Abstract
The endangered yellow-spotted river turtle (Podocnemis unifilis) has experienced a dramatic population decline in the Ecuadorian Amazonia, mainly due to overexploitation of its eggs. To reverse this trend, the Wildlife Conservation Society has developed a head-start program in Yasuní National Park since 2008, but the potential risk that microbes associated with its eggs might represent for hatching success has not been evaluated yet. Members of the Fusarium solani species complex (FSSC) are involved in egg failure in sea turtles under natural and hatchery conditions, but their role in infecting the eggs of P. unifilis is unknown. In this study, we collected eggshells of P. unifilis and obtained 50 fungal and bacterial isolates. Some potentially pathogenic fungi of the genera Fusarium, Penicillium and Rhizopus were identified based on molecular data. Most importantly, the sea turtle pathogenic species F. keratoplasticum not only was present, but it was the most frequently found. Conversely, we have also isolated other microorganisms, such as Pseudomonas or Phoma-like species, producing a wide spectrum of antifungal compounds that may have a protective role against fungal diseases. Our survey provides useful information on potential pathogens found in P. unifilis eggshells, upon which the success of conservation programs may depend.
Keywords: bacteria, chelonians, conservation, endangered species, fungi, FSSC, pathogens, Yasuní National Park, yellow-spotted river turtle
1. Introduction
Turtles (class Reptilia, order Testudines) are some of the most endangered vertebrates in the world. Indeed, according to International Union for the Conservation of Nature (IUCN) Red List of Threatened Species, most species of this order are threatened, with only 18% being assigned the “least concern” category. The yellow-spotted river turtle (Podocnemis unifilis Troschel, 1848; fam. Podocnemididae), native to several South American countries, is currently classified as vulnerable [1], and listed in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora [2].
In the Ecuadorian Amazonia, P. unifilis inhabits the northwest region of the Yasuní National Park (YNP), where it has experienced a dramatic population decline mainly due to the overexploitation of its eggs for human consumption. Besides, the sale of turtle meat at local illegal markets and the collection of juveniles for pet trade also have a negative influence [3,4,5,6,7]. In order to reverse this trend towards local extinction, the Wildlife Conservation Society (WCS) established a conservation program for P. unifilis, commonly referred to as “taricaya” or “charapa”, back in 2008. This ongoing project, self-managed by Kichwa and Waorani Indigenous communities, consists of transplanting nests threatened by egg poachers and floods to hatcheries, and rearing hatchlings in captivity during the first year of life, when mortality reaches a maximum [8,9,10,11].
So far, this collaborative effort has allowed the release of thousands of juveniles into the wild, in exchange for sustainable harvest of eggs and economic alternatives to reduce communities’ dependence on such a resource [9]. However, the real outcome of this, and any other long-term conservation program [12,13,14,15], depends on controlling potential threats that could compromise hatchlings’ survival. To this regard, incubating egg clutches in hatcheries and rearing juveniles in captivity, which involve high densities of eggs and individuals, respectively, could potentially lead to fungal, bacterial, viral and protozoan disease outbreaks [16,17,18,19,20,21].
Previous studies have revealed a broad variety of potentially pathogenic bacteria on sea turtle eggs [22,23,24,25,26]. Moreover, a detailed metagenomic analysis on the microbial community of the eggs of the hawksbill turtle, Eretmochelys imbricata, confirmed the presence of some potentially pathogenic bacteria, but also several strains with antifungal activity [27]. In addition, it has been shown that ubiquitous soil fungi belonging to the so-called Fusarium solani species complex (hereafter, FSSC), such as Fusarium falciforme and Fusarium keratoplasticum, can seriously affect hatching success [28]. These species are involved in “sea turtle egg fusariosis” (STEF), an emergent fungal disease linked to egg mortality in endangered sea turtle nests worldwide [29]. Thus, they could compromise the performance of conservation practices based on ex situ incubations.
As for the freshwater turtle P. unifilis, several pathogens, such as parasitic nematodes [30,31,32,33], protozoans [34,35], yeasts [36] and bacteria [35,37,38], are known to affect adults. However, little is known about potentially pathogenic or beneficial microorganisms associated with their eggs. In this study we aimed to (1) confirm the presence of Fusarium spp. in eggs of P. unifilis showing symptoms of Fusarium infection, (2) determine the presence of additional fungal and bacterial species, and (3) identify those microorganisms that could either represent a risk or play a protective role, during embryonic development.
2. Materials and Methods
2.1. Sample Collection
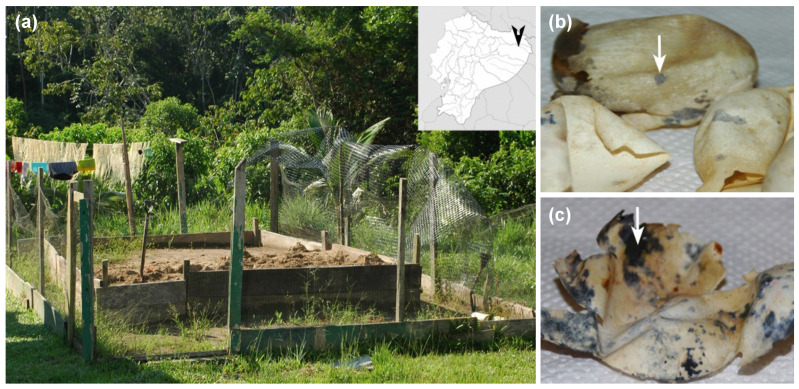
We collected 17 eggshells of P. unifilis from several nests relocated in three different hatcheries (Figure 1a), at the end of 2011 nesting season. After egg eclosion, eggshells showing typical symptoms of Fusarium infection (Figure 1b,c) were manipulated using sterile gloves and placed individually in plastic sealed bags. Samples were maintained in an ice box at 4 °C and transported to the laboratory for processing.
Figure 1.
Sampling location at Yasuní National Park (Ecuador) and eggshells of P. unifilis with macroscopic signs of Fusarium infection (colored spots). The hatcheries were located in the territories of Guiyero, Nueva Providencia and Sani Isla Communities (distributed along the Napo and Tiputini rivers). (a) Artificial hatchery located at Guiyero; (b) eggshells showing early symptoms of fusariosis (indicated with an arrow); (c) eggshells with advanced symptoms (arrow).
2.2. Fungal Isolation and DNA Extraction
Several fragments (ca. 0.5 cm2) with obvious signs of infection were excised from each eggshell using a sterile scalpel and plated on peptone glucose agar (PGA) supplemented with ampicillin (100 mg/L). We sub-cultured the resulting fungal colonies to obtain pure cultures, inoculated on potato dextrose agar (PDA) slants in 15 mL tubes. Anexic cultures were incubated at 25 °C for 24 to 72 h, and then permanently stored at 4 °C in the culture collection of the Real Jardín Botánico-CSIC (Madrid, Spain), (Table S1). Total genomic DNA was extracted from pure cultures using a DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA), according to the manufacturer’s instructions.
2.3. PCR Amplification and BLAST-Based Identification of Fungal Isolates
For molecular identification of fungal isolates, we obtained sequences of the nuclear ribosomal ITS region (ITS1, 5.8S and ITS2), using universal primers (Table 1). PCR reactions were performed using Ready-To-Go PCR Beads™ (GE Healthcare Life Sciences, Little Chalfont, UK). Samples were subjected to an initial denaturation step at 94 °C for 5 min, to ensure complete denaturation of the DNA template, as recommended by the manufacturer. After that, the PCR cycling protocol included 5 cycles at 94 °C for 30 s, 54 °C for 30 s and 72 °C for 1 min, followed by 33 cycles at 94 °C for 30 s, 48 °C for 30 s and 72 °C for 1 min, with a final extension step at 72 °C for 10 min [39]. Amplicons were separated by electrophoresis on 2% agarose gels and visualized with an UV transilluminator. The bands of interest were excised from the gels, purified using a QIAquick gel extraction kit (Qiagen, Germantown, MD, USA), and sequenced in both directions by Macrogen (Seoul, Korea) with the same primers used for amplification. Raw sequence edition and consensus assembly were done in Geneious v. 7.1.9 [40]. Sequences were trimmed to exclude low quality and primer-binding sites.
Table 1.
Primers used to amplify different molecular regions analyzed in this study.
| Organism/ Target Gene |
Primer Name a |
Primer Sequence (5′→3′) | Reference |
|---|---|---|---|
| Fungi | |||
| ITS | ITS5 (f) | GGAAGTAAAAGTCGTAACAAGG | [41] |
| ITS4 (r) | TCCTCCGCTTATTGATATGC | ||
| EF-1α | EF-1 (f) | ATGGGTAAGGA(A/G)GACAAGAC | [42] |
| EF-2 (r) | GGA(G/A)GTACCAGT(G/C)ATCATGTT | ||
| LSU | LR0R (f) | ACCCGCTGAACTTAAGC | [43,44] |
| LR5 (r) | ATCCTGAGGGAAACTTC | ||
| Bacteria | |||
| 16S | fD2 | AGAGTTTGATCATGGCTCAG | [45] |
| rP1 | ACGGTTACCTTGTTACGACTT | ||
a If not indicated by the primer name itself, forward and reverse primers are marked with (f) and (r), respectively.
In an attempt to improve the resolution provided by a single molecular region (ITS), we obtained data for two additional molecular regions, i.e., elongation factor-1 alpha (EF-1α) and the nuclear ribosomal large subunit (LSU), from 17 isolates belonging to the FSSC (see Figure S1). Both regions were amplified by PCR using primers previously designed (Table 1).
Cycling parameters for EF-1α amplification were: initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 90 s, and extension at 68 °C for 2 min, with a final extension step at 76 °C for 5 min. For LSU, the PCR cycling profile consisted of an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 45 s, extension at 72 °C for 1 min, and a final extension step at 72 °C for 7 min [43].
PCR product purification and sequence edition were done as described above. All 62 new fungal sequences generated in this study were submitted to GenBank (Table 2 and Table S1).
Table 2.
FSSC Clade 3 isolates obtained from the eggshells of P. unifilis analyzed in this study.
| Isolate | Geographic Origin a | Species ID b | GenBank Accession No. | ||
|---|---|---|---|---|---|
| ITS | LSU | EF-1α | |||
| 143 FUS | Guiyero | F. keratoplasticum | MW390926 | MW390975 | MW389342 |
| 144 FUS | Guiyero | F. keratoplasticum | MW390927 | MW390976 | MW389343 |
| 145 FUS | Guiyero | F. suttonianum | MW390928 | MW390977 | MW389356 |
| 151 FUS | Guiyero | F. keratoplasticum | MW390930 | MW390978 | MW389344 |
| 152 FUS | Nueva Providencia | F. suttonianum | MW390931 | MW390979 | MW389357 |
| 153 FUS | Sani Isla | F. cf. crassum | MW390932 | MW390980 | MW389341 |
| 154 FUS | Sani Isla | F. keratoplasticum | MW390933 | MW390981 | MW389345 |
| 156 FUS | Sani Isla | F. keratoplasticum | MW390935 | MW390982 | MW389346 |
| 157 FUS | Sani Isla | F. keratoplasticum | MW390936 | MW390983 | MW389347 |
| 158 FUS | Undetermined | F. keratoplasticum | MW390937 | MW390984 | MW389348 |
| 160 FUS | Undetermined | F. keratoplasticum | MW390939 | MW390985 | MW389349 |
| 161 FUS | Undetermined | F. keratoplasticum | MW390940 | MW390986 | MW389350 |
| 162 FUS | Undetermined | F. keratoplasticum | MW390941 | MW390987 | MW389351 |
| 163 FUS | Undetermined | F. keratoplasticum | MW390942 | MW390988 | MW389352 |
| 197 FUS | Guiyero | F. keratoplasticum | MW390949 | MW390989 | MW389353 |
| 198 FUS | Sani Isla | F. keratoplasticum | MW390950 | MW390990 | MW389354 |
| 200 FUS | Sani Isla | F. keratoplasticum | MW390952 | MW390991 | MW389355 |
a Guiyero: 00°36′14″ S, 76°28′03” W; Nueva Providencia: 00°29′35′′ S, 76°28′21″ W; Sani Isla: 00°28′30″ S, 76°18′37′′ W. b Species identification based on the multigene phylogenetic tree presented in this study.
2.4. Sequence Alignment and Phylogenetic Analyses of the FSSC Isolates
To better place our 17 FSSC isolates in a phylogenetic context, we selected 130 GenBank specimens representing different Fusarium species, for which data of two or all three regions analyzed here were available (Table S2). These included, at least, one specimen from each of the subgroups within the FSSC Clade 3 [46], and 20 specimens designated as type material, used to delineate the species in the resulting clusters (Table S2).
Homologous sequences were automatically aligned with MAFFT v. 7.017 [47] with the E-INS-i algorithm and default settings, as implemented in Geneious. Obvious alignment errors were manually adjusted. All alignments used in this study are available under request.
Both Maximum Likelihood (ML) and Bayesian Inference (BI) analyses were first done separately for each region, using resources available in CIPRES [48]. Specifically, individual ML trees were estimated using IQ-TREE v. 1.6.12 [49]. The best-fit model of nucleotide substitution for each dataset was selected with the integrated version of ModelFinder [50]. It was also used to determine the optimal partitioning scheme for each region (ITS was partitioned into ITS1, 5.8S and ITS2, EF-1α was divided into six partitions corresponding to three introns and each codon position of four exons, and LSU was not partitioned). We used the “complete bootstrap” option with 1000 non-parametric bootstrap replicates to assess nodal bootstrap support (BS).
BI single-gene analyses were carried out using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method, as implemented in MrBayes v. 3.2.7 [51]. For each dataset, the best-fit substitution model was estimated using the reversible jumping model choice [52], allowing a gamma distributed rate heterogeneity across sites, and a proportion of invariant sites. In the case of the EF-1α and ITS analyses, we used the partitioning schemes selected by ModelFinder, unlinking model parameters across different partitions. In all cases, we used four independent runs of 100 million generations, each with six chains. Trees were sampled every 1000 generations, with the first 25% discarded as burn-in and the posterior probabilities (PP) being calculated from the remaining ones. The Bayesian analyses automatically stopped when the average standard deviation of split frequencies fell below 0.01. Additionally, we assessed run convergence in Tracer v. 1.7.2 [53] by checking the effective sampling size (ESS) values for all parameters (>200).
Single-gene trees were visualized in FigTree v. 1.4.3 [54]. For each locus, both ML and BI analyses yielded very similar topologies, so only the Bayesian tree, showing PP and BS values, is provided (Figures S2–S4). By comparing these trees, we found an incongruent sample, i.e., 153 FUS, that was removed prior to subsequent concatenated analyses.
We predefined 10 partitions for the concatenated dataset (LSU, ITS1, 5.8S, ITS2, each codon position of EF-1α and three EF-1α introns). Again, IQ-TREE was used to choose the best partition scheme and substitution models, and to estimate the concatenated ML tree. The combined Bayesian analysis was performed considering the best partition scheme selected by ModelFinder and the same settings used for the individual analyses. Both ML and BI concatenated trees were visualized and edited in FigTree, and further processed using Adobe Illustrator CS5 (Adobe Systems Inc., San Jose, CA, USA). Only the Bayesian tree showing node supports from both phylogenetic approaches is presented. Members of the FSSC Clade 1 were selected as outgroup, based on a recent phylogenomic study [46].
2.5. Bacterial Isolation and DNA Extraction
We used a method previously described for isolating bacterial DNA from other turtle eggs [27], with some modifications. In short, each eggshell fragment was suspended in 10 mL of sterile tap water, vortexed for 2 min and incubated at room temperature for 24 h. Suspensions were diluted (1:10) and then a volume of 50 µL of each dilution was plated on PGA and incubated at 25 °C for five days. The resulting colonies were repeatedly transferred into fresh plates to obtain pure cultures. We prepared bacterial suspensions for long-term storage by inoculating single colonies in 1 mL of sterile tap water into 2 mL tubes, incubating them at 25 °C for 24 h. After that, we added 1 mL of 80% glycerol to the tubes. All suspensions were deposited at −20 °C in the culture collection of the Real Jardín Botánico-CSIC (Table 3). For bacterial DNA extraction, we collected ca. 2 mg of single colonies in 1.5 µL sterile tubes, and followed the same protocol mentioned above.
Table 3.
Bacterial isolates obtained from the eggshells of P. unifilis analyzed in this study.
| Isolate | Geographic Origin | Genus ID a | Phylum | 16S GenBank Accession No. |
|---|---|---|---|---|
| B13 | Nueva Providencia | Bacillus | Firmicutes | MW391108 |
| B14 | Nueva Providencia | Bacillus | Firmicutes | MW391109 |
| B15 | Nueva Providencia | Tsukamurella | Actinobacteria | MW391110 |
| B16 | Guiyero | Paracoccus | Proteobacteria | MW391111 |
| B17 | Guiyero | Nocardioides | Actinobacteria | MW391112 |
| B18 | Sani Isla | Nocardioides | Actinobacteria | MW391113 |
| B19 | Sani Isla | Gordonia | Actinobacteria | MW391114 |
| B20 | Sani Isla | Cupriavidus or Ralstonia | Proteobacteria | MW391115 |
| B21 | Sani Isla | Pseudoxanthomonas | Proteobacteria | MW391116 |
| B22 | Sani Isla | Pseudomonas | Proteobacteria | MW391117 |
| B23 | Sani Isla | Pseudomonas | Proteobacteria | MW391118 |
| B24 | Sani Isla | Delftia | Proteobacteria | MW391119 |
| B25 | Undetermined | Chryseobacterium | Bacteroidetes | MW391120 |
| B26 | Undetermined | Achromobacter | Proteobacteria | MW391121 |
| B27 | Undetermined | Elizabethkingia | Bacteroidetes | MW391122 |
| B28 | Undetermined | Stenotrophomonas | Proteobacteria | MW391123 |
| B29 | Undetermined | Pseudomonas | Proteobacteria | MW391124 |
| B30 | Undetermined | Stenotrophomonas | Proteobacteria | MW391125 |
| B31 | Undetermined | Delftia | Proteobacteria | MW391126 |
| B32 | Undetermined | Stenotrophomonas | Proteobacteria | MW391127 |
| B33 | Undetermined | Stenotrophomonas | Proteobacteria | MW391128 |
| B34 | Undetermined | Pseudomonas | Proteobacteria | MW391129 |
a Genus identification based on the 16S phylogenetic tree presented in this study.
2.6. PCR Amplification, BLAST-Based Identification of Bacterial Isolates, and 16S Phylogeny
The universal primer pair fD2/rP1 (Table 1) was selected to amplify the 16S rDNA gene using Ready-to-Go PCR Beads™ (GE Healthcare Life Sciences, Little Chalfont, UK). PCR reactions were performed in a final volume of 25 µL containing 23 µL of DNA template and 1 µL of each primer (10 µM). The cycling parameters were: initial denaturalization step at 94 °C for 5 min; 35 cycles at 94 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min; and a final extension step at 72 °C for 7 min.
Amplicon purification, sequencing and sequence edition were carried out as previously described. All bacterial 16S sequences newly generated were submitted to GenBank (Table 3).
To roughly identify the bacterial strains isolated here, we compared our edited 16S sequences to GenBank database (Table S3). To establish their phylogenetic relationships and better determine their identity, we conducted phylogenetic analyses as previously described. A set of 137 GenBank 16S sequences from related bacteria, including the type of several genera, was used in these analyses (Table S4). Taking into account that our isolates belong to different phyla and that high levels of sequence divergence between ingroup and outgroup can potentially lead to odd topologies [55,56], we rooted the tree at the mid-point of its longest path, instead of including an outgroup.
3. Results
3.1. Fungal Isolation and Phylogenetic Analyses of the FSSC Isolates
We obtained 28 fungal isolates from the 17 eggshells of P. unifilis analyzed here. According to BLAST searches, 20 of these isolates corresponded to the genus Fusarium (BLAST similarity values > 99.5%; Table S1).
To determine whether these isolates belonged to the sea turtle pathogenic FSSC, we run a preliminary phylogenetic analysis based on ITS data (Figure S1). This included most Fusarium complexes, and members of the genus Neonectria as outgroup according to previous results [46]. Most of the Fusarium strains isolated here (17 out of 20) nested within the FSSC, with only three isolates forming part of other major groups, i.e., Fusarium oxysporum species complex (FOSC) and, most probably, Fusarium fujikuroi species complex (FFSC).
To further investigate their phylogenetic relationships, two other molecular regions were analyzed. Individually, no region was able to fully resolve all relationships within the FSSC, as evidenced by the presence of several polytomies and the generalized low support values recovered for most branches in our single-gene trees (Figures S2–S4).
The isolate 153 FUS appeared sister with full support to the reference sequence of F. crassum and one unidentified species in the ITS tree (Figure S2). However, in the EF-1α tree, it nested within a moderately supported clade including the type material of F. keratoplasticum and many other samples of this species (Figure S4). Therefore, 153 FUS was removed from the data matrices prior to analyzing them together.
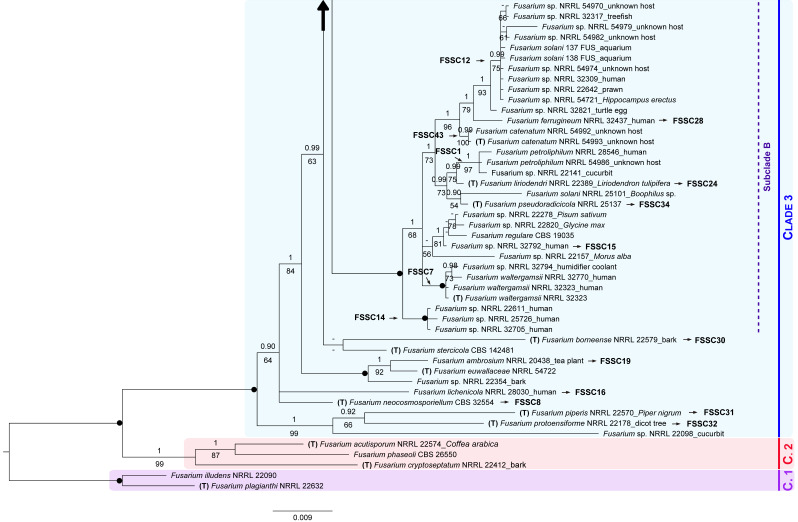
The resulting combined dataset consisted of 1732 characters. Only 277 were parsimony informative given the scarcity of informative characters in each individual data matrix. Out of the 146 isolates analyzed, 144 unique three-locus haplotypes were identified. The corresponding phylogenetic reconstruction (Figure 2) was neither totally resolved. Still, three main monophyletic groups were recovered, all with high support: FSSC Clade 1 (PP = 1, BS = 100%), Clade 2 (PP = 1, BS = 99%), and Clade 3 (PP = 1, BS = 100%).
Figure 2.
Combined Bayesian tree of the Fusarium solani species complex Clade 3, based on 1732 nucleotide positions from 146 isolates, with two members of the FSSC Clade 1 as outgroup. For each isolate, the species name is followed by the corresponding herbarium code, and host if available. Specimens corresponding to F. keratoplasticum initially identified as F. solani are marked with an asterisk. GenBank specimens corresponding to type material are marked with a “T”. Isolates for which we obtained sequences are in blue bold (153 FUS is not included in this concatenated tree). Clades 1, 2 and 3 represent designations previously proposed by O’Donnell [57]. Subclades A, B and C follow Sarmiento-Ramírez et al. [28]. Different lineages within the Clade 3 follow the nomenclature by O’Donnell et al. [58]. Numbers above and below the branches correspond to Bayesian posterior probability (PP) and maximum likelihood bootstrap values (BS), respectively (shown if PP ≥ 0.90 and BS ≥ 50%). Black solid dots indicate full support in both analyses. The scale bar represents the average number of substitutions per site. A large vertical black arrow indicates that the tree continues along the corresponding branch.
Within the FSSC Clade 3, some specimens occupied an undefined phylogenetic position. Besides, several minor clades and three recognizable large monophyletic groups were recovered, i.e., Subclade A (PP = 0.79, BS = 33%), Subclade B (PP = 1, BS = 100%), and Subclade C (PP = 1, BS = 93%).
All 17 FSSC isolates were linked to a species reference sequence (i.e., generated from type material). Specifically, 14 isolates clustered in the Subclade C along with the type strain F. keratoplasticum FRC S-2477 and additional specimens of this species. Sister to the Subclade A with no support (PP = 0.70, BS = 14%), we found a clade formed by 145 FUS, 152 FUS, F. suttonianum NRRL 32858 (type material), and other representatives of this species (PP = 1, BS = 99%).
The remaining internal relationships within the FSSC Clade 3 are not detailed for the sake of brevity.
Considering the phylogenetic relationships among the isolates of the FSSC Clade 3 and those corresponding to type material (retrieved from GenBank) in this multigene phylogeny, but also in the ITS tree (Figure S1), most isolates were identified as F. keratoplasticum (Table 2).
3.2. Bacterial Isolates and Phylogenetic Analyses
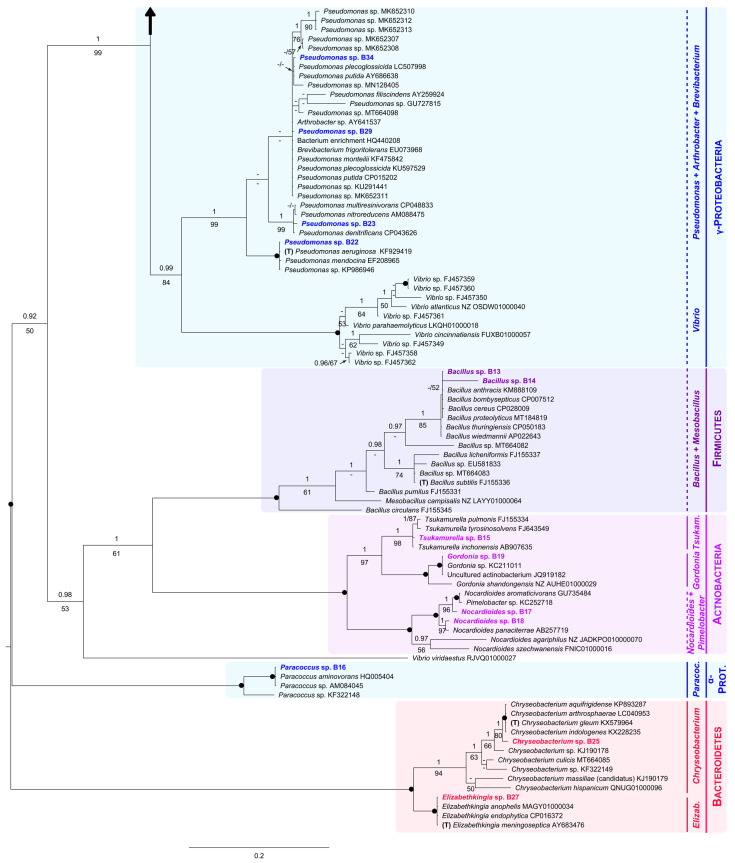
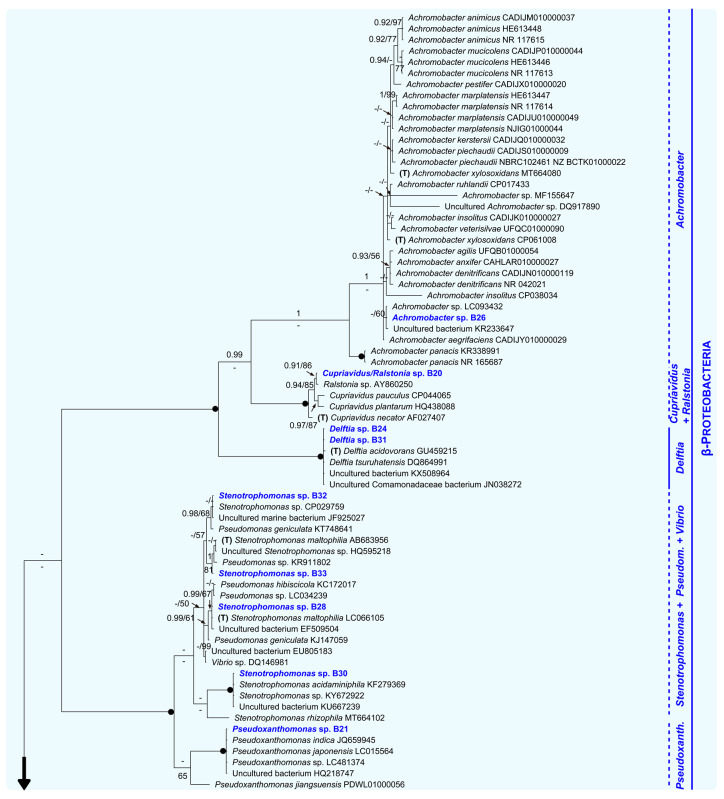
We obtained 22 bacterial isolates and analyzed them in a phylogenetic context (Figure 3). Specifically, in our 16S phylogenetic tree, six main clades were recovered: (1) subphylum Beta-Proteobacteria, (2) subphylum Gamma-Proteobacteria, (3) phylum Firmicutes, (4) phylum Actinobacteria, (5) subphylum Alfa-Proteobacteria, and (6) phylum Bacteroidetes (Figure 3).
Figure 3.
Unrooted Bayesian tree of the bacteria isolated from eggshells of P. unifilis, and allies. For each isolate, the species name is followed by the GenBank accession number. Sequences obtained in this study are in bold. Specimens representing the type of a genus are marked with a “T”. Numbers above and below the branches correspond to Bayesian posterior probability (PP) and maximum likelihood bootstrap values (BS), respectively (shown if PP ≥ 0.90 and BS ≥ 50%). Black solid dots indicate full support in both analyses. The scale bar represents the average number of substitutions per site. Discontinuous vertical lines indicate either unsupported or non-monophyletic genera. Vertical lines correspond to well-supported monophyletic genera. A large vertical black arrow indicates that the tree continues along the corresponding branch.
Within the Beta-Proteobacteria clade, the isolate B26 and all representatives of the genus Achromobacter analyzed here, including the type (A. xylosoxidans), constituted a group only supported in the Bayesian analysis (PP = 1, BS = 33%). A fully supported multispecies group (PP = 1, BS = 100%) comprised the isolate B20, the type of the genus Cupriavidus, and one member of Ralstonia. Sister with full support to both mentioned assemblages, we found another highly supported monophyletic group (PP = 1, BS = 100%). This was formed by B24, B31, two species of the genus Delftia (including the type, D. acidovorans), and two unidentified specimens. Additionally, within Beta-Proteobacteria, B28, B30, B32 and B33 formed an unsupported group with several species of the genus Pseudomonas, a single representative of Vibrio, and the type of Stenotrophomonas (S. maltophilia, recovered as polyphyletic). Closely related to this unsupported group, we found a small robust clade constituted by the isolate B21 and several species of Pseudoxanthomonas (PP = 1, BS = 100%).
Within the Gamma-Proteobacteria clade (PP = 0.99, BS = 84%), B22, B23, B29 and B34 nested within a strongly supported group (PP = 1, BS = 99%) that comprised most Pseudomonas analyzed here (including the type, P. aeruginosa), and two representatives of the genera Arthrobacter and Brevibacterium.
As for the Firmicutes clade (PP = 1, BS = 100%), it contained both isolates B13 and B14, all sequences of Bacillus analyzed here, including that of the type (B. subtilis), and one representative of Mesobacillus.
Sister to Firmicutes, with moderate support (PP = 1, BS = 61%), we found the Actinobacteria as monophyletic, receiving full support. It was further divided into three groups: (1) the well-supported Tsukamurella clade (PP = 1, BS = 98%), including the isolate B15; (2) the Gordonia clade (PP = 1, BS = 100%), holding the isolate B19; and (3) the Nocardiodes/Pimelobacter clade, also fully supported (PP = 1, BS = 100%), harboring B17 and B18.
The Alfa-Proteobacteria clade (PP = 1, BS = 100%) appeared in a distant position to both subphyla Gamma and Beta-Proteobacteria. It was formed by the isolate B16 and three members of the genus Paracoccus.
Finally, within the Bacteroidetes clade (PP = 1, BS = 100%), the isolates B25 and B27 were part of two robust clades constituted by members of the genera Chryseobacterium (PP = 1, BS = 94%) and Elizabethkingia (PP = 1, BS = 100%), respectively.
To summarize, the bacteria isolated from the eggshells of P. unifilis corresponded to 13 different genera belonging to four phyla (Table 3).
4. Discussion
The yellow-spotted river turtle, Podocnemis unifilis, is one of the most threatened reptiles in the Ecuadorian Amazonia. This has made it necessary to implement conservation actions through a collaborative effort among WCS and several Indigenous communities historically linked to the YNP. Specifically, the head start program developed by WCS has focused its strategy on egg translocations and their incubation in protected hatcheries. However, so far, the presence of microorganisms representing a potential risk for hatching success has not been evaluated under hatchery conditions.
Here, we describe, for the first time, the fungal and microbial communities associated with eggs of P. unifilis apparently colonized by Fusarium spp. Given the difficulties to collect samples from threatened species, a limited number of eggshells could be analyzed. Therefore, our results provide a glimpse into the fungi and bacteria associated with its eggs.
The multigene tree shown in Figure 2 was substantially congruent with that presented in a recent phylogenomic study [46]. Based on our tree, several fungi isolated from the eggshells of P. unifilis were revealed to be members of the FSSC Clade 3. Specifically, most isolates corresponded to F. keratoplasticum (lineage FSSC 2), a soil-borne species globally distributed. Notably, previous studies have reported that this and other species of the FSSC constitute a real threat to sea turtle nests worldwide, especially to those subject to environmental stressors, among others, inundation and clay/silt composition of nests [28,59,60,61].
Under natural conditions, the nests of P. unifilis are often exposed to flooding, which might favor the development of Fusarium in the eggs. Besides, clutches incubated under hatchery conditions are defenseless against additional stressors leading to the accumulation of pathogen spores and the spread of contaminants (e.g., manipulation, high density of nests, and reusing the substrate and the same wood frame for several seasons). These factors might exacerbate Fusarium development and eggs contamination by this and other microorganisms that could seriously affect hatching success [62,63].
While we cannot conclude whether F. keratoplasticum directly contributes to P. unifilis embryo death, this species has been not only isolated from unhatched loggerhead eggs, but it has been proved responsible for sea turtle mass mortalities related to STEF [27,28,60,61,64]. Thus, research efforts should focus on further characterizing this fungus and its potential pathogenicity in P. unifilis. Likewise, considering that F. keratoplasticum is less frequently isolated in in situ than in relocated sea turtle nests [60,65], it should also be investigated whether common management practices, such as bare-hand contact with the eggs or reusing the hatchery structure, increase the risk of fungal outbreaks for P. unifilis.
Apart from causing STEF, F. keratoplasticum is one of the most frequent etiological agents of mycotic keratitis, onychomycosis and disseminated infections in immunocompromised persons [66,67,68], who can get infected by inhalation of microconidia and/or skin penetration [69,70]. Considering the potential vulnerability of Ecuadorian Indigenous people, exposed to untreated toxic wastes from oil industries [71,72,73], and in close contact with the eggs, every attempt to prevent F. keratoplasticum infection from occurring and spreading should be made. We recommend implementing basic preventive measures, such as using protective masks and single-use gloves, hand washing and disinfection when handling eggs and/or hatchlings.
Although predominant within the FSSC Clade 3, F. keratoplasticum was not the only fungal species found in the eggshells of P. unifilis. We also recovered three isolates phylogenetically related to F. suttonianum (Figure 2) and, possibly, F. crassum (Figures S1 and S2). The former is an uncommon human pathogenic species, reported from blood samples, that can cause human keratitis [58], while F. crassum has been isolated from numerous hosts, including human clinical samples [74]. However, to the best of our knowledge, none of these species has been previously isolated from turtle eggs.
Additionally, three samples were identified as members of other complexes. More in detail, although ITS data from the type material of F. oxysporum could not be included in our analyses, several specimens of this species appeared sister to 199 FUS. For this reason, this isolate is considered here as a member of the FOSC, and it could correspond to F. oxysporum, or a closely related species (Figure S1). Fusarium oxysporum has been repeatedly found in both nests and failed eggs of several sea turtles [75,76,77,78,79,80]. Additionally, its presence in Dermochelys coriacea eggs negatively affects the size of the hatchlings [81], which could have undesirable consequences on their survival. The remaining two Fusarium isolates, 168 FUS and 170 FUS, form part of the FFSC, which is recovered as paraphyletic in our preliminary ITS tree by including members of the FOSC and other complexes (Figure S1). The relationships among F. concentricum, F. fujikuroi, F. proliferatum and both isolates are unclear, but they share the same ITS sequence with a specimen of the first mentioned species, while only one base change differentiates their ITS sequence from those obtained from the type material of F. fujikuroi and several specimens of F. proliferatum. Some members of the FFSC cause severe diseases in economically important plants [82,83,84,85,86]. Moreover, F. fujikuroi is known to act as an entomopathogenic fungi but, to our knowledge, only F. proliferatum has been previously associated to mycotic infections in turtle eggs [87]. Additional data from more informative loci are needed to firmly establish the identity of this pair of isolates.
Other than Fusarium spp., the fungi isolated from our samples also included the species Rhizopus microsporus, and one member of the genus Penicillium. To our knowledge, R. microsporus has not been previously reported from reptiles. In contrast, its close relatives R. stolonifer and R. oryzae have been isolated from eggs and nests of green turtles, Chelonia mydas [75,78,88,89]. Rhizopus stolonifer has been also found in soft-shell turtles, Apalone ferox, affected by cutaneous mycosis [90]. Interestingly, several members of the genus Rhizopus produce mycotoxins [75,91] that might be harmful for the turtle embryonic development, although this remains to be proved. As for Penicillium, some species of this genus have been identified in nests, eggs and skin lesions of numerous chelonians, including terrapins, tortoises and sea turtles [75,78,89,92,93,94]. They are known for their mycotoxigenic properties, thought to be detrimental for developing eggs under hatchery conditions [95]. Furthermore, some Penicillium species have been related to bronchopneumonia in sea turtles [96] and so, caution is recommended to prevent the risk of respiratory tract allergies for people handling both eggs and hatchlings of P. unifilis.
Members of the family Didymellaceae (possibly Allophoma, Didymella, and Phoma and allied genera) also formed part of the microflora of P. unifilis eggs. These taxa comprise plant pathogenic, saprobic and endophytic species associated with a wide range of hosts, including crops [97,98,99,100]. More interestingly, the species Phoma multirostrata has been isolated from eggs of C. mydas [64], and several congenerics synthetize antifungal compounds with broad-spectrum activity [101,102]. Hence, it would be interesting to investigate the role of Phoma spp. and its relatives on pathogenicity or mitigation of Fusarium infections in sea and freshwater turtle eggs.
While this constitutes the first report of fungi associated with eggshells of P. unifilis, we also provide novel data on the accompanying bacteria. We could not identify most of them at the species level, but members of the phyla Proteobacteria and Actinobacteria prevailed over other species, as occurs in sea turtle eggs [27]. In particular, the bacterial community associated with the eggs of P. unifilis seems to be dominated by Gram-negative aerobic bacteria normally present in the environment, or as part of the turtle microbiota, that may act as opportunistic pathogens under stressful conditions [24,103]. Among such bacteria we found Pseudomonas and Stenotrophomonas, both previously isolated from chelonians [24,104,105,106]. The genus Pseudomonas is known to be part of the normal microbiota of the mouth and cloaca of several turtles [22,25,103,104,105], including P. unifilis [37]. Besides, Pseudomonas spp. have been also isolated from eggs, and their presence has been linked to low hatching success [22,23,24,25,104]. The only bacterium identified at the species level, Stenotrophomonas maltophilia, has been isolated from several diseased adult animals [107,108], and also from unhatched sea turtle eggs [24,106,107]. Notably, infections caused by Pseudomonas spp. and Stenotrophomonas spp. are especially difficult to control because of their high resistance to most antibiotics [25,58,103,109,110,111,112]. So, apart from being a potential reproductive hazard to P. unifilis, they may have an important role on the dissemination of antimicrobial resistance, an increasingly concerning issue, and may also pose a health risk for immunocompromised people [26,105,113]. Therefore, it is recommendable to take certain precautions while handling eggs of P. unifilis.
On the other hand, some Pseudomonas species contribute to the natural soil suppressiveness against several fungal pathogens, including Fusarium [114,115,116]. Thus, the potential role of Pseudomonas as a pathogen and/or as an antagonistic of Fusarium disease on P. unifilis eggs needs investigation.
The second most abundant phylum, Actinobacteria, was represented by Gordonia, Nocardioides and Tsukamurella. Species of these genera have been described as pathogens in snakes [117,118] and tortoises [119]. However, there is no evidence of their presence in turtle eggs. Other bacteria found in P. unifilis eggs include Bacillus spp. (phylum Firmicutes), well-known human pathogens that probably account for developmental arrest in turtle embryos [26]. We have also identified some members of Bacteroidetes, one of the three most abundant phyla affecting both wild-captured and stranded green turtles [120]. Among them, we found the genus Elizabethkingia, which has also been regarded as potentially pathogenic for reptiles [121]. Consequently, in-depth studies are needed to further characterize these strains, which could eventually represent a serious hazard. Despite the limitations derived from the absence of a microbiological assessment, our survey provides useful information on the bacteria found in eggs of P. unifilis upon which the success of WCS conservation program depends.
5. Conclusions
This is the first molecular study on the microbiota associated to P. unifilis eggshells. It significantly contributes to the existing literature on fungal and bacterial contamination of freshwater turtle eggs. Most importantly, this study points to a potential major problem for the conservation of P. unifilis that is the extended presence of the pathogenic fungus F. keratoplasticum in its eggs. If it were proved that this fungal species causes a disease, then, by analogy with STEF, it could be referred to as “freshwater turtle egg fusariosis” (FTEF). On the other hand, we have also identified other fungi and bacteria that might have antagonistic activity against Fusarium. These findings have direct application on the WCS conservation program since feasible measures could be easily implemented to prevent Fusarium disease development in eggs of P. unifilis, and to protect individuals working in hatcheries. Further studies on the microbiota of P. unifilis eggs are necessary for a better understanding of their pathogenic or beneficial effects, and their role on the conservation of the yellow-spotted river turtle.
Acknowledgments
We express our deep gratitude to all members of the Indigenous Communities of Guiyero, Nueva Providencia and Sani Isla (Ecuadorian Amazonia) for allowing us to conduct this study in their territories. Special thanks are extended to our collaborators from the Wildlife Conservation Society (Ecuador), in particular R. Cuevas and W. Prado, for their kind assistance and logistical support during sampling. We also thank two anonymous reviewers for their time, effort, and constructive comments on earlier versions of this article.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7090742/s1, Figure S1: Preliminary Bayesian ITS tree of all 20 Fusarium isolated from P. unifilis eggshells, with Neonectria as outgroup, Figure S2: Bayesian ITS tree of FSSC isolates, based on 499 nucleotides, with members of the FSSC Clade 1 as outgroup, Figure S3: Bayesian LSU tree of FSSC isolates, based on 520 nucleotides, with members of the FSSC Clade 1 as outgroup, Figure S4: Bayesian EF-1α tree of FSSC isolates, based on 713 nucleotides, with members of the FSSC Clade 1 as outgroup, Table S1: Fungal isolates obtained from P. unifilis eggshells, and putative identity based on ITS data, Table S2: GenBank sequences from 130 fungal specimens used in phylogenetic analyses, Table S3: Bacteria isolated from P. unifilis eggshells and putative identity based on 16S Blast results, Table S4: GenBank 16S sequences from related bacterial species used in phylogenetic analyses.
Author Contributions
Conceptualization, J.M.G.-M., J.M.S.-R. and J.D.-U.; methodology, J.M.G.-M. and J.M.S.-R.; software, J.M.G.-M.; validation, J.M.G.-M.; formal analysis, J.M.G.-M.; investigation, J.M.G.-M.; resources, J.D.-U.; data curation, J.M.G.-M.; writing—original draft preparation, J.M.G.-M.; writing—review and editing, J.M.G.-M., J.M.S.-R. and J.D.-U.; visualization, J.M.G.-M.; supervision, J.D.-U.; project administration, J.D.-U.; funding acquisition, J.D.-U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Real Jardín Botánico (Spanish National Research Council, CSIC), and the MS program “Biodiversity and its Conservation in Tropical Areas” of the Menéndez Pelayo International University-CSIC.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the findings of this study are presented within this article and online Supplementary Materials. Sequences generated here are deposited in GenBank. Any additional data are available on request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.IUCN Tortoise & Freshwater Turtle Specialist Group Podocnemis unifilis (Errata Version Published in 2016). The IUCN Red List of Threatened Species 1996: E.T17825A97397562. [(accessed on 7 August 2021)]. Available online: [DOI]
- 2.CITES Convention on International Trade In Endangered Species of Wild Fauna and Flora. Appendices I, II and III. [(accessed on 7 August 2021)]. Available online: https://cites.org/sites/default/files/eng/app/2020/E-Appendices-2020-08-28.pdf.
- 3.Caputo F.P., Canestrelli D., Boitani L. Conserving the terecay (Podocnemis unifilis, Testudines: Pelomedusidae) through a community-based sustainable harvest of its eggs. Biol. Conserv. 2005;126:84–92. doi: 10.1016/j.biocon.2005.05.004. [DOI] [Google Scholar]
- 4.Wildlife Conservation Society Programa Ecuador, Boletín 2. El Tráfico de Carne Silvestre en el Parque Nacional Yasuní: Caracterización de un Mercado Creciente en la Amazonía Norte del Ecuador. [(accessed on 7 August 2021)]. Available online: http://s3.amazonaws.com/WCSResources/file_20110823_035823_ecu_pub_ProgramaEcuadorBoletin2_2007_oAuB.pdf.
- 5.Norris D., Michalski F. Socio-economic and spatial determinants of anthropogenic predation on yellow-spotted river turtle, Podocnemis unifilis (Testudines: Pelomedusidae), nests in the Brazilian Amazon: Implications for sustainable conservation and management. Zoologia. 2013;30:482–490. doi: 10.1590/S1984-46702013000500003. [DOI] [Google Scholar]
- 6.Norris D., Peres C.A., Michalski F., Gibbs J.P. Prospects for freshwater turtle population recovery are catalyzed by pan-Amazonian community-based management. Biol. Conserv. 2019;233:51–60. doi: 10.1016/j.biocon.2019.02.022. [DOI] [Google Scholar]
- 7.Casal A.C., Fornelino M.M., Restrepo M.F.G., Torres M.A.C., Velasco F.G. Uso histórico y actual de las tortugas charapa (Podocnemis expansa) y terecay (Podocnemis unifilis) en la Orinoquia y la Amazonia. Biota Colomb. 2013;14:45–64. [Google Scholar]
- 8.Cueva R., Utreras V., Muñoz I. Manejo Comunitario de Tortugas Charapas en la Comunidades Kichwa y Waorani del Parque Nacional Yasuní. Wildlife Conservation Society; Quito, Ecuador: 2010. [Google Scholar]
- 9.Wildlife Conservation Society Especies. Programas de Manejo. [(accessed on 7 August 2021)]. Available online: https://ecuador.wcs.org/Especies/Especies-semiacuaticas/Tortugas-charapa/Programa-de-manejo.aspx.
- 10.Soini P. Estudio, reproducción y manejo de los quelonios acuáticos del género Podocnemis (Charapa, Cupiso y Taricaya) en la Cuenca del Rio Pacaya. In: Gonzales G., editor. Seminario Sobre Proyectos de Investigación Ecológica Para el Bosque Tropical Húmedo. Instituto de Investigaciones de la Amazonia Peruana, Dirección General Forestal y de Fauna; Lima, Peru: 1981. pp. 124–143. [Google Scholar]
- 11.Iverson J.B. Patterns of survivorship in turtles (order Testudines) Can. J. Zool. 1991;69:385–391. doi: 10.1139/z91-060. [DOI] [Google Scholar]
- 12.Norris D., Michalski F., Gibbs J.P. Community based actions save yellow-spotted river turtle (Podocnemis unifilis) eggs and hatchlings flooded by rapid river level rises. PeerJ. 2020;8:e9921. doi: 10.7717/peerj.9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fantin C., Machado-Andrade P.C., Pires-Farias I., Pereira D.I. Mirroring-in nature? Comparison of kinship analysis in clutches of the endangered giant Amazon river turtle, Podocnemis expansa (Chelonia: Podocnemididae) in both captivity and natural habitat. Lat. Am. J. Aquat. Res. 2018;46:346–354. doi: 10.3856/vol46-issue2-fulltext-10. [DOI] [Google Scholar]
- 14.Chen P.N. Conservation of the southern river terrapin Batagur affinis (Reptilia: Testudines: Geoemydidae) in Malaysia: A case study involving local community participation. J. Threat. Taxa. 2017;9:10035–10046. doi: 10.11609/jott.3267.9.4.10035-10046. [DOI] [Google Scholar]
- 15.Townsend W.R., Borman A.R., Yiyoguaje E., Mendua L. Cofán Indians’ monitoring of freshwater turtles in Zábalo, Ecuador. Biodivers. Conserv. 2005;14:2743–2755. doi: 10.1007/s10531-005-8410-1. [DOI] [Google Scholar]
- 16.Burke R.L. Head-starting turtles: Learning from experience. Herpetol. Conserv. Biol. 2015;10:299–308. [Google Scholar]
- 17.Boede E., Hernández O. Diseases in arrau sideneck turtle, Podocnemis expansa, kept in Venezuelan farms. Rev. Cient. Fac. Cienc. Vet. 2004;14:395–403. [Google Scholar]
- 18.Arpini C.M., Nóbrega Y.C., Castheloge V.D., Neves D.S., Tadokoro C.E., Costa G.L.d., Oliveira M.M.E., Santos M.R.d.D. Purpuriocillium lilacinum infection in captive loggerhead sea turtle hatchlings. Med. Mycol. Case Rep. 2019;23:8–11. doi: 10.1016/j.mmcr.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitura A., Niemczuk K., Zaręba K., Zając M., Laroucau K., Szymańska-Czerwińska M. Free-living and captive turtles and tortoises as carriers of new Chlamydia spp. PLoS ONE. 2017;12:e0185407. doi: 10.1371/journal.pone.0185407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfitzer S., Boustead K.J., Vorster J.H., du Plessis L., la Grange L.J. Adenoviral hepatitis in two Nile crocodile (Crocodylus niloticus) hatchlings from South Africa. J. S. Afr. Vet. Assoc. 2019;90:1–4. doi: 10.4102/jsava.v90i0.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardi E., Noviello E., Hofmannová L. Protozoa and protozoal infections in chelonians. J. Exot. Pet Med. 2019;31:5–12. doi: 10.1053/j.jepm.2019.06.006. [DOI] [Google Scholar]
- 22.Wyneken J., Burke T., Salmon M., Pedersen D. Egg failure in natural and relocated sea turtle nests. J. Herpetol. 1988;22:88–96. doi: 10.2307/1564360. [DOI] [Google Scholar]
- 23.Girondot M., Fretey J., Prouteau I., Lescure J. Hatchling success for Dermochelys coriacea in a French Guiana hatchery; Proceedings of the Tenth Annual Workshop on Sea Turtle Biology and Conservation; Hilton Head Island, SC, USA. 20–24 February 1990; pp. 229–232. NOAA Technical Memorandum NMFS-SEFC-278. [Google Scholar]
- 24.Craven K.S., Awong-Taylor J., Griffiths L., Bass C., Muscarella M. Identification of bacterial isolates from unhatched loggerhead (Caretta caretta) sea turtle eggs in Georgia, USA. Mar. Turt. Newsl. 2007;115:9–11. [Google Scholar]
- 25.Al-Bahry S., Mahmoud I., Elshafie A., Al-Harthy A., Al-Ghafri S., Al-Amri I., Alkindi A. Bacterial flora and antibiotic resistance from eggs of green turtles Chelonia mydas: An indication of polluted effluents. Mar. Pollut. Bull. 2009;58:720–725. doi: 10.1016/j.marpolbul.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Soslau G., Spotila J.R., Chun A., Yi S., Weber K.T. Potentially lethal bacteria in leatherback turtle eggs in the wild threaten both turtles and conservationists. J. Exp. Mar. Biol. Ecol. 2011;410:101–106. doi: 10.1016/j.jembe.2011.10.018. [DOI] [Google Scholar]
- 27.Sarmiento-Ramírez J.M., van der Voort M., Raaijmakers J.M., Diéguez-Uribeondo J. Unravelling the microbiome of eggs of the endangered sea turtle Eretmochelys imbricata identifies bacteria with activity against the emerging pathogen Fusarium falciforme. PLoS ONE. 2014;9:e95206. doi: 10.1371/journal.pone.0095206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarmiento-Ramírez J.M., Abella-Perez E., Phillott A.D., Sim J., Van West P., Martín M.P., Marco A., Diéguez-Uribeondo J. Global distribution of two fungal pathogens threatening endangered sea turtles. PLoS ONE. 2014;9:e85853. doi: 10.1371/journal.pone.0085853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth C.W., Sarmiento-Ramírez J.M., Short D.P., Diéguez-Uribeondo J., O’donnell K., Geiser D.M. Unraveling the ecology and epidemiology of an emerging fungal disease, sea turtle egg fusariosis (STEF) PLoS Path. 2019;15:e1007682. doi: 10.1371/journal.ppat.1007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons L.M., Khalil L., Marinkelle C. A new nematode genus, Podocnematractis, for Atractis ortleppi Thapar, 1925 (Cosmocercoidea: Atractidae) and the description of another new species, P. colombiaensis from turtles Podocnemis spp. in Colombia. Syst. Parasitol. 1995;30:47–56. doi: 10.1007/BF00009244. [DOI] [Google Scholar]
- 31.Khalil L., Gibbons L.M. Two nematodes, Paratractis hystrix (Diesing, 1851) and Buckleyatractis marinkelli n. g., n. sp. (Atractidae: Cosmocercoidea) from Podocnemis spp. in Colombia. Syst. Parasitol. 1988;12:187–198. doi: 10.1007/BF00007767. [DOI] [Google Scholar]
- 32.Bursey C.R., Reavill D., Greiner E. Pneumoatractis podocnemis n. gen., n. sp. (Nematoda: Atractidae) from the yellow-spotted Amazon River Turtle, Podocnemis unifilis (Testudines: Pelomedusidae) Comp. Parasitol. 2009;76:149–153. doi: 10.1654/4379.1. [DOI] [Google Scholar]
- 33.Jesus R.F., Cardoso E.L., Willkens Y., Futado A.P., dos Santos J.N., Melo F.T. “Long time no see”: Redescription of Orientatractis leiperi (Nematoda: Atractidae) parasite of Podocnemis unifilis (Testudines: Podocnemididae) from Pará State, Brazil. Rev. Mex. Biodivers. 2020;91:913464. [Google Scholar]
- 34.Bradford C.M., Denver M.C., Cranfield M.R. Development of a polymerase chain reaction test for Entamoeba invadens. J. Zoo Wildl. Med. 2008;39:201–207. doi: 10.1638/2007-0145.1. [DOI] [PubMed] [Google Scholar]
- 35.Úngari L.P., Santos A.L.Q., Paiva G.C.M., Mota K.C.P., de Almeida Borges L.F., Cury M.C. Concomitant infection of Haemogregarina sp. and Staphylococcus aureus in free-living yellow-spotted river turtle (Podocnemis unifilis): Case report. Arch. Vet. Sci. 2018;23:50–55. doi: 10.5380/avs.v23i3.59102. [DOI] [Google Scholar]
- 36.Morais P.B.d., Pimenta R.S., Tavares I.B., de Garcia V., Rosa C.A. Yeasts occurring in surface and mouth cavity of two chelonian species, Podocnemis expansa Schweigger and P. unifilis Troschel (Reptilia: Chelonia: Pelomedusidae), in the Javaés River Border of Araguaia National Park in Brazil. Int. J. Microbiol. 2010;2010:504524. doi: 10.1155/2010/504524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morais P.B.d., Souza D.R.d., Sousa F.M.P.d., Oliveira K.W.d., Pimenta R.S. Enterobacteriaceae in mouth and cloaca of Podocnemis expansa and P. unifilis (Testudines: Chelonia) populations of National Park of Araguaia Plains, Brazil. Braz. J. Microbiol. 2011;42:526–530. doi: 10.1590/S1517-83822011000200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miranda J.M.S., Rocha K.d.S., Monteiro L.H., Baia I.W.M., Monteiro T.R.M., Brito J.d.S., Mesquita E.Y.E., Moraes C.C.G.d. Presence of anti-Leptospira spp. antibodies in captive yellow-spotted river turtles (Podocnemis unifilis) in the Eastern Amazon. Cienc. Rural. 2020;50:e20190088. doi: 10.1590/0103-8478cr20190088. [DOI] [Google Scholar]
- 39.Martín M.P., Winka K. Alternative methods of extracting and amplifying DNA from lichens. Lichenologist. 2000;32:189–196. doi: 10.1006/lich.1999.0254. [DOI] [Google Scholar]
- 40.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White T., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press Limited; London, UK: 1990. pp. 315–322. [Google Scholar]
- 42.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cubeta M., Echandi E., Abernethy T., Vilgalys R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology. 1991;81:1395–1400. doi: 10.1094/Phyto-81-1395. [DOI] [Google Scholar]
- 45.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geiser D.M., Al-Hatmi A., Aoki T., Arie T., Balmas V., Barnes I., Bergstrom G.C., Bhattacharyya M.K.K., Blomquist C.L., Bowden R., et al. Phylogenomic analysis of a 55.1 kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology. 2020 doi: 10.1094/PHYTO-08-20-0330-LE. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K., Misawa K., Kuma K.i., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 49.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyaanamoorthy S., Minh B.Q., Wong T.K., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huelsenbeck J.P., Larget B., Alfaro M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. [DOI] [PubMed] [Google Scholar]
- 53.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut A. FigTree v. 1.4.3. 2016. [(accessed on 21 November 2020)]. Available online: http://tree.bio.ed.ac.uk/software/figtree.
- 55.Wheeler W.C. Nucleic acid sequence phylogeny and random outgroups. Cladistics. 1990;6:363–367. doi: 10.1111/j.1096-0031.1990.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 56.Wade T., Rangel L.T., Kundu S., Fournier G.P., Bansal M.S. Assessing the accuracy of phylogenetic rooting methods on prokaryotic gene families. PLoS ONE. 2020;15:e0232950. doi: 10.1371/journal.pone.0232950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Donnell K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 2000;92:919–938. doi: 10.2307/3761588. [DOI] [Google Scholar]
- 58.O’Donnell K., Sutton D.A., Fothergill A., McCarthy D., Rinaldi M.G., Brandt M.E., Zhang N., Geiser D.M. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gleason F.H., Allerstorfer M., Lilje O. Newly emerging diseases of marine turtles, especially sea turtle egg fusariosis (STEF), caused by species in the Fusarium solani complex (FSSC) Mycology. 2020;11:184–194. doi: 10.1080/21501203.2019.1710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey J.B., Lamb M., Walker M., Weed C., Craven K.S. Detection of potential fungal pathogens Fusarium falciforme and F. keratoplasticum in unhatched loggerhead turtle eggs using a molecular approach. Endanger. Species Res. 2018;36:111–119. doi: 10.3354/esr00895. [DOI] [Google Scholar]
- 61.Sarmiento-Ramírez J.M., Abella E., Martín M.P., Tellería M.T., López-Jurado L.F., Marco A., Diéguez-Uribeondo J. Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. FEMS Microbiol. Lett. 2010;312:192–200. doi: 10.1111/j.1574-6968.2010.02116.x. [DOI] [PubMed] [Google Scholar]
- 62.Clusella Trullas S., Paladino F.V. Micro-environment of olive ridley turtle nests deposited during an aggregated nesting event. J. Zool. 2007;272:367–376. doi: 10.1111/j.1469-7998.2006.00277.x. [DOI] [Google Scholar]
- 63.Özdemir B., Türkozan O. Hatching success of original and hatchery nests of the green turtle, Chelonia mydas, in northern Cyprus. Turk. J. Zool. 2006;30:377–381. [Google Scholar]
- 64.Sarmiento-Ramírez J.M., Sim J., Van West P., Dieguez-Uribeondo J. Isolation of fungal pathogens from eggs of the endangered sea turtle species Chelonia mydas in Ascension Island. J. Mar. Biol. Assoc. UK. 2017;97:661–667. doi: 10.1017/S0025315416001478. [DOI] [Google Scholar]
- 65.Hoh D.Z., Lin Y.-F., Liu W.-A., Sidique S.N.M., Tsai I.J. Nest microbiota and pathogen abundance in sea turtle hatcheries. Fungal Ecol. 2020;47:100964. doi: 10.1016/j.funeco.2020.100964. [DOI] [Google Scholar]
- 66.Short D.P., O’Donnell K., Thrane U., Nielsen K.F., Zhang N., Juba J.H., Geiser D.M. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet. Biol. 2013;53:59–70. doi: 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 67.Peinado-Acevedo J.S., Ramírez-Sánchez I.C. Endocarditis by Fusarium keratoplasticum. Mycopathologia. 2020;186:131–133. doi: 10.1007/s11046-020-00502-4. [DOI] [PubMed] [Google Scholar]
- 68.Boutati E.I., Anaissie E.J. Fusarium, a significant emerging pathogen in patients with hematologic malignancy: Ten years’ experience at a cancer center and implications for management. Blood J. Am. Soc. Hematol. 1997;90:999–1008. [PubMed] [Google Scholar]
- 69.Guarro J. Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. Eur. J. Clin. Microbiol. Infect. Dis. 2013;32:1491–1500. doi: 10.1007/s10096-013-1924-7. [DOI] [PubMed] [Google Scholar]
- 70.Dallé da Rosa P., Ramirez-Castrillón M., Borges R., Aquino V., Meneghello Fuentefria A., Zubaran Goldani L. Epidemiological aspects and characterization of the resistance profile of Fusarium spp. in patients with invasive fusariosis. J. Med. Microbiol. 2019;68:1489–1496. doi: 10.1099/jmm.0.001059. [DOI] [PubMed] [Google Scholar]
- 71.Hurtig A.-K., Sebastián M.S. Incidence of childhood leukemia and oil exploitation in the Amazon Basin of Ecuador. Int. J. Occup. Environ. Health. 2004;10:245–250. doi: 10.1179/oeh.2004.10.3.245. [DOI] [PubMed] [Google Scholar]
- 72.San Sebastián M., Karin Hurtig A. Oil exploitation in the Amazon basin of Ecuador: A public health emergency. Rev. Panam. Salud Pública. 2004;15:205–211. doi: 10.1590/S1020-49892004000300014. [DOI] [PubMed] [Google Scholar]
- 73.Maurice L., López F., Becerra S., Jamhoury H., Le Menach K., Dévier M.-H., Budzinski H., Prunier J., Juteau-Martineau G., Ochoa-Herrera V., et al. Drinking water quality in areas impacted by oil activities in Ecuador: Associated health risks and social perception of human exposure. Sci. Total Environ. 2019;690:1203–1217. doi: 10.1016/j.scitotenv.2019.07.089. [DOI] [PubMed] [Google Scholar]
- 74.Sandoval-Denis M., Lombard L., Crous P.W. Back to the roots: A reappraisal of Neocosmospora. Pers. Mol. Phylogeny Evol. Fungi. 2019;43:90–185. doi: 10.3767/persoonia.2019.43.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elshafie A., Al-Bahry S.N., AlKindi A.Y., Ba-Omar T., Mahmoud I. Mycoflora and aflatoxins in soil, eggshells, and failed eggs of Chelonia mydas at Ras Al-Jinz, Oman. Chelonian Conserv. Biol. 2007;6:267–270. doi: 10.2744/1071-8443(2007)6[267:MAAISE]2.0.CO;2. [DOI] [Google Scholar]
- 76.Phillott A.D., Parmenter C.J., Limpus C.J. Occurrence of mycobiota in eastern Australian sea turtle nests. Mem. Qld. Mus. 2004;49:701–703. [Google Scholar]
- 77.Gambino D., Persichetti M.F., Gentile A., Arculeo M., Visconti G., Currò V., Caracappa G., Crucitti D., Piazza A., Mancianti F. First data on microflora of loggerhead sea turtle (Caretta caretta) nests from the coastlines of Sicily. Biol. Open. 2020;9:bio045252. doi: 10.1242/bio.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Güçlü Ö., Bıyık H., Şahiner A. Mycoflora identified from loggerhead turtle (Caretta caretta) egg shells and nest sand at Fethiye beach, Turkey. Afr. J. Microbiol. Res. 2010;4:408–413. [Google Scholar]
- 79.Neves M.S.C., de Melo Moura C.C., de Oliveira L.G. Mycobiota from the eggs, nests and stillbirths of Eretmochelys imbricata Linneus 1766 (Testudines: Cheloniidae) in Pernambuco State, Brazil. Afr. J. Microbiol. Res. 2015;9:1195–1199. [Google Scholar]
- 80.Marco A., Diéguez-Uribeondo J., Abella Pérez E., Martín M.P., Tellería M.T., López-Jurado L.F. Natural colonization of loggerhead turtle eggs by the pathogenic fungus Fusarium oxysporum; Proceedings of the Twenty Sixth Annual Symposium on Sea Turtle Biology and Conservation; Athens, Greece. 3–8 April 2006. [Google Scholar]
- 81.Patiño-Martínez J., Marco A., Quiñones L., Abella E., Abad R.M., Diéguez-Uribeondo J. How do hatcheries influence embryonic development of sea turtle eggs? Experimental analysis and isolation of microorganisms in leatherback turtle eggs. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2012;317:47–54. doi: 10.1002/jez.719. [DOI] [PubMed] [Google Scholar]
- 82.Spadaro D., Gullino M., Garibaldi A. Recent findings on the disease of rice caused by Fusarium fujikuroi. Prot. Delle Colt. 2016;2:11–17. [Google Scholar]
- 83.Sunani S., Bashyal B., Rashmi A., Prakash G. Conidial germination study of Fusarium fujikuroi causing bakanae disease of rice. Environ. Ecol. 2017;35:2790–2794. [Google Scholar]
- 84.Amatulli M.T., Spadaro D., Gullino M.L., Garibaldi A. Conventional and real-time PCR for the identification of Fusarium fujikuroi and Fusarium proliferatum from diseased rice tissues and seeds. Eur. J. Plant Pathol. 2012;134:401–408. doi: 10.1007/s10658-012-9998-0. [DOI] [Google Scholar]
- 85.Borrero C., Capote N., Gallardo M., Avilés M. First report of vascular wilt caused by Fusarium proliferatum on strawberry in Spain. Plant Dis. 2019;103:581. doi: 10.1094/PDIS-09-18-1544-PDN. [DOI] [Google Scholar]
- 86.Xiao R., Wang J., Zheng M., Su H., Zhu Y., Liu B. First report of Fusarium concentricum causing stem rot disease on the medicinal plant Paris polyphylla var. chinensis in China. Plant Dis. 2019;103:1418. doi: 10.1094/PDIS-10-18-1810-PDN. [DOI] [Google Scholar]
- 87.Sidique S.N.M., Azuddin N.F., Joseph J. First report of Fusarium species at nesting sites of endangered sea turtles in Terengganu and Melaka, Malaysia. Malays. Appl. Biol. 2017;46:195–205. [Google Scholar]
- 88.Phillott A.D., Parmenter C.J., Limpus C.J. Mycoflora identified from failed green (Chelonia mydas) and loggerhead (Caretta caretta) sea turtle eggs at Heron Island, Australia. Chelonian Conserv. Biol. 2001;4:170–172. [Google Scholar]
- 89.Candan E.D. Molecular identification of fungal isolates and hatching success of green turtle (Chelonia mydas) nests. Arch. Microbiol. 2018;200:911–919. doi: 10.1007/s00203-018-1496-0. [DOI] [PubMed] [Google Scholar]
- 90.Jacobson E.R., Cheatwood J.L., Maxwell L.K. Mycotic diseases of reptiles. Semin. Avian Exot. Pet Med. 2000;9:94–101. doi: 10.1053/AX.2000.4621. [DOI] [Google Scholar]
- 91.Pedras M.C., Biesenthal C.J. HPLC analyses of cultures of Phoma spp.: Differentiation among groups and species through secondary metabolite profiles. Can. J. Microbiol. 2000;46:685–691. doi: 10.1139/w00-048. [DOI] [PubMed] [Google Scholar]
- 92.Phillott A.D., Parmenter C.J., Limpus C.J., Harrower K. Mycobiota as acute and chronic cloacal contaminants of female sea turtles. Aust. J. Zool. 2002;50:687–695. doi: 10.1071/ZO01057. [DOI] [Google Scholar]
- 93.Nardoni S., Papini R., Marcucci G., Mancianti F. Survey on the fungal flora of the cloaca of healthy pet reptiles. Rev. Med. Vet. 2008;3:159–165. [Google Scholar]
- 94.Oros J., Ramirez A., Poveda J., Rodriguez J., Fernandez A. Systemic mycosis caused by Penicillium griseofulvum in a Seychelles giant tortoise (Megalochelys gigantea) Vet. Rec. 1996;139:295–296. doi: 10.1136/vr.139.12.295. [DOI] [PubMed] [Google Scholar]
- 95.Rosado-Rodríguez G., Maldonado-Ramírez S.L. Mycelial fungal diversity associated with the leatherback sea turtle (Dermochelys coriacea) nests from western Puerto Rico. Chelonian Conserv. Biol. 2016;15:265–272. doi: 10.2744/CCB-1217.1. [DOI] [Google Scholar]
- 96.Glazebrook J., Campbell R., Thomas A. Studies on an ulcerative stomatitis-obstructive rhinitis-pneumonia disease complex in hatchling and juvenile sea turtles Chelonia mydas and Caretta caretta. Dis. Aquat. Org. 1993;16:133–147. doi: 10.3354/dao016133. [DOI] [Google Scholar]
- 97.Chen Q., Jiang J.R., Zhang G.Z., Cai L., Crous P.W. Resolving the Phoma enigma. Stud. Mycol. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barilli E., Cobos M.J., Rubiales D. Clarification on host range of Didymella pinodes the causal agent of pea Ascochyta blight. Front. Plant Sci. 2016;7:592. doi: 10.3389/fpls.2016.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pearce T.L., Wilson C.R., Gent D.H., Scott J.B. Multiple mutations across the succinate dehydrogenase gene complex are associated with boscalid resistance in Didymella tanaceti in pyrethrum. PLoS ONE. 2019;14:e0218569. doi: 10.1371/journal.pone.0218569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larki R., Mehrabi-Koushki M., Farokhinejad R. Ectophoma iranica sp. nov. and new hosts and records of Allophoma spp. in Iran. J. Phytopathol. 2019;167:538–545. doi: 10.1111/jph.12840. [DOI] [Google Scholar]
- 101.Herath K., Harris G., Jayasuriya H., Zink D., Smith S., Vicente F., Bills G., Collado J., González A., Jiang B. Isolation, structure and biological activity of phomafungin, a cyclic lipodepsipeptide from a widespread tropical Phoma sp. Bioorg. Med. Chem. 2009;17:1361–1369. doi: 10.1016/j.bmc.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 102.Pignati M.T., Fernandes L.F., Miorando P.S., Ferreira P.D., Pezzuti J.C.B. Effects of the nesting environment on embryonic development, sex ratio, and hatching success in Podocnemis unifilis (Testudines: Podocnemididae) in an area of Várzea floodplain on the Lower Amazon River in Brazil. Copeia. 2013;2013:303–311. doi: 10.1643/CE-11-146. [DOI] [Google Scholar]
- 103.Pace A., Dipineto L., Fioretti A., Hochscheid S. Loggerhead sea turtles as sentinels in the western mediterranean: Antibiotic resistance and environment-related modifications of gram-negative bacteria. Mar. Pollut. Bull. 2019;149:110575. doi: 10.1016/j.marpolbul.2019.110575. [DOI] [PubMed] [Google Scholar]
- 104.Santoro M., Orrego C.M., Hernández Gómez G. Flora bacteriana cloacal y nasal de Lepidochelys olivacea (Testudines: Cheloniidae) en el pacífico norte de Costa Rica. Rev. Biol. Trop. 2006;54:43–48. doi: 10.15517/rbt.v54i1.13990. [DOI] [PubMed] [Google Scholar]
- 105.Santoro M., Hernandéz G., Caballero M., García F. Potential bacterial pathogens carried by nesting leatherback turtles (Dermochelys coriacea) in Costa Rica. Chelonian Conserv. Biol. 2008;7:104–108. doi: 10.2744/CCB-0666.1. [DOI] [Google Scholar]
- 106.Candan O., Candan E.D. Bacterial diversity of the green turtle (Chelonia mydas) nest environment. Sci. Total Environ. 2020;720:137717. doi: 10.1016/j.scitotenv.2020.137717. [DOI] [PubMed] [Google Scholar]
- 107.Albini. Abril. Franchini. Hüssy. Filioussis Stenotrophomonas maltophilia isolated from the airways of animals with chronic respiratory disease. Schweiz. Arch. Für Tierheilkd. 2009;151:323–328. doi: 10.1024/0036-7281.151.7.323. [DOI] [PubMed] [Google Scholar]
- 108.Martínez-Silvestre A., Verdaguer I., Vidal F., Fortuño L., Franch M.A., Soler J., Velarde R. High mortality associated with thyroid hyperplasia in European pond turtles, Emys orbicularis (L., 1758) (Emydidae) in a breeding facility at the Ebro Delta, NE Spain. Acta Zool. Bulg. Suppl. 2017;10:85–89. [Google Scholar]
- 109.Díaz M.A., Cooper R.K., Cloeckaert A., Siebeling R.J. Plasmid-mediated high-level gentamicin resistance among enteric bacteria isolated from pet turtles in Louisiana. Appl. Environ. Microbiol. 2006;72:306–312. doi: 10.1128/AEM.72.1.306-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petridou E., Filioussis G., Karavanis E., Kritas S.K. Stenotrophomonas maltophilia as a causal agent of pyogranulomatous hepatitis in a buffalo (Bubalus bubalis) J. Vet. Diagn. Investig. 2010;22:772–774. doi: 10.1177/104063871002200522. [DOI] [PubMed] [Google Scholar]
- 111.Mercier-Darty M., Royer G., Lamy B., Charron C., Lemenand O., Gomart C., Fourreau F., Madec J.-Y., Jumas-Bilak E., Decousser J.-W. Comparative whole-genome phylogeny of animal, environmental, and human strains confirms the genogroup organization and diversity of the Stenotrophomonas maltophilia complex. Appl. Environ. Microbiol. 2020;86:e02919. doi: 10.1128/AEM.02919-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delli Paoli Carini A., Ariel E., Picard J., Elliott L. Antibiotic resistant bacterial isolates from captive green turtles and in vitro sensitivity to bacteriophages. Int. J. Microbiol. 2017;2017:5798161. doi: 10.1155/2017/5798161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Warwick C., Arena P.C., Steedman C. Health implications associated with exposure to farmed and wild sea turtles. JRSM Short Rep. 2013;4:1–7. doi: 10.1177/2042533313475574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mendes R., Kruijt M., De Bruijn I., Dekkers E., van der Voort M., Schneider J.H., Piceno Y.M., DeSantis T.Z., Andersen G.L., Bakker P.A., et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 115.Weller D.M., Raaijmakers J.M., Gardener B.B.M., Thomashow L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002;40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 116.Haas D., Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 117.Tang Y., Teng J.L., Cheung C.L., Ngan A.H., Huang Y., Wong S.S., Yip E.K., Ng K.H., Que T.-L., Lau S.K. Tsukamurella serpentis sp. nov., isolated from the oral cavity of Chinese cobras (Naja atra) Int. J. Syst. Evol. Microbiol. 2016;66:3329–3336. doi: 10.1099/ijsem.0.001187. [DOI] [PubMed] [Google Scholar]
- 118.Maeda Y., Stanley T., Stirling J., Griffiths M., Calvert A., Stuart Elborn J., Cherie Millar B., Goldsmith C., Rendall J., Loughrey A. No evidence of transmission of bacteria between reptiles and a CF patient—A case report of a young adult CF patient and reptiles. Zoonoses Public Health. 2010;57:e47–e53. doi: 10.1111/j.1863-2378.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- 119.Martinez-Silvestre A., Mateu-de Antonio E. Bacteriological features of rhinitis in captive Greek tortoises, Testudo graeca. Bull. Assoc. Reptil. Amphib. Vet. 1997;7:12–15. doi: 10.5818/1076-3139.7.2.12. [DOI] [Google Scholar]
- 120.Ahasan M.S., Waltzek T.B., Huerlimann R., Ariel E. Fecal bacterial communities of wild-captured and stranded green turtles (Chelonia mydas) on the Great Barrier Reef. FEMS Microbiol. Ecol. 2017;93:fix139. doi: 10.1093/femsec/fix139. [DOI] [PubMed] [Google Scholar]
- 121.Heynol V., Heckers K.O., Behncke H., Heusinger A., Marschang R.E. Detection of bacteria in oral swabs from healthy commonmusk turtles (Sternotherus odoratus) and West African mud turtles (Pelusios castaneus) J. Herpetol. Med. Surg. 2015;25:33–39. doi: 10.5818/1529-9651-25.1.33. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are presented within this article and online Supplementary Materials. Sequences generated here are deposited in GenBank. Any additional data are available on request to the corresponding author.