Abstract
While studies show that nutrient pollution shifts reef trophic interactions between fish, macroalgae, and corals, we know less about how the microbiomes associated with these organisms react to such disturbances. To investigate how microbiome dynamics are affected during nutrient pollution, we exposed replicate Porites lobata corals colonized by the fish Stegastes nigricans, which farm an algal matrix on the coral, to a pulse of nutrient enrichment over a two-month period and examined the microbiome of each partner using 16S amplicon analysis. We found 51 amplicon sequence variants (ASVs) shared among the three hosts. Coral microbiomes had the lowest diversity with over 98% of the microbiome dominated by a single genus, Endozoicomonas. Fish and algal matrix microbiomes were ~20 to 70× more diverse and had higher evenness compared to the corals. The addition of nutrients significantly increased species richness and community variability between samples of coral microbiomes but not the fish or algal matrix microbiomes, demonstrating that coral microbiomes are less resistant to nutrient pollution than their trophic partners. Furthermore, the 51 common ASVs within the 3 hosts indicate microbes that may be shared or transmitted between these closely associated organisms, including Vibrionaceae bacteria, many of which can be pathogenic to corals.
Keywords: coral reefs, Stegastes nigricans, turf algae, 16S, nutrient pollution, marine bacteria, microbial symbiosis
1. Introduction
The role of bottom-up forcing [1] on trophic interactions and nutrient-dependent symbioses is profound, particularly in ecosystems that are oligotrophic, such as coral reefs. It is well established that nutrient enrichment can increase macroalgal growth in the absence of fish herbivory, resulting in corals becoming overgrown, shaded, and/or diseased [2,3,4,5]. These combined negative effects on coral physiological traits (inhibition of coral recruitment and growth) can shift a reef from a coral-dominated to algal-dominated state which makes corals less resilient to disturbances such as bleaching, disease, and hurricanes [5,6,7,8,9].
In this study, we examine coral-associated algal farming fish in the genus Stegastes. These fish farms alter algal assemblages on coral by removing fleshy macroalgae and cultivating filamentous turf algae, which the fish find more palatable [10,11]. Stegastes defend these food resources by demonstrating aggression towards other reef fish such as corallivores, herbivores, and egg predators, while ignoring most carnivores and omnivores [12]. Stegastes territories are dominated by the epilithic algal matrix (EAM), a conglomeration of turf algae, juvenile macroalgae, detritus, invertebrates, and bacterial assemblages [13,14]. Previous studies have documented numerous detrimental effects of EAM on corals including inhibition of coral recruitment [15,16], decreases in coral growth [17,18,19], and coral physiological stress [20,21,22]. Further, evidence suggests that territorial Stegastes farming may promote the development of reservoirs of potential coral disease pathogens (genera Geitlerinema, Leptolyngbya, Oscillatoria, and Sphingomonas) within the EAM [23] and within Stegastes territories (genera Leptolyngbya and Oscillatoria) [10]. Additionally, the presence of territorial Stegastes nigricans increases the rate of coral mortality [23] and may further increase algal growth by enhancing nutrient supply via recycled waste products [24]. While we understand many macro-scale aspects of the trophic relationship between corals, fish, and turf algae under nutrient enrichment, we know much less about the interactions and dynamics among the microbiomes of these hosts under nutrient enrichment.
Studying multiple hosts at the microbial scale gives new insights into the interactions between coral reef organisms. Microbial sampling of the coral-algal interface uncovered unique microbial assemblages characterized by higher microbial abundances and larger microbial cells in the interface compared to the coral or the turf algae [25]. We also previously showed that fishes have the capability to share and transfer microbiome members with corals. This includes the potential coral pathogens Vibrio vulnificus and Photobacterium rosenbergii which significantly changed in abundance in corals exposed to Surgeonfish feces and thermal stress [26]. Parrotfish predation on corals also resulted in increased bacterial alpha diversity and the detection of both beneficial and opportunistic bacteria was only found on corals post predation, indicating direct transmission of bacteria from fish mouth to coral mucus and tissue, or indirectly facilitating bacterial growth or invasion within the coral or from the surrounding environment [27].
At the same time, nutrient pollution in oligotrophic coral reef ecosystems can alter the structure and function of coral holobionts (host, symbiont, microbiome, and virome), mediating changes in the symbiotic interactions among its members [28,29]. Corals rely on their symbiotic dinoflagellate partners, family Symbiodiniaceae, for carbon production [30,31,32] and the breakdown of this symbiosis is known as coral bleaching, which can lead to coral starvation and death [33,34]. Research on inorganic nutrient amendment has revealed the depletion of phosphate can stress coral Symbiodiniaceae and lead to bleaching [35]. Additionally, nitrogen addition can counteract the nitrogen limitation necessary for a stable coral-algal symbiosis and can also lead to bleaching [36,37]. Corals also rely on their bacterial partners, or their microbiomes, to produce antimicrobial compounds that can defend the coral from pathogens [38], and nutrient cycling including carbon, nitrogen, sulfur, and phosphorous [39,40]. The combined effects of overfishing and nutrient enrichment are known to destabilize coral microbiomes and increase putative pathogen loads [29]. Coral microbiomes undergoing stressors such as nutrient enrichment demonstrate higher microbiome variability, particularly when exposed to nitrate or ammonia [41] or the combination of nitrogen and phosphorus [42,43]. In addition, the combination of stressors such as nutrients, predation, and increased temperatures have an antagonistic effect on microbial diversity [44].
Ultimately, shifts in the abundance of limiting nutrients can impact both the trophic interactions among corals, fish, and algae and their microbiomes in ways that negatively impact coral health, resilience, and recovery [45,46,47]. Yet, while much research has focused on the effects of nutrients on the benthic community and coral microbiome dynamics, few, if any, have explored how nutrient pollution might affect microbial interactions among these members of the benthic community. Additionally, it is unclear how the destabilization of the coral microbiome under nutrient enrichment compares to the microbiomes of other hosts in close association with corals. These microbiome comparisons can help us understand whether corals are particularly susceptible or robust to nutrient pollution.
To test how nutrient enrichment might alter microbiome interactions among members of an ecological symbiosis, we compared Porites lobata coral microbiomes to the microbiomes of their resident fishes, Stegastes nigricans, and their farmed EAM gardens of turf algae or the ‘algal matrix’. This in situ 8-week nutrient pulse experiment allowed us (1) to test which microbes are shared among or are unique to the 3 partners, (2) to identify microbes that are variably sensitive to nutrient amendment, and (3) to compare microbiome resistance and resilience across the 3 different hosts that represent unique trophic levels. We hypothesized that coral microbiomes would be uniquely susceptible to nutrient pollution because of their low microbiome diversity [48] while fish and turf algae would be more resistant due to their high microbiome diversity [49,50,51]. Additionally, we hypothesized that fish and algal matrix microbiomes would reflect similar changes during the experiment because of the close trophic symbiosis between the two (e.g., algal farming and consumption by the fish). We also hypothesized that microbiomes would generally increase in diversity and become more variable under nutrient enrichment by supporting bacterial communities with high variability and increased presence and abundance of opportunistic bacterial taxa.
2. Materials and Methods
2.1. Experimental Setup and Sampling
Porites lobata colonies inhabited by Stegastes nigricans fish that were actively farming turf algae were used for this experiment. Colonies were randomly selected along Maharepa reef, in Mo’orea, French Polynesia (17.483194 S, 149.814056 W). Control colonies (n = 7) were left undisturbed and nutrient treated colonies (n = 7) were exposed to slow-release nutrient diffusers with 200 g Osmocote © classic (19-6-12 N-P-K) in 15.25 cm PVC pipes covered in mesh (Figure 1A). Sampling for both treatments began (Time 0 or T0) following nutrient diffuser installation. Coral mucus, tissue, and skeleton were sampled with bonecutters, and stored in 6 mL DNA/RNA Shield (Zymo Research, Irvine, CA, USA) with sterilized matrix A (MP Biomedicals, Santa Ana, CA, USA). Turf algae or the ‘algal matrix’, was sampled with different bonecutters and stored in 2 mL Zymo DNA/RNA shield lysing tubes. Coral and algal matrix samples were taken for all timepoints. We could only obtain fish samples for the first (T0) and last (T4) timepoints because most colonies were inhabited by ~3–5 fish; as sampling required sacrificing the fish, if fish were sampled at each timepoint the original cohort would have been removed and a new cohort that would not have undergone the entire pulse treatment would have inhabited the colony.
Figure 1.
Experimental nutrient enrichment set up. (A) In situ Porites lobata colonies inhabited by turf algae farming Stegastes nigricans, on a northern fringing reef in Mo’orea French Polynesia, were left untreated (n = 7) (top image), or exposed to Osmocote © slow-release nutrient diffusers (n = 7) (bottom image). (B) Sampling of P. lobata, algal matrix, and S. nigricans started on 7/02/18, and continued over an 8-week period, sampling every two weeks. Local rainfall and water temperature was recorded over the experimental period (B top graph). The fish S. nigricans were only sampled on the first and last timepoints (T0 and T4), while the coral P. lobata and turf algae were sampled for every timepoint (T0 to T4) (B bottom images).
For T0, S. nigricans were caught via spear-gun, and for T4 they were anesthetized with a 1:5 clove oil to ethanol solution (Jedwards International, Inc. Braintree, MA, USA) and collected with hand nets. For both time-points S. nigricans were euthanized on the reef in whirl-paks with MS222 (100 mg of MS222 into 0.5 L seawater, balanced to a pH of ~7.5 w/500 mg NaCO3) and kept on ice until dissected (~1 h). In the lab, the S. nigricans were dissected with a sterile scalpel, and organ pieces (kidney, liver, gut) were stored in 2 mL Zymo DNA/RNA shield lysing tubes. All S. nigricans samples were collected under approval of the Institutional Animal Care and Use Committee (IACUC) at Oregon State University (Animal Care and Use Protocol #5056). All host samples stored in DNA/RNA Shield tubes were bead-beaten for 20 min before aliquots of host tissue slurry were taken for DNA extraction.
2.2. DNA Extraction, 16S Library Preparation, Sequencing
To extract DNA from samples from each host, 250 μL of slurry preserved in DNA/RNA Shield (Zymo) was input into the Qiagen DNeasy Power-Soil kit and DNA was extracted according to the kit protocol. Next, the V4 region of the 16S rRNA gene was amplified via 2-step PCR coupling forward and reverse primers 515F (5′-GTG YCA GCM GCC GCG GTA A-3′) [52] and 806R (5′-GGA CTA CNV GGG TWT CTA AT-3′) [53]. First-step PCR was conducted according to the reaction and thermocycler protocol described in Maher et al. 2020 [54]. Second-step PCR was conducted according to the methods described in Ezzat et al. 2021 [26]. Briefly, each 16S band visualized on a 1.5% agarose gel was poked with a pipette tip and swirled into a second-step barcoding master mix solution which was then run on a thermocycler for barcoding. Lastly, amplicons were pooled into equivolume ratios in a single pool and cleaned using Agencourt® AMPure XP beads. Libraries were then sequenced at Oregon State University (OSU) by the Center for Quantitative Life Sciences (CQLS) with v.3 reagent 2 × 300 bp read chemistry on an Illumina MiSeq.
2.3. Data Processing
Demultiplexed reads from the CQLS were trimmed of primers, adapters, and barcodes using Cutadapt (v 3.1). Reads were then processed separately for each host using DADA2 (v 1.16.0) [55] in R (v 4.0.0) [56]. Forward and reverse reads were truncated at their 3’ end at 260 and 210 base pairs, respectively. Sequences were truncated at the first position having a quality score less than or equal to 2, and reads with a total expected error >2 or with the presence of Ns were discarded. Error rates were then learned independently on filtered forward and reverse reads, followed by dereplication and sample inference. Next, forward and reverse reads were merged and an amplicon sequence table was constructed. Two-parent chimeras (bimeras) were removed and taxonomy was assigned at 100% sequence identity using the Silva reference database (v132) [57]. The resulting unique ASVs for each host were imported into phyloseq (v 1.32.0) [58]. ASVs that were annotated as mitochondrial or chloroplast sequences as well as ASVs with a Kingdom classification of “NA” were removed. Two algae samples were determined to have insufficient sequencing depth (<5000 reads) based on the distribution of sequencing depths for algae samples (Figure S1). The removal of these algae samples did not impact the total number of microbial taxa. ASVs were then agglomerated to a genus level classification via phylogenetic distance (tip_glom, h = 0.05) and rarefied to the lowest sequence number for each host (Coral: 2308, Fish: 10,097, Algae: 5973). Changes in read numbers per sample throughout data processing are recorded in the Supplemental Files (Coral: File S1, Algae: File S2, Fish: File S3). Changes in the number of taxa for each host throughout phyloseq filtering is outlined in Table S1. A phylogenetic tree was made from the resulting ASVs for each host using QIIME 2 2019.10 [59]. Briefly, ASVs were aligned with mafft [60] via the q2-alignment plugin and used to construct a phylogeny with fasttree2 [61] via the q2-phylogeny plugin.
2.4. Statistical Analyses
Alpha diversity metrics including observed richness and Simpson’s Diversity index and beta diversity statistics were run separately for each host. To improve the normality of observed richness for each host, observed richness was square-root transformed. Simpson’s index was arcsine-transformed to improve normality for coral samples, while transformation of this metric was not necessary for fish or algae samples. Experimental group effects on each alpha diversity metric were assessed with linear mixed effect models (LMM) using lme4 (v.1.1.23) [62] with time, treatment, and their interaction as fixed effects and individual coral colony as a random effect. Multiple comparisons were performed with estimated marginal means (EMMs) using the emmeans (v.1.4.8) package. For beta diversity statistics, Bray-Curtis dissimilarities were first calculated in phyloseq. Next, Permutational Multivariate Analyses of Variance (PERMANOVA) [63] were conducted to test for differences in bacterial community compositions between treatment groups or across time and between group factorial interactions (Treatment*Time). In addition, Permutational Analyses of Multivariate Dispersions (PERMDISP) [64] were used to test for homogeneity of multivariate dispersions between groups and to calculate the distance to centroid for each sampling group. PERMANOVA and PERMDISP were performed using the functions adonis and betadisper in the package vegan (v.2.5.6) followed by a pairwise analysis of variance with the pairwise.adonis.dm function and permutest in vegan, respectively, with FDR adjusted p-values.
Additionally, changes in the abundance of different bacterial genera across time and treatment for each host were assessed with Analysis of Composition of Microbiomes (ANCOM) with controls for false discovery rate [65]. For each host, an unrarefied ASV table agglomerated to the genus-level (as described above) was used as input into ANCOM. Fish and algae samples were further filtered to only include ASVs with at least 3 counts in 20% of the samples. For each host, ANCOM was run with a model including time, treatment, and their interaction as fixed effects and the individual coral colony as a random effect. A significance level of W = 0.7 was used in which the null hypothesis for a given taxon was rejected in 70% of the tests and p-values were corrected with Benjamini-Hochberg FDR [65,66].
2.5. Shared ASVs
The ASV phyloseq object prior to genus agglomeration, was rarefied to the minimum number of sequences for each host (coral: 2308, fish: 10,097, algae: 5973). Shared ASV’s were found via the Reduce and intersect functions in base R. The relative abundance of the shared ASV’s was calculated via the transform function in the microbiome (v.1.10.0) package and visualized via the plot_heatmap function in phyloseq (v.1.32.0).
2.6. Water Nutrient Analysis
Water samples for nutrient analysis were taken at time-points T0, T2, and T4 from 6 random colonies (n = 3 for each treatment). Water was collected in whirl-paks and stored on ice until filtered (5 mL through 0.2 um). Precipitation and water temperature data was accessed via the Mo’orea Coral Reef LTER data repository [67,68]. At the CEOAS Chemical Analysis Lab (OSU) the continuous segmented flow systems were utilized to determine inorganic nutrients in seawater. Technicon AutoAnalyzer II™ components were used to measure phosphate [69]; and Alpkem RFA 300™ components were used for nitrate plus nitrite and nitrite [70,71]. A detailed description of the continuous segmented flow procedures used can be found in Gordon et al. (1994) [72].
3. Results
3.1. Environmental Conditions Significantly Varied over 8-Week Experimental Period
Throughout the experimental time period, coral health and algal matrix growth were visually monitored during each sampling timepoint. No significant changes in phenotype were observed throughout the experiment for both control and nutrient treated coral colonies. Environmental conditions, such as water temperature and rainfall in the area were recorded, with the greatest increase in rainfall and decrease in temperature occurring between T0 and T1 (Figure 1B). Later timepoints (T2–T4) had steadier changes in temperature and rainfall. Changes in concentration (micromole/liter) of measured phosphate (PO4), nitrate and nitrite (N + N), and nitrite (NO2) over the three sampled time-points (T0, T2, and T4) are plotted in Figure S2. For each nutrient, no significant changes occurred over treatment or time (Kruskal-Wallis rank sum test, Nutrient ~ Treatment, Nutrient ~ Time).
3.2. Corals Have Increased Sensitivity to Nutrient Enrichment Compared to Their Resident Fish and Algal Matrix
Although control corals were left undisturbed, observed species richness changed across the course of the experiment with a transient increase at T3 (6 weeks), which disappeared 2 weeks later at T4 (Figure 2A, blue bars). For nutrient treated corals, within observed species richness increased until T2, stayed similar at T3, and then decreased between T3 and T4 (Figure 2A, red bars). When examined over the whole experiment, nutrient treatment showed significantly increased coral microbial diversity (p = 0.045, Fstat = 4.99) (Figure 2B).
Figure 2.
Alpha diversity results for coral, fish, and algal matrix microbiomes. Only statistically significant results are shown in graphs on the right (B,D,F). (A) Coral microbiome observed richness increases for nutrient treated colonies over time (T0–T2), and then becomes more similar between treatments from timepoint T3–T4. (B) Only treatment had a significant effect on coral microbiome observed richness, with increased diversity for nutrient treated colonies. (C) Algal matrix microbiome observed richness changed more over time than between treatments. (D) Only time had a significant effect on algal matrix observed richness, with diversity decreasing over time. (E) Fish microbiome observed richness increased over time for control colonies and remained similar for nutrient treated colonies. (F) Only the Time*Treatment interaction had a significant effect on fish microbiome Simpson diversity. Lower-case letters indicate statistical significance, i.e. boxes that do not have any letters in common are significantly different from one another (B,D).
In contrast, observed species richness for algal matrix microbiomes decreased over time, with the largest decrease occurring between T1 and T2 (Figure 2C). Algal matrix microbiomes changed more across timepoints than across treatment (Figure 2C) resulting in only ‘time’ having a significant effect on algal matrix microbiome diversity (p = 3.72 × 10−6 Fstat = 10.53) (Figure 2D). Additionally, while within sample diversity (observed and Simpson) increased for control fish microbiomes over time, there was a decrease in diversity for nutrient treated fish microbiomes across time (Figure 2E,F). The treatment*time interaction had the only significant effect on within sample Simpson diversity of fish microbiomes (p = 0.0094, Fstat = 9.52) (Figure 2F). Alpha-diversity statistics for all hosts across all variables tested are recorded in Table S2.
3.3. Between Sample Diversity Changed Significantly for Each Host
We analyzed beta-diversity in two ways: (1) a shift in between sample diversity or the community structure between samples (PERMANOVA) and (2) the dispersion or variability of between sample diversity (PERMDISP; how dissimilar samples were from one another). Coral microbiome between sample diversity was significantly affected by nutrients, while only time had a significant effect on the between sample diversity of algal matrix and fish microbiomes (Figure 3). Between sample diversity was significantly different for control and nutrient treated coral microbiomes (p = 0.006**, R2 = 0.041) (Figure 3A) and dispersion significantly increased for nutrient treated coral microbiomes (p = 0.023*) (Figure 3B). In other words, coral microbiomes became more dissimilar from one another after nutrient exposure. In contrast, between sample diversity significantly changed over time for algal matrix microbiomes (p = 0.006**, R2 = 0.079) (Figure 3C) and dispersion significantly decreased for algal matrix microbiomes over time (p = 0.0027**) (Figure 3D). Like algal matrix microbiomes, between sample diversity was significantly different for fish microbiomes over time (p = 0.001***, R2 = 0.21) (Figure 3E), and dispersion significantly decreased from T0 to T4 (p = 4.93 × 10−5***) (Figure 3F). The treatment*time interaction had a significant effect on algal matrix microbiome dispersion (p = 0.0068) as well as the between sample diversity (p = 0.025*, R2 = 0.069) and dispersion/variability (p = 1.76 × 10−5) of fish microbiomes. Beta-diversity statistics for all hosts across all variables tested are recorded in Tables S3 and S4.
Figure 3.
Significant beta diversity results for coral, fish, and algal matrix microbiomes. Bray-Curtis dissimilarity was significantly different between control and nutrient treated coral microbiomes (A), and was significantly different across time for algal matrix microbiomes ((C), T0–T4) and fish microbiomes ((E), T0 and T4). Dispersion/variability, or distance to centroid, was significantly increased for coral microbiomes under nutrient enrichment compared to coral microbiomes under control conditions (B). Dispersion significantly increased over time for algal matrix microbiomes (D) and decreased over time for fish microbiomes (F). Lower-case letters indicate statistical significance, i.e. boxes that do not have any letters in common are significantly different from one another (B,D,F).
3.4. Coral Microbiomes Are Dominated by One Bacterial Genus While Fish and Algae Microbiomes Are More Even
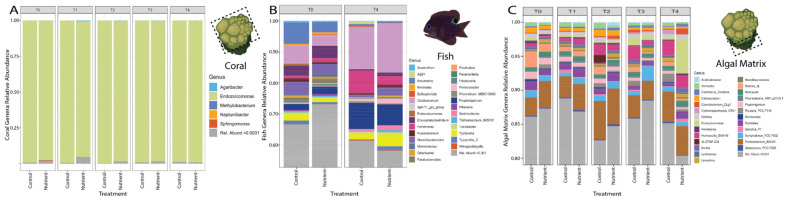
Each host had a unique microbial community (Figure S3), characterized by dominant or minor taxa. Coral microbiomes, across time and treatment, were dominated by the Endozoicomonas bacterial genus, which made up over 90–99% of every sample (Figure 4A). In contrast, both fish and algal matrix microbiomes were dominated by minor taxa. For fish microbiomes (1095 total taxa), ~70% of taxa in T0 had a relative abundance of less than 0.001, and ~60% of taxa in T4 had a relative abundance of less than 0.001 (Figure 4B). Some of the more abundant taxa (rel. abund. > 0.001) in fish T0 microbiomes include Brevinema, Cetobacterium, Erysipelatoclostridium, and Macellibacteroides, and for fish T4 microbiomes include Cetobacterium, Ferrimonas, Propionigenium, and Tyzzerella. Across time and treatment, ~80–85% of taxa in algal matrix microbiomes (6179 total taxa) had a relative abundance of less than 0.001 (Figure 4C). Some more abundant (rel. abund. > 0.001) algal matrix microbiome taxa include Hormoscilla_SI04-45, Moorea_3L, Propionigenium, Symphothece_PCC-7002, and Trichodesmium_IMS101.
Figure 4.
Relative abundance of bacterial genera across the three host microbiomes. (A) Over 90–99% of coral microbiomes were dominated by one genera Endozoicomonas. (B) Approximately 60–70% of fish microbiomes were dominated by genera with a relative abundance of less than 0.001 and changes in taxa are more apparent across time than across treatment. (C) Approximately 80–85% of algal matrix microbiome genera had a relative abundance of less than 0.001 and again, changes in taxa are more apparent across time than across treatment. The range of the y-axis of graphs (B,C) have been altered to highlight the more abundant taxa.
3.5. Fish and Algae Microbiomes Had Several Taxa Significantly Varying in Abundance over Time
For fish microbiomes, 8 bacterial genera (named to the lowest identifiable taxonomy) showed significant changes (ANCOM detection = 0.7) in relative abundance over time (Figure 5A) and all of these, with the exception of the Family Erysipelotrichaceae, were significant for both time and the treatment*time interaction. Family Erysipelotrichaceae and Marinomonas both decreased in abundance over time. Propionigenium, Persicobacter, Sediminitomix, Phormidium_MBIC10003, Family Vibrionaceae (BLAST nr similarities to different Vibro species, Table S5), and Family Shewanellaceae (BLAST nr similarity to Paraferrimonas haliotis, which is in Family Ferrimonadaceae) increased in abundance over time. For fish microbiomes, the taxa identified to genera with significant differential abundance across time (Marinomonas, Propionigenium, Persicobacter, Sediminitomix, Phormidium_MBIC10003) (Figure 5A) were all part of the more abundant genera (abundance >0.001) (Figure 4B).
Figure 5.
Fish and algal matrix bacterial taxa with significant changes in abundance. (A) Seven bacterial taxa (Propionigenium, Persicobacter, Sediminitomix, Marinomonas, Phormidium_MBIC10003, Family Vibrionaceae, and Family Shewanellaceae) within fish microbiomes changed significantly over time and one taxa (Family Erysipelotrichaceae) changed significantly over time and the treatment*time interaction. (B) Four bacterial taxa (Family Flavobacteriaceae, Salinirepens, Kingdom Bacteria, and Class Gammaproteobacteria) within algal matrix microbiomes changed significantly over time and the treatment*time interaction. Coral microbiomes did not have any taxa that significantly changed over treatment, time, or the interaction of these variables. If not identified to genus, the next higher classification taxonomic assignment is used. Darker colors indicate higher abundance, and no color indicates an absence of that taxa.
For algal matrix microbiomes, 4 genera had significant changes (ANCOM detection = 0.7) in relative abundance over time (Figure 5B), and time and the treatment*time interaction had a significant effect on all of these genera (named to the lowest identifiable taxonomy): Family Flavobacteriaceae, Salinirepens, Kingdom Bacteria, and Class Gammaproteobacteria. Family Flavobacteriaceae decreased in abundance from T3 to T4. Salinirepens, Kingdom Bacteria (BLAST nr similarity to uncultured Phylum Planctomycete and uncultured Class Gammaproteobacteria), and Class Gammaproteobacteria (one BLAST nr similarity to Saccharophagus degradans), all increased from T0 to T1, decreased from T1 to T2 and stayed similar to T4. For algal matrix microbiomes, the one taxon identified to genus with significant differential abundance across time (Salinirepens) is part of the minor taxa (abundance <0.001). No genera with significant changes in relative abundance were detected for coral microbiomes.
3.6. Despite a Strong Trophic Interaction Few Microbes Are Shared among the Three Hosts
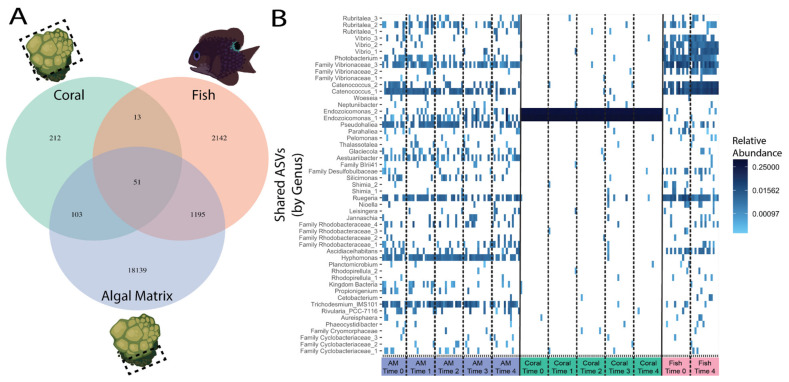
All hosts shared 51 ASVs in common. Fish microbiomes had ~10X and algal matrix microbiomes had ~100× more ASVs than coral microbiomes (Figure 6A). Fish microbiomes shared more ASVs with algal matrix microbiomes than coral microbiomes (Figure 6A). Coral microbiomes shared more ASVs with algal microbiomes than fish microbiomes (Figure 6A). The 51 shared ASVs changed in relative abundance over time, depending on host (Figure 6B). The Endozoicomonas ASVs shared between the 3 hosts were the most dominant out of all 30 Endozoicomonas ASVs found in coral microbiomes (Figure S4). Apart from these dominant Endozoicomonas, all other 49 shared ASVs were generally present in the corals in low abundance, and most increased in abundance at T3 but then decreased at T4. Several shared ASVs were more highly abundant in both fish and algal matrix microbiomes: Rubritalea (_2), Family Vibrionacae (_3), Catenococcus (_1), and Ruegeria. ASVs that were highly abundant in only algal matrix microbiomes include: Pseudohaliea, Aestuariibacter, Hyphomonas, and Trichodesmium_IMS101. ASVs that were highly abundant in only fish microbiomes include: 3 Vibrio ASVs, Catenocccus (_1), and Ascidiaceihabitans. BLAST nr similarities: Kingdom Bacteria ASV to uncultured bacterium, Family Vibrionaceae_2 ASV to Vibrio sagamiensis, Family Cryomorphaceae ASV to uncultured Owenweeksia sp., Family Cyclobacteriaceae_1 and Family Cyclobacteriaceae_3 ASVs to Family Flammeovirgaceae, Family Rhodobacteraceae_3 to Epibacterium sp., Family Rhodobacteraceae_4 to Pseudoruegeria lutimaris and Maritimibacter alkaliphilus.
Figure 6.
Unique and common ASVs across the three host microbiomes. (A) Venn-diagram of unique and shared ASVs between the 3 hosts shows that all hosts had 51 ASVs in common. (B) Relative abundance of these 51 ASVs for each host across time (‘AM’ = algal matrix). Darker colors indicate higher abundance, and no color indicates an absence of that taxa.
4. Discussion
Anthropogenic sources of nutrients contribute to coral reef decline [45,46]. Numerous studies have looked at how nutrient pollution in oligotrophic coral reef ecosystems alters both coral-fish-algae trophic dynamics and the coral holobiont (host, symbiont, microbiome, and virome) structure and function [11,28,29]. For example, previous studies show that Stegastes exclusion and nutrient enrichment increase turf algae cover and alter trophic dynamics in reef ecosystems [11], but the interactions among the microbiomes of the corals, fish, and algae under such experiments are yet to be researched. It is well-established that coral disease, such as tissue loss and decay, is correlated with the destabilization of mutualistic coral microbial communities [73]. However, it was recently shown that overfishing and nutrient pollution surprisingly interact to increase the susceptibility of corals to mortality from typically unharmful fish predation [28,29,47]. Given this link to nutrient enrichment and increased disease and mortality from fish predation, we posited that symbioses between algal matrix farming fishes and their coral hosts would be negatively affected by the addition of nitrogen and phosphorus into highly oligotrophic habitats such as those on the island of Mo’orea. To evaluate both which microbes are shared within this unique trophic interaction and determine how nutrients might alter this relationship, we exposed several Stegastes nigricans colonized Porites lobata corals to nutrient enrichment over 8 weeks and sampled the fishes, farmed algal matrix, and corals at different frequencies.
4.1. Coral Microbiomes Respond Uniquely to Nutrient Enrichment
In this study, nutrient enrichment significantly changed the coral microbiome within and between sample diversity, while enrichment on its own did not have a significant impact on the within and between sample diversity of fish and algal matrix microbiomes (Figure 2 and Figure 3). This indicates that coral microbiomes may be less resistant to nutrient enrichment than algal matrix and fish microbiomes. However, changes in coral microbiome observed richness over time and treatment (Figure 2A) also indicates that coral microbiomes may be resilient to nutrient enrichment, as richness levels between treatments became more similar by T3, which would be the end of the nutrient pulse, and stay similar till the last time point (T4). We previously saw similar patterns in coral microbiomes after exposure to a natural temperature anomaly in which species richness peaked after peak temperatures, and declined following a recovery period [54].
Coral microbiome dispersion appears to be most significantly impacted by nutrient enrichment as it significantly increased in nutrient treated corals, indicating that coral microbiomes become more dissimilar under nutrient enrichment. This significant increase in dispersion is most likely also driving the significant shift we calculated in the community structure between samples (Figure 3A). When calculating significance in community shifts (PERMANOVA) we assume dispersion is equal across our samples, but since this was not the case, the significant changes in dispersion may be influencing the significant shifts in community structure between samples. There does not appear to be as notable a shift in communities in coral samples between treatments (Figure 3A) and algal matrix samples across time (Figure 3C) as there is in fish samples over time (Figure 3E). Therefore, the statistically significant results we see in coral and algal matrix microbiome shifts may be driven by the significant change in dispersion we find in each host (Figure 3B,D).
The unique effect of nutrient enrichment on coral microbiomes may have occurred because coral microbiomes are less diverse compared to fish and algal matrix microbiomes. Our coral microbiome samples were dominated by Endozoicomonas, a common coral-associated bacteria [74]. There were 30 Endozoicomonas ASVs present in coral samples with varying abundances (Figure S4) and the richness of Endozoicomonas ASVs changed more over time rather than treatment (Figure S5A) with a significant increase in richness between T1 and T3 (Figure S5B). Therefore, even though Endozoicomonas accounted for approximately 98% of the coral microbiome for each sample, the diversity of this genera significantly changed over time and not treatment, so it is most likely not a main driver of the increase in diversity we detected for the whole coral microbiome under nutrient enrichment (Figure 2B). This made us question whether less abundant coral microbial taxa may be driving the changes we see in alpha- and beta-diversity. However, no taxa in coral microbiomes were found to be differentially abundant across treatment, time, or the treatment*time interaction.
It was difficult to discern what was driving changes in coral diversity between treatments, however, our ability to compare coral microbiomes to fish and algal matrix microbiomes gave us some more insights. Compared to the coral microbiomes, the more even and diverse fish and algal matrix microbiomes may be contributing to their resistance to change under nutrient enrichment. It is still unclear whether microbial richness and evenness contribute to resistance and resilience [75], yet some microbial experiments have showed that higher evenness and richness support functional stability and contribute to resistance and resilience to environmental stress [76,77]. Fish and algal matrix microbiomes are more diverse and dominated by minor taxa with an average abundance <0.001 across identified genera. We detected differentially abundant taxa in fish and algal matrix microbiomes, and time and the treatment*time interaction were the only variables significantly impacting their microbial diversity. In comparison, no differentially abundant taxa were found in coral microbiomes, even though treatment had an effect on coral microbiome alpha-diversity and dispersion. These results give an initial indication that overall microbiome diversity and evenness may play a role in buffering any changes driven by nutrient enrichment.
4.2. Cryptic Environmental and/or Biological Factors May Be Altering Fish and Algal Matrix Microbiomes
While nutrient enrichment appears to have the most significant effect on coral microbiome dispersion, fish and algal matrix microbiomes were most consistently impacted by time and the treatment*time interaction. We hypothesized that fish and algal matrix microbiomes would respond similarly and would become more variable, or increase in dispersion, over the experimental period. While we saw this trend with algal matrix microbiomes, fish microbiome dispersion decreased over time. This indicates that while Stegastes nigricans eat the turf algae, their microbiomes do not behave similarly. Most research suggests that S. nigricans are exclusively herbivores [78,79] that eat the turf algae they farm, however their gut contents are paradoxically not dominated by turf algae [13,14]. It was suggested that detritus within the algal matrix was the main source of nitrogen within the fish diet, while the algae, invertebrates, and sediment found in algal matrices were underrepresented [14]. This may explain the differences we found in the fish and algal matrix microbiomes. If S. nigricans diet benefits more from the detritus within the algal matrix than any other component, their microbiomes will not be as similar to the algal matrix as we would expect.
Because fish and algal matrix microbiomes did not significantly change in response to nutrient amendment alone, we examined whether the significant changes we see in their microbiomes over time (Figure 2 and Figure 3) correlated with other environmental changes, particularly rainfall, which was measured over the experimental period (Figure 1B). Based on the results of differential abundance (Figure 5) it does not appear that fish and algal matrix changes in taxonomic abundance match these few environmental patterns. Increased rainfall would indicate a decrease in water salinity and a potential decrease in halophilic bacteria. However, microbial taxa identified as Salinirepens, a halophilic bacteria, within the algal matrix microbiomes increase at the same time we observed the largest amount of rainfall (T1, Figure S6A). We also see an absence of the halophilic Marinomonas genera from fish microbiomes at T4, which was preceded with little rainfall (Figure S6B). Again, the high microbial diversity and evenness we found in fish and algal matrix microbiomes may be buffering or complicating microbial changes due to other uncontrolled environmental variables. In addition, we did not measure other variables such as water compounds other than N and P, amount of light exposure, or hormonal/growth changes in the fish, which could be contributing to the changes in differentially abundant bacteria. Additionally, a caveat of our experiment is our inability to know the composition of and changes within fish microbiomes from T1 to T3, since we could only sample at T0 and T4. Microbial samples at these timepoints would have further clarified how fish samples changed over the entire nutrient pulse experiment.
Another caveat of our experiment is our ability to confirm increased nutrient levels from our water samples. Based on the results of changes in nutrient concentration over time (Figure S2), we cannot confirm that nutrient levels actually increased for nutrient treated coral colonies. Our Osmocote © diffusers were set up in a method similar to Vega Thurber et al., 2014, which confirmed an increase in nutrient levels in the water column, however our method for collecting the water was not the same (we did not use syringes), which could be contributing to our results [47]. We also could have sampled Turbinaria growing around our coral colonies to measure increases in nutrient concentrations [80]. This method may provide more accurate measures of nutrient concentrations, which are difficult to detect in oligotrophic waters.
4.3. Shared ASVs Indicate Microbial Transmission Amongst Members of This Trophic Symbiosis
Although we did not directly test the transmission of microbes across the three hosts, examining the shared microbes between the hosts gives us insights into which microbes are potentially being transmitted. The most abundant Endozoicomonas ASV (Figure S4, Endo_Seq1, Endo_Seq2, for sequence see File S4), is the only Endozoicomonas ASV shared among all three hosts (Figure 6, Endozoicomonas_1, Endozoicomonas_2). Its high abundance in corals may lead to its transfer to the algal matrix and then algae to fish, and/or coral directly to fish through consumption during grazing of the coral for algal matrix farming. However, fish and algal microbiomes also contain other Endozoicomonas ASVs with lower abundances and this makes it hard to infer which host is transmitting which Endozoicomonas strain, or whether each strain is endemic to each host. In addition, we expected to see more bacteria shared between fish and algal matrix samples since S. nigricans farm and exclusively eat the turf algal matrix. However, algal matrix microbiomes were the most diverse but only shared about 6% of their bacteria with fish microbiomes, and only 0.5% of their bacteria with coral microbiomes.
From the ASVs that were more abundant in both fish and algal matrix microbiomes, ASVs within the Family Vibrionacae are of interest because they have been shown to proliferate and be pathogenic to corals by forming tissue lesions that affect coral-algal symbiont photosynthetic efficiency (Vibrio coralliilyticus) [81,82,83,84]. In addition, Ruegeria was more abundant in fish and algal matrix microbiomes, and certain strains of this bacteria can inhibit growth of Vibrio species in corals or may be opportunistic pathogens [85]. It is unclear whether these ASVs are being transmitted from either or both the algal matrix and fish to the coral, but their higher abundance in fish and algal matrix microbiomes indicates these hosts as potential vectors of these coral pathogens [26,27,81].
5. Conclusions
Our study is first to examine the microbiomes of three closely-associated hosts in the coral reef ecosystem under one experimental nutrient-pulse period. We found that the three host, Porites lobata, Stegastes nigricans, and its farmed algal matrix, microbiomes respond uniquely to changes across time and/or treatment. The most striking difference between the host microbiomes and their response to nutrient enrichment over time was the diversity and evenness of the microbial communities. Fish and algal matrix microbiomes were more diverse and even than coral microbiomes. This variability may have contributed to fish and algal microbiome resistance to change under nutrient enrichment alone, but does not explain the significant changes we see over time or the treatment*time interaction. Since we only manipulated one environmental variable, it is unclear whether other environmental factors were contributing to these changes or compounding the effect of nutrient enrichment over time. This indicates that while increased microbial diversity may prevent major changes in response to one stressor, such as nutrient pollution, it does not guarantee a stabilized microbiome across time. Conversely coral microbiomes did not change significantly over time, which may be due to their highly uneven microbiome dominated by one genus Endozoicomonas, suggesting it provides a mechanism of stability. However, nutrient enrichment alone had a significant effect on coral microbiome richness and dispersion. The more noticeable effect of nutrient enrichment on coral microbiomes may have to do with the susceptibility of change in the coral-algal symbiosis under nutrient stress [35,36,37,86,87] which leads to a marked change in the overall coral microbiome and coral physiology [88,89].
Our experiment has shown that although these three organisms have tightly linked trophic interactions, their microbiomes tell a different story. While corals provide a structure for algae to grow and for fish to find shelter in, their microbiomes remain unique but susceptible to change under nutrient enrichment. Because the fish actively eat and farm the turf algae, they have a more direct contact with one another, yet their microbiomes are dissimilar and respond differently to temporal changes, in ways that are not intuitively expected. Our study does show that under a stressor such as nutrient enrichment, host microbiomes will respond differently regardless of their proximity or ecological role in a trophic dynamic. In other words, the microbial communities of these hosts do not exactly mirror their macro-scale interactions as the microbiomes respond to nutrient pollution in more complex ways.
The ability to examine each host microbiome under a single experiment also allowed us to uncover which bacteria were shared among the three hosts. The 51 shared ASVs give us the first insights into how linked the microbiomes of these coral reef organisms are. While we were unable to specify the role of mechanism behind bacterial sharing, the number of ASVs shared between the physically linked microbiomes reflected trophic dynamics, (i.e., fish, which directly touch/feed on the and algal matrix shared more ASVs with the algal matrix than with coral microbiomes) while also highlighting how unique each microbiome is despite the interconnectedness of these hosts. More work needs to be done to understand the mechanisms and pathways of microbial transmission among these organisms as well to understand the functional nature of these relationships. Nevertheless, our study shows that microbiomes provide innovative insights into how anthropogenic stressors are impacting vulnerable but essential marine ecosystem members via their smallest members—the microbes.
Acknowledgments
Thank you to the University of California Gump Station Staff for their aid and resources: Hinano Teavai-Murphy, Jacques You Sing, Irma You Sing, Tony You Sing, Valentine Brotherson, and Neil Davies. Thank you to the Mo’orea Coral Reef LTER Network for their aid, advice, and resources: Russell J. Schmitt, Sally Holbrook, Andrew Brooks, and Sam Degregori. And thank you to Joe Jennings Jr. at the CEOAS Chemical Analysis Lab (OSU) for conducting the water nutrient analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9091873/s1, Figure S1: Histogram of the number of read counts within algal matrix microbiome samples, Figure S2: Water nutrient concentrations over time, Figure S3: Community structures of the ASVs found in each host, Figure S4: Relative abundance of coral microbiome Endozoicomonas ASVs across time, Figure S5: Changes in coral Endozoicomonas within sample diversity, Figure S6: Changes in algal matrix Salinirepens and fish Marinomonas abundance compared to changes in rainfall across time, Table S1: Changes in taxa numbers throughout data processing pipeline, Table S2: Effects of treatment, time, and treatment*time interaction on microbial community alpha diversity metrics, Table S3: Effects of treatment, time, and treatment*time interaction on microbial community dissimilarity, Table S4: Effects of treatment, time, and treatment*time interaction on microbial community group dispersion, Table S5: BLASTn similarities (top 5 based on Max Score) of differentially abundant genera identified within Family Vibrionaceae in fish microbiome samples, File S1: coral_read_nums.txt, File S2: algae_read_nums.txt, File S3: fish_read_nums.txt, File S4: Coral_Endo_ASV_seq.txt.
Author Contributions
Conceptualization, A.M., S.S.M. and R.V.T.; methodology, A.M., S.S.M., R.L.M.; validation, A.M., R.L.M. and R.V.T.; formal analysis, A.M. and R.L.M.; resources, A.M. and R.V.T.; data curation, A.M.; writing—original draft preparation, A.M.; writing—review and editing, R.L.M., S.S.M. and R.V.T.; visualization, A.M.; funding acquisition, R.V.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the NSF Ocean Sciences grant #1635913 to RV, used for sample collection, and the NSF Ocean Sciences grant #2023424 to RV, used for sequencing data generation.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Oregon State University (Animal Care and Use Protocol #5056 approved 19 June 2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw read data can be found at NCBI SRA, BioProject ID: PRJNA752131, which will be made public once the manuscript is published.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stock C.A., Powell T.M., Levin S.A. Bottom–up and Top–down Forcing in a Simple Size-Structured Plankton Dynamics Model. J. Mar. Syst. 2008;74:134–152. doi: 10.1016/j.jmarsys.2007.12.004. [DOI] [Google Scholar]
- 2.Williams I., Polunin N. Large-Scale Associations between Macroalgal Cover and Grazer Biomass on Mid-Depth Reefs in the Caribbean. Coral Reefs. 2001;19:358–366. doi: 10.1007/s003380000121. [DOI] [Google Scholar]
- 3.Friedlander A., Brown E., Monaco M. Defining Reef Fish Habitat Utilization Patterns in Hawaii: Comparisons between Marine Protected Areas and Areas Open to Fishing. Mar. Ecol.-Prog. Ser. 2007;351:221–233. doi: 10.3354/meps07112. [DOI] [Google Scholar]
- 4.Tebbett S.B., Goatley C.H.R., Bellwood D.R. The Effects of Algal Turf Sediments and Organic Loads on Feeding by Coral Reef Surgeonfishes. PLoS ONE. 2017;12:e0169479. doi: 10.1371/journal.pone.0169479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith J.E., Brainard R., Carter A., Grillo S., Edwards C., Harris J., Lewis L., Obura D., Rohwer F., Sala E., et al. Re-Evaluating the Health of Coral Reef Communities: Baselines and Evidence for Human Impacts across the Central Pacific. Proc. R. Soc. B Biol. Sci. 2016;283:20151985. doi: 10.1098/rspb.2015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkepile D.E., Hay M.E. Herbivore vs. Nutrient Control of Marine Primary Producers: Context-Dependent Effects. Ecology. 2006;87:3128–3139. doi: 10.1890/0012-9658(2006)87[3128:HVNCOM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Hughes T.P., Rodrigues M.J., Bellwood D.R., Ceccarelli D., Hoegh-Guldberg O., McCook L., Moltschaniwskyj N., Pratchett M.S., Steneck R.S., Willis B. Phase Shifts, Herbivory, and the Resilience of Coral Reefs to Climate Change. Curr. Biol. 2007;17:360–365. doi: 10.1016/j.cub.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 8.Bellwood D.R., Pratchett M.S., Morrison T.H., Gurney G.G., Hughes T.P., Álvarez-Romero J.G., Day J.C., Grantham R., Grech A., Hoey A.S., et al. Coral Reef Conservation in the Anthropocene: Confronting Spatial Mismatches and Prioritizing Functions. Biol. Conserv. 2019;236:604–615. doi: 10.1016/j.biocon.2019.05.056. [DOI] [Google Scholar]
- 9.Tebbett S.B., Bellwood D.R. Algal Turf Sediments on Coral Reefs: What’s Known and What’s Next. Mar. Pollut. Bull. 2019;149:110542. doi: 10.1016/j.marpolbul.2019.110542. [DOI] [PubMed] [Google Scholar]
- 10.Casey J.M., Ainsworth T.D., Choat J.H., Connolly S.R. Farming Behaviour of Reef Fishes Increases the Prevalence of Coral Disease Associated Microbes and Black Band Disease. Proc. R. Soc. B Biol. Sci. 2014;281:20141032. doi: 10.1098/rspb.2014.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gobler C., Gutierrez D., Davis T., Peterson B., Liddle L. Algal Assemblages Associated with Stegastes Sp. Territories on Indo-Pacific Coral Reefs: Characterization of Diversity and Controls on Growth. J. Exp. Mar. Biol. Ecol. 2006;336:135–145. doi: 10.1016/j.jembe.2006.04.012. [DOI] [Google Scholar]
- 12.Johnson M.K., Holbrook S.J., Schmitt R.J., Brooks A.J. Fish Communities on Staghorn Coral: Effects of Habitat Characteristics and Resident Farmerfishes. Environ. Biol. Fish. 2011;91:429–448. doi: 10.1007/s10641-011-9802-6. [DOI] [Google Scholar]
- 13.Wilson S., Bellwood D., Choat J., Furnas M. Detritus in the Epilithic Algal Matrix and Its Use by Coral Reef Fishes. Oceanogr. Mar. Biol. Annu. Rev. 2003;41:279–309. [Google Scholar]
- 14.Wilson S., Bellwood D.R. Cryptic Dietary Components of Territorial Damselfishes (Pomacentridae, Labroidei) Mar. Ecol. Prog. Ser. 1997;153:299–310. doi: 10.3354/meps153299. [DOI] [Google Scholar]
- 15.Birrell C.L., McCook L.J., Willis B.L. Effects of Algal Turfs and Sediment on Coral Settlement. Mar. Pollut. Bull. 2005;51:408–414. doi: 10.1016/j.marpolbul.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Vermeij M.J.A., Smith J.E., Smith C.M., Vega Thurber R., Sandin S.A. Survival and Settlement Success of Coral Planulae: Independent and Synergistic Effects of Macroalgae and Microbes. Oecologia. 2009;159:325–336. doi: 10.1007/s00442-008-1223-7. [DOI] [PubMed] [Google Scholar]
- 17.Birrell C., Mccook L., Willis B., Diaz-Pulido G. Effects of Benthic Algae on the Replenishment of Corals and the Implications for the Resilience of Coral Reefs. Oceanogr. Mar. Biol. 2008;46:25–51. [Google Scholar]
- 18.River G.F., Edmunds P.J. Mechanisms of Interaction between Macroalgae and Scleractinians on a Coral Reef in Jamaica. J. Exp. Mar. Biol. Ecol. 2001;261:159–172. doi: 10.1016/S0022-0981(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 19.Titlyanov E.A., Yakovleva I.M., Titlyanova T.V. Interaction between Benthic Algae (Lyngbya Bouillonii, Dictyota Dichotoma) and Scleractinian Coral Porites Lutea in Direct Contact. J. Exp. Mar. Biol. Ecol. 2007;342:282–291. doi: 10.1016/j.jembe.2006.11.007. [DOI] [Google Scholar]
- 20.Barott K.L., Rodriguez-Mueller B., Youle M., Marhaver K.L., Vermeij M.J.A., Smith J.E., Rohwer F.L. Microbial to Reef Scale Interactions between the Reef-Building Coral Montastraea Annularis and Benthic Algae. Proc. R. Soc. B Biol. Sci. 2012;279:1655–1664. doi: 10.1098/rspb.2011.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barott K., Smith J., Dinsdale E., Hatay M., Sandin S., Rohwer F. Hyperspectral and Physiological Analyses of Coral-Algal Interactions. PLoS ONE. 2009;4:e8043. doi: 10.1371/journal.pone.0008043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas A., El-Zibdah M., Wild C. Seasonal Monitoring of Coral–Algae Interactions in Fringing Reefs of the Gulf of Aqaba, Northern Red Sea. Coral Reefs. 2010;29:93–103. doi: 10.1007/s00338-009-0556-y. [DOI] [Google Scholar]
- 23.Casey J., Connolly S., Ainsworth T. Coral Transplantation Triggers Shift in Microbiome and Promotion of Coral Disease Associated Potential Pathogens. Sci. Rep. 2015;5:11903. doi: 10.1038/srep11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchette A., Ely T., Zeko A., Sura S.A., Turba R., Fong P. Damselfish Stegastes Nigricans Increase Algal Growth within Their Territories on Shallow Coral Reefs via Enhanced Nutrient Supplies. J. Exp. Mar. Biol. Ecol. 2019;513:21–26. doi: 10.1016/j.jembe.2019.02.001. [DOI] [Google Scholar]
- 25.Roach T.N.F., Little M., Arts M.G.I., Huckeba J., Haas A.F., George E.E., Quinn R.A., Cobián-Güemes A.G., Naliboff D.S., Silveira C.B., et al. A Multiomic Analysis of in Situ Coral–Turf Algal Interactions. Proc. Natl. Acad. Sci. USA. 2020;117:13588–13595. doi: 10.1073/pnas.1915455117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezzat L., Merolla S., Clements C.S., Munsterman K.S., Landfield K., Stensrud C., Schmeltzer E.R., Burkepile D.E., Vega Thurber R. Thermal Stress Interacts With Surgeonfish Feces to Increase Coral Susceptibility to Dysbiosis and Reduce Tissue Regeneration. Front. Microbiol. 2021;12:620458. doi: 10.3389/fmicb.2021.620458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ezzat L., Lamy T., Maher R.L., Munsterman K.S., Landfield K.M., Schmeltzer E.R., Clements C.S., Vega Thurber R.L., Burkepile D.E. Parrotfish Predation Drives Distinct Microbial Communities in Reef-Building Corals. Anim. Microbiome. 2020;2:5. doi: 10.1186/s42523-020-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaver E., Shantz A., McMinds R., Burkepile D., Vega Thurber R., Silliman B. Effects of Predation and Nutrient Enrichment on the Success and Microbiome of a Foundational Coral. Ecology. 2016;98:830–839. doi: 10.1002/ecy.1709. [DOI] [PubMed] [Google Scholar]
- 29.Zaneveld J.R., Burkepile D.E., Shantz A.A., Pritchard C.E., McMinds R., Payet J.P., Welsh R., Correa A.M.S., Lemoine N.P., Rosales S., et al. Overfishing and Nutrient Pollution Interact with Temperature to Disrupt Coral Reefs down to Microbial Scales. Nat. Commun. 2016;7:11833. doi: 10.1038/ncomms11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearse V.B., Muscatine L. Role of Symbiotic Algae (Zooxanthellae) in Coral Calcification. Biol. Bull. 1971;141:350–363. doi: 10.2307/1540123. [DOI] [Google Scholar]
- 31.Rinkevich B., Loya Y. Oriented Translocation of Energy in Grafted Reef Corals. Coral Reefs. 1983;1:243–247. doi: 10.1007/BF00304422. [DOI] [Google Scholar]
- 32.Rinkevich B., Loya Y. Coral Illumination through an Optic Glass-Fiber: Incorporation of 14C Photosynthates. Mar. Biol. 1984;80:7–15. doi: 10.1007/BF00393121. [DOI] [Google Scholar]
- 33.Hayes R.L., Bush P.G. Microscopic Observations of Recovery in the Reef-Building Scleractinian Coral, Montastrea Annularis, after Bleaching on a Cayman Reef. Coral Reefs. 1990;8:203–209. doi: 10.1007/BF00265012. [DOI] [Google Scholar]
- 34.Hughes T.P., Baird A.H., Bellwood D.R., Card M., Connolly S.R., Folke C., Grosberg R., Hoegh-Guldberg O., Jackson J.B.C., Kleypas J., et al. Climate Change, Human Impacts, and the Resilience of Coral Reefs. Science. 2003;301:929–933. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- 35.D’Angelo C., Wiedenmann J. Impacts of Nutrient Enrichment on Coral Reefs: New Perspectives and Implications for Coastal Management and Reef Survival. Curr. Opin. Environ. Sustain. 2014;7:82–93. doi: 10.1016/j.cosust.2013.11.029. [DOI] [Google Scholar]
- 36.Wooldridge S. A New Conceptual Model for the Warm-Water Breakdown of the Coral-Algae Endosymbiosis. Mar. Freshw. Res. 2009;60:483–496. doi: 10.1071/MF08251. [DOI] [Google Scholar]
- 37.Wooldridge S.A., Done T.J. Improved Water Quality Can Ameliorate Effects of Climate Change on Corals. Ecol. Appl. 2009;19:1492–1499. doi: 10.1890/08-0963.1. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie K.B. Regulation of Microbial Populations by Coral Surface Mucus and Mucus-Associated Bacteria. Mar. Ecol. Prog. Ser. 2006;322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 39.Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F. Metagenomic Analysis of the Microbial Community Associated with the Coral Porites Astreoides. Environ. Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 40.Robbins S.J., Singleton C.M., Chan C.X., Messer L.F., Geers A.U., Ying H., Baker A., Bell S.C., Morrow K.M., Ragan M.A., et al. A Genomic View of the Reef-Building Coral Porites Lutea and Its Microbial Symbionts. Nat. Microbiol. 2019;4:2090–2100. doi: 10.1038/s41564-019-0532-4. [DOI] [PubMed] [Google Scholar]
- 41.Rice M.M., Maher R.L., Vega Thurber R., Burkepile D.E. Different Nitrogen Sources Speed Recovery from Corallivory and Uniquely Alter the Microbiome of a Reef-Building Coral. PeerJ. 2019;7:e8056. doi: 10.7717/peerj.8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaneveld J.R., McMinds R., Vega Thurber R. Stress and Stability: Applying the Anna Karenina Principle to Animal Microbiomes. Nat. Microbiol. 2017;2:1–8. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Shantz A.A., Payet J.P., Sharpton T.J., Foster A., Burkepile D.E., Vega Thurber R. Corals and Their Microbiomes Are Differentially Affected by Exposure to Elevated Nutrients and a Natural Thermal Anomaly. Front. Mar. Sci. 2018;5:1–14. doi: 10.3389/fmars.2018.00101. [DOI] [Google Scholar]
- 44.Maher R.L., Rice M.M., McMinds R., Burkepile D.E., Vega Thurber R. Multiple Stressors Interact Primarily through Antagonism to Drive Changes in the Coral Microbiome. Sci. Rep. 2019;9:6834. doi: 10.1038/s41598-019-43274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szmant A. Nutrient Enrichment on Coral Reefs: Is It a Major Cause of Coral Reef Decline? Estuaries. 2002;25:743–766. doi: 10.1007/BF02804903. [DOI] [Google Scholar]
- 46.Fabricius K. Coral Reefs: An Ecosystem in Transition. Springer; Dordrecht, The Netherlands: 2011. Factors Determining the Resilience of Coral Reefs to Eutrophication: A Review and Conceptual Model; pp. 493–505. [Google Scholar]
- 47.Thurber R.L.V., Burkepile D.E., Fuchs C., Shantz A.A., McMinds R., Zaneveld J.R. Chronic Nutrient Enrichment Increases Prevalence and Severity of Coral Disease and Bleaching. Glob. Chang. Biol. 2014;20:544–554. doi: 10.1111/gcb.12450. [DOI] [PubMed] [Google Scholar]
- 48.McDevitt-Irwin J.M., Baum J.K., Garren M., Vega Thurber R.L. Responses of Coral-Associated Bacterial Communities to Local and Global Stressors. Front. Mar. Sci. 2017;4:262. doi: 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- 49.Ghanbari M., Kneifel W., Domig K.J. A New View of the Fish Gut Microbiome: Advances from next-Generation Sequencing. Aquaculture. 2015;448:464–475. doi: 10.1016/j.aquaculture.2015.06.033. [DOI] [Google Scholar]
- 50.Parris D.J., Brooker R.M., Morgan M.A., Dixson D.L., Stewart F.J. Whole Gut Microbiome Composition of Damselfish and Cardinalfish before and after Reef Settlement. PeerJ. 2016;4:e2412. doi: 10.7717/peerj.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hester E.R., Barott K.L., Nulton J., Vermeij M.J., Rohwer F.L. Stable and Sporadic Symbiotic Communities of Coral and Algal Holobionts. ISME J. 2016;10:1157–1169. doi: 10.1038/ismej.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parada A.E., Needham D.M., Fuhrman J.A. Every Base Matters: Assessing Small Subunit RRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 53.Apprill A., McNally S., Parsons R., Weber L. Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 54.Maher R.L., Schmeltzer E.R., Meiling S., McMinds R., Ezzat L., Shantz A.A., Adam T.C., Schmitt R.J., Holbrook S.J., Burkepile D.E., et al. Coral Microbiomes Demonstrate Flexibility and Resilience Through a Reduction in Community Diversity Following a Thermal Stress Event. Front. Ecol. Evol. 2020;8:356. doi: 10.3389/fevo.2020.555698. [DOI] [Google Scholar]
- 55.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R: The R Project for Statistical Computing. [(accessed on 5 August 2021)]. Available online: https://www.r-project.org/
- 57.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurdie P.J., Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Author Correction: Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019;37:1091. doi: 10.1038/s41587-019-0252-6. [DOI] [PubMed] [Google Scholar]
- 60.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bates D., Mächler M., Bolker B., Walker S. Fitting Linear Mixed-Effects Models Using Lme4. arXiv. 20141406.5823 [Google Scholar]
- 63.Anderson M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral. Ecol. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 64.Anderson M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics. 2006;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 65.Mandal S., Van Treuren W., White R.A., Eggesbø M., Knight R., Peddada S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 67.Leichter J., Seydel K., Gotschalk C. MCR LTER: Coral Reef: Benthic Water Temperature, Ongoing Since 2005 Ver 11. [(accessed on 2 April 2021)]. Available online: [DOI]
- 68.Washburn L., Brooks A.J. MCR LTER: Coral Reef: Gump Station Meteorological Data, Ongoing since 2006 Ver 45. [(accessed on 2 April 2021)]. Available online: [DOI]
- 69.Bernhardt H., Wilhelms A. The Continuous Determination of Low Level Iron, Soluble Phosphate and Total Phosphate with the AutoAnalyzer TM. Technicon Symp. 1967;1:386. [Google Scholar]
- 70.Armstrong F.A.J., Stearns C.R., Strickland J.D.H. The Measurement of Upwelling and Subsequent Biological Process by Means of the Technicon Autoanalyzer® and Associated Equipment. Deep Sea Res. A. 1967;14:381–389. doi: 10.1016/0011-7471(67)90082-4. [DOI] [Google Scholar]
- 71.Patton C.J. Ph.D. Thesis. Michigan State University; East Lansing, MI, USA: 1983. Design, Characterization and Applications of a Miniature Continuous Flow Analysis System. [Google Scholar]
- 72.Gordon L., Jennings J., Ross A., Krest J. A Suggested Protocol for Continuous Flow Automated Analysis of Seawater Nutrients (Phosphate, Nitrate, Nitrite and Silicic Acid) in the WOCE Hydrographic Program and the Joint Global Ocean Fluxes Study. 1993. [(accessed on 2 August 2021)]. p. 91. Available online: http://www.ioccp.org/images/06Nutrients/WOCE_nutrients-manual_1993.pdf.
- 73.Vega Thurber R., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R.A., Angly F., Dinsdale E., Kelly L., Rohwer F. Metagenomic Analysis of Stressed Coral Holobionts. Environ. Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 74.Neave M.J., Apprill A., Ferrier-Pagès C., Voolstra C.R. Diversity and Function of Prevalent Symbiotic Marine Bacteria in the Genus Endozoicomonas. Appl. Microbiol. Biotechnol. 2016;100:8315–8324. doi: 10.1007/s00253-016-7777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shade A., Peter H., Allison S.D., Baho D., Berga M., Buergmann H., Huber D.H., Langenheder S., Lennon J.T., Martiny J.B., et al. Fundamentals of Microbial Community Resistance and Resilience. Front. Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Griffiths B.S., Ritz K., Bardgett R.D., Cook R., Christensen S., Ekelund F., Sørensen S.J., Bååth E., Bloem J., Ruiter P.C.D., et al. Ecosystem Response of Pasture Soil Communities to Fumigation-Induced Microbial Diversity Reductions: An Examination of the Biodiversity–Ecosystem Function Relationship. Oikos. 2000;90:279–294. doi: 10.1034/j.1600-0706.2000.900208.x. [DOI] [Google Scholar]
- 77.Wittebolle L., Marzorati M., Clement L., Balloi A., Daffonchio D., Heylen K., De Vos P., Verstraete W., Boon N. Initial Community Evenness Favours Functionality under Selective Stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 78.Ceccarelli D.M., Jones G.P., McCook L.J. Territorial Damselfishes as Determinants of the Structure of Benthic Communities on Coral Reefs. Oceanogr. Mar. Biol. Annu. Rev. 2001;39:355–389. [Google Scholar]
- 79.Meekan M.G., Steven A.D.L., Fortin M.J. Spatial Patterns in the Distribution of Damselfishes on a Fringing Coral Reef. Coral Reefs. 1995;14:151–161. doi: 10.1007/BF00367233. [DOI] [Google Scholar]
- 80.Bender-Champ D., Diaz-Pulido G., Dove S. Effects of Elevated Nutrients and CO2 Emission Scenarios on Three Coral Reef Macroalgae. Harmful Algae. 2017;65:40–51. doi: 10.1016/j.hal.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 81.Gibbin E., Gavish A., Krueger T., Kramarsky-Winter E., Shapiro O., Guiet R., Jensen L., Vardi A., Meibom A. Vibrio Coralliilyticus Infection Triggers a Behavioural Response and Perturbs Nutritional Exchange and Tissue Integrity in a Symbiotic Coral. ISME J. 2019;13:989–1003. doi: 10.1038/s41396-018-0327-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pollock F.J., Morris P.J., Willis B.L., Bourne D.G. Detection and Quantification of the Coral Pathogen Vibrio Coralliilyticus by Real-Time PCR with TaqMan Fluorescent Probes. Appl. Environ. Microbiol. 2010;76:5282–5286. doi: 10.1128/AEM.00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sussman M., Mieog J.C., Doyle J., Victor S., Willis B.L., Bourne D.G. Vibrio Zinc-Metalloprotease Causes Photoinactivation of Coral Endosymbionts and Coral Tissue Lesions. PLoS ONE. 2009;4:e4511. doi: 10.1371/journal.pone.0004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de O Santos E., Alves N., Dias G.M., Mazotto A.M., Vermelho A., Vora G.J., Wilson B., Beltran V.H., Bourne D.G., Le Roux F., et al. Genomic and Proteomic Analyses of the Coral Pathogen Vibrio Coralliilyticus Reveal a Diverse Virulence Repertoire. ISME J. 2011;5:1471–1483. doi: 10.1038/ismej.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo D., Wang X., Feng X., Tian M., Wang S., Tang S.-L., Ang P., Yan A., Luo H. Population Differentiation of Rhodobacteraceae along with Coral Compartments. ISME J. 2021;15:1–17. doi: 10.1038/s41396-021-01009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fabricius K.E. Effects of Terrestrial Runoff on the Ecology of Corals and Coral Reefs: Review and Synthesis. Mar. Pollut. Bull. 2005;50:125–146. doi: 10.1016/j.marpolbul.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 87.Stambler N., Popper N., Dubinsky Z., Stimson J. Effects of Nutrient Enrichment and Water Motion on the Coral Pocillopora Damicornis. 1991. [(accessed on 6 June 2021)]. Available online: https://scholarspace.manoa.hawaii.edu/bitstream/10125/1396/v45n3-299-307.pdf.
- 88.Dougan K.E., Ladd M.C., Fuchs C., Vega Thurber R., Burkepile D.E., Rodriguez-Lanetty M. Nutrient Pollution and Predation Differentially Affect Innate Immune Pathways in the Coral Porites Porites. Front. Mar. Sci. 2020;7:742–751. doi: 10.3389/fmars.2020.563865. [DOI] [Google Scholar]
- 89.Morris L.A., Voolstra C.R., Quigley K.M., Bourne D.G., Bay L.K. Nutrient Availability and Metabolism Affect the Stability of Coral–Symbiodiniaceae Symbioses. Trends Microbiol. 2019;27:678–689. doi: 10.1016/j.tim.2019.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw read data can be found at NCBI SRA, BioProject ID: PRJNA752131, which will be made public once the manuscript is published.