Abstract
The effective control of rodent populations on farms is crucial for food safety, as rodents are reservoirs and vectors for several zoonotic pathogens. Clear links have been identified between rodents and farm-level outbreaks of pathogens throughout Europe and Asia; however, comparatively little research has been devoted to studying the rodent–agricultural interface in the USA. Here, we address this knowledge gap by metabarcoding bacterial communities of rodent pests collected from Minnesota and Wisconsin food animal farms. We leveraged the Oxford Nanopore MinION sequencer to provide a rapid real-time survey of putative zoonotic foodborne pathogens, among others. Rodents were live trapped (n = 90) from three dairy and mixed animal farms. DNA extraction was performed on 63 rodent colons along with 2 shrew colons included as outgroups in the study. Full-length 16S amplicon sequencing was performed. Our farm-level rodent-metabarcoding data indicate the presence of multiple foodborne pathogens, including Salmonella spp., Campylobacter spp., Staphylococcus aureus, and Clostridium spp., along with many mastitis pathogens circulating within five rodent species (Microtus pennsylvanicus, Mus musculus, Peromyscus leucopus, Peromyscus maniculatus, and Rattus norvegicus) and a shrew (Blarina brevicauda). Interestingly, we observed a higher abundance of enteric pathogens (e.g., Salmonella) in shrew feces compared to the rodents analyzed in our study. Knowledge gained from our research efforts will directly inform and improve farm-level biosecurity efforts and public health interventions to reduce future outbreaks of foodborne and zoonotic disease.
Keywords: agriculture, 16S amplicon sequencing, metabarcoding, nanopore sequencing, dairy cattle, One Health, Peromyscus leucopus, Mus musculus, Blarina brevicauda, Rattus norvegicus
1. Introduction
Rodents are the largest group of mammals in the world, and they are well known for harboring a plethora of zoonotic pathogens of concern for human and animal health [1]. Both native and invasive species of mice and rats benefit from human activities, especially agricultural systems. Rodents are a common hindrance of food production systems globally and they are known to transmit zoonotic pathogens to food animals and raw produce by contaminating the overall farm environment [2,3,4,5]. This transmission is largely due to the amplification of foodborne pathogens through the daily deposition of urine and fecal pellets into the production environment. For example, a single rodent within a barn or food-production facility can introduce upwards of 23 million Salmonella bacteria into production pipelines within 24 h [6,7]. However, the functional role that peridomestic (i.e., living in and around human habitations) rodents serve in the amplification and transmission of various zoonoses is likely underappreciated. Clear links have been identified between rodent pests and outbreaks of zoonotic diseases throughout Europe and Asia [8,9,10,11,12]; yet, little research has been devoted to studying this relationship in the United States [4,13]. Specifically, regional studies focused on specific rodent species and their pathogen reservoir status across the diverse agricultural landscapes of the United States are lacking. Hence, our overarching research goal was to investigate the role of rodent pests on food animal farms as reservoirs or carriers of zoonotic pathogens, especially with respect to species-specific patterns.
Emerging genomic technologies are providing exciting new opportunities for the surveillance of zoonotic pathogens in diverse settings and environments. Next-generation sequencing platforms allow for the metabarcoding of complex bacterial communities using taxonomically informative genes (e.g., the 16S rRNA gene). 16S rRNA sequence data are particularly useful as a molecular marker for bacterial identification, including for pathogens with clinical relevance [14,15]. The 16S rRNA gene has nine hypervariable regions (V1-V9) with varying levels of phylogenetic signal, of which the V3 and V4 regions are particularly useful for resolving genus and species-level relationships [16]. Second-generation sequencing platforms frequently used for bacterial metabarcoding experiments (e.g., Illumina MiSeq and NextSeq) provide high per-base accuracy and sequencing throughput; however, the resulting data consist of relatively short (~300 bp) reads, often permitting the analysis of particular subregions of the full-length (~1550 bp) 16S rRNA gene [17]. Alternatively, the Oxford Nanopore Technologies (ONT) MinION sequencer is a third-generation single-molecule sequencing platform that can sequence exceptionally long DNA fragments (i.e., thousands to millions of bases in length) [18,19]. For this reason, the MinION can sequence the entire ~1550 bp 16S rRNA gene, thus providing two to five times greater coverage of the 16S rRNA gene when compared to sequencing data originating from second-generation technologies. Full-length 16S sequence data provide a greater number of phylogenetically informative characters, thus enhancing downstream bacterial taxonomic assignment. This approach is important given that bacterial pathogenicity is typically considered a species or strain level phenomenon [20]. Although per-base accuracy of nanopore sequencing is lower (~98%) than that of more commonly used next-generation sequencing platforms (e.g., Illumina and Pacific Biosciences HiFi (i.e., circular consensus sequencing); both at >99.9% accuracy) similar or even greater taxonomic resolution is still achieved with the ONT MinION [21,22,23,24]. Furthermore, with continued technological and bioinformatic advancements, ONT-based DNA sequencing will continually improve over time [25,26].
With respect to metabarcoding, the ONT MinION platform has been successfully applied in several studies, including the characterization of bacterial mock communities [25,27,28]; microbiota profiling of species and tissues such as dog skin [29], canine feces [30], equine gut [31], water buffalo milk [32], sea louse [33], and microalgae [34]; identification of fungi [35]; and characterization of plastic-associated species in the Mediterranean sea [36]. Additionally, metagenetic analyses of environmental samples obtained from glacial regions [37], aquatic environments (e.g., ocean water column [38], river water [39], wastewater [40], and freshwater [41]), building dust [22], and the International Space Station [42] demonstrate the potential and applicability of nanopore sequencing for microorganism detection across diverse environments and field settings. Notably, nanopore sequencing has been used to describe human gut [43], and nasal microbiota [44], as well as those associated with colorectal cancer tumors [45], and thrombus samples [46]. Nanopore-based pathogen surveillance of EMS vehicles [15], prosthetic devices [47], hospitals [48], and antibiotic resistance markers in clinical samples [49,50,51,52] clearly demonstrate the potential of the MinION platform as a pathogen surveillance tool.
In light of the growing number of studies showing the utility of the ONT MinION for a diverse range of biosurveillance applications and, given the lack of research on the rodent–agriculture interface in the USA, we set out to examine the potential of the MinION for metabarcoding rodent-borne zoonoses. Here, we show how the ONT MinION can be used to taxonomically characterize fecal bacterial communities of farm-dwelling rodents. Our study area included farms in the Upper Midwest of the United States (i.e., Minnesota and Wisconsin), regions where studies focused on the rodent–farm interface are lacking [1]. Our intent was twofold: (1) to elucidate the farm-level rodent diversity in our study area and (2) to use full-length 16S metabarcoding to identify rodent-borne zoonoses of agricultural concern (Figure 1).
Figure 1.
Workflow of the overall study design performed herein. (A): Rodents effectively serve as amplifiers of bacterial pathogens across a given farm environment through fecal deposition, including possible transmission to resident farm animals. (B): DNA extraction from rodent colon contents (i.e., feces) and quantification. (C): Laboratory workflow to monitor bacterial communities from rodent samples using nanopore sequencing.
2. Results
2.1. Rodent Trapping on Farms
We live-trapped 90 rodents during the course of our study; 29 from Farm A, 43 from Farm B and 18 from Farm C (Table 1). We additionally captured two shrews (Blarina brevicauda) on Farm C and included those in our analyses. We identified five rodent species across our study sites, including three native (Peromyscus maniculatus, P. leucopus, and Microtus pennsylvanicus) and two invasive species (Mus musculus and Rattus norvegicus). The captured shrew species, B. brevicauda, is native to the Midwest. The majority of rodent captures were centered around feed bunks, grain storage sites, and within cow barns.
Table 1.
Index of all captured animals from Farm A, B and C.
| Rodent Species | Farm A | Farm B | Farm C |
|---|---|---|---|
| House mouse (Mus musculus) | 27 | 21 | 0 |
| White-footed mouse (Peromyscus leucopus) |
0 | 13 | 11 |
| Deer mouse (Peromyscus maniculatus) |
1 | 4 | 0 |
| Meadow vole (Microtus pennsylvanicus) |
0 | 5 | 1 |
| Norway rat (Rattus norvegicus) | 1 | 0 | 6 |
| Shrew Species | |||
| Northern short-tailed shrew (Blarina brevicauda) | 0 | 0 | 2 |
| Total = 92 | 29 | 43 | 20 |
2.2. Nanopore Sequencing Workflow of Full-Length 16S rRNA for Rodent Microbiome Analysis
We generated full-length 16S amplicon sequencing data comprising more than 33 million reads from 63 farm-caught rodent and 2 shrew colon extracts. Run 1 and Run 3 included 22 rodent colon samples from the large conventional Farm (A), Run 2 and Run 4 included 23 samples from the med-sized Farm (B), while Run 5 included all 20 samples from the small family Farm (C; Table 2). Each of the MinION sequencing runs included 12 molecularly barcoded samples except Run 5, which included 20 barcoded samples that were collected from individual rodent and shrew colons (Table 2). Average raw 16S rRNA reads generated across the five sequencing runs ranged from 3.5 to 9.4 million reads and mean quality scores of filtered reads ranged from 8.2 to 10.1 (Table 2). The per-base error rate of our MinION sequencing was approximately 1 in 11 bases, a result that is consistent with other nanopore studies [22,53]. However, read depths of full-length 16S amplicons averaged 16× coverage, resulting in high-quality consensus sequences and reducing concerns of per-base sequencing error [54]. To sort high-quality reads, pass fast5 reads were generated from raw data using the Guppy base calling program and FAST model setting. Although Run 3 data (~3.5 million reads) showed fewer sequencing reads than other sequencing runs, the mean Q score (8.2), mean read length (1625.40 bp) and read length N50 (1593 bp) were comparable (Table 2). For all sequencing runs, read lengths had a narrow distribution, with mean read lengths ranging from 1132.70 to 1625.40 bp, which was close to the full-length of the 16S rRNA gene (about 1550 bp). A filtering program (Cutadapt) was used to discard reads out of a 1200 to 1800 bp length range. Collectively, our quality control measures (see Methods) filtered out approximately 20% of initial raw reads from all runs.
Table 2.
Summary statistics of 16S nanopore sequencing of rodent colon contents performed herein. N = number of rodent samples barcoded and pooled on each MinION sequencing experiment. BP = Base Pairs.
| MinION Sequencing Run | Farm (N) | Active Pores avg. | Read Count (Unit Million Reads) | Mean Read Length | Mean Q Score | Read Length N50 | Total bp | QC > Q7 |
|---|---|---|---|---|---|---|---|---|
| 1 | A (12) | 509 | 5.0 | 1132.70 | 9.3 | 1564 | 5.7 × 109 | 85.80% |
| 2 | B (11) | 500 | 4.5 | 1178.30 | 8.3 | 1587 | 5.3 × 109 | 85.50% |
| 3 | A (11) | 509 | 3.5 | 1625.40 | 8.2 | 1593 | 5.7 × 109 | 68.50% |
| 4 | B (11) | 500 | 9.4 | 1477.10 | 10.1 | 1576 | 13.9 × 109 | 85.40% |
| 5 | C (20) | 504 | 5.8 | 1472.30 | 8.4 | 1564 | 8.6 × 109 | 77.70% |
2.3. Rodent Fecal Core Microbiome and Microbial Diversity
After taxonomic classification, we obtained 96.25% of classified reads and 3.75% of unclassified reads. Total reads corresponding to bacteria were at 96.25%, and 0% reads were assigned to virus, fungi and protozoa. The microbial classifications were obtained at different taxonomic levels (e.g., division, phylum, class, order, family, genus, and species) for all of the 65 colon extract samples. Overall, the most abundant phylum for all five rodent species was Firmicutes (~75% of total reads), followed in abundance by Bacteroidetes (12.5%) and Proteobacteria (~10%), and other phyla comprised less than ~2.5% of total classified phyla (n = 80; Figure 2).
Figure 2.
Core rodent fecal microbiome observed herein (n = 63). Numbers above nodes are reads assigned to each taxon.
At the rodent species level, the house mouse (Mus musculus) was the most captured species, with a total of 48 animals trapped from Farms A and B. After taxonomic classification of the combined house mouse reads, 77 phyla, 886 genera, and 473 bacterial species were obtained. At the genus and species levels, a filtering step was applied that included a threshold of 100 reads binned to the respective taxonomic level, which retained 261 genera and 181 species passing the threshold. For M. musculus, the most abundant genera were Lactobacillus (27.7%), Ruminococcus (10%), Helicobacter (8%), Bacteroides (7.6%), and Blautia (6.8%) (see Supplementary Table S1). Other abundant genera included fecal or mammalian gut microbiota (e.g., Coprococcus, Faecalibacterium, Dorea, Roseburia, Oscillospira) and potential human pathogens and bovine mastitis-causing pathogens such as Staphylococcus, Streptococcus, Bacillus, and Enterococcus, each constituting more than 1% of the total classified reads.
The second most dominant rodents in across our study area consisted of Peromyscus spp., with a total of 29 captured from Farms A, B, and C. After combining and taxonomically classifying all Peromyscus 16S rRNA reads, 58 phyla, 619 genera, and 344 species were obtained. After threshold filtering, 139 genera and 89 species were retained. The most abundant genus was Lactobacillus (37.6%), followed in abundance by Ruminococcus (16.5%), Blautia (10.6%), Dorea (4.6%), Helicobacter (4%), and Streptococcus (3.4%). Other fecal-related genera formed less than ~3% of the total (see Supplementary Table S1).
Brown rats (Rattus norvegicus) were captured from Farms A and C with a total of seven animals. The combined nanopore reads were taxonomically classified into 47 phyla, 583 genera and 357 species. Threshold filtering retained 119 genera and 88 species, where Lactobacillus (22%), Blautia (17.3%), Ruminococcus (11.5%), Streptococcus (7.8%), and Dorea (6.5%) were the most abundant genera (see Supplementary Table S1). Other mammalian gut microbiota (e.g., Oscillospira, Coprococcus, Faecalibacterium, Roseburia) and potential human pathogenic genera (Helicobacter, Prevotella, Staphylococcus, Clostridium, Bacteroides) compiled more than 23% of the bacterial genera.
We captured a total of six meadow voles (Microtus pennsylvanicus) from Farms B and C. Taxonomic classification of vole 16S rRNA reads revealed 52 phyla, 542 genera and 302 species. Subsequent filtering retained 121 genera and 61 species, with the most prominent genera in the meadow vole feces consisting of Lactobacillus (24%), Ruminococcus (19.6%), Blautia (11.7%), Oscillospira (7.6%), and Coprococcus (5.3%) (see Supplementary Table S1).
While examining species level composition, several bacterial species were abundant and observed across all five rodent species (Figure 3). For example, abundant Lactobacillus species included L. reuteri, L. zeae, L. salivarius, L. delbrueckii, L. brevis, L. helveticus, L. ruminis, and L. iners. Another dominant genus Ruminococcus consisted of R. gnavus, R. torques, R. flavefaciens, R. bromii, and R. callidus. Blautia species included B. producta and B. obeum. The following species were dominant across all rodent samples in our study: Dorea formicigenerans, Roseburia faecis, Prevotella copri, Faecalibacterium prausnitzii, Oscillospira guilliermondii, Clostridium perfringens, Helicobacter pylori, and Coprococcus eutactus. The most abundant Staphylococcus species included S. aureus, S. epidermidis, S. haemolyticus, and S. sciuri. The most abundant Streptococcus species included S. luteciae, S. anginosus, S. alactolyticus, and S. infantis.
Figure 3.
Heatmap of the most abundant (>1%) bacterial genera, across rodent species, identified by mapping 16S rRNA gene amplicons against the GreenGenes reference database. Genera having low relative abundance are light in color, while those with high abundance are dark. Genera that are pathogenic to humans appear in red font.
2.4. Shrew Fecal Core Microbiome and Microbial Diversity
Two shrews from the same species, Blarina brevicauda, were captured from a small family farm (Farm C). Taxonomic classification analysis of the shrew 16S sequencing data revealed 28 phyla, 281 genera, and 178 species. The most abundant phylum for both shrews was Proteobacteria (~91% of total reads), followed in abundance by Firmicutes (8%), and other phyla comprised less than ~1% of total classified phyla (n = 28; Figure 4).
Figure 4.
Shrew (B. brevicauda) fecal microbiome observed herein (n = 2). Numbers above nodes are reads assigned to each taxon.
At the genus level, a total of 281 genera were identified and 36 genera were retained after applying a threshold of 100 reads binned to each genus. The shrew fecal microbiome was rich in Klebsiella (18.8%), followed in abundance by Salmonella (16.8%), Serratia (15.7%), Erwinia (12.6%), and Citrobacter (6.2%) (see Supplementary Table S1). Moreover, it contained other fecal-related and potential pathogenic genera, including Providencia, Enterococcus, Morganella, Yersinia, Enterobacter, Proteus, Clostridium, Plesiomonas, Vibrio, Bacillus, Pseudomonas, Staphylococcus, and Streptococcus, representing less than 5% each of the total bacterial composition. The relative abundance of the 18 most abundant taxa determined at the genus level is shown using bar graphs in Figure 5.
Figure 5.
Shrew (B. brevicauda) microbiota representing > 1% relative abundance at genus (A) and species (B) levels. Potential pathogenic genera (A) and species (B) to humans appear in red font.
Metabarcoding analyses at the species level revealed a total of 178 species, with 29 retained after applying a threshold of 100 reads binned to each species. Salmonella enterica was the most abundant species with 21.5% reads assigned, followed in abundance by Serratia marcescens (17.3%), Klebsiella oxytoca (16%), Erwinia soli (7.4%), Staphylococcus sciuri (6.2%), and Trabulsiella farmeri (6%). Other abundant species included putative human and plant pathogens such as Morganella morganii, Providencia stuartii, Enterobacter cowanii, Staphylococcus aureus, and Brenneria quercina, each >1% of the total bacterial species composition.
3. Discussion
We used MinION nanopore sequencing to metabarcode fecal microbial communities in peridomestic small mammals (i.e., rodents, shrews). Rodents and shrews within our study were collected from three dairy and mixed animal farms over a two-year period. Our nanopore-based metabarcoding pipeline (Figure 1) demonstrates the utility of the MinION technology for the surveillance of pathogenic organisms in peridomestic pests [55,56,57,58,59] and, when used for farm-level surveillance, the methodology can be leveraged to inform and strengthen biosecurity practices. The long read length, depth of coverage, and rapid results make MinION sequencing a robust option for pathogen surveillance. Although nanopore sequencing has a higher per-base accuracy error rate than other next-generation sequencing methods [53], we based our taxonomic classification on read depths of over 30 million full-length 16S (~1.5 kb) reads. We acknowledge that a high number of sequencing reads for putative pathogens does not necessarily indicate the absolute presence of the organism; thus, standard culturing techniques and related molecular methods are still valuable tools for the confirmation of putative pathogens. Nevertheless, for our purpose of a primary screening tool, MinION 16S sequencing was a cost effective and rapid method that achieved our surveillance goals. Regarding the usage of 16S rRNA gene sequence data for the taxonomic classification of bacteria, we note that shotgun metagenomic approaches (e.g., deep sequencing DNA isolates with second-generation Illumina technologies) can provide a more accurate depiction of the diversity of a given microbial community than single-gene sequence data. Moreover, such approaches can also recover detailed profiling of antimicrobial-resistant genes within a sample. Despite these observations, there are clear benefits with respect to MinION 16S rRNA sequencing, as the method can be performed within individual labs (or in the field for real-time surveillance) and can effectively identify bacteria that are abundant within a given sample.
To the best of our knowledge, this is the first study to reveal and compare the composition of bacterial communities in gastrointestinal tracts of wild-caught rodents and shrews (e.g., M. musculus, Peromyscus spp., M. pennsylvanicus, R. norvegicus and B. brevicauda) using a third-generation sequencing technology. Rodents are considered to be among the most commensal synanthropic animals, especially within the context of food-production systems. Contrastingly, shrews (B. brevicauda) naturally live in woodlands, cultivated fields, vegetable gardens and mainly feed on invertebrates [60,61]. Both rodents and shrews retreat into barns, cellars and sheds during fall and winter months (both for shelter and foraging), thus providing opportunities for direct and indirect (i.e., with rodent/shrew feces and urine) contacts with humans, pets, livestock, and poultry. Such interactions have the potential for the transmission of rodent-borne zoonoses, especially pathogenic bacteria [1,62].
For all five rodent species, phylum level gut microbiomes were similar to that of other rodent gut microbiomes reported in the scientific literature, with Firmicutes, Bacteroidetes and Proteobacteria comprising more than 97% of the gut microbiota [63,64]. Firmicutes was the most abundant phylum in all rodent species, ranging from 64% to 91.5%. In shrews, the most abundant phylum was Proteobacteria, covering 91.7% of the total shrew microbiota. At the genus level, Lactobacillus was the top genus for all the rodent species in our study, which is in line with the findings of a previously reported study on laboratory rodent microbiomes (see Supplementary Table S1) [65]. This finding implies that the core fecal bacterial compositions of wild and laboratory rodents share a degree of similarity. Lactobacillus also constitutes a significant component of the human gut microbiome [66]. Furthermore, Ruminococcus was the second most prominent genus in all mouse species (M. musculus, Peromyscus spp., M. pennsylvanicus), whereas Blautia was prominent in rats (R. norvegicus). Alternatively, Klebsiella was the most prominent genus in the northern short-tailed shrew feces, followed by Salmonella, Serratia and Erwinia. Of note is that metabarcoding data are scant across all shrews, thus limiting genus or species-level comparisons. A single study originating from China included the Asian house shrew (Suncus murinus) in their 16S rRNA metabarcoding analyses and identified Clostridium as the most abundant genus in their sample [67].
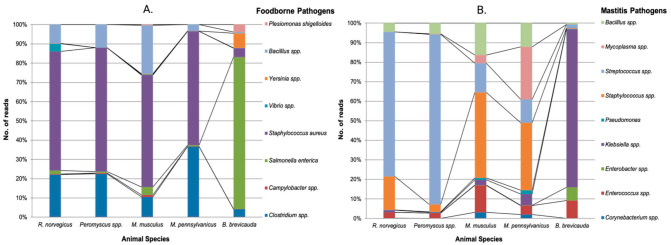
We identified a higher relative abundance of potential human and animal pathogens in shrew fecal samples than in rodent fecal samples. In brief, the resulting data indicate the presence of multiple putative foodborne pathogens from the Enterobacteriales order, including Salmonella spp., Shigella spp., Plesiomonas shigelloides, Yersinia spp., and Escherichia coli, in all small mammal species (Figure 6). Other top foodborne pathogens observed in our analysis included Listeria monocytogenes, Campylobacter spp., Clostridium perfringens, Vibrio spp., and Staphylococcus aureus. These are identified as major bacterial pathogens that cause foodborne illness and hospitalization in the USA and globally each year [68].
Figure 6.
Krona plot showing overall rodent and shrew-associated (n = 65) bacterial species abundance within the order Enterobacteriales.
Interestingly, Salmonella enterica was the most abundant species (~21.5%) in the shrew feces but was much lower across our rodent samples (Figure 7). On the contrary, Clostridium perfringens, Bacillus spp., and Staphylococcus aureus were abundant within rodents when compared to the two shrew samples. Within our rodent sample, Vibrio spp. and Campylobacter spp. were observed in comparatively higher abundance in R. norvegicus and M. musculus, respectively. These bacteria can be transmitted by contaminated food and water, and, within our study, they had a greater relative abundance in shrew fecal samples than in those of rodents.
Figure 7.
Abundance of foodborne (A) and mastitis (B) pathogens in all small mammal species.
Mastitis is a leading cause of cow culling and causes substantial economic losses to the dairy industry [69]. Because a majority of our rodent samples were collected from large-sized (Farm A; ~20,000 cattle) and medium-sized (Farm B; ~600 cattle) dairy farms, we investigated the presence and abundance of pathogens causing bovine mastitis in the fecal samples of the farm-dwelling rodents. Many important mastitis-causing pathogens, including Streptococcus spp., Staphylococcus spp., Klebsiella spp., Enterococcus spp., Enterobacter spp., Mycoplasma spp., and Corynebacterium spp., were observed in varying abundance across our rodent and shrew samples. E. coli, Streptococcus spp., Staphylococcus spp., and Corynebacterium spp. are well known environmental mastitis pathogens [70], and the presence of these pathogens in the resident rodent population of each dairy farm is a putative health risk to the resident cattle, especially when considering the fecal output of commensal rodent species into the farm environment [1]. Our metabarcoding data indicate that the rodents sampled during our trapping events are possible reservoirs of mastitis pathogens and have the potential to continuously introduce these pathogens into the dairy farm environments. Moreover, they can amplify and mechanically vector these pathogens from sick to susceptible animals through fecal-based pathogen amplification [1]. When performing rodent species comparisons of the relative abundance of mastitis pathogens across Farms A, B, and C (Figure 7B), the house mouse (M. musculus) exhibited the highest number of mastitis-associated pathogens. Given this observation and in light of our farm-level observations, we hypothesize that because M. musculus cohabitates with cows inside barns (i.e., tunneling and nesting extensively in bedding material and wall insulation), they are exposed to a greater overall amount of mastitis pathogens by direct and indirect interactions with resident dairy cow herds.
Rodent and shrew-borne bacterial pathogens can cause human diseases through various routes, especially by urine and fecal output into water and food resources. In our study, rodent fecal samples contained a variety of zoonoses of concern to human health in high abundance, including: Helicobacter pylori, known to cause chronic gastritis, gastric ulcers, and stomach cancer [71]; Prevotella copri, associated with the pathogenesis of rheumatoid arthritis [72]; pathogens related to nosocomial infections such as Morganella morganii, Serratia marcescens, and Providencia stuartii [73,74]; and opportunistic pathogens associated with splenic abscess (Parabacteroides distasonis) and anaerobic peritoneal infections (Bacteroides fragilis) [75,76].
Rodents are well-known across the globe for their commensal nature, living in close proximity to populations of both humans and domesticated species. An increasing number of studies suggest that rodents may serve as potential sources of infectious zoonotic diseases via pathogen amplification and cross-species transmission [1]. Surprisingly little attention has been paid to rodents associated with food-production systems in the United States, despite the fact that they occupy food animal farms, fresh produce lands, and processing facilities across the country. In our study, a substantial number of sequences obtained from the northern short-tailed shrew were identified as potential foodborne pathogens. It is possible that B. brevicauda is a reservoir of bacterial foodborne pathogens; however, shrews are not considered peridomestic species and thus transmission risk is likely quite low. Alternatively, invasive M. musculus and R. norvegicus and native Peromyscus spp. readily adapt to farm environments and have the capacity to establish abundant on-farm populations. We recorded evidence of large on-farm populations of both M. musculus and R. norvegicus consisting of subfloor tunneling in barns and outbuildings, nesting within wall insulation and livestock bedding, and accessing livestock food storage areas. Despite observing a large population of R. norvegicus on Farm A, our live-trapping methods were not ideal for collecting R. norvegicus. Enhanced species-specific trapping efforts with pre-baiting measures to acclimate the rats and the use of multiple trapping methods might overcome trap avoidance behaviors exhibited by R. norvegicus [77]. Our personal observations during farm-level trapping and subsequent bacterial metabarcoding data indicate that farm-dwelling rodents are potential reservoirs of many putative zoonotic foodborne and mastitis-associated pathogens. In light of these results, we strongly recommend additional research focused on the rodent–agricultural interface across the United States.
4. Materials and Methods
4.1. Rodent Trapping and Sample Collection on Farms
During the summer (2019) and fall (2019, 2020), we collected rodents from one dairy cattle farm (A), one mixed animal (dairy cattle and hog) farm (B) and another mixed animal (cattle and horse) farm (C). Farms A and B are located in Nicollet and Stevens counties of Minnesota, respectively, and Farm C is in southeastern Wisconsin in Sauk County. Farm A is a large dairy operation (~20,000 dairy cattle), Farm B is a medium-sized operation (600 dairy cattle and 400 hogs), and Farm C is a small-sized family farm (cattle and horses < 100 animals). Rodent activity was elevated on farms per observations by farm managers, particularly on Farm A where we observed hundreds of Norway rats (R. norvegicus) actively foraging around compost piles during daylight hours. All farms had poison bait stations, kill traps and cats as rodent control measures during the time of our visits. Four nights of rodent trapping were conducted at each study site using 150 Sherman live traps baited with oats. Decontamination of all traps was performed using a 10% sodium hypochlorite solution (10 min soak) before and after each trapping event. All trapped animals were humanely euthanized following approved UMN IACUC protocols (protocol number 1809-36374A). Small mammals were collected with approval by the Minnesota Department of Natural Resources under Special Permit No. 23896. Two shrews (Blarina brevicauda) were collected during the course of our research and were included in our analyses. Standard morphological techniques were used to identify rodents and shrews to species-level and metadata (e.g., species, age, weight, sex, body measurements) were collected for each individual animal. Biological samples (e.g., feces, colon) were collected and preserved (e.g., liquid nitrogen, freezer) for metabarcoding analysis and further quantification of pathogens of interest. A schematic workflow of the overall study design is shown in Figure 1 and specimens examined are provided in Table S2.
4.2. DNA Extraction
DNA was extracted with a QIAamp PowerFecal Pro DNA Kit (QIAGEN, Hilden, Germany). Snap-frozen rodent and shrew feces and colon extracts were stored at −80 °C and were used for DNA extraction. Briefly, 250 mg of colon contents were added to PowerBead Pro tubes and 800 μL of solution CD1 was mixed by vortexing. A bench top PowerLyzer 24 Homogenizer (QIAGEN, Hilden, Germany) was used for homogenizing the samples at 2000 rpm for 30 s, pausing for 30 s, then homogenizing again at 2000 rpm for 30 s to enhance cell lysis. PowerBead Pro tubes were centrifuged at 15,000× g for 1 min, and the resulting supernatant was transferred to clean microcentrifuge tubes. We used fully automated QIAcube connect instruments (QIAGEN, Hilden, Germany) for DNA extraction following the manufacturer’s instructions. DNA concentrations were measured by fluorescence in a Qubit 4 fluorometer (Thermofisher Scientific, Waltham, MA, USA) using the Qubit dsDNA BR Assay Kit (Thermofisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions.
4.3. Nanopore Library Construction and Sequencing
The 16S Barcoding Kit (SQK-RAB204; Oxford Nanopore Technologies, Oxford, UK) was used to prepare the amplicon library, following the manufacturer’s instructions for 1D sequencing strategy. The 16S region (1.5 kb) of bacteria was amplified using specific primers (27F-1492R) and subsequently barcoded. This approach enables targeted sequencing of multiple samples and provides genus-level resolution. Five sequencing runs were performed with a total of 65 samples, including 11 (run 1, 3, 4), 12 (run 2), and 20 (run 5) barcoded samples from individual rodents and shrews. Briefly, genomic DNA samples were diluted to 100 ng/μL and amplification of the full-16S rRNA gene was performed by PCR with reaction volume of 50 μL, using the primers 27F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R 5′-GGTTACCTTGTTACGACTT-3′, and Taq DNA polymerase LongAmp (NewEngland Biolabs, Ipswich, MA, USA). Amplification was performed using Bio-Rad Laboratories PCR Thermal Cycler T100™ (Bio-Rad Laboratories, Hercules, CA, USA) with the following PCR conditions: initial denaturation at 95 °C for 1 min, 25 cycles of 95 °C for 20 s, 55 °C for 30 s, and 65 °C for 2 min, followed by a final extension at 65 °C for 5 min.
PCR products (50 μL each) were purified with 30 μL Agencourt AMPure XP beads and incubated in a HulaMixer for 5 min at room temperature. After a magnetic bead washing step, purified products were eluted in 10 μL of elution buffer (10 mM Tris-HCl pH8.0 with 50 mM NaCl). The amount and purity of the sequencing library was quantified using a Qubit 4 fluorometer (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Libraries were pooled in multiplex mode following the addition of 1 μL of rapid adapter (Oxford Nanopore Technologies, Oxford, UK) and incubated at room temperature for 5 min. The amplicon library (11 μL) was then diluted with a running buffer (35 μL) containing 3.5 μL of nuclease-free water and 25.5 μL of loading beads. Five nanopore sequencing libraries were separately run on FLO-MIN106 R9.4 (run 1, 2, 4, 5) and FLO-MIN111 R10.3 (run 3) flow cells (Oxford Nanopore Technologies, Oxford, UK). Sequencing runs were performed for 48 h. using the MinION control software, MinKNOW 4.0.20 (Oxford Nanopore Technologies, Oxford, UK).
4.4. Bioinformatic Analyses
After the completion of each sequencing run, raw signals in nanopore fast5 files were base-called using Guppy (version3.2.2, Oxford Nanopore Technologies, Oxford, UK), and a quality filter step was applied to retain only sequences with a mean Q-score ≥ 7. De-multiplexing of the barcoded samples was conducted using Porechop [78]. Adapter trimming and a second round of de-multiplexing were performed using Cutadapt 1.91 [79]. Only reads between 1200 and 1800 bp were selected for further analysis using Cutadapt. Read statistics for each sequencing run were obtained using Nanostat and NanoPlot [80]. For taxonomic assignments, Kraken2 [81] and Bracken [82] were used with the Greengenes (GG) database (https://benlangmead.github.io/aws-indexes/k2, accessed on 26 March 2021). While generating the Bracken classification report, a threshold of >100 reads was applied for higher confidence at the genus and species levels. For visualization Krona tools and Pavian interactive applications were used to generate taxonomic charts and flow diagrams [83,84]. The ggplot2 package (version 3.2.1) in RStudio software (version 3.3.3) was used to create a heatmap [85]. BioRender was used for illustrations and diagrams (Created with BioRender.com). Base-called data were uploaded to the EPI2ME interface, a platform for cloud-based analysis of MinION data, and WIMP (ONT; “What’s in my pot” software) analysis was performed in parallel to compare results.
Acknowledgments
We thank the Minnesota Supercomputing Institute for computational and data storage resources. Suzanne Stone kindly provided logistical assistance within the molecular lab. We thank all farm owners and associated staff for their assistance throughout our work.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091183/s1, Table S1: Percentages of full-length 16S rRNA reads taxonomically assigned to bacterial genera per rodent and shrew species. Listed genera represent the top 15 of each rodent and shrew species, collectively; Table S2: specimens examined.
Author Contributions
Conceptualization, P.A.L. and N.A.J.; methodology, N.A.J., L.L.L., A.R., E.J.K., P.A.L., A.M.R.; data analysis, N.A.J.; validation, N.A.J., L.L.L.; formal analysis, N.A.J., L.L.L.; investigation, N.A.J., L.L.L., E.J.K., A.R., B.J.H., A.M.R., P.A.L.; resources, P.A.L., B.J.H., A.M.R.; data curation, N.A.J., L.L.L.; writing—original draft preparation, N.A.J., P.A.L.; writing—review and editing, N.A.J., L.L.L., E.J.K., A.R., B.J.H., A.M.R., P.A.L.; visualization, N.A.J.; supervision, P.A.L., L.L.L.; project administration, P.A.L.; funding acquisition, P.A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by start-up research funds awarded to PAL through the Agricultural Research, Education, Extension and Technology Transfer (AGREETT) program at the University of Minnesota.
Institutional Review Board Statement
The study was conducted according to the guidelines of the IACUC and approved by the Ethics Committee of the University of Minnesota (protocol number 1809-36374A).
Informed Consent Statement
Not applicable.
Data Availability Statement
All nanopore sequence data are available on the National Center for Biotechnology Information Sequence Read Archive website under project accession number PRJNA759117.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jahan N.A., Lindsey L.L., Larsen P.A. The Role of Peridomestic Rodents as Reservoirs for Zoonotic Foodborne Pathogens. Vector Borne Zoonotic Dis. 2020;21:133–148. doi: 10.1089/vbz.2020.2640. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez A., Pangloli P., Richards H.A., Mount J.R., Draughon F.A. Prevalence of Salmonella in Diverse Environmental Farm Samples. J. Food Prot. 2006;69:2576–2580. doi: 10.4315/0362-028X-69.11.2576. [DOI] [PubMed] [Google Scholar]
- 3.Meerburg B.G. Rodents Are a Risk Factor for the Spreading of Pathogens on Farms. Vet. Microbiol. 2010;142:464–465. doi: 10.1016/j.vetmic.2009.06.038. [DOI] [PubMed] [Google Scholar]
- 4.Kilonzo C., Li X., Vivas E.J., Jay-Russell M.T., Fernandez K.L., Atwill E.R. Fecal Shedding of Zoonotic Food-Borne Pathogens by Wild Rodents in a Major Agricultural Region of the Central California Coast. Appl. Environ. Microbiol. 2013;79:6337–6344. doi: 10.1128/AEM.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backhans A., Fellström C. Rodents on Pig and Chicken Farms—A Potential Threat to Human and Animal Health. Infect. Ecol. Epidemiol. 2012;2:17093. doi: 10.3402/iee.v2i0.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies R.H., Wray C. Mice as Carriers of Salmonella Enteritidis on Persistently Infected Poultry Units. Vet. Rec. 1995;137:337–341. doi: 10.1136/vr.137.14.337. [DOI] [PubMed] [Google Scholar]
- 7.Trampel D.W., Holder T.G., Gast R.K. Integrated Farm Management to Prevent Salmonella Enteritidis Contamination of Eggs. J. Appl. Poult. Res. 2014;23:353–365. doi: 10.3382/japr.2014-00944. [DOI] [Google Scholar]
- 8.Berndtson E., Emanuelson U., Engvall A., Danielsson-Tham M.-L. A 1-Year Epidemiological Study of Campylobacters in 18 Swedish Chicken Farms. Prev. Vet. Med. 1996;26:167–185. doi: 10.1016/0167-5877(95)01008-4. [DOI] [Google Scholar]
- 9.Burt S.A., Siemeling L., Kuijper E.J., Lipman L.J.A. Vermin on Pig Farms Are Vectors for Clostridium Difficile PCR Ribotypes 078 and 045. Vet. Microbiol. 2012;160:256–258. doi: 10.1016/j.vetmic.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Lapuz R.R.S.P., Umali D.V., Suzuki T., Shirota K., Katoh H. Comparison of the Prevalence of Salmonella Infection in Layer Hens from Commercial Layer Farms with High and Low Rodent Densities. Avian Dis. 2012;56:29–34. doi: 10.1637/9704-030711-Reg.1. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa L., Gray A., Duffy G., Fanning S., McMahon B.J. A Scoping Review on the Prevalence of Shiga-Toxigenic Escherichia Coli in Wild Animal Species. Zoonoses Public Health. 2018;65:911–920. doi: 10.1111/zph.12508. [DOI] [PubMed] [Google Scholar]
- 12.Camba S.I., Del Valle F.P., Umali D.V., Soma T., Shirota K., Katoh H., Sasai K. The Expanded Role of Roof-Rats (Rattus Rattus) in Salmonella Spp. Contamination of a Commercial Layer Farm in East Japan. Avian Dis. 2020;64:46–52. doi: 10.1637/0005-2086-64.1.46. [DOI] [PubMed] [Google Scholar]
- 13.Henzler D.J., Opitz H.M. The Role of Mice in the Epizootiology of Salmonella Enteritidis Infection on Chicken Layer Farms. Avian Dis. 1992;36:625–631. doi: 10.2307/1591757. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan R., Karaoz U., Volegova M., MacKichan J., Kato-Maeda M., Miller S., Nadarajan R., Brodie E.L., Lynch S.V. Use of 16S RRNA Gene for Identification of a Broad Range of Clinically Relevant Bacterial Pathogens. PLoS ONE. 2015;10:e0117617. doi: 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan T., Hakstol R., Kailasam S., Glaister G.D., Hudson A.J., Wieden H.-J. Rapid Metagenomics Analysis of EMS Vehicles for Monitoring Pathogen Load Using Nanopore DNA Sequencing. PLoS ONE. 2019;14:e0219961. doi: 10.1371/journal.pone.0219961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters W.A., Caporaso J.G., Lauber C.L., Berg-Lyons D., Fierer N., Knight R. PrimerProspector: De Novo Design and Taxonomic Analysis of Barcoded Polymerase Chain Reaction Primers. J. Bioinform. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin S., McPherson J.D., McCombie W.R. Coming of Age: Ten Years of Next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016;17:333. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyler A.D., Mataseje L., Urfano C.J., Schmidt L., Antonation K.S., Mulvey M.R., Corbett C.R. Evaluation of Oxford Nanopore’s MinION Sequencing Device for Microbial Whole Genome Sequencing Applications. Sci. Rep. 2018;8:123. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T., et al. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrd A.L., Belkaid Y., Segre J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018;16:143. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 21.Nygaard A.B., Tunsjø H.S., Meisal R., Charnock C. A Preliminary Study on the Potential of Nanopore MinION and Illumina MiSeq 16S RRNA Gene Sequencing to Characterize Building-Dust Microbiomes. Sci. Rep. 2020;10:3209. doi: 10.1038/s41598-020-59771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J., Lee S., Go M.-J., Lee S.Y., Kim S.C., Lee C.-H., Cho B.-K. Analysis of the Mouse Gut Microbiome Using Full-Length 16S RRNA Amplicon Sequencing. Sci. Rep. 2016;6:29681. doi: 10.1038/srep29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilianski A., Haas J.L., Corriveau E.J., Liem A.T., Willis K.L., Kadavy D.R., Rosenzweig C.N., Minot S.S. Bacterial and Viral Identification and Differentiation by Amplicon Sequencing on the MinION Nanopore Sequencer. Gigascience. 2015;4:12. doi: 10.1186/s13742-015-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oikonomopoulos S., Wang Y.C., Djambazian H., Badescu D., Ragoussis J. Benchmarking of the Oxford Nanopore MinION Sequencing for Quantitative and Qualitative Assessment of CDNA Populations. Sci. Rep. 2016;6:31602. doi: 10.1038/srep31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Chng K.R., Boey E.J.H., Ng A.H.Q., Wilm A., Nagarajan N. INC-Seq: Accurate Single Molecule Reads Using Nanopore Sequencing. Gigascience. 2016;5:34. doi: 10.1186/s13742-016-0140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calus S.T., Ijaz U.Z., Pinto A.J. NanoAmpli-Seq: A Workflow for Amplicon Sequencing for Mixed Microbial Communities on the Nanopore Sequencing Platform. Gigascience. 2018;7:giy140. doi: 10.1093/gigascience/giy140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benítez-Páez A., Portune K.J., Sanz Y. Species-Level Resolution of 16S RRNA Gene Amplicons Sequenced through the MinIONTM Portable Nanopore Sequencer. Gigascience. 2016;5:4. doi: 10.1186/s13742-016-0111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown B.L., Watson M., Minot S.S., Rivera M.C., Franklin R.B. MinIONTM Nanopore Sequencing of Environmental Metagenomes: A Synthetic Approach. Gigascience. 2017;6:gix007. doi: 10.1093/gigascience/gix007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuscó A., Viñes J., D’Andreano S., Riva F., Casellas J., Sánchez A., Francino O. Using MinIONTM to Characterize Dog Skin Microbiota through Full-Length 16S RRNA Gene Sequencing Approach. Biorxiv. 2017:167015. [Google Scholar]
- 30.Cuscó A., Pérez D., Viñes J., Fàbregas N., Francino O. Long-Read Metagenomics Retrieves Complete Single-Contig Bacterial Genomes from Canine Feces. BMC Genom. 2021;22:330. doi: 10.1186/s12864-021-07607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinoshita Y., Hidekazu N., Uchida-Fujii E., Nukada T. Establishment and Assessment of An Amplicon Sequencing Method Targeting The 16S-ITS-23S RRNA Operon for Analysis of The Equine Gut Microbiome. Sci. Rep. 2021;11:11884. doi: 10.1038/s41598-021-91425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catozzi C., Ceciliani F., Lecchi C., Talenti A., Vecchio D., De Carlo E., Grassi C., Sánchez A., Francino O., Cuscó A. Milk Microbiota Profiling on Water Buffalo with Full-Length 16S RRNA Using Nanopore Sequencing. J. Dairy Sci. 2020;103:2693–2700. doi: 10.3168/jds.2019-17359. [DOI] [PubMed] [Google Scholar]
- 33.Gonçalves A.T., Collipal-Matamal R., Valenzuela-Muñoz V., Nuñez-Acuña G., Valenzuela-Miranda D., Gallardo-Escárate C. Nanopore Sequencing of Microbial Communities Reveals the Potential Role of Sea Lice as a Reservoir for Fish Pathogens. Sci. Rep. 2020;10:2895. doi: 10.1038/s41598-020-59747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin H., Lee E., Shin J., Ko S.-R., Oh H.-S., Ahn C.-Y., Oh H.-M., Cho B.-K., Cho S. Elucidation of the Bacterial Communities Associated with the Harmful Microalgae Alexandrium Tamarense and Cochlodinium Polykrikoides Using Nanopore Sequencing. Sci. Rep. 2018;8:5323. doi: 10.1038/s41598-018-23634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Andreano S., Cuscó A., Francino O. Rapid and Real-Time Identification of Fungi up to the Species Level with Long Amplicon Nanopore Sequencing from Clinical Samples. Biol. Methods Protoc. 2020;6:bpaa026. doi: 10.1093/biomethods/bpaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidov K., Iankelevich-Kounio E., Yakovenko I., Koucherov Y., Rubin-Blum M., Oren M. Identification of Plastic-Associated Species in the Mediterranean Sea Using DNA Metabarcoding with Nanopore MinION. Sci. Rep. 2020;10:17533. doi: 10.1038/s41598-020-74180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards A., Debbonaire A.R., Nicholls S.M., Rassner S.M., Sattler B., Cook J.M., Davy T., Soares A., Mur L.A., Hodson A.J. In-Field Metagenome and 16S RRNA Gene Amplicon Nanopore Sequencing Robustly Characterize Glacier Microbiota. BioRxiv. 2019:073965. doi: 10.1101/073965. [DOI] [Google Scholar]
- 38.Hamner S., Brown B.L., Hasan N.A., Franklin M.J., Doyle J., Eggers M.J., Colwell R.R., Ford T.E. Metagenomic Profiling of Microbial Pathogens in the Little Bighorn River, Montana. Int. J. Environ. Res. Public Health. 2019;16:1097. doi: 10.3390/ijerph16071097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddington K., Eccles D., O’Grady J., Drown D.M., Hansen L.H., Nielsen T.K., Ducluzeau A.-L., Leggett R.M., Heavens D., Peel N. Metagenomic Analysis of Planktonic Riverine Microbial Consortia Using Nanopore Sequencing Reveals Insight into River Microbe Taxonomy and Function. Gigascience. 2020;9:giaa053. doi: 10.1093/gigascience/giaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X., Stachler E., Bibby K. Evaluation of Oxford Nanopore MinIONTM Sequencing for 16S RRNA Microbiome Characterization. BioRxiv. 2017:099960. [Google Scholar]
- 41.Urban L., Holzer A., Baronas J.J., Hall M.B., Braeuninger-Weimer P., Scherm M.J., Kunz D.J., Perera S.N., Martin-Herranz D.E., Tipper E.T. Freshwater Monitoring by Nanopore Sequencing. Elife. 2021;10:e61504. doi: 10.7554/eLife.61504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl-Rommel S., Jain M., Nguyen H.N., Arnold R.R., Aunon-Chancellor S.M., Sharp G.M., Castro C.L., John K.K., Juul S., Turner D.J. Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing. Genes. 2021;12:106. doi: 10.3390/genes12010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuo Y., Komiya S., Yasumizu Y., Yasuoka Y., Mizushima K., Takagi T., Kryukov K., Fukuda A., Morimoto Y., Naito Y. Full-Length 16S RRNA Gene Amplicon Analysis of Human Gut Microbiota Using MinIONTM Nanopore Sequencing Confers Species-Level Resolution. BMC Microbiol. 2021;21:35. doi: 10.1186/s12866-021-02094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heikema A.P., Horst-Kreft D., Boers S.A., Jansen R., Hiltemann S.D., de Koning W., Kraaij R., de Ridder M.A., van Houten C.B., Bont L.J. Comparison of Illumina versus Nanopore 16S RRNA Gene Sequencing of the Human Nasal Microbiota. Genes. 2020;11:1105. doi: 10.3390/genes11091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor W.S., Pearson J., Miller A., Schmeier S., Frizelle F.A., Purcell R.V. MinION Sequencing of Colorectal Cancer Tumour Microbiomes—A Comparison with Amplicon-Based and RNA-Sequencing. PLoS ONE. 2020;15:e0233170. doi: 10.1371/journal.pone.0233170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vajpeyee A., Chauhan P.S., Pandey S., Tiwari S., Yadav L.B., Shroti A.K., Vajpeyee M. Metagenomics Analysis of Thrombus Samples Retrieved from Mechanical Thrombectomy. Neurointervention. 2021;16:39. doi: 10.5469/neuroint.2020.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderson N.D., Street T.L., Foster D., Swann J., Atkins B.L., Brent A.J., McNally M.A., Oakley S., Taylor A., Peto T.E. Real-Time Analysis of Nanopore-Based Metagenomic Sequencing from Infected Orthopaedic Devices. BMC Genom. 2018;19:714. doi: 10.1186/s12864-018-5094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quick J., Ashton P., Calus S., Chatt C., Gossain S., Hawker J., Nair S., Neal K., Nye K., Peters T. Rapid Draft Sequencing and Real-Time Nanopore Sequencing in a Hospital Outbreak of Salmonella. Genome Biol. 2015;16:114. doi: 10.1186/s13059-015-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S., Wain J., O’grady J. MinION Nanopore Sequencing Identifies the Position and Structure of a Bacterial Antibiotic Resistance Island. Nat. Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 50.Bradley P., Gordon N.C., Walker T.M., Dunn L., Heys S., Huang B., Earle S., Pankhurst L.J., Anson L., De Cesare M. Rapid Antibiotic-Resistance Predictions from Genome Sequence Data for Staphylococcus Aureus and Mycobacterium Tuberculosis. Nat. Commun. 2015;6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Judge K., Harris S.R., Reuter S., Parkhill J., Peacock S.J. Early Insights into the Potential of the Oxford Nanopore MinION for the Detection of Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2015;70:2775–2778. doi: 10.1093/jac/dkv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemon J.K., Khil P.P., Frank K.M., Dekker J.P. Rapid Nanopore Sequencing of Plasmids and Resistance Gene Detection in Clinical Isolates. J. Clin. Microbiol. 2017;55:3530–3543. doi: 10.1128/JCM.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ip C.L.C., Loose M., Tyson J.R., de Cesare M., Brown B.L., Jain M., Leggett R.M., Eccles D.A., Zalunin V., Urban J.M., et al. MinION Analysis and Reference Consortium: Phase 1 Data Release and Analysis. F1000Res. 2015;4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikheyev A.S., Tin M.M.Y. A First Look at the Oxford Nanopore MinION Sequencer. Mol. Ecol. Resour. 2014;14:1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 55.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A., et al. Real-Time, Portable Genome Sequencing for Ebola Surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro-Wallace S.L., Chiu C.Y., John K.K., Stahl S.E., Rubins K.H., McIntyre A.B.R., Dworkin J.P., Lupisella M.L., Smith D.J., Botkin D.J., et al. Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Sci. Rep. 2017;7:18022. doi: 10.1038/s41598-017-18364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goordial J., Altshuler I., Hindson K., Chan-Yam K., Marcolefas E., Whyte L.G. In Situ Field Sequencing and Life Detection in Remote (79°26′N) Canadian High Arctic Permafrost Ice Wedge Microbial Communities. Front. Microbiol. 2017;8:2594. doi: 10.3389/fmicb.2017.02594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson S.S., Zaikova E., Goerlitz D.S., Bai Y., Tighe S.W. Real-Time DNA Sequencing in the Antarctic Dry Valleys Using the Oxford Nanopore Sequencer. J. Biomol. Tech. 2017;28:2–7. doi: 10.7171/jbt.17-2801-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pomerantz A., Peñafiel N., Arteaga A., Bustamante L., Pichardo F., Coloma L.A., Barrio-Amorós C.L., Salazar-Valenzuela D., Prost S. Real-Time DNA Barcoding in a Rainforest Using Nanopore Sequencing: Opportunities for Rapid Biodiversity Assessments and Local Capacity Building. Gigascience. 2018;7:giy033. doi: 10.1093/gigascience/giy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamilton W.J., Whitaker J.O., Jr. Mammals of the Eastern United States. 3rd ed. Cornell University Press; New York, NY, USA: 1998. [Google Scholar]
- 61.Rue L.L. Pictorial Guide to the Mammals of North America. Crowell University Press; New York, NY, USA: 1967. [Google Scholar]
- 62.Rahman M., Islam S., Masuduzzaman M., Alam M., Chawdhury M.N.U., Ferdous J., Islam M.N., Hassan M.M., Hossain M.A., Islam A. Prevalence and Diversity of Gastrointestinal Helminths in Free-Ranging Asian House Shrew (Suncus Murinus) in Bangladesh. Vet. World. 2018;11:549. doi: 10.14202/vetworld.2018.549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X., Zhao Y., Zhang M., Pang X., Xu J., Kang C., Li M., Zhang C., Zhang Z., Zhang Y. Structural Changes of Gut Microbiota during Berberine-Mediated Prevention of Obesity and Insulin Resistance in High-Fat Diet-Fed Rats. PLoS ONE. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Everard A., Lazarevic V., Gaïa N., Johansson M., Ståhlman M., Backhed F., Delzenne N.M., Schrenzel J., François P., Cani P.D. Microbiome of Prebiotic-Treated Mice Reveals Novel Targets Involved in Host Response during Obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D., Chen H., Mao B., Yang Q., Zhao J., Gu Z., Zhang H., Chen Y.Q., Chen W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci. Rep. 2017;7:45840. doi: 10.1038/srep45840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldstein E.J.C., Tyrrell K.L., Citron D.M. Lactobacillus Species: Taxonomic Complexity and Controversial Susceptibilities. Clin. Infect. Dis. 2015;60:S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 67.He W., Xiong Y., Ge J., Chen Y., Chen X., Zhong X., Ou Z., Gao Y., Cheng M., Mo Y., et al. Composition of Gut and Oropharynx Bacterial Communities in Rattus Norvegicus and Suncus Murinus in China. BMC Vet. Res. 2020;16:413. doi: 10.1186/s12917-020-02619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.-A., Roy S.L., Jones J.L., Griffin P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halasa T., Huijps K., Østerås O., Hogeveen H. Economic Effects of Bovine Mastitis and Mastitis Management: A Review. Vet. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- 70.Jahan N.A., Godden S.M., Royster E., Schoenfuss T.C., Gebhart C., Timmerman J., Fink R.C. Evaluation of the Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) System in the Detection of Mastitis Pathogens from Bovine Milk Samples. J. Microbiol. Methods. 2021;182:106168. doi: 10.1016/j.mimet.2021.106168. [DOI] [PubMed] [Google Scholar]
- 71.Kusters J.G., An Liet H., Kuipers E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drago L. Prevotella Copri and Microbiota in Rheumatoid Arthritis: Fully Convincing Evidence? J. Clin. Med. 2019;8:1837. doi: 10.3390/jcm8111837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singla N., Kaistha N., Gulati N., Chander J. Morganella morganii Could Be an Important Intensive Care Unit Pathogen. Indian J. Crit. Care Med. 2010;14:154. doi: 10.4103/0972-5229.74176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donkor E.S. Nosocomial Pathogens: An in-Depth Analysis of the Vectorial Potential of Cockroaches. Trop. Med. Int. Dis. 2019;4:14. doi: 10.3390/tropicalmed4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunalan A., Biswas R., Sridharan B., Elamurugan T.P. Pathogenic Potential of Parabacteroides Distasonis Revealed in a Splenic Abscess Case: A Truth Unfolded. BMJ Case Rep. 2020;13:e236701. doi: 10.1136/bcr-2020-236701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brook I. Pathogenicity of the Bacteroides Fragilis Group. Ann. Clin. Lab. Sci. 1989;19:360–376. [PubMed] [Google Scholar]
- 77.Himsworth C.G., Zabek E., Desruisseau A., Parmley E.J., Reid-Smith R., Jardine C.M., Tang P., Patrick D.M. Prevalence and Characteristics of Escherichia coli and Salmonella spp. in the feces of wild urban Norway and Black rats (Rattus norvegicus and Rattus rattus) from an inner-city neighborhood of Vancouver, Canada. J. Wildl. Dis. 2015;51:589–600. doi: 10.7589/2014-09-242. [DOI] [PubMed] [Google Scholar]
- 78.Wick R.R. Porechop. 2017. [(accessed on 26 March 2021)]. Available online: https://github.com/rrwick/Porechop.
- 79.Martin M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 80.De Coster W., D’Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood D.E., Salzberg S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ Comput. Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 83.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Breitwieser F.P., Salzberg S.L. Pavian: Interactive Analysis of Metagenomics Data for Microbiome Studies and Pathogen Identification. Bioinformatics. 2020;36:1303–1304. doi: 10.1093/bioinformatics/btz715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.RStudio Team . RStudio: Integrated Development for R. PBC; Boston, MA, USA: 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All nanopore sequence data are available on the National Center for Biotechnology Information Sequence Read Archive website under project accession number PRJNA759117.