for patients aged 70–79 years, and 54.6% (315/577) for patients aged 80–89 years (Table 4).
Table 4. Number and Proportion (%) of Four Mycobacterium Species according to Age and Sex of Patients.
| Age (yr) | Mycobacterium intracellulare | Mycobacterium avium | Mycobacterium fortuitum complex | Mycobacterium abscessus | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Subtotal | Female | Male | Subtotal | Female | Male | Subtotal | Female | Male | Subtotal | |
| 0–9 | 0 (0) | 2 (100) | 2 (0.1) | 0 (0) | 1 (100) | 1 (0.2) | 0 (0) | 1 (100) | 1 (0.7) | 0 (0) | 0 (0) | 0 (0) |
| 10–19 | 1 (11.1) | 8 (88.9) | 9 (0.6) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (0.7) | 0 (0) | 0 (0) | 0 (0) |
| 20–29 | 4 (50.0) | 4 (50.0) | 8 (0.5) | 2 (50.0) | 2 (50.0) | 4 (0.7) | 0 (0) | 1 (100) | 1 (0.7) | 0 (0) | 2 (100) | 2 (1.5) |
| 30–39 | 14 (48.3) | 15 (51.7) | 29 (1.9) | 9 (75.0) | 3 (25.0) | 12 (2.2) | 3 (60.0) | 2 (40.0) | 5 (3.4) | 3 (100) | 0 (0) | 3 (2.2) |
| 40–49 | 38 (52.8) | 34 (47.2) | 72 (4.8) | 31 (77.5) | 9 (22.5) | 40 (7.3) | 3 (37.5) | 5 (62.5) | 8 (5.5) | 8 (100) | 0 (0) | 8 (5.9) |
| 50–59 | 101 (49.8) | 102 (50.2) | 203 (13.5) | 68 (68.7) | 31 (31.3) | 99 (18.1) | 2 (22.2) | 7 (77.8) | 9 (6.2) | 20 (64.5) | 11 (35.5) | 31 (23.0) |
| 60–69 | 119 (43.8) | 153 (56.3) | 272 (18.0) | 57 (42.9) | 76 (57.1) | 133 (24.4) | 11 (27.5) | 29 (72.5) | 40 (27.6) | 11 (40.7) | 16 (59.3) | 27 (20.0) |
| 70–79 | 229 (40.0) | 344 (60.0) | 573 (38.0) | 67 (41.1) | 96 (58.9) | 163 (29.9) | 22 (40.7) | 32 (59.3) | 54 (37.2) | 21 (50.0) | 21 (50.0) | 42 (31.1) |
| 80–89 | 149 (47.3) | 166 (52.7) | 315 (20.9) | 26 (31.3) | 57 (68.7) | 83 (15.2) | 5 (19.2) | 21 (80.8) | 26 (17.9) | 8 (38.1) | 13 (61.9) | 21 (15.6) |
| ≥90 | 13 (50.0) | 13 (50.0) | 26 (1.7) | 7 (63.6) | 4 (36.4) | 11 (2.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (0.7) |
| Total | 668 (44.3) | 841 (55.7) | 1509 (100) | 267 (48.9) | 279 (51.1) | 546 (100) | 47 (32.4) | 98 (67.6) | 145 (100) | 71 (52.6) | 64 (47.4) | 135 (100) |
Data are presented as n (%).
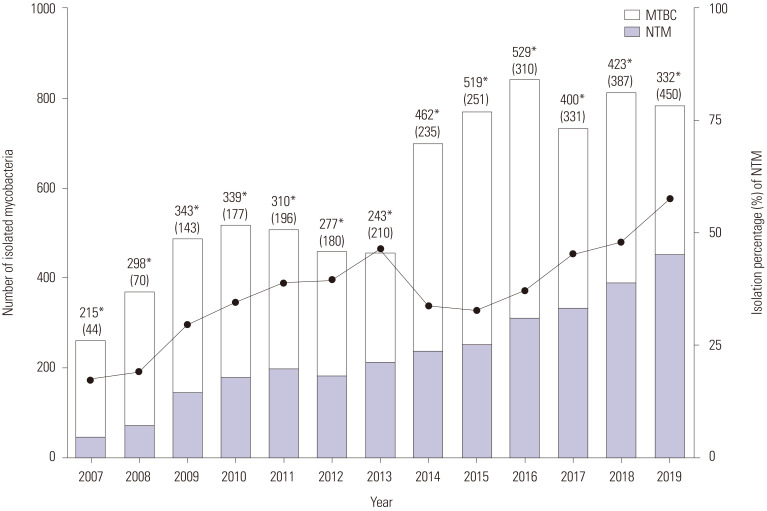
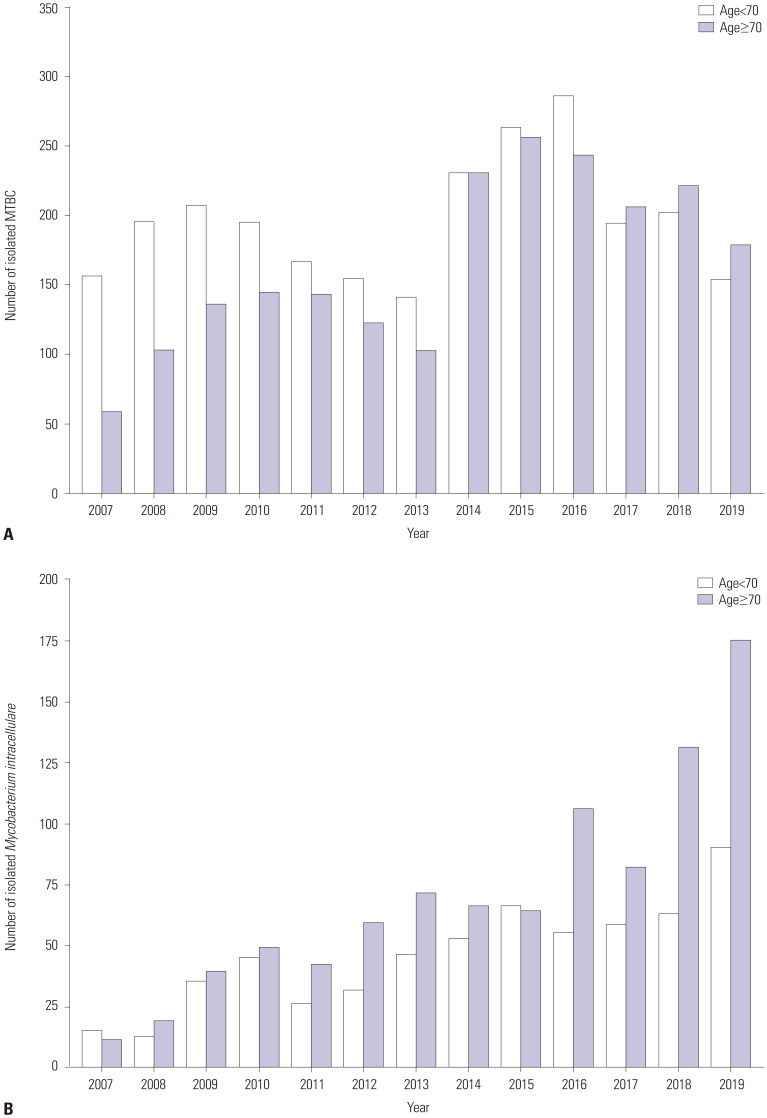
The proportion of NTM among all mycobacteria showed an upward trend over the 13-year study period and increased from 17.0% in 2007 to 57.5% in 2019 (Fig. 1). Moreover, in patients aged over 70 years, the proportion of M. intracellulare isolates among all mycobacterial isolates continuously increased from 13.9% (11/79) in 2007 to 37.4% (175/468) in 2019 (Fig. 2).
Fig. 1. Continued upward trend in the proportion of NTM isolates over the 13-year study period. Filled circles (●) denote the percentage of NTM isolates each year, while asterisks (*) represent the number of MTBC isolates each year. The number in brackets is the number of NTM isolates. MTBC, Mycobacterium tuberculosis complex; NTM, nontuberculous mycobacteria.
Fig. 2. The number of MTBC (A) and Mycobacterium intracellulare (B) isolates over the 13-year study period. The height of left and right bars are the number of isolated mycobacteria in patients aged less than 70 years and more than 70 years, respectively. MTBC, Mycobacterium tuberculosis complex.
DISCUSSION
There are several factors that need to be considered when estimating and comparing the prevalence of NTM pulmonary disease (NTM-PD). Unlike TB, which is a notifiable disease in most regions due to management policies, NTM-PD is not a notifiable disease. Its diagnosis depends on the integration of clinical, radiological, and microbiological findings, as summarized in the American Thoracic Society/Infectious Diseases Society of America 2007 criteria, which have become the accepted definition for this disease.15 In the genus Mycobacterium, there are more than 200 species and 13 subspecies.16 Many NTM species show similar phenotypic characteristics17 and high DNA homology18; therefore, they have been sorted into groups or complexes, each consisting of two to seven species, although some species remain ungrouped. The prevalence of NTM-PD and the proportions of different NTM species may vary based on the following: country,3 study period,19 and methods employed for isolation20,21 and identification.19 The AdvanSure Mycobacteria GenoBlot assay (LG Chem) used in this study has been reported to be comparable to the GenoType Mycobacterium CM/AS assay (Hain Life-science, Nehren, Germany) in terms of performance and may be useful as a routine method for NTM identification in clinical settings, especially in locations where MAC is the main cause of NTM infection.22 The NTM species most commonly isolated worldwide belong to the MAC, comprising 34%–61% of isolates, depending on the continent, with the highest proportions reported in North America and Oceania.23 A nationwide survey in Japan showed that the distribution of the two major MAC species, M. avium and M. intracellulare, followed a clear gradient from the northern to southern regions.24 The relative ratio of M. avium to M. intracellulare (M. avium/M. intracellulare) in different parts of the world, in decreasing order, is North America (9.8)>South America (4.9)> Asia (2.8)>Europe (1.5)>South Africa (0.27)>Australia (0.25).5 In Korea, the prevalence of M. avium was high mainly in the capital city of Seoul,2,25,26,27 whereas the prevalence of M. intracellulare was high in southern cities, such as Busan, Yangsan, and Ulsan.28,29 Ambient temperature, heavy rainfall, flooding, and drought are likely to influence the prevalence of environmental microorganisms.30 Although NTM are found in soil and water, the factors that influence their transmission from the environment to humans are largely unknown.6 The environmental reservoirs reported to be predominantly associated with M. avium are water and soil, while those associated with M. intracellulare include dust and soil.6 M. intracellulare infection was shown to be more common in agricultural regions and was associated with shallow soil,31 and there was no significant association between the incidence of M. intracellulare and temperature or rainfall.32 Shallow soil depth, which is associated with poor crop yields, as well as the process of “deep ripping” to improve yields, have been hypothesized to result in aerosolization of mycobacteria from the soil.32 However, there may be other aspects of soil quality or other yet unknown environmental factors that favor M. intracellulare growth in these regions.
Han, et al.10 reported that M. intracellulare was frequently isolated from postmenopausal women with estrogen deficiency. However, in the present study, both M. intracellulare and M. avium were frequently isolated in men over 60 years of age. M. intracellulare was isolated from extrapulmonary specimens, such as wounds (7%), whereas M. avium was isolated only from respiratory specimens. In addition, 50% of NTM isolates from synovial fluid were M. intracellulare. Although the exact reason for the geographic diversity in distribution of mycobacterial species is unknown, the degree of human exposure to certain endemic NTM species is considered to be the most important factor. We postulated that, depending on environmental factors such as occupation, residence, or participation in activities associated with water and/or soil, a deficiency in cellular immunity may be a risk factor for infection when exposed to M. avium. Underlying lung disease or prior lung injury resulting from bronchiectasis and smoking may be a risk factor for M. intracellulare infection.
Further research is needed to identify the major exposure routes and environmental sources of NTM infections. This information would then enable the design of effective control strategies for diseases associated with NTM.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Young Uh.
- Data curation: Young Uh.
- Formal analysis: Kwangjin Ahn and Young Uh.
- Investigation: Kwangjin Ahn and Young Uh.
- Methodology: Gyu Yel Hwang and Hyunmi Cho.
- Project administration: Young Uh.
- Resources: Kwangjin Ahn and Young Uh.
- Software: Kwangjin Ahn.
- Supervision: Young Uh.
- Validation: Young Keun Kim.
- Visualization: Kwangjin Ahn and Young Uh.
- Writing—original draft: Kwangjin Ahn.
- Writing—review & editing: all authors.
- Approval of final manuscript: all authors.
References
- 1.World Health Organization. Global tuberculosis report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Yoon HJ, Choi HY, Ki M. Nontuberculosis mycobacterial infections at a specialized tuberculosis treatment centre in the Republic of Korea. BMC Infect Dis. 2017;17:432. doi: 10.1186/s12879-017-2532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowman S, van Ingen J, Griffith DE, Loebinger MR. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J. 2019;54:1900250. doi: 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Nontuberculous mycobacteria (NTM) infections [Internet] [accessed on 2020 August 12]. Available at: https://www.cdc.gov/hai/organisms/nontuberculous-mycobacteria.html.
- 5.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. 2013;42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 6.Halstrom S, Price P, Thomson R. Review: environmental mycobacteria as a cause of human infection. Int J Mycobacteriol. 2015;4:81–91. doi: 10.1016/j.ijmyco.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Burns EA, Goodwin JS. Immunodeficiency of aging. Drugs Aging. 1997;11:374–397. doi: 10.2165/00002512-199711050-00005. [DOI] [PubMed] [Google Scholar]
- 8.Guthertz LS, Damsker B, Bottone EJ, Ford EG, Midura TF, Janda JM. Mycobacterium avium and Mycobacterium intracellulare infections in patients with and without AIDS. J Infect Dis. 1989;160:1037–1041. doi: 10.1093/infdis/160.6.1037. [DOI] [PubMed] [Google Scholar]
- 9.Inderlied CB, Kemper CA, Bermudez LE. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han XY, Tarrand JJ, Infante R, Jacobson KL, Truong M. Clinical significance and epidemiologic analyses of Mycobacterium avium and Mycobacterium intracellulare among patients without AIDS. J Clin Microbiol. 2005;43:4407–4412. doi: 10.1128/JCM.43.9.4407-4412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkinham JO, 3rd, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol. 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirschner RA, Jr, Parker BC, Falkinham JO., 3rd Epidemiology of infection by nontuberculous mycobacteria. Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum in acid, brown-water swamps of the southeastern United States and their association with environmental variables. Am Rev Respir Dis. 1992;145:271–275. doi: 10.1164/ajrccm/145.2_Pt_1.271. [DOI] [PubMed] [Google Scholar]
- 13.Wang HY, Kim H, Kim S, Bang H, Kim DK, Lee H. Evaluation of PCR-reverse blot hybridization assay for the differentiation and identification of Mycobacterium species in liquid cultures. J Appl Microbiol. 2015;118:142–151. doi: 10.1111/jam.12670. [DOI] [PubMed] [Google Scholar]
- 14.Cho H, Kim JB, Uh Y. Frequency of Mycobacterium tuberculosis among M. tuberculosis complex strains isolated from clinical specimen. Ann Clin Microbiol. 2020;23:21–31. [Google Scholar]
- 15.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 16.Parte AC. LPSN–list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 17.Bhalla GS, Sarao MS, Kalra D, Bandyopadhyay K, John AR. Methods of phenotypic identification of non-tuberculous mycobacteria. Pract Lab Med. 2018;12:e00107. doi: 10.1016/j.plabm.2018.e00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, Shin JH. Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J Clin Lab Anal. 2018;32:e22184. doi: 10.1002/jcla.22184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donohue MJ. Increasing nontuberculous mycobacteria reporting rates and species diversity identified in clinical laboratory reports. BMC Infect Dis. 2018;18:163. doi: 10.1186/s12879-018-3043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotcheewaphan S, Odusanya OE, Henderson CM, Stephenson D, Olivier KN, Perry JD, et al. Performance of RGM medium for isolation of nontuberculous mycobacteria from respiratory specimens from non-cystic fibrosis patients. J Clin Microbiol. 2019;57:e01519-18. doi: 10.1128/JCM.01519-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Ingen J. Diagnosis of nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34:103–109. doi: 10.1055/s-0033-1333569. [DOI] [PubMed] [Google Scholar]
- 22.Yang M, Huh HJ, Kwon HJ, Kim JY, Song DJ, Koh WJ, et al. Comparative evaluation of the AdvanSure Mycobacteria GenoBlot assay and the GenoType Mycobacterium CM/AS assay for the identification of non-tuberculous mycobacteria. J Med Microbiol. 2016;65:1422–1428. doi: 10.1099/jmm.0.000376. [DOI] [PubMed] [Google Scholar]
- 23.Zweijpfenning SMH, Ingen JV, Hoefsloot W. Geographic distribution of nontuberculous mycobacteria isolated from clinical specimens: a systematic review. Semin Respir Crit Care Med. 2018;39:336–342. doi: 10.1055/s-0038-1660864. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto K, Hasegawa N, Izumi K, Namkoong H, Uchimura K, Yoshiyama T, et al. A laboratory-based analysis of nontuberculous mycobacterial lung disease in Japan from 2012 to 2013. Ann Am Thorac Soc. 2017;14:49–56. doi: 10.1513/AnnalsATS.201607-573OC. [DOI] [PubMed] [Google Scholar]
- 25.Park Y, Kim CY, Park MS, Kim YS, Chang J, Kang YA. Age- and sexrelated characteristics of the increasing trend of nontuberculous mycobacteria pulmonary disease in a tertiary hospital in South Korea from 2006 to 2016. Korean J Intern Med. 2020;35:1424–1431. doi: 10.3904/kjim.2019.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko RE, Moon SM, Ahn S, Jhun BW, Jeon K, Kwon OJ, et al. Changing epidemiology of nontuberculous mycobacterial lung diseases in a tertiary referral hospital in Korea between 2001 and 2015. J Korean Med Sci. 2018;33:e65. doi: 10.3346/jkms.2018.33.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joo YS, Kwak NE, Kim GH, Yoon EJ, Jeong SH. Prevalence and species spectrum of pulmonary nontuberculous mycobacteria isolates at a tertiary care center. Ann Clin Microbiol. 2019;22:71–76. [Google Scholar]
- 28.Lee MY, Lee T, Kim MH, Byun SS, Ko MK, Hong JM, et al. Regional differences of nontuberculous mycobacteria species in Ulsan, Korea. J Thorac Dis. 2014;6:965–970. doi: 10.3978/j.issn.2072-1439.2014.07.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim N, Yi J, Chang CL. Recovery rates of non-tuberculous mycobacteria from clinical specimens are increasing in Korean tertiarycare hospitals. J Korean Med Sci. 2017;32:1263–1267. doi: 10.3346/jkms.2017.32.8.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda JR, Bernhard JN, Chan ED. Natural disasters and nontuberculous mycobacteria: a recipe for increased disease? Chest. 2015;147:304–308. doi: 10.1378/chest.14-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou MP, Clements AC, Thomson RM. A spatial epidemiological analysis of nontuberculous mycobacterial infections in Queensland, Australia. BMC Infect Dis. 2014;14:279. doi: 10.1186/1471-2334-14-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson RM, Furuya-Kanamori L, Coffey C, Bell SC, Knibbs LD, Lau CL. Influence of climate variables on the rising incidence of nontuberculous mycobacterial (NTM) infections in Queensland, Australia 2001-2016. Sci Total Environ. 2020;740:139796. doi: 10.1016/j.scitotenv.2020.139796. [DOI] [PubMed] [Google Scholar]