Abstract
Two sorts of proteins bind to, and mediate the developmental and homeostatic effects of, retinoic acid (RA): the RAR and RXR nuclear receptors, which act as ligand-dependent transcriptional regulators, and the cellular RA binding proteins (CRABPI and CRABPII). CRABPs are generally known to be implicated in the synthesis, degradation, and control of steady-state levels of RA, yet previous and recent data have indicated that they could play a role in the control of gene expression. Here we show for the first time that, both in vitro and in vivo, CRABPII is associated with RARα and RXRα in a ligand-independent manner in mammalian cells (HL-60, NB-4, and MCF-7). In the nucleus, this protein complex binds the RXR-RAR-specific response element of an RA target gene (RARE-DR5). Moreover, in the presence of retinoids that bind both the nuclear receptors and CRABPII, enhancement of transactivation by RXRα-RARα heterodimers is observed in the presence of CRABPII. Thus, CRABPII appears to be a novel transcriptional regulator involved in RA signaling.
The vitamin A metabolite retinoic acid (RA) is a potent modulator of cell growth and differentiation. It plays a central role in development processes, controls adult tissue homeostasis, and is, clinically, a novel tool for the treatment of skin disorders, the prevention of epidermal cancer, and the treatment of acute promyelocytic leukemia (APL) (12).
The effects of RA are mediated by at least two sorts of proteins, the nuclear receptors and cellular RA binding proteins (CRABPs). The nuclear receptors belong to the steroid/thyroid hormone superfamily, members of which act as ligand-dependent transcription factors (9, 34). CRABPI and CRABPII are small-molecular-size proteins (15 kDa) which belong to a family of proteins, the β-clamp protein family, members of which bind small hydrophobic ligands (39).
So far the function which has been attributed to CRABPs is to protect retinoids in vivo from other cellular proteins, transform bound retinoids into specific biological compounds, and modulate the concentration of free RA available to the nuclear receptors (19). While CRABPI is widely expressed and has been extensively studied, CRABPII has been less thoroughly characterized, due to its low abundance in most tissues. CRABPI and CRABPII have 75% amino acid sequence similarity and are the same size (19, 24). Distinct features of CRABPII such as differential expression in certain tissues (16) and direct control by an RA-responsive element (RARE) (2, 17) indicate that CRABPII may have a function different from that of CRABPI. Of the natural isomers, all-trans RA binds CRABPII with a stronger affinity than 9-cis RA, with Kds of 10 to 20 and 50 to 70 nM, respectively (19).
To date these binding proteins, known to be cytosolic, have not been implicated in nuclear events, although in vitro data in different tissues demonstrate that the presence of CRABPs influences RA efficacy and gene expression (6). Direct control of gene expression by CRABPI has been ruled out previously (48), yet we and others have suggested that a nuclear function of CRABPII may be expected (15, 22, 30). Having observed an increase in RA receptor alpha (RARα) and CRABPII proteins during all-trans RA differentiation therapy in APL (12–14), we investigated the potential role of CRABPII in RAR signaling both in vitro and in vivo. The results strongly place CRABPII as a novel ligand-dependent transcription regulator of the RAR signaling pathway in eucaryotic cells.
MATERIALS AND METHODS
Plasmids.
The human RARβ2 (hRARβ2)–luciferase (Luc) (−5 kb to +155 bp) and RARE3–thymidine kinase (TK)-Luc reporter genes have been described previously (10, 43, 47). Expression vectors for hRARα (pSG5-hRARα), human retinoid X receptor alpha (hRXRα) (pSG5-hRXRα), murine CRABPII (mCRABPII) (pTL1-mCRABPII), and mCRABPI (pSG5-mCRABPI) have been described previously (22, 43). The Gal4 fusion protein expression vector Gal4-RAR(DEF) (36) and the 17-mer ERE-G-chloramphenicol acetyltransferase (CAT) reporter gene (45) have already been described. The Gal4-CRABPII chimeric expression vector was constructed by replacing the human estrogen receptor (ER) exon 7 from the vector Gal4-exon7-F (52) with full-length mCRABPII. For in vitro binding assays, the cDNAs for full-length RARα and RXRα (as well as those for the vitamin D3 receptor [VDR], c-Jun, and ER) were fused to glutathione S-transferase (GST) in the pGEX2T plasmid (Pharmacia) (50). Full-length mCRABPII was cloned into the pET15b plasmid, which directs the synthesis of six-His-tagged fusion protein in Escherichia coli. His-mCRABPII was expressed in E. coli and purified on HiTrap chelating columns (Pharmacia Biotech).
Antibodies.
Mouse monoclonal antibodies (MAbs) against the F region of RARα [MAb 9α(F)], the DE region of RXRα (MAb 4RX3A2 and MAb 4RX1D12), CRABPI (3CRA10F5), or CRABPII (5CRA3B3 and 1CRA4C9) and rabbit polyclonal antibodies against the F region of RARα [RPα(F)] or the A region of RXRα [RPRXα(A)] were described previously (22, 41).
Cells.
HL-60, NB4, MCF-7, and Cos-1 cells were cultured as previously described (11, 30, 43).
Retinoids.
All-trans RA and 9-cis RA were supplied by Hoffmann-La Roche (Basel, Switzerland). CD336, CD2307, and CD582 were provided by Cird-Galderma (Sophia Antipolis, France); Am80 and Ch55 were provided by K. Shudo (Tokyo, Japan).
Transfections and luciferase assays.
HL-60 cells were electroporated as previously described (43) with the CRABPII expression vector (2 μg) and the luciferase reporter gene (hRARβ2-Luc) or RARE3-TK-Luc (5 μg) in the presence or absence of all-trans RA. All transfections were performed with 1 μg of the β-galactosidase expression vector (pCH110) as an internal standard. Cells were harvested 48 h after transfection, and a luciferase assay was performed by a standard procedure. Cos-1 cells were transfected with the same vectors by the calcium phosphate precipitation technique as previously described (43). All the results are expressed as fold induction based on the basal activity of the reporter gene (arbitrarily set at 1) observed in the absence of any receptor expression vector and in the absence of any ligand.
Immunofluorescence.
Cytospun NB4 cells and transiently transfected Cos-1 cells were fixed in 4% paraformaldehyde and incubated overnight at 4°C, with the MAbs 3CRA10F5 (dilution, 1/100), 5CRA3B3 (dilution, 1/50) and 9α(F) and/or the polyclonal antibody RPRXα(A) (dilution, 1/100) or with purified normal mouse immunoglobulin G (IgG) (dilution, 1/50) (Dako, Glostrüp, Denmark) as a control. Then the cells were incubated with antibodies specific for mouse or rabbit immunoglobulin subclasses conjugated to fluorochromes (fluorescein, cyanine 3, or cyanine 5) (dilution, 1/100) (Caltag, San Francisco, Calif.). Nuclei were counterstained with Hoechst 33258. Cells were analyzed by fluorescence microscopy using a confocal laser scanning microscope. The scanning conditions and exposure times in all subsequent photographic processes were identical for all cells from a given experiment.
EMSA, immunoblotting, and immunoprecipitation.
The electrophoretic mobility shift assay (EMSA) procedures used were similar to those previously described (7). In addition to the extracts (2 to 5 μg), reaction mixtures contained 20 μl of binding buffer and the double-stranded DR5 oligonucleotide probe (37 bp) (30 ng) corresponding to the RARE of the RARβ2 natural promoter. Where indicated, extracts of Cos-1 CRABPII-transfected cells or purified bacterially expressed His-mCRABPII was added (at 2 μg or in increasing concentrations). In experiments performed in the presence of RA, Cos-1 cells were cultured in medium with charcoal-dextran prior to transfection. Proteins were resolved on nondenaturing polyacrylamide gels and autoradiographed. For immunoprecipitation, nuclear extracts were incubated with protein A-Sepharose beads cross-linked with MAb 9α(F) or MAb 4RX3A2. The immunoprecipitated proteins were detected by immunoblotting and chemiluminescence.
In vitro binding assays.
In vitro binding assays were performed as previously described (50). Briefly, GST or GST fusion proteins were expressed in E. coli and purified on glutathione-Sepharose beads (Pharmacia). Purified proteins were quantified by a Bradford protein assay and by Coomassie staining after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Purified recombinant His-mCRABPII (500 ng) was incubated at 4°C for 1 h with 20 μg of each of the different GST fusion proteins bound to glutathione-Sepharose beads in a 100-μl total volume of binding buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 0.3 mM dithiothreitol [DTT], 5% glycerol, 0.1% Nonidet P-40). Reactions were performed in the absence or presence of all-trans RA or 9-cis RA (10−7 M). Beads were then washed four times with the same buffer, and bound proteins were eluted with 30 μl of SDS loading buffer, resolved by SDS-PAGE, and analyzed by Western blotting.
RA binding assay.
Extracts (200 μg of protein) were incubated with increasing concentrations of [3H]all-trans RA (2, 4, 10, 30, and 60 nM) in binding buffer (50 mM Tris-HCl [pH 8]–150 mM NaCl–1 mM EDTA–1 mM DTT) in the presence or absence of a 200-fold excess of unlabeled all-trans RA. After 18 h of incubation at 4°C, 0.1 ml of a chilled charcoal-dextran suspension (3% NortiA–0.3% dextran T70 in 50 nM Tris-HCl [pH 8]–10 mM KCl–1 mM DTT) was added to 0.2 ml of the incubated mixture, mixed vigorously, and left for 15 min at 4°C. The tubes were then centrifuged at 5,000 × g for 10 min, and 0.15 ml of supernatant samples was counted for radioactivity. At each retinoid concentration, the number of molecules bound was determined. Radioactivity bound in the presence of a 200-fold molar excess of unlabeled all-trans RA (nonspecific binding) was subtracted from the total binding to obtain the specific binding.
RESULTS
CRABPII is a nuclear protein which enhances RA-mediated gene transcription.
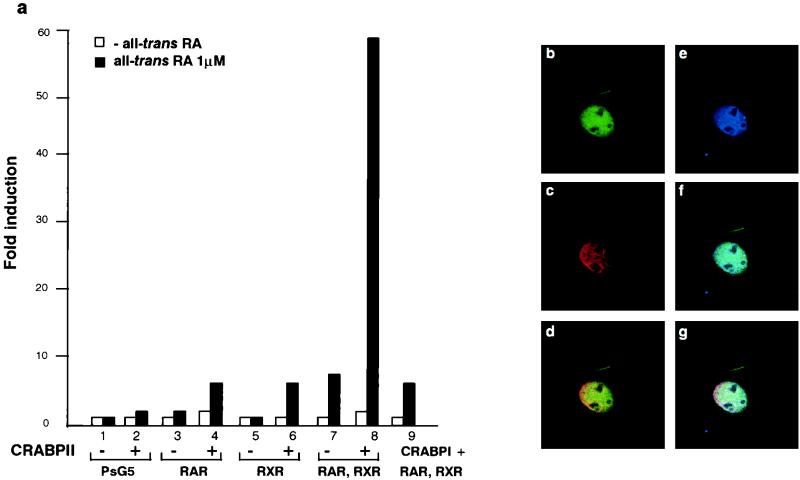
When CRABPII was coexpressed with RARα and RXRα in transiently transfected Cos-1 cells, the RA-dependent activation of the hRARβ2 promoter that contains a DR5 RARE (10, 47) was enhanced ∼10-fold (Fig. 1a, columns 7 and 8). A lower level of stimulation was noted when CRABPII was expressed with either RARα or RXRα (Fig. 1a, columns 3 to 6). Under similar conditions, overexpression of CRABPI did not affect transactivation by RARα-RXRα (Fig. 1a, column 9) (47). Transfection experiments performed with a synthetic RARE3–TK–Luc reporter gene yielded similar results (data not shown), implying that the observed stimulation could be mediated by the DR5 RARE sequence.
FIG. 1.
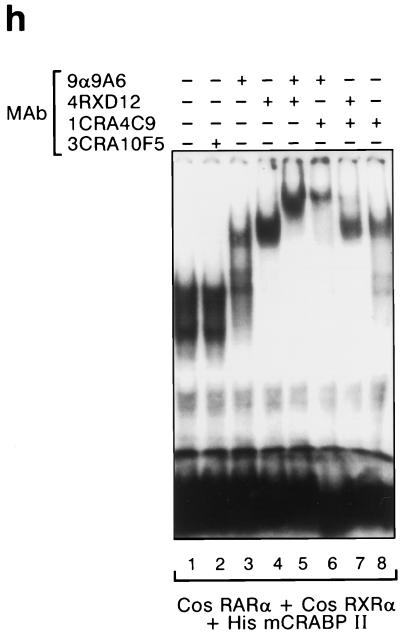
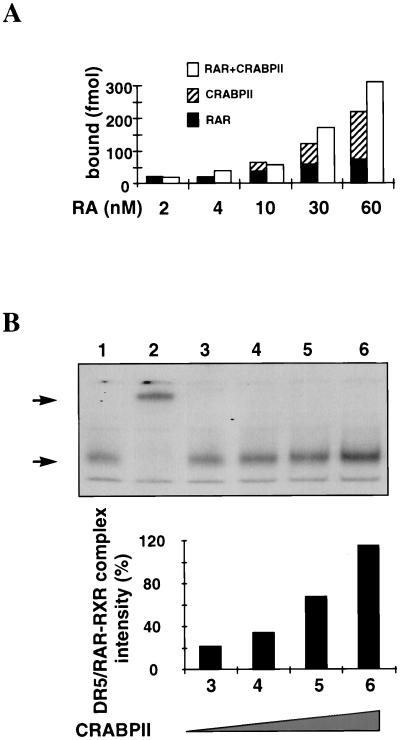
CRABPII is a nuclear protein which participates with the DR5-bound complex to enhance transcription. (a) Cos-1 cells were cotransfected with the RAREβ-Luc reporter gene containing the native RARβ promoter region (−5 kb to +155 bp) (5 μg) and pSG5-hRARα and/or pSG5-hRXRα expression vectors (0.5 μg each) with or without pTL1-CRABPII (2 μg) (columns 1 to 8) or pSG5-mCRABPI (2 μg) (column 9) as previously reported (43). Cells were or were not incubated with all-trans RA (1 μM). The results shown correspond to a representative experiment among at least five. All experiments were normalized to β-galactosidase (1 μg). The results are expressed as fold induction compared to the basal activity of the RAREβ-Luc reporter. (b to g) Immunofluorescence and confocal analysis of Cos-1 cells cotransfected with RARα, RXRα, and CRABPII. (b) Green, 5CRA3B3 (anti-CRABPII). (c) Red, MAb 9α(F) (anti-RARα). (d) Yellow, overlapping red and green fluorescence. CRABPII is localized as RARα in the nucleus. (e) Blue, RPRXα (anti-RXRα). (f) Turquoise, overlapping blue and green fluorescence. CRABPII is in the nucleus with RXRα. (g) White, overlapping green, red, and blue fluorescence. CRABPII is in the nucleus along with RARα and RXRα. (h) EMSA performed with a DR5 probe and nuclear extracts (2 μg) from Cos-1 cells overexpressing RARα and RXRα with added bacterially purified recombinant His-mCRABPII (2 μg). A supershift was obtained with the CRABPII antibody (lane 8), and a super-supershift was obtained when this antibody was combined with the RARα MAb (compare lane 6 with lanes 3 and 8) and, to a lesser extent, with the RXRα MAb.
The use of specific antibodies and confocal microscopy immunofluorescence analysis of Cos-1 cells expressing CRABPII, RARα, and RXRα showed that the distribution of CRABPII in the nucleus (Fig. 1b) was similar to those of RARα (Fig. 1c and d), RXRα (Fig. 1e and f), or both RARα and RXRα (Fig. 1g). Therefore, we hypothesized that CRABPII could be associated with transcriptional DR5 RARE-receptor complexes.
CRABPII is part of the protein complex which binds to RARE.
EMSAs were performed by using purified recombinant mouse CRABPII, extracts from Cos-1 cells transfected with both RARα and RXRα expression vectors (Fig. 1h), and a labeled DR5 RAREβ2 oligonucleotide probe. Upon the addition of recombinant CRABPII to RARα and RXRα, the DR5-bound complex (Fig. 1h, lane 1) was clearly shifted by the CRABPII MAb (Fig. 1h, lane 8) and supershifted upon the further addition of RARα antibody (Fig. 1h, lane 6); addition of the RXRα antibody was less efficient in this supershifting, perhaps reflecting steric hindrance problems (Fig. 1h, lane 7).
CRABPII interacts directly with RARα and RXRα.
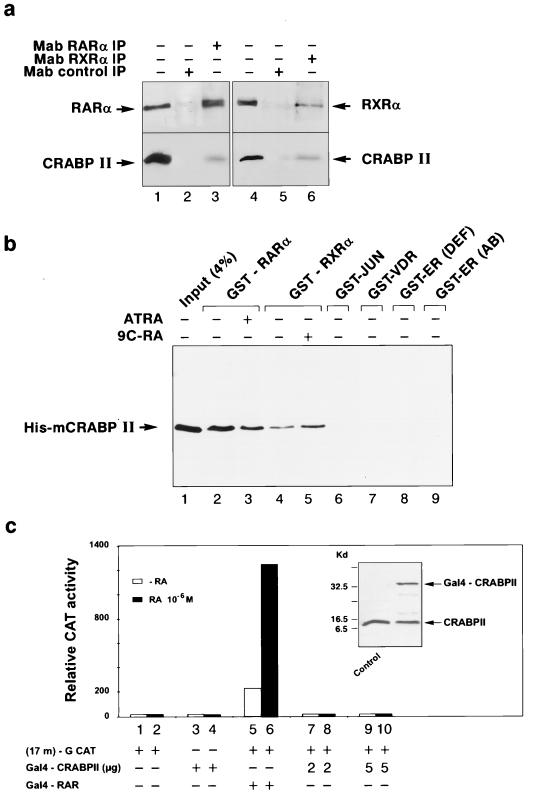
To further investigate the interaction of CRABPII with RARα and RXRα, extracts from Cos-1 cells expressing CRABPII with either RARα or RXRα were immunoprecipitated with MAbs directed against RARα or RXRα. SDS-PAGE, electrotransfer onto a nitrocellulose filter, and immunoblotting with a CRABPII MAb showed that CRABPII could be immunoprecipitated with RARα and RXRα (Fig. 2a, lanes 3 and 6). This interaction was a direct one, as shown by the fact that, irrespective of the presence of ligand, purified bacterially expressed His-tagged mCRABPII was specifically pulled down by the bacterially expressed fusion proteins GST-RARα and GST-RXRα bound to glutathione-Sepharose beads (Fig. 2b, lanes 2 through 5). This interaction was specific to the RA nuclear receptors, as evidenced by the fact that no binding was noted with either the GST-cJun fusion protein (Fig. 2b, lane 6), a GST-VDR protein (Fig. 2b, lane 7), or either of the GST-ER fusions (Fig. 2b, lanes 8 and 9).
FIG. 2.
CRABPII interacts directly with RARα and RXRα in the absence of ligand. (a) Coimmunoprecipitation of CRABPII with RARα and RXRα. Whole-cell extracts from Cos-1 cells (1 mg) cotransfected with CRABPII and RARα or RXRα were immunoprecipitated with MAb 9α(F) (lane 3) or MAb 4RX3A2 (lane 6) and then immunoprobed with RPα(F) (upper panel, lanes 1 to 3), RPRXα(A) (upper panel, lanes 4 to 6), or 5CRA3B3 (lower panels, lanes 1 to 6). Each extract was also immunoprecipitated with nonimmune antibodies (MAb control IP) (lanes 2 and 5). Lanes 1 and 4, unprecipitated extracts from the different cell lines. CRABPII interacts with RARα and RXRα. (b) Binding of GST-RARα (lanes 2 and 3), GST-RXRα (lanes 4 and 5), or other GST fusion proteins (GST-Jun [lane 6], GST-VDR [lane 7], and GST-ER [lanes 8 and 9]) to purified bacterially expressed His-mCRABPII (500 ng) was assessed in a GST pull-down assay as indicated (50). Bound CRABPII was detected by Western blotting with MAb 1CRA4C9. Lane 1 represents 4% of input His-mCRABPII fusion protein. Addition of all-trans RA (ATRA) or 9-cis RA (9C-RA) (0.1 μM) does not affect the direct specific binding (lanes 3 and 5). (c) Cos-1 cells were cotransfected with the (17m)-G-CAT reporter plasmid (36) (1 μg) and chimeric expression vectors encoding the DNA-binding domain of the yeast transcription factor Gal4 fused to either CRABPII (Gal4–CRABPII) (2 or 5 μg) or the DEF regions of RARα (Gal4–RAR) (45) (50 ng). Fold inductions compared to the activity of the control Gal4 expression vector are indicated. No significant activation was observed with Gal4-CRABPII even in the presence of the ligand and despite the confirmed presence of the protein by Western blotting. However, the Gal4–RAR(DEF) (52) expression vector induced an expected increase of CAT activity in the presence of RA.
Altogether, the above results support the existence of a protein-DR5 RARE transcriptional complex in which CRABPII directly interacts with the receptors in a ligand-independent manner to further enhance the transcriptional activity of the RXR-RAR heterodimer in the presence of ligand.
How could CRABPII enhance the transcriptional activity of RARα-RXRα heterodimers in transfected Cos-1 cells? CRABPII did not exhibit any transcriptional activity in Cos-1 cells in the transient transfection assay shown in Fig. 1a, and we further confirmed that when it was fused to the DNA-binding domain of the yeast transcription factor Gal4 (inset in Fig. 2c), no transactivating activity was noted, irrespective of the presence of the ligand (Fig. 2c, columns 7 to 10). Under similar conditions, a control Gal4-RAR(DEF) expression vector induced the expected ligand-dependent increase in CAT activity (Fig. 2c, column 6). Therefore, it seems obvious that CRABPII is not, by itself, a transcriptional factor and that the enhancement of transactivation observed in its presence is linked to its association with RARα and RXRα.
Enhanced transcription through CRABPII requires ligand binding.
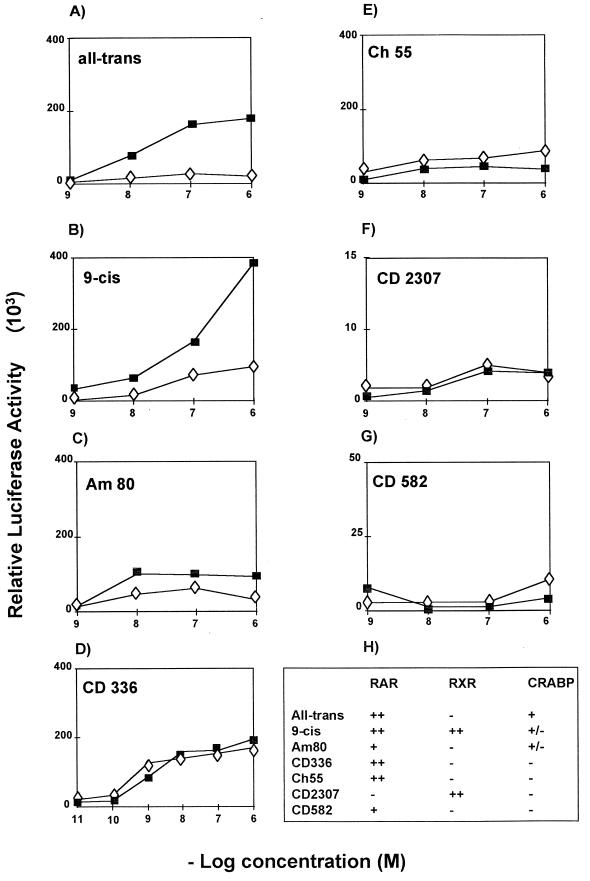
Since, in contrast to other known nuclear receptor transcriptional cofactors (25), CRABPII binds the ligand, we investigated whether its transcriptional stimulatory activity required RA binding. The effect of CRABPII on the transactivation of the RARβ2 promoter-based reporter by RXRα-RARα heterodimers was studied in the presence of RA and synthetic retinoids known to possess different binding affinities for retinoid receptors and CRABPII (Fig. 3H) (1, 7, 20, 23, 44). Enhanced transcription in the presence of CRABPII was observed only with retinoids which bind both CRABPII and the receptors: all-trans RA and 9-cis RA (Fig. 3A and B), Am80 (Fig. 3C), and CD270 (data not shown). Incubation with retinoids reported not to bind CRABPII (Fig. 3D through F), such as the RXRα agonists Ch55 and CD582 or an RARα-specific agonist (CD336), did not elicit transcription enhancement and even reduced it. Thus, enhancement of transcription by CRABPII requires ligand binding to both CRABPII and the nuclear receptors.
FIG. 3.
CRABPII is a ligand-dependent transcriptional regulator. Cos-1 cells were transfected with the luciferase reporter gene hRARβ2-Luc (5 μg) and the RARα and RXRα expression vectors (0.5 μg each) with (■) or without (◊) cotransfection of the CRABPII expression vector (2 μg). The cells were then treated with different retinoids: all-trans RA (A), 9-cis RA (B), Am80 (C), and retinoids which do not bind CRABPII (D through G). (H) The reported relative binding of the different retinoids to the receptors and CRABPII (1, 7, 20, 23, 44) is summarized. Results of one experiment representative of at least three are shown. All experiments were normalized to β-galactosidase (1 μg).
The exact mechanism(s) through which CRABPII induces this enhancement is unknown. As preliminary indications of research, we studied whether the presence of CRABPII enhanced the interaction of all-trans RA with the receptors. Increasing concentrations of [3H]all-trans RA were incubated with either Cos-1 RARα, Cos-1 CRABPII, or a mixture of Cos-1 RARα and Cos-1 CRABPII extracts. The same quantity of proteins was added to each reaction mixture. Figure 4A shows that when CRABPII and RARα are both incubated with increasing concentrations of [3H]all-trans RA, the number of bound molecules is greater than the sum of molecules bound to each protein incubated separately with [3H]all-trans RA. This suggests a positive cooperative effect between CRABPII and RARα.
FIG. 4.
The presence of CRABPII increases the binding of all-trans RA to the receptors and that of the receptors to DR5. (A) RA binding assay. Increasing concentrations of [3H]all-trans RA (2, 4, 10, 30, and 60 nM) were incubated with Cos-1 RARα, Cos-1 CRABPII, or a mixture of Cos-1 RARα and Cos-1 CRABPII extracts. Two hundred micrograms of proteins was added to each reaction. Results are expressed as the number of specifically bound all-trans RA molecules. (B) EMSA performed with a DR5 probe and nuclear extracts (2 μg) from Cos-1 cells overexpressing RARα and RXRα, with increasing concentrations of Cos-1 cells overexpressing CRABPII (1.25, 2.5, 5, and 7.25 μg [lanes 3 to 6, respectively]) added in the presence of 10−9 M all-trans RA. The DR5 bound complex obtained in the presence of 10−9 M all-trans RA (lane 1, arrow), which contains RARα and RXRα, as shown by the supershift (lane 2, arrow) in the presence of anti-RARα and anti-RXRα antibodies, increases with the concentration of CRABPII added (compare lane 3 to lane 4). The relative increase was quantified and expressed in relation to the binding intensity obtained in the absence of CRABPII (lane 1).
We further show that incubation of increasing concentrations of CRABPII (Fig. 4B, lanes 3 to 6) drastically increases the binding of the DR5-bound complex (Fig. 4B, lane 1) which contains RARα and RXRα, as shown by the supershift obtained in the presence of anti-RARα and anti-RXRα antibodies (Fig. 4B, lane 2). CRABPII could facilitate the delivery or accessibility of all-trans RA to the receptors, thereby increasing the binding or stability of the RA nuclear complex (RANC) receptors to the promoters of their target genes.
Transactivation of myeloid cells is enhanced by CRABPII.
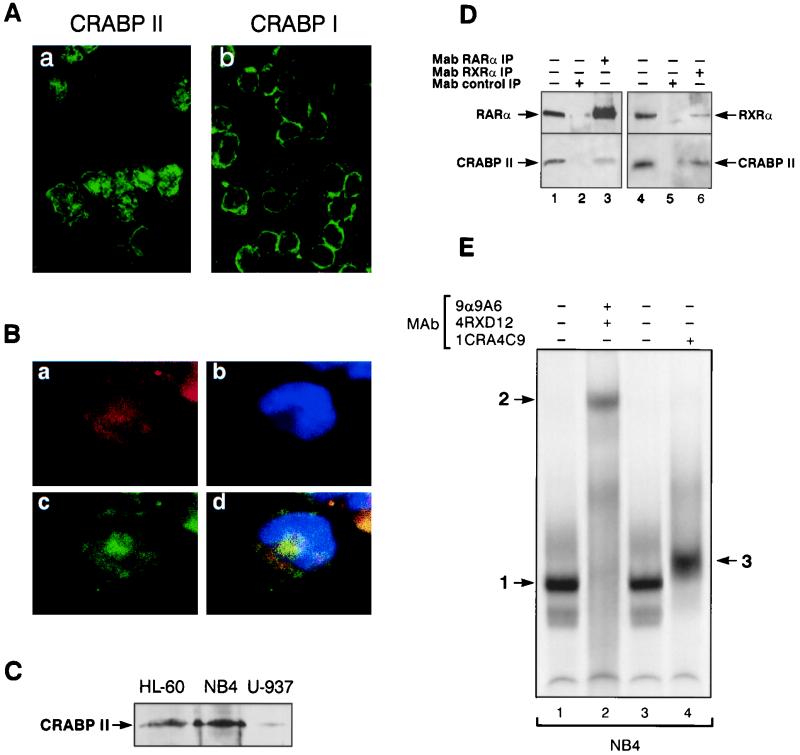
By Western blotting and immunoblotting with specific CRABPII antibodies, we were able to detect CRABPII in myeloid cells. Although CRABPII is found in limited amounts, confocal microscopy analysis showed the presence of CRABPII in both the nucleus and the cytoplasm (Fig. 5 Aa and Bc), whereas CRABPI remained cytoplasmic (Fig. 5Ab). The proportion of CRABPII found in the nucleus or in the cytoplasm was found to vary from one cell to another and from one sample to another, further indicating that CRABPII shuttles between the cytoplasm and the nucleus. Immunolabeling with an RARα antibody (Fig. 5Ba) and fluorochrome bound to a different IgG subtype offered evidence that CRABPII could be found in the same cellular compartments as RARα (Fig. 5Bd). A Western blotting assay of myeloid cell line extracts showed that different amounts of CRABPII were detected in different cell types (Fig. 5C). Last, coimmunoprecipitation using nuclear extracts from NB4 cells, which express the highest levels of CRABPII among myeloid cells (Fig. 5C), allowed us to confirm that indeed endogenous CRABPII interacted with both RARα and RXRα (Fig. 5D, lanes 3 and 6).
FIG. 5.
CRABPII is present in the nuclei of RA-sensitive myeloid cells. (A and B) Confocal microscopy. CRABPII is localized in both the nucleus and the cytoplasm. (A) Immunofluorescence of NB4 cells with MAbs for CRABPII (5CRA3B3) (a) and CRABPI (3CRA10F5) (b). (B) Immunofluorescence of an NB4 cell labeled with MAbs for RARα [MAb 9α(F)] (a, red fluorescence) and CRABPII (5CRA3B3) (c, green fluorescence). (d) Yellow, overlapping red and green fluorescence. (b) Nuclei counterstained with Hoechst 33258. CRABPII is localized with RARα in both the nucleus and the cytoplasm. (C) Western blot analysis of extracts from NB4 (75 μg), HL-60 (150 μg), and U-937 (150 μg) cells with 5CRA3B3, showing the nuclear localization of CRABPII and the different levels of CRABPII in the different cell types. (D) Coimmunoprecipitation of CRABPII with RARα and RXRα. Nuclear extracts from NB4 (1.5 mg) cells were immunoprecipitated with MAb 9α(F) (lane 3) or MAb 4RX3A2 (lane 5) and then immunoprobed with either RPα(F) (upper panel, lanes 1 to 3), RPRXα(A) (upper panel, lanes 4 and 5), or 5CRA3B3 (lower panels, lanes 1 to 5). Unprecipitated extracts were used as controls (lanes 1 and 5). Extracts were immunoprecipitated with nonimmune antibodies (MAb control IP, lanes 2 and 4). CRABPII interacts in vivo with RARα and RXRα. (E) EMSA performed with a DR5 probe and nuclear extracts (2 μg) from NB4 cells. The CRABPII antibody (lane 4) induces a supershift (arrow 3) of the DR5-bound complex (lane 1 or 3, arrow 1), which is shown to comprise RARα and RXRα (lane 2, arrow 2).
The presence of CRABPII in the nucleus and its specific binding to RARα and RXRα strongly suggest that, as observed in Cos-1 transfected cells, CRABPII may participate in the transcription of RA target genes. EMSA studies performed with extracts of NB4 cells show that, as observed in Cos-1 transfected cells, CRABPII is part of the complex which binds to the RAREβ-DR5 oligonucleotide which contains RARα and RXRα (Fig. 5E, lane 1), as shown by the supershift in the presence of anti-RARα and anti-RXRα antibodies (lane 2). Indeed upon incubation with an anti-CRABPII antibody, the complex is clearly shifted (Fig. 5E; compare lanes 3 and 4). CRABPII could facilitate the delivery or accessibility of all-trans RA to the receptors, thereby increasing the binding or stability of the RANC receptors to the promoters of their target genes.
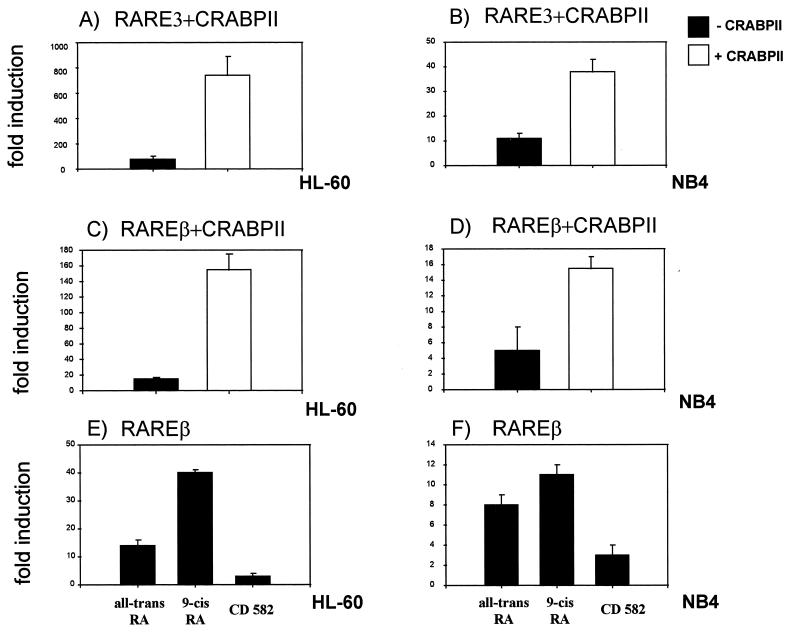
When the same RAREβ reporter gene and the CRABPII expression vector are both transfected in HL-60 or NB4 cells, enhancement of transcription from the endogenous RARs is induced 10- or 2-fold, respectively, with the RAREβ reporter gene (Fig. 6C and D). When similar experiments were performed with a synthetic reporter gene bearing only the nucleotides of a RARE direct repeat (RARE3-TK-Luc) (43), enhancement of transcription was equally observed, confirming that the RAREs of the RARβ promoter are involved (Fig. 6A and B). Spontaneous high levels of CRABPII in NB4 cells may be responsible for a less striking enhancement. In the absence of overexpressed CRABPII, myeloid cells respond to induction of transactivation only with ligands which bind both the nuclear receptors and CRABPII. Thus, in myeloid cells, CRABPII binds the RA nuclear receptors.
FIG. 6.
CRABPII participates in RA-mediated transactivation in myeloid cells. (A through D) HL-60 cells and NB4 cells were cotransfected with either of the reporter genes RARE3-TK-Luc (5 μg) (A and B) or hRARβ2-Luc (5 μg) (C and D) and the CRABPII expression vector (2 μg) and were treated with all-trans RA at 1 μM. (E and F) HL-60 cells and NB4 cells were transfected with the hRARβ2-Luc reporter gene and treated with different retinoids: all-trans RA, 9-cis RA, and a retinoid which does not bind CRABPII (CD582) at 1 μM. In all cases, similar results were obtained in at least five independent experiments, and the results of a representative experiment are shown. All experiments were normalized to β-galactosidase (1 μg). Results are expressed as fold induction compared to the activity of the reporter gene alone.
CRABPII is immunoprecipitated by RARα and RXRα in mammary and teratocarcinoma cells.
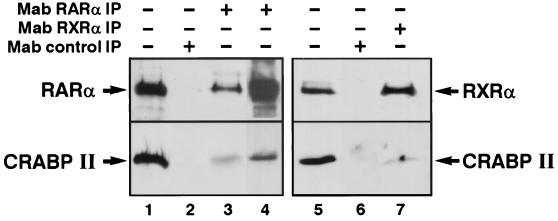
Because RA plays a pivotal role in the control of differentiation and proliferation in other tissues (6, 42), we studied the expression of CRABPII in the nuclear extracts of nonhematopoietic cells, such as mammary and teratocarcinoma cells, known to respond to RA (26). Indeed, in MCF-7 cells, we observe that CRABPII is expressed in the nucleus (Fig. 7, lane 1) and is coimmunoprecipitated with RARα and RXRα. Interestingly, the addition of labeled DR5 increases the amount of bound CRABPII (Fig. 7, lane 4).
FIG. 7.
Coimmunoprecipitation of endogenous CRABPII with RARα and RXRα in mammary cells. Nuclear extracts from MCF-7 cells (1 mg) were immunoprecipitated with MAb 9a(F) (lane 3) or MAb 4RX3A2 (lane 7) and then immunoprobed with either RPα(F) (upper panel, lanes 1 to 4), RPRXa(A) (upper panel, lanes 5 to 7), or 5CRA3B3 (lower panels, lanes 1 to 7). Addition of a DR5 oligonucleotide increases the efficiency of RARα and CRABPII recovery (lane 4). Unprecipitated extracts were used as controls (lanes 1 and 5).
DISCUSSION
In this study, we have confirmed for the first time that CRABPII interacts in vitro and in vivo with the RARs (RARα and RXRα) and participates in the RANC that transactivates RA target genes. Overexpression of CRABPII in transfected Cos-1 cells, as in myeloid cells and in breast cancer cells, as recently reported (30), suggests that these interactions could be implicated in RA-mediated transcription. Indeed, we provide evidence that physical interactions of CRABPII with the receptors are observed in both myeloid and breast cancer cells, bringing strong arguments that these observations may be of physiological importance.
To date the mechanism of action of retinoids in a given cell has been shown to result from the integration of a certain number of signals resulting from specific interactions with proteins of the transcriptional machinery (4, 28, 29), coactivators or corepressors (37, 46), heterodimeric configuration (53), protein phosphorylations (40), and ligand structure and concentration, to name a few more widely studied parameters (9, 25, 51). In this report, we present evidence of a novel level of regulation. CRABPII, a small RA binding protein, which to date was assumed to be only cytoplasmic and linked to the control of the intracellular concentration of RA, is shown to be involved in the enhancement of RA-mediated transactivation and to participate in the DNA-bound nuclear receptor complex. As such, CRABPII could be included in the already defined nuclear receptor coregulator family. Indeed, it binds to RARα and RXRα; its positive control of transcription requires the presence of the RXR-RAR heterodimer, as it cannot on its own bind to the RARE; and it has by itself no transcriptional effect when recruited close to a transcription initiating site.
However, specific characteristics distinguish CRABPII from the coactivators previously described (3, 8, 29, 33, 49). First, it is the sole identified coregulator which binds the ligand. Second, it does not show any structural homology with any of the known activators and does not have an LXXLL sequence (27). Third, it is likely to be specific to the RANC, as it does not bind to any other members of the nuclear receptor family, such as the ER or VDR. Nevertheless, CRABPII may share some features with the other RA nuclear receptor cofactors, such as binding to RXR and RAR in the presence of ligand, with the interaction involving the D and E domains of RARα (unpublished data). To date, most of the coregulators of the nuclear receptors have been assigned specific functions (helicase, protein kinase, histone acetylase, or chromatin activation) (5, 21, 31, 35, 38, 50). CRABPII is the first example of a novel function for a coregulator of the nuclear receptor transcriptional complex, as it binds the ligand and the receptor equally.
These findings raise numerous questions related to our already complex understanding of RA-regulated transcription. It will be interesting to elucidate the specific role of CRABPII in relation to the other nuclear receptor-bound proteins, the transport of the nuclear receptors to the RARE, or any other unknown functions. A hypothesis could be that since CRABPII is an RA-binding and -metabolizing protein, it could bring further local control of target genes at the promoter level and exquisitely coordinate the nuclear signaling of RXR-RAR-mediated transcription in specific conditions. The existing model of mice in which the CRABPII gene has been disrupted will offer us a valuable tool to address these questions.
Indeed, although it may at first appear surprising that despite the novel function of CRABPII, the CRABPII knockout mice we and others have studied show no major abnormalities (18, 32), it is now more frequently observed that disruptions of genes encoding proteins implicated in crucial cell control mechanisms (and even proteins implicated in nuclear receptor complexes such as PML or SRC-1) have not always proved lethal, and their functional consequences have often required rigorous studies (51, 54). In this respect, it should be kept in mind that the role of CRABPII might be observed only under certain conditions which have not yet been addressed.
Numerous features of CRABPs, such as conservation during evolution, coexpression with RARs, regulation of gene expression, and direct control by RA of the CRABPII gene were already indicators that CRABPII played a major role in RA signaling (2, 16, 17). Our results identify a novel level of specific receptor control via nuclear in situ ligand regulation which should be integrated with the already identified interacting factors of nuclear signaling.
ACKNOWLEDGMENTS
We are indebted to V. Giguère, H. de Thé, R. M. Evans, C. K. Glass, and M. P. Gaub for providing plasmids. We thank E. E. Baulieu and R. Losson for a critical analysis of the manuscript, M. P. Gaub for her participation, L. Penna for help in some experiments, G. Linares-Cruz for excellent technical advice on immunocytofluorescence, M. Schmidt and J. Vassy for assistance with confocal microscopy analysis, and the members of the Photography Laboratory of the Institute of Hematology for photography and artworks. We also thank Marie Hélène Schlageter and Philippe Lefebvre for their help in the binding studies and analysis.
This work was supported by grants from the Association pour la Recherche sur le Cancer and from the Fondation sur la Recherche contre la Leucémie. L.D. benefited from a grant from the Association pour la Recherche sur le Cancer. This work was also supported by funds from the CNRS, INSERM, the Hôpital Universitaire de Strasbourg (HUS), and the Collège de France.
L.D. and J.-N.B. contributed equally to this work.
REFERENCES
- 1.Allenby G, Bocquel M T, Saunders M, Kazmer S, Speck J, Rosenberg M, Lovey A, Kastner P, Grippo J F, Chambon P, Levin A A. Retinoic acid receptors and retinoic X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aström A, Pettersson U, Chambon P, Voorhees J J. Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J Biol Chem. 1994;269:22334–22339. [PubMed] [Google Scholar]
- 3.Baniahmad C, Nawaz Z, Baniahmad A, Gleeson M A G, Tsai M, O’Malley B W. Enhancement of human estrogen receptor activity by SPT6: a potential coactivator. Mol Endocrinol. 1995;9:34–39. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- 4.Berkenstam A, del Mar Vivanco Ruiz M, Barettino D, Horikoshi M, Stunnenberg H G. Cooperativity in transactivation between retinoic acid receptor and TFIID requires an activity analogous to E1A. Cell. 1992;69:401–412. doi: 10.1016/0092-8674(92)90443-g. [DOI] [PubMed] [Google Scholar]
- 5.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan J F, Gudas L J. Overexpression of the cellular retinoic acid binding protein-I (CRABPI) results in differentiation-specific gene expression in F9 teratocarcinoma cells. J Cell Biol. 1991;112:965–979. doi: 10.1083/jcb.112.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpentier A, Balitrand N, Rochette-Egly C, Shroot B, Degos L, Chomienne C. Distinct sensitivity of neuroblastoma cells for retinoid receptor agonists: evidence for functional receptor heterodimers. Oncogene. 1997;15:1805–1813. doi: 10.1038/sj.onc.1201335. [DOI] [PubMed] [Google Scholar]
- 8.Cavaillès V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 10.Chen Z, Guidez F, Rousselot P, Agadir A, Chen S J, Wang Z Y, Degos L, Zelent A, Waxman S, Chomienne C. PLZF-RARα fusion proteins generated from the variant t(11;17)(q23;21) translocation in acute promyelocytic leukemia inhibit ligand-dependent transactivation of wild-type retinoic acid receptors. Proc Natl Acad Sci USA. 1994;91:1178–1182. doi: 10.1073/pnas.91.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomienne C, Balitrand N, Ballerini P, Castaigne S, de Thé H, Degos L. All-trans retinoic acid modulates the retinoic acid receptor alpha in promyelocytic cells. J Clin Investig. 1991;88:2150–2154. doi: 10.1172/JCI115547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomienne C, Fenaux P, Degos L. Retinoid differentiation therapy in promyelocytic leukemia. FASEB J. 1996;10:1025–1030. doi: 10.1096/fasebj.10.9.8801163. [DOI] [PubMed] [Google Scholar]
- 13.Cornic M, Delva L, Balitrand N, Guidez F, Micléa J M, Delmer A, Teillet F, Fenaux P, Castaigne S, Degos L, Chomienne C. Induction of retinoic acid-binding protein in normal and malignant human myeloid cells by retinoic acid in acute promyelocytic leukemia patients. Cancer Res. 1992;52:3329–3334. [PubMed] [Google Scholar]
- 14.Delva L, Cornic M, Balitrand N, Guidez F, Micléa J M, Delmer A, Teillet F, Fenaux P, Castaigne S, Degos L, Chomienne C. Resistance to all-trans retinoic acid (ATRA) therapy in relapsing acute promyelocytic leukemia: study of in vitro ATRA sensitivity and cellular retinoic acid binding protein levels in leukemic cells. Blood. 1993;82:2175–2181. [PubMed] [Google Scholar]
- 15.Delva L, Bastie J N, Kraiba R, Guidez F, Balitrand N, Gaub M P, Chambon P, Rochette-Egly C, Chomienne C. CRABPII is part of a nuclear complex which binds to retinoic acid response elements in hematopoietic cells. Blood. 1996;88:48a. . (Abstract 180-I.) [Google Scholar]
- 16.Dollé P, Ruberte E, Kastner P, Petkovich M, Stoner C M, Gudas L J, Chambon P. Differential expression of genes encoding α, β, and γ retinoic acid receptors and CRABP in the developing limbs of the mouse. Nature. 1989;342:702–705. doi: 10.1038/342702a0. [DOI] [PubMed] [Google Scholar]
- 17.Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of mouse CRABPII gene transcription is mediated by RAR/RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- 18.Fawcett D, Pasceri P, Fraser R, Colbert M, Rossant J, Giguère V. Postaxial polydactyly in forelimbs of CRABP-mutant mice. Dev Suppl. 1995;121:671–679. doi: 10.1242/dev.121.3.671. [DOI] [PubMed] [Google Scholar]
- 19.Fiorella P D, Giguère V, Napoli J L. Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli: characterization and comparison to cellular retinoic acid-binding protein (type I) J Biol Chem. 1993;268:21545–21552. [PubMed] [Google Scholar]
- 20.Fogh K, Voorhees J J, Aström A. Expression, purification, and binding properties of human cellular retinoic acid binding protein type I and type II. Arch Biochem Biophys. 1993;300:751–755. doi: 10.1006/abbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- 21.Fraser R A, Heard D J, Adam S, Lavigne A C, Le Douarin B, Tora L, Losson R, Rochette-Egly C, Chambon P. The putative cofactor TIF1α is a protein kinase that is hyperphosphorylated upon interaction with liganded nuclear receptors. J Biol Chem. 1998;273:16199–16204. doi: 10.1074/jbc.273.26.16199. [DOI] [PubMed] [Google Scholar]
- 22.Gaub M P, Lutz Y, Ghyselinck N P, Scheuer I, Pfister V, Chambon P, Rochette-Egly C. Nuclear detection of cellular retinoic acid binding proteins I and II with new antibodies. J Histochem Cytochem. 1998;46:1103–1111. doi: 10.1177/002215549804601002. [DOI] [PubMed] [Google Scholar]
- 23.Gazith J, Eustache J, Watts O, Cavey M T, Shroot B. An improved assay procedure and a new chemically stable ligand for retinoic acid binding protein. Anal Biochem. 1998;88:238–247. doi: 10.1016/0003-2697(88)90481-2. [DOI] [PubMed] [Google Scholar]
- 24.Giguère V. Retinoic acid receptors and cellular retinoic acid binding proteins: complex interplay in retinoid signaling. Endocr Rev. 1994;15:61–79. doi: 10.1210/edrv-15-1-61. [DOI] [PubMed] [Google Scholar]
- 25.Glass C, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 26.Guilbaud N F, Gas N, Dupont M A, Valette A. Effects of differentiation-inducing agents on maturation of human MCF-7 breast cancer cells. J Cell Physiol. 1990;145:162–172. doi: 10.1002/jcp.1041450122. [DOI] [PubMed] [Google Scholar]
- 27.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 28.Ing N H, Beekman J M, Tsai S Y, Tsai M J, O’Malley B W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 29.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 30.Jing Y, Waxman S, Mira-y-Lopez R. The cellular retinoic acid binding protein II is a positive regulator of retinoic acid signalling in breast cancer cells. Cancer Res. 1997;57:1668–1672. [PubMed] [Google Scholar]
- 31.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 32.Lampron C, Rochette-Egly C, Gorry P, Dollé P, Mark M, Lufkin T, LeMeur M, Chambon P. Mice deficient in cellular retinoic acid binding protein II (CRABPII) or in both CRABPI and CRABPII are essentially normal. Development. 1995;121:539–548. doi: 10.1242/dev.121.2.539. [DOI] [PubMed] [Google Scholar]
- 33.Le Douarin B, Zechel C, Garnier J, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand dependent activation function (AF2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangesldorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Ong D, Newcomer M, Chytil F. Cellular retinoid-binding proteins. In: Sporn M B, et al., editors. Retinoids. 2nd ed. New York, N.Y: Raven; 1994. pp. 283–317. [Google Scholar]
- 40.Rochette-Egly C, Adam S, Rossignol M, Egly J M, Chambon P. Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by cdk7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 41.Rochette-Egly C, Lutz Y, Pfister V, Heyberger S, Scheuer I, Chambon P, Gaub M P. Detection of retinoid X receptors using specific monoclonal and polyclonal antibodies. Biochem Biophys Res Commun. 1994;204:525–536. doi: 10.1006/bbrc.1994.2491. [DOI] [PubMed] [Google Scholar]
- 42.Roman S D, Clarke C L, Hall R E, Alexander I E, Sutherland R L. Expression and regulation of retinoic acid receptors in human breast cancers. Cancer Res. 1992;52:2236–2242. [PubMed] [Google Scholar]
- 43.Rousselot P, Hardas B, Patel A, Guidez F, Gäken J, Castaigne S, Dejean A, de Thé H, Degos L, Farzaneh F, Chomienne C. The PML-RARα gene product of the t(15;17) translocation inhibits retinoic acid-induced granulocytic differentiation and mediated transactivation in human myeloid cells. Oncogene. 1994;9:545–551. [PubMed] [Google Scholar]
- 44.Shibakura M, Koyama T, Saito T, Shudo K, Miyasaka N, Kamiyama R, Hirosawa S. Anticoagulant effects of synthetic retinoids mediated via different receptors on human leukemia and umbilical vein endothelial cells. Blood. 1997;90:1545–1551. [PubMed] [Google Scholar]
- 45.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 46.Torchia J, Glass C K, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 47.Umesono K, Giguère V, Glass C, Rosenfeld M G, Evans R. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:185–188. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 48.Venepally P, Reddy L G, Sani B. Analysis of the effects of CRABPI expression on the RA-induced transcription mediated by retinoid receptors. Biochemistry. 1996;35:9974–9982. doi: 10.1021/bi9603276. [DOI] [PubMed] [Google Scholar]
- 49.Voegel J J, Heine J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.vom Baur E, Zechel C, Heery D, Heine M J, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1993;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 52.Webster N J, Green S, Tasset D, Ponglikitmongkol M, Chambon P. The transcriptional activation function located in the hormone-binding domain of the human oestrogen receptor is not encoded in a single exon. EMBO J. 1989;8:1441–1446. doi: 10.1002/j.1460-2075.1989.tb03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O’Malley B W. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]