Abstract
Besides Mycoplasma hyopneumoniae (M. hyopneumoniae), many other viruses and bacteria can concurrently be present in pigs. These pathogens can provoke clinical signs, known as porcine respiratory disease complex (PRDC). A sampling technique on live animals, namely tracheobronchial swab (TBS) sampling, was applied to detect different PRDC pathogens in pigs using PCR. The objective was to determine prevalence of different PRDC pathogens and their variations during different seasons, including correlations with local weather conditions. A total of 974 pig farms and 22,266 pigs were sampled using TBS over a 5-year period. TBS samples were analyzed using mPCR and results were categorized and analyzed according to the season of sampling and local weather data. In samples of peri-weaned and post-weaned piglets, influenza A virus in swine (IAV-S), porcine reproductive and respiratory syndrome virus—European strain (PRRSV1), and M. hyopneumoniae were found as predominant pathogens. In fattening pigs, M. hyopneumoniae, porcine circovirus type 2 (PCV-2) and PRRSV1 were predominant pathogens. Pathogen prevalence in post-weaned and finishing pigs was highest during winter, except for IAV-S and A. pleuropneumoniae, which were more prevalent during autumn. Associations between prevalence of several PRDC pathogens, i.e., M. hyopneumoniae, PCV-2 and PRRSV, and specific weather conditions could be demonstrated. In conclusion, the present study showed that many respiratory pathogens are present during the peri-weaning, post-weaning, and fattening periods, which may complicate the clinical picture of respiratory diseases. Interactions between PRDC pathogens and local weather conditions over the 5-year study period were demonstrated.
Keywords: PRDC, tracheobronchial swabs, prevalence, swine
1. Introduction
Porcine respiratory disease complex (PRDC) is a multifactorial and complex disease in nursery and growing pigs [1], provoked by a combination of several infectious viral and bacterial pathogens, environmental stressors, differences in production systems, and management practices [2,3,4]. The disease, characterized by pneumonia and reduced growth performance, is an economically significant respiratory disorder of weaned piglets and finishing pigs, and remains a challenge to the swine industry worldwide. Multiple agents were reported to be associated with PRDC, including the major pathogens porcine reproductive and respiratory syndrome virus (PRRSV), Mycoplasma hyopneumoniae (M. hyopneumoniae), influenza A virus in swine (IAV-S), and porcine circovirus type 2 (PCV-2) [5,6]. Other pathogens associated with PRDC are porcine cytomegalovirus (PCMV), porcine respiratory coronavirus (PRCV), and Actinobacillus pleuropneumoniae (A. pleuropneumoniae) [2,5,6]. Infection with each single pathogen does not necessarily result in appearance of symptoms, but complex infections with a variety of pathogens can develop severe conditions.
Mycoplasma hyopneumoniae plays a major role within PRDC as the etiological agent of enzootic pneumonia, a chronic respiratory disease that mainly affects finishing pigs [7,8]. Mycoplasma infections are related to a chronic, non-productive cough. This results in economic losses due to reduced growth rate, poorer feed conversion, increased medication use and a higher susceptibility to secondary pathogens, such as Pasteurella multocida (P. multocida) and A. pleuropneumoniae [7]. Moreover, M. hyopneumoniae may act as a facilitator to other primary pathogens such as PRRSV [9,10], IAV-S [2,11,12], and PCV-2 [13,14]. The respiratory form of PRRSV primarily affects growing and finishing pigs, causing interstitial pneumonia, which induces respiratory signs [15]. PRRSV increases the susceptibility of pigs to secondary bacterial and viral infection [16,17,18,19,20,21]. Concurrent infections with PRRSV, PCV-2, and M. hyopneumoniae have been associated with more severe disease and higher mortality [17,22,23,24]. Swine influenza is mainly caused by influenza type A viruses and several subtypes of IAV-S have become enzootic in the pig population. Indeed, three IAV-S subtypes, namely H1N1, H2N1, and H3N2, currently circulate among pigs worldwide [25,26]. The enzootic within-farm persistence of IAV-S has recently been described as consecutive waves of diverse intensity in some Spanish farrow-to-finish operations [27]. Recently, pigs with passive immunity to IAV-S have been identified as potential disseminators of IAV-S, despite a potential reduction in clinical disease implied by this immunity [28]. Porcine circovirus type 2 is also responsible for considerable economic losses in the swine industry worldwide [29]. PCV-associated disease (PCVAD) can manifest as enteric, respiratory, reproductive, and systemic disease [30]. PCVAD is characterized by lymphoid depletion, which is considered the hallmark lesion [31]. This is thought to induce immunosuppression or immunomodulation in the host [32], leading to secondary infections with other viral or bacterial pathogens [33,34,35]. A field study in Spain confirmed detection of PCV-2 in several types of respiratory samples [36]. Porcine respiratory coronavirus (PRCV) is a naturally occurring respiratory variant of transmissible gastroenteritis virus in pigs, leading to fever and atypical pneumonia [37]. Seroprevalence of PRCV in young fattening pigs in Belgium varied from 34% during winter to 50% during late summer and autumn [38]. Dual infections involving PRCV and PRRSV or IAV-S did only identify little to no interactions between pathogens [39]. Porcine cytomegalovirus infection is endemic in the pig population [40]. Virus transmission occurs horizontally through nasal and ocular secretions, milk, and urine. Actinobacillus pleuropneumoniae, the etiological agent of pleuropneumonia in pigs, is the most important bacterial pulmonary pathogen in pigs worldwide. In its most virulent form, A. pleuropneumoniae induces severe, rapidly fatal pleuropneumonia in naïve pigs of all ages [41]. Virulence of A. pleuropneumoniae strains varies remarkably, ranging from acute disease with high mortality to more chronic respiratory problems without significant mortality. Animals may be carriers of A. pleuropneumoniae at the level of the tonsils and in chronic lung lesions.
Using conventional necropsy findings or serology is a relevant first step towards diagnosis of the complex combination of several PRDC pathogens. However, due to the absence of pathognomonic lesions involved and the variable interval between infection and seroconversion for each of these pathogens, it is frequently not easy to obtain a conclusive diagnosis. Therefore, direct detection of pathogens present through PCR techniques is currently used [5,6,42,43]. A validated sampling technique using tracheobronchial sampling [43,44,45,46] applied for early detection of M. hyopneumoniae in pigs [47] was used to collect samples in pigs of different age categories, with clinical signs of respiratory diseases, for subsequent analysis with a multiplex PCR (M. hyopneumoniae, PRRSV (PRRSV1, European strain; PRRSV2, North-American strain; PRRSV1,2, combined European and North-American strain), IAV-S, PCV-2, PRCV, PCMV), and a supplementary bacterial PCR (A. pleuropneumoniae), to detect seven different PRDC pathogens.
The aim of the present study was to detect the prevalence of several respiratory pathogens in pigs with clinical signs of respiratory disease using the TBS sampling technique and to gain insights into seasonal variation and correlation with local weather conditions.
2. Results
2.1. Prevalence of M. hyopneumoniae and Other PRDC Pathogens among Age Categories
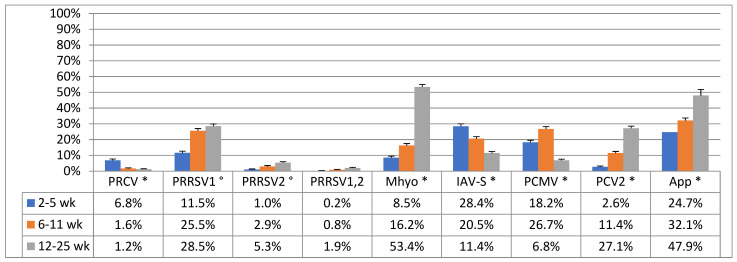
The prevalence data of M. hyopneumoniae and all other PRDC pathogens among the different age categories are given in Figure 1. At 3–5 weeks of age, 8.5% of piglets were already M. hyopneumoniae-positive, increasing to 16.2% at 6–11 weeks of age. In the fattening period, sampled pigs with clinical signs of respiratory disease were 53.4% M. hyopneumoniae-positive.
Figure 1.
Prevalence (expressed as % positive samples ± SEM) of different single PRDC pathogens at different age categories, namely peri-weaned piglets (3–5 weeks of age), post-weaned piglets (6–11 weeks of age), and fattening pigs (12–25 weeks of age). In total, 22,266 pigs were sampled. PRCV, porcine respiratory coronavirus; PRRSV1, European strain of porcine reproductive and respiratory syndrome virus; PRRSV2, North-American strain of PRRSV; PRRSV1,2, combined European and North-American strain of PRRSV; Mhyo, Mycoplasma hyopneumoniae; IAV-S, influenza A virus in swine; PCMV, porcine cytomegalovirus; PCV-2, porcine circovirus type 2; App, Actinobacillus pleuropneumoniae. Significant differences (p < 0.05) in pathogen prevalence are indicated by superscript: * differences between all three age categories significant; ° differences between 2–5 weeks and 6–11 weeks of age, and between 2–5 weeks and 12–25 weeks.
During the peri-weaning period (3–5 weeks of age), most prevalent PRDC pathogens were IAV-S (28.4%), PCMV (18.2%), PRRSV1 (11.5%), and PRCV (6.8%). Other pathogens, such as PRRSV2, PRRSV1,2, and PCV-2 were much less prevalent. The secondary pathogen A. pleuropneumoniae occupied an intermediate position with a prevalence of 24.7% at 3–5 weeks of age.
During the post-weaning period (6–11 weeks of age), predominant PRDC pathogens were PCMV (26.7%), PRRSV1 (25.5%), IAV-S (20.5%), and PCV-2 (11.4%). Other pathogens, such as PRRSV2, PRRSV1,2, and PRCV had a rather low prevalence (<3.0%). Prevalence of A. pleuropneumoniae rose from 24.7 to 32.1% during the post-weaning period.
During the fattening period (12–25 weeks of age), prevalence of several PRDC pathogens showed a clear shift. PRRSV1 prevalence remained stable at 28.5%, although other PRRSV strain types also gained importance with PRRSV2 at 5.3% and PRRSV1,2 at 1.9% prevalence. PRCV and PCMV prevalence further decreased to 1.2% and 6.8%, respectively. Prevalence of IAV-S decreased to 11.4%, whereas PCV-2 increased to 27.1%. Prevalence of A. pleuropneumoniae further increased towards 47.9% during the fattening period.
Prevalence of different double and triple PRDC major pathogen interactions are given in Table 1. Most prevalent pathogen combinations among the different age categories are PRRSV–M. hyopneumoniae, PRRSV–IAV-S, PRRSV–PCV-2 and M. hyopneumoniae–IAV-S within the double infections and PRRSV–M. hyopneumoniae–IAV-S and PRRSV–M. hyopneumoniae–PCV-2 within the triple infections.
Table 1.
Prevalence (expressed as % positive samples ± SEM) of different double and triple PRDC major pathogen interactions in different age categories, namely peri-weaned piglets (3–5 weeks of age), post-weaned piglets (6–11 weeks of age), and fattening pigs (12–25 weeks of age). In total, 22,266 pigs were sampled. PRRSV1, European strain of porcine reproductive and respiratory syndrome virus; PRRSV2, North-American strain of PRRSV; PRRSV1,2, combined European and North-American strain of PRRSV; Mhyo, Mycoplasma hyopneumoniae; IAV-S, influenza A virus in swine; PCV-2, porcine circovirus type 2; App, Actinobacillus pleuropneumoniae.
| Pathogen Combination | Age Category | ||
|---|---|---|---|
| 3–5 w | 6–11 w | 12–25 w | |
| Double infections | |||
| PRRSV–Mhyo | 1.9 ± 0.5% a | 6.1 ± 0.8% b | 21.0 ± 1.3% c |
| PRRSV1–Mhyo | 1.8 ± 0.5% a | 5.2 ± 0.7% b | 15.9 ± 1.1% c |
| PRRSV2–Mhyo | 0.1 ± 0.1% a | 0.7 ± 0.3% a | 3.7 ± 0.6% a |
| PRRSV1,2–Mhyo | 0.0 ± 0.1% a | 0.2 ± 0.2% a | 1.3 ± 0.4% a |
| PRRSV–IAV-S | 3.3 ± 0.6% a | 5.1 ± 0.7% b | 2.8 ± 0.5% ab |
| PRRSV1–IAV-S | 3.1 ± 0.6% a | 4.5 ± 0.7% a | 2.4 ± 0.5% a |
| PRRSV2–IAV-S | 0.2 ± 0.2% a | 0.5 ± 0.3% a | 0.4 ± 0.2% a |
| PRRSV1,2–IAV-S | 0.0 ± 0.1% a | 0.2 ± 0.2% a | 0.0 ± 0.0% a |
| PRRSV–PCV-2 | 1.0 ± 0.3% a | 5.4 ± 0.8% a | 9.5 ± 0.9% a |
| PRRSV–PCV-2 | 0.9 ± 0.3% a | 4.4 ± 0.7% a | 7.7 ± 0.8% a |
| PRRSV2–PCV-2 | 0.1 ± 0.1% a | 0.6 ± 0.3% a | 1.2 ± 0.3% a |
| PRRSV1,2–PCV-2 | 0.0 ± 0.0% a | 0.3 ± 0.2% a | 0.7 ± 0.3% a |
| PRRSV–App | 1.0 ± 0.9% a | 2.7 ± 1.7% a | 4.8 ± 1.7% a |
| PRRSV1–App | 0.9 ± 0.9% a | 2.3 ± 1.3% b | 3.7 ± 1.5% c |
| PRRSV2–App | 0.0 ± 0.2% a | 0.3 ± 0.3% a | 0.6 ± 0.6% a |
| PRRSV1,–App | 0.0 ± 0.2% a | 0.1 ± 0.2% a | 0.5 ± 0.5% a |
| Mhyo–IAV-S | 2.7 ± 0.6% a | 2.5 ± 0.5% a | 5.4 ± 0.7% b |
| Mhyo–PCV-2 | 0.8 ± 0.3% a | 3.3 ± 0.6% a | 17.1 ± 1.2% a |
| Mhyo–App | 0.7 ± 0.7% a | 1.9 ± 0.7% b | 7.6 ± 1.2% c |
| Triple infections | |||
| PRRSV–Mhyo–IAV-S | 0.4 ± 0.2% a | 0.8 ± 0.3% a | 1.6 ± 0.4% a |
| PRRSV1–Mhyo–IAV-S | 0.3 ± 0.2% a | 0.8 ± 0.3% ac | 1.5 ± 0.4% bc |
| PRRSV2–Mhyo–IAV-S | 0.0 ± 0.1% a | 0.0 ± 0.1% a | 0.1 ± 0.1% a |
| PRRSV1,2–Mhy –IAV-S | 0.0 ± 0.1% a | 0.0 ± 0.0% a | 0.0 ± 0.0% a |
| PRRSV–Mhyo–PCV-2 | 0.3 ± 0.2% a | 1.7 ± 0.4% a | 6.9 ± 0.8% a |
| PRRSV –Mhyo–PCV-2 | 0.3 ± 0.2% a | 1.3 ± 0.4% b | 5.4 ± 0.7% c |
| PRRSV2–Mhy –PCV-2 | 0.0 ± 0.1% a | 0.2 ± 0.2% a | 0.9 ± 0.3% a |
| PRRSV1,2–Mhy–PCV-2 | 0.0 ± 0.0% a | 0.1 ± 0.1% a | 0.7 ± 0.3% a |
| PRRSV–IAV-S–PCV-2 | 0.3 ± 0.2% a | 0.7 ± 0.3% a | 0.5 ± 0.2% a |
| PRRSV1–IAV-S–PCV-2 | 0.2 ± 0.2% a | 0.7± 0.3% a | 0.5 ± 0.2% a |
| PRRSV2–IAV-S–PCV-2 | 0.0 ± 0.1% a | 0.0 ± 0.1% a | 0.0 ± 0.0% a |
| PRRSV1,2–IAV-S–PCV-2 | 0.0 ± 0.0% a | 0.0 ± 0.1% a | 0.0 ± 0.0% a |
Significant differences (p < 0.05) in pathogen prevalence are indicated by different letters (a–c) in superscript.
2.2. Seasonal Variation in Prevalence of M. hyopneumoniae and Other PRDC Pathogens at Piglet Level
Effect of season on prevalence of different PRDC pathogens or combinations of pathogens in different age categories are given in Table 2.
Table 2.
Effect of season on prevalence of different PRDC pathogens or PRDC pathogen combinations in different age categories. Pathogen prevalences are given in %. Significant differences (p < 0.05) are indicated with different letters in superscript. PRRSV1, European strain of porcine reproductive and respiratory syndrome virus; PRCV, porcine respiratory coronavirus; PCMV, porcine cytomegalovirus; M. hyopneumoniae, Mycoplasma hyopneumoniae; IAV-S, influenza A virus in swine; PCV-2, porcine circovirus type 2; A. pleuropneumoniae, Actinobacillus pleuropneumoniae. Seasons with the highest pathogen prevalence are in red.
| Age Category | Pathogen | Season | |||
|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | ||
| 6–11 w | PRRSV1 | 14.2 b | 10.3 ab | 8.2 a | 13.0 ab |
| IAV-S | 23.5 a | 32.8 b | 21.3 a | 33.9 b | |
| PCMV | 21.4 b | 18.4 ab | 14.8 a | 18.3 ab | |
| A. pleuropneumoniae | 20.2 ab | 31.0 b | 32.6 b | 15.0 a | |
| 12–25 w | PRCV | 1.8 ab | 2.2 b | 0.7 a | 1.5 ab |
| PRRSV1 | 35.5 b | 24.3 a | 22.8 a | 21.5 a | |
| PCMV | 25.6 ab | 26.9 bc | 21.9 a | 31.1 c | |
| PCV-2 | 16.3 c | 12.8 bc | 9.2 ab | 8.2 a | |
| A. pleuropneumoniae | 30.5 b | 37.1 bc | 45.2 c | 18.0 a | |
| PRRSV1/M. hyopneumoniae | 7.2 b | 4.8 ab | 5.5 ab | 3.7 a | |
| PRRSV1/PCV-2 | 8.1 b | 3.9 a | 3.9 a | 3.6 a | |
| PRRSV1/A. pleuropneumoniae | 2.2 ab | 2.3 ab | 4.0 b | 1.2 a | |
| M. hyopneumoniae/IAV-S | 3.7 b | 2.0 ab | 1.5 a | 2.8 ab | |
| M. hyopneumoniae/PCV-2 | 5.7 b | 3.5 ab | 2.2 a | 2.3 a | |
| PRRSV1/M. hyopneumoniae/PCV-2 | 3.3 b | 0.7 a | 1.0 a | 0.6 a | |
| PRRSV1/IAV-S/PCV-2 | 1.4 b | 0.6 ab | 0.6 ab | 0.3 a | |
Significant differences (p < 0.05) in pathogen prevalence are indicated by different letters (a–c) in superscript.
At 3–5 weeks of age, no seasonal effect on prevalence of M. hyopneumoniae and other PRDC pathogens could be observed. At 6–11 weeks of age, several pathogens had a seasonal variation in occurrence. Actinobacillus pleuropneumoniae had the highest prevalence during summer (S3), whereas IAV-S was most prevalent during autumn (S4). For the viral agents PRRSV1 and PCMV, the highest prevalence occurred during winter (S1).
During the fattening period, both single PRDC pathogens and combined infections, including M. hyopneumoniae, showed seasonal variations. These combined infections included M. hyopneumoniae–PRRSV1 (7.2%), M. hyopneumoniae–PCV-2 (5.7%), M. hyopneumoniae–IAV-S (3.7%) and M. hyopneumoniae–PRRSV1–PCV-2 (3.3%). For most pathogens, highest prevalence could be observed in winter (S1), except for PCMV (autumn, S4), A. pleuropneumoniae, and the combined infection of PRRSV1–A. pleuropneumoniae (summer, S3).
2.3. Impact of Climatological Parameters on Piglet Positivity for M. hyopneumoniae and Other PRDC Pathogens
The most relevant associations (expressed as odds ratio (OR) with the 95% confidence intervals between brackets) between prevalence of different pathogens during the 5-year study in Belgium and the Netherlands and local weather parameters are presented in Table 3.
Table 3.
Associations (OR—odds ratio) between overall pathogen prevalence during the five-year study in Belgium and the Netherlands and specific weather parameter registered at the weather stations located nearby the sampled farm included in the study. Only significant associations (Q < 0.001) for pathogens with an overall prevalence in all three age categories >5% are given. PRRSV, porcine reproductive and respiratory syndrome virus; PRRSV1, European strain of PRRSV; Mhyo, Mycoplasma hyopneumoniae; PCV-2, porcine circovirus type 2; App, Actinobacillus pleuropneumoniae. WS—wind speed; WSavg—average measured wind speed; Tdiff—difference between maximal and minimal temperature measured over the day; Sdur—duration of sunshine; RHmin—minimal relative humidity and WD—wind direction. Co-notations associated with the weather parameter indicated the period: 10 w—10-week rolling average. Positive associations have an OR > 1, negative associations have an OR < 1.
| Pathogen | WS.10w | WSavg.10w | Tdiff.10w | Sdur.10w | RHmin.10w | WD.10w |
|---|---|---|---|---|---|---|
| App | 1.164 | 0.940 | ||||
| (1.063; 1.274) | (0.911; 0.970) | |||||
| PRRSV1 | 1.280 | 1.267 | 0.921 | 0.911 | 1.020 | 0.981 |
| (1.126; 1.456) | (1.116; 1.440) | (0.880; 0.965) | (0.861; 0.964) | (1.009; 1.032) | (0.970; 0.992) | |
| PCV-2 | 1.836 | 1.832 | 0.834 | 1.030 | ||
| (1.516; 2.225) | (1.517; 2.214) | (0.777; 0.896) | (1.012; 1.049) | |||
| M. hyopneumoniae—PCV2 | 1.918 | 1.947 | 0.818 | 1.026 | ||
| (1.460; 2.520) | (1.484; 2.553) | (0.736; 0.911) | (1.012; 1.040) | |||
| M. hyopneumoniae—PRRSV | 1.404 | |||||
| (1.150; 1.714) | ||||||
| PRRSV—PCV-2 | 1.753 | 1.738 | 0.799 | 0.817 | 1.048 | |
| (1.327; 2.316) | (1.317; 2.293) | (0.718; 0.889) | (0.721; 0.925) | (1.021; 1.075) |
Wind speed in the 10 weeks prior to sampling (WS.10w) and average wind speed in the 10 weeks prior to sampling (WSavg.10w) were only positively associated with all pathogen prevalence with OR ranging from 1.280 (PRRSV1) to 1.918 (PCV-2) for WS.10w and OR ranging from 1.267 (PRRSV1) to 1.947 (M. hyopneumoniae–PCV-2) for WSavg.10w.
The difference between minimum and maximum outside temperature in the 10 weeks prior to sampling (Tdiff.10w) was negatively associated with PRRSV1 (OR = 0.921), PCV-2 (OR = 0.834), M. hyopneumoniae–PCV-2 (OR = 0.818), and PRRSV–PCV-2 (OR = 0.799).
Duration of sunshine in the 10 weeks prior to sampling (Sdur.10w) was positively associated with A. pleuropneumoniae (OR = 1.164) and negatively associated with PRRSV1 (OR = 0.911) and PRRSV–PCV-2 (OR = 0.817).
Minimal relative humidity in the 10 weeks prior to sampling (RHmin.10w) was negatively associated with A. pleuropneumoniae (OR = 0.940), and positively associated with PRRSV1 (OR = 1.020), PCV-2 (OR = 1.030), and PRRSV–PCV-2 (OR = 1.048).
Wind direction in the 10 weeks prior to sampling (WD.10w) was negatively associated with PRRSV1 (OR = 0.981) and positively associated with M. hyopneumoniae–PCV-2 (OR = 1.026).
3. Discussion
Porcine respiratory disease complex remains one of the most important health concerns with a high economic impact for pig producers worldwide. The disease involves multiple viral and bacterial pathogens together with several non-infectious factors, such as ventilation, housing conditions, and management, leading to respiratory distress in pigs ranging from the peri-weaning period (3–5 weeks of age) over the post-weaning period (6–11 weeks of age) to the finishing stage (12–25 weeks of age). Interaction between both infectious (viral and bacterial agents) and non-infectious factors may all contribute to the development and severity of the respiratory disease [23]. The most commonly identified pathogens are PRRSV, IAV-S, PCV-2, and M. hyopneumoniae, besides other pathogens associated with PRDC, such as Streptococcus suis, A. pleuropneumoniae, P. multocida, DNT-positive P. multocida, Glaeserella parasuis (G. parasuis), Mycoplasma hyorhinis (M. hyorhinis), Mycoplasma hyosynoviae, PRCV, and PCMV [3,23,48].
Detection of the etiologic agents of PRDC has long been difficult, especially due to the wide variety of diagnostic approaches applied in practice. Diagnosis of M. hyopneumoniae could be performed using clinical signs, slaughterhouse checks of affected lungs [49,50], serological examination of relevant age groups [49,50], direct pathogen identification through bacteriological culture [44] or PCR techniques [51,52]. As for other respiratory pathogens involved in PRDC, more or less the same diagnostic approach has been applied, mainly due to lack of diagnostic tests able to simultaneously detect multiple respiratory pathogens in a single-reaction method [6]. Although these single pathogen detection techniques may be reliable and sensitive, they remain time-consuming, labor-intensive and, therefore, quite expensive. Moreover, for bacterial pathogens detection typically depends on culture-based methods that can take up to several days to obtain the final results.
Polymerase chain reactions and real-time PCR tests have been developed for several pathogens involved in PRDC and are characterized by their high sensitivity and ease of use. In combination with a reliable sampling technique, such as TBS, these detection methods based on PCR have been able to detect M. hyopneumoniae at an early age [45,46] and in an early stage of infection [53,54].
The results from the current study in 974 pig farms clearly demonstrate that pigs can be infected at an early stage with M. hyopneumoniae, which is in accordance with previous reports applying the same sampling technique [46]. However, besides M. hyopneumoniae, several other PRDC pathogens may be involved in the clinical picture of coughing piglets at 3–5 weeks of age, such as IAV-S, PRRSV1, PRCV, and PCMV, whereas the prevalence of PCV-2 is rather low at that stage. From a bacterial perspective, A. pleuropneumoniae is present in a quarter of the pigs sampled. These observations are in accordance with Sunaga et al. (2020) [6] who observed mixed infection with several bacterial (A. pleuropneumoniae, Bordetella bronchiseptica, P. multocida, M. hyopneumoniae, M. hyorhinis), and viral agents (PCV-2, PCMV, PRRSV US strain, PRCV) in some of the farms included in the study. Comparison of prevalence data is however not possible due to the low sample number (n = 6 farms, n = 30 samples) in this study [6] as compared to our study (n = 974 farms, n = 22,266 samples).
In contrast to previous studies [45,46,53,54], where sampling was focused on M. hyopneumoniae detection and prevalence only, the current study clearly demonstrates that M. hyopneumoniae is in many cases combined with other pathogens related to PRDC. The demonstrated presence of M. hyopneumoniae at an early stage (pre-weaning or post-weaning) might significantly impact the clinical course of other PRDC pathogens such as PRRSV [9,10], IAV-S [2,11,12], or PCV-2 [14,15].
In the post-weaned piglets (6-11 weeks of age), some clear differences in PRDC pathogen prevalence could be observed. M. hyopneumoniae and both PRRSV1 and PRRSV2 doubled in prevalence as compared to peri-weaned piglets (3–5 weeks of age), together with PCMV and A. pleuropneumoniae that showed a more moderate increase. In contrast, PRCV rapidly decreased during the post-weaning period. The prevalence of PCV-2 increased over four times during the post-weaning phase, indicating that PCVAD became more prominently involved in respiratory problems during this phase. Besides single infections with one of these PRDC pathogens, several combined double and triple infections (Table 1) could also be detected involving the major pathogens PRRSV, M. hyopneumoniae, PCV-2, and IAV-S and A. pleuropneumoniae as the most prevalent bacterial agent. This implies that piglets in the post-weaning phase are exposed to mixed infections of several viral pathogens known as immunosuppressive, leading to a compromised immune response towards vaccination or other concurrent bacterial infections such as post-weaning diarrhea due to enterotoxigenic Escherichia coli.
During the fattening phase, the most prominent PRDC pathogens in pigs with clinical signs of respiratory disease were M. hyopneumoniae, PRRSV1, PRRSV2, and PCV-2, together with A. pleuropneumoniae and to a lesser extent IAV-S. The high prevalence of M. hyopneumoniae is of particular interest and may be partially explained by the long duration of pathogen persistence in the respiratory tract of the infected pigs. Previous studies have detected M. hyopneumoniae up to 254 days post-infection in deep laryngeal swabs using a PCR technique [55]. Recent research into specific risk factors influencing M. hyopneumoniae infection status revealed that both age and type of production system had an impact on infection rate and pathogen prevalence [56]. Infection rates were higher in older animals and the prevalence was higher in the one- and two-site systems than in the three-site systems. Dynamics of infection by RT-PCR showed that M. hyopneumoniae infection on one-site farms occurs earlier, while on two- and three-site farms occurs later but spreads faster, suggesting that contact between animals of different age favors the transmission. However, in the present study, data on the type of production system were not recorded. Nevertheless, the impact of age was similar in our study, since the prevalence of M. hyopneumoniae is substantially higher in the fattening pigs as compared to peri-weaned or post-weaned piglets with clinical signs of respiratory disease [56]. Therefore, piglets infected with M. hyopneumoniae during the post-weaning period or the early fattening period can excrete the pathogen for the entire fattening period, thus, increasing the probability of detection when pigs are sampled during an episode of respiratory problems. This is in contrast with other pathogens such as IAV-S that have a rather short period (max. 6–8 days) of detection at the level of the respiratory tract [57,58]. The results obtained concerning PCV-2 dynamics in the current study are in accordance with a recent study on serological and viral dynamics of PCV-2 carried out in PCV-2 infected pig herds in Taiwan [59]. As reported previously, we also observed an increased prevalence during the grow (11.2%) and finish phase (27.1%) [59].
In the peri-weaned piglets, no effect of season on prevalence of PRDC pathogens could be observed. Presence of PRDC pathogens at that age is mainly determined by the infection status of their dam and outside conditions have very little impact on infection status at that age. In contrast, in post-weaned piglets, single PRDC pathogens are affected in their seasonal prevalence, with A. pleuropneumoniae at the highest level in summer (S3), IAV-S in autumn (S4) and both PRRSV1 and PCMV in winter (S1). In practice, it is well-known that respiratory problems related with A. pleuropneumoniae frequently occur at the change of seasons, especially with warmer days and colder nights, as might happen in our region at start of spring (April/May; S2) and the end of summer (August/September; S3), which explains the higher prevalence in these two seasons in our study. On the contrary, IAV-S occurs later in the year, when weather conditions turn cold and wet, which is the case in autumn (S4). In the colder winter months, PRRSV1 and PCMV seem to circulate more pronouncedly. The same pattern continues in the older pigs during the fattening period (12–25 weeks of age), where most PRDC pathogens and several combined respiratory infections are highly prevalent during winter (S1). Only exceptions from this pattern are PCMV (S4) and infections involving A. pleuropneumoniae, which are again more prominent during late summer (S3), as previously discussed.
Several climatological parameters were significantly associated with PRDC pathogen prevalence. The most prominent parameters are relative humidity (RHmin.10w), temperature difference between minimum–maximum temperature (Tdiff.10w), wind speed (WS.10w), average wind speed (WSavg.10w), wind direction (WD.10w) and duration of sunshine (Sdur.10w). Apparently, the weather conditions in the 10 weeks prior to sampling have the largest impact on pathogen prevalence. Another interesting observation is that from all PRDC pathogens detected in prevalences higher than 5%, prevalence of M. hyopneumoniae and PRRSV is most frequently impacted by weather conditions. In a Spanish study [60], the influence of climatological parameters on M. hyopneumoniae dynamics was also explored. This study revealed the higher the precipitation rate, the higher the probability of being M. hyopneumoniae nPCR-positive on nasal swabs, whereas the lower the temperature, the higher the probability of being M. hyopneumoniae seropositive. From a seasonal perspective, animals born in autumn and reaching slaughter in spring had the highest probability of being infected by M. hyopneumoniae and the highest probability of being M. hyopneumoniae seropositive [60]. Moreover, bio-aerosols, containing M. hyopneumoniae and PRRSV, were capable of spreading pathogens between herds via airborne route. Conditions common to both pathogens include cool temperature and specific wind direction, and more specifically low sunlight levels, low wind velocity in combination with rising humidity and air pressure [61].
The present study clearly shows that M. hyopneumoniae and different viral and bacterial pathogens responsible for PRDC may be present during the peri-weaning, post-weaning, and fattening period. Following analysis of seasonal variation, it can be concluded that depending on the pathogen, a clear variation in seasonal impact on the prevalence of PRDC pathogens is present. Moreover, several climatological parameters may influence the prevalence of the detected PRDC pathogens.
4. Materials and Methods
4.1. Selection of Study Herds
The study was conducted from September 2011 to September 2016 in Belgium and the Netherlands. Closed pig herds were selected through regular contacts with local veterinary practices. Inclusion criteria were as following: at least 200 sows, at least two age groups (3–5 weeks of age, 6–11 weeks of age, and 12–25 weeks of age) available for sampling, presence of clinical signs of respiratory disease (coughing), and no use of antimicrobials active against M. hyopneumoniae in piglets less than 3 weeks of age and during the last 2 weeks prior to sampling in the post-weaning or fattening phase. Piglets or fattening pigs eligible for sampling were marked up by the swine farmer or herd veterinarian prior to sampling. In total, 974 closed pig herds were included in the study, equally distributed over different seasons of the year (Table 4). In total, 22,266 pigs with clinical signs of respiratory disease were sampled. On average, 22.8 pigs were sampled per herd with a minimum of 10 pigs and a maximum of 30 pigs, distributed over the different age categories that suffered from clinical signs of respiratory disease. Therefore, samples were collected during the peri-weaning (3–5 weeks of age), post-weaning (6–11 weeks of age), and fattening (12–25 weeks of age) period. The latter group was only sampled in case of presence of clinical signs of respiratory disease in that age group. Within each herd, we sampled piglets affected with clinical signs of respiratory disease from as many different compartments and pens in the nursery as possible. Sampling was always performed by the same veterinarian trained on TBS sampling.
Table 4.
Number of swine farms in Belgium and the Netherlands sampled per year and per season during the entire study period from S4 2011 until S3 2016, including total number of sampled farms per year and per season. S1, winter; S2, spring; S3, summer; and S4, autumn.
| Season | Year | Total/Season | |||||
|---|---|---|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | ||
| S1 | 51 | 53 | 44 | 34 | 45 | 227 | |
| S2 | 40 | 40 | 53 | 45 | 69 | 247 | |
| S3 | 31 | 41 | 47 | 32 | 62 | 213 | |
| S4 | 47 | 50 | 70 | 69 | 51 | 287 | |
| Total/Year | 47 | 172 | 204 | 213 | 162 | 176 | 974 |
4.2. Tracheobronchial Swab (TBS) Sampling Procedure
TBS sampling was performed as previously described [45,46]. Briefly, TBS were obtained through thorough fixation of the piglets with a nose snare, followed by use of a mouth opener. The TBS (aspiration tube, 50 cm, 12CH; Medinorm GmbH, Spiesen-Elversberg, Germany) was subsequently inserted through the mouth, through the glottis down to the tracheobronchial split. Mucus was collected through gentle movement of the swab at the level of the tracheobronchial split and the swab was subsequently retrieved. The tip of the swab was collected in a sterile tube (MLS, Menen, Belgium) with 1 mL of sterile saline solution (Saline Solution 0.9%; Eurovet, Heusden-Zolder, Belgium) and kept cool at 3 °C until analysis within 48 h after sampling.
4.3. Analysis of TBS Swabs
The material collected by the TBS was processed according to Strait et al. (2008) [62]. A multiplex PCR (mPCR) analysis was performed according to the standard operating procedure of the laboratory (IVD GmbH, Hannover, Germany). The mPCR included analysis of M. hyopneumoniae, PRRSV (including differentiation in European (PRRSV1) and North-American (PRRSV2) or combined European and North-American strain type (PRRSV1,2)), PCV-2, IAV-S, PCMV, and PRCV. A second PCR for bacterial species (A. pleuropneumoniae) was run on the same samples. The test was an App apxIV-PCR (IVD GmbH, Germany; in-house test; Strutzberg-Minder, 2009). It was a specific 414 bp-target, which had been tested for the detection of App serotype 1 to 19. Specificity was tested against the following species: Glaesserella parasuis serotypes 1–15, Actinobacillus porcitonsillarum, Actinobacillus suis, Actinobacillus minor, Actinobacillus indolicus, Actinobacillus porcinum, Actinobacillus equuli, Actinobacillus lignieresii, Mannheimia haemolytica, Pasteurella multocida, and Streptococcus suis. Analytical sensitivity was 1000 GE/mL. PCR results were reported as negative or positive for the presence of the different PRDC pathogens.
4.4. Data Categorization for Seasonality
In order to assess the association among season, as defined by calendar months, and infection dynamics of M. hyopneumoniae and other PRDC pathogens sampled, herds were categorized for seasonality based on date of sampling. Seasonality was implemented as the following: S1—winter (21/12–20/03; n = 227 herds), S2—spring (21/03–20/06; n = 247 herds), S3—summer (21/06–20/09; n = 213 herds), and S4—autumn (21/09–20/12; n = 287 herds) (Table 4).
4.5. Climatologic Data
Climatologic data collected are presented in Table 5. These data were collected from July 2011 to September 2016. Climatologic data were provided by the local meteorological institute (KNMI, Koninklijk Nederlands Meteorologisch Instituut; http://www.knmi.nl/klimatologie (accessed on 16 October 2016)).
Table 5.
List of climatological observation parameters with their abbreviations used and their specific units.
| Parameter | Abbreviation | Units |
|---|---|---|
| WD | Wind direction | ° |
| WS | Wind speed | 0.1 m/s |
| WSavg | Average wind speed over 12 h | 0.1 m/s |
| T | Average temperature over 12 h | 0.1 °C |
| Tmin | Minimum temperature | 0.1 °C |
| Tmax | Maximum temperature | 0.1 °C |
| Tdiff | Difference maximal–minimal temperature measured over the day | 0.1 °C |
| Sdur | Duration of sunshine | 0.1 h |
| Pdur | Duration of precipitation | 0.1 h |
| P | Total precipitation in 12 h | 0.1 mm |
| AP | Average air pressure at sea level over 12 h | 0.1 Pa |
| RH | Relative humidity | % |
| RHmax | Maximal relative humidity | % |
| RHmin | Minimal relative humidity | % |
Besides the climatologic data observed on the day of sampling, a rolling average of the data over a 1-, 2-, 4-, and 10-week period were calculated for all sampling days throughout the 5-year study period.
4.6. Statistical Analysis
The effect of season on the prevalence of PRDC pathogens was investigated with logistic regression mixed models with a random effect for herd. The overall effect of season was tested with the likelihood ratio test, and all pairwise comparisons between seasons were performed on the log odds ration scale with Wald tests [63] and with Bonferroni adjustment for controlling the familywise error rate (FWER) at 5%. These analyses were repeated for all pathogens and within the three age categories (3–5, 6–11, and 12–25 weeks of age).
The effects of the climate parameters on the prevalence were investigated by fitting logistic regression mixed models with herd as random effect. The models were fitted for each combination of pathogen and climate parameter. The significance of the effects was tested by means of Wald tests. Given the large number of tests, false positives were controlled with the method of Storey (2003) [64] for controlling the false discovery rate (FDR) at 5%.
All statistical analyses were performed in the statistical software R [65] (R Core Team, 2021) and the R packages lme4, emmeans, and qvalue. All hypothesis tests were performed at the 5% level of significance, and multiple testing procedures were performed at the 5% FWER or the 5% FDR level.
5. Conclusions
In conclusion, the present study showed that many respiratory pathogens are present during the peri-weaning, post-weaning, and fattening period, which may complicate the clinical picture of respiratory disease. Moreover, interactions between these PRDC pathogens, season, and local weather conditions could be demonstrated over the five-year study period.
Acknowledgments
The authors acknowledge the swine veterinarians and swine farmers cooperating in this field study for their help in identifying eligible farms and for their assistance in sample collection.
Author Contributions
Conceptualization, F.A.C.J.V. and O.T.; methodology, F.A.C.J.V.; formal analysis, F.A.C.J.V.; investigation, F.A.C.J.V.; resources, F.A.C.J.V.; data curation, F.A.C.J.V. and O.T.; writing—original draft preparation, F.A.C.J.V.; writing—review and editing, F.A.C.J.V. and O.T.; visualization, F.A.C.J.V.; supervision, F.A.C.J.V.; project administration, F.A.C.J.V.; funding acquisition, F.A.C.J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The manuscript has been reviewed internally and was approved for disclosure under the ID number SDR_502.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maes D., Deluyker H., Verdonck M., Castryck F., Miry C., Vrijens B., de Kruif A. Herd factors associated with the seroprevalences of four major respiratory pathogens in slaughter pigs from farrow-to-finish pig herds. Vet. Res. 2000;31:313–327. doi: 10.1051/vetres:2000122. [DOI] [PubMed] [Google Scholar]
- 2.Thacker E.L. Immunologyof the porcine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2001;17:551–565. doi: 10.1016/S0749-0720(15)30006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen M.S., Pors S.E., Jensen H.E., Bille-Hansen V., Bisgaard M., Flachs E.M., Nielsen O.L. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J. Comp. Pathol. 2010;143:120–131. doi: 10.1016/j.jcpa.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Garcia J., Robben N., Magnée D., Eley T., Dennis I., Kayes S.M., Thomson J.R., Tucker A.W. The use of oral fluids to monitor key pathogens in porcine respiratory disease complex. Porc. Health Manag. 2017;3:7–12. doi: 10.1186/s40813-017-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fablet C., Marois-Créhan C., Simon G., Grasland B., Jestin A., Kobisch M., Madec F., Rose N. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: A cross-sectional study. Vet. Microbiol. 2012;157:152–163. doi: 10.1016/j.vetmic.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Sunaga F., Tsuchiaka S., Kishimoto M., Aoki H., Kakinoki M., Kure K., Okumura H., Okumura M., Okumura A., Nagai M., et al. Development of a one-run real-time PCR detection system for pathogens associated with porcine respiratory diseases. J. Vet. Med. Sci. 2020;82:217–223. doi: 10.1292/jvms.19-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibila M., Pieters M., Molitor T., Maes D., Haesebrouck F., Segalés J. Current perspectives on the diagnosis and epidemiology of Mycoplasma hyopneumoniae infection. Vet. J. 2009;181:221–231. doi: 10.1016/j.tvjl.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maes D., Sibila M., Kuhnert P., Segalés J., Haesebrouck F., Pieters M. Update on Mycoplasma hyopneumoniae infections in pigs: Knowledge gaps for improved disease control. Transbound. Emerg. Dis. 2017;62:1–15. doi: 10.1111/tbed.12677. [DOI] [PubMed] [Google Scholar]
- 9.Fano E., Pijoan C., Dee S. Infection dynamics of porcine reproductive and respiratory syndrome virus in a continuous-flow population of pigs also infected with Mycoplasma hyopneumoniae. Vet. Rec. 2007;161:515–520. doi: 10.1136/vr.161.15.515. [DOI] [PubMed] [Google Scholar]
- 10.Park S.-J., Seo H.W., Park C., Chae C. Interaction between single-dose Mycoplasma hyopneumoniae and porcine reproductive and respiratory syndrome virus vaccines on dually infected pigs. Res. Vet. Sci. 2014;96:516–522. doi: 10.1016/j.rvsc.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Deblanc C., Gorin S., Quéguiner S., Gautier-Bouchardon A.V., Ferré S., Amenna N., Cariolet R., Simon G. Pre-infection of pigs with Mycoplasma hyopneumoniae modifies outcomes of infection with European swine influenza virus of H1N1, but not H1N2 subtype. Vet. Microbiol. 2012;157:96–105. doi: 10.1016/j.vetmic.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deblanc C., Robert F., Pinard T., Gorin S., Quéguiner S., Gautier-Bouchardon A.V., Ferré S., Garraud J.M., Cariolet R., Brack M., et al. Pre-infection of pigs with Mycoplasma hyopneumoniae induces oxidative stress that influences outcomes of a subsequent infection with a swine influenza virus of H1N1 subtype. Vet. Microbiol. 2013;162:643–651. doi: 10.1016/j.vetmic.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Kim D., Kim C.H., Han K., Seo H.W., Oh Y., Park C., Kang I., Chae C. Comparative efficacy of commercial Mycoplasma hyopneumoniae and porcine circovirus 2 (PCV2) vaccines in pigs experimentally infected with M. hyopneumoniae and PCV2. Vaccine. 2011;29:3206–3212. doi: 10.1016/j.vaccine.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Opriessnig T., Thacker E.L., Yu S., Fenaux M., Meng X.-J., Halbur P.G. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet. Pathol. 2004;41:624–640. doi: 10.1354/vp.41-6-624. [DOI] [PubMed] [Google Scholar]
- 15.Rossow K.D. Porcine reproductive and respiratory syndrome. Vet. Pathol. 1998;35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- 16.Labarque G., Van Reeth K., Van Gucht S., Nauwynck H., Pensaert M. Porcine reproductive-respiratory syndrome virus infection predisposes pigs for respiratory signs upon exposure to bacterial lipopolysaccharide. Vet. Microbiol. 2002;88:1–12. doi: 10.1016/S0378-1135(02)00104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thacker E.L., Halbur P.G., Ross R.F., Thanawongnuwech R., Thacker B.J. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J. Clin. Microbiol. 1999;37:620–627. doi: 10.1128/JCM.37.3.620-627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockmeier S.L., Palmer M.V., Bolin S.R. Effects of intranasal inoculation of porcine reproductive and respiratory syndrome virus, Bordetella bronchiseptica, or a combination of both organisms in pigs. Am. J. Vet. Res. 2000;61:892–899. doi: 10.2460/ajvr.2000.61.892. [DOI] [PubMed] [Google Scholar]
- 19.Drew T.W. A review of evidence for immunosuppression due to porcine reproductive and respiratory syndrome virus. Vet. Res. 2000;31:27–39. doi: 10.1051/vetres:2000106. [DOI] [PubMed] [Google Scholar]
- 20.Halbur P.G., Thanawongnuwech R., Brown G., Kinyon J., Roth J., Thacker E., Thacker B. Efficacy of antimicrobial treatments and vaccination regimens for control of porcine reproductive and respiratory syndrome virus and Streptococcus suis coinfection of nursery pigs. J. Clin. Microbiol. 2000;38:1156–1160. doi: 10.1128/JCM.38.3.1156-1160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renukaradhya G.J., Alekseev K., Jung K., Fang Y., Saif L.J. Porcine reproductive and respiratory syndrome virus-induced immunosuppression exacerbates the inflammatory response to porcine respiratory coronavirus in pigs. Viral Immunol. 2010;23:457–466. doi: 10.1089/vim.2010.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segalés J., Calsamiglia M., Rosell C., Soler M., Maldonado J., Martin M., Domingo M. Porcine reproductive and respiratory syndrome virus (PRRSV) infection status in pigs naturally infected with post-weaning multisystemic wasting syndrome (PMWS) in Spain. Vet. Microbiol. 2002;85:23–30. doi: 10.1016/S0378-1135(01)00474-6. [DOI] [PubMed] [Google Scholar]
- 23.Opriessnig T., Giménez-Lirola L.G., Halbur P.G. Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 2011;12:133–148. doi: 10.1017/S1466252311000120. [DOI] [PubMed] [Google Scholar]
- 24.Rovira A., Balasch M., Segalés J., Garcia L., Plana-Duran J., Rosell C., Ellerbrok H., Mankertz A., Domingo M. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J. Virol. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen C.W., Brown I.H., Easterday B.C., Van Reeth K. Swine Influenza. In: Straw B., Zimmerman W., D’Allaire S., Taylor D., editors. Diseases of Swine. 9th ed. Iowa State University Press; Ames, IA, USA: 2006. pp. 469–482. [Google Scholar]
- 26.Kuntz-Simon G., Madec F. Genetic and antigenic evolution of swine influenza viruses in Europe and evaluation of their zoonotic potential. Zoonoses Public Health. 2009;56:310–325. doi: 10.1111/j.1863-2378.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 27.Simon-Grife M., Martin-Valls G.E., Vilar M.J., Busquets N., Mora-Salvatierra M., Besteboer T.M., Fouchier R.A., Martin M., Mateu E., Casal J. Swine influenza virus infection dynamics in two pigs farms: Results of a longitudinal assessment. Vet. Res. 2012;43:24. doi: 10.1186/1297-9716-43-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allerson M., Deen J., Detmer S.E., Gramer M.R., Joo H.S., Romagosa A., Torremorell M. The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine. 2013;31:500–505. doi: 10.1016/j.vaccine.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segalès J., Allan G.M., Domingo M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005;6:119–142. doi: 10.1079/AHR2005106. [DOI] [PubMed] [Google Scholar]
- 30.Opriessnig T., Meng X.J., Halbur P.G. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007;19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 31.Segalés J., Rosell C., Domingo M. Pathological findings associated with naturally acquired porcine circovirus type 2 associated disease. Vet. Microbiol. 2004;98:137–149. doi: 10.1016/j.vetmic.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Darwich L., Pie S., Rovira A., Segalés J., Domingo M., Oswald I.P., Mateu E. Cytokine mRNA expression profiles in lymphoid tissues of pigs naturally affected by postweaning multisystemic wasting syndrome. J. Gen. Virol. 2003;84:2117–2125. doi: 10.1099/vir.0.19124-0. [DOI] [PubMed] [Google Scholar]
- 33.Pallarés F.J., Halbur P.G., Opriessnig T., Sorden S.D., Villar D., Janke B.H., Yaeger M.J., Larsen D.J., Schwartz K.J., Yoon K.J., et al. Porcine circovirus type 2 (PCV-2) coinfections in US field cases of postweaning multisystemic wasting syndrome (PMWS) J. Vet. Diagn. Investig. 2002;14:515–519. doi: 10.1177/104063870201400614. [DOI] [PubMed] [Google Scholar]
- 34.Kim J., Chung H.K., Chae C. Associations of porcine circovirus 2 with porcine respiratory disease complex. Vet. J. 2003;166:251–256. doi: 10.1016/S1090-0233(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 35.Dorr P.M., Baker R.B., Almond G.W., Wayne S.R., Gebreyes W.A. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. J. Am. Vet. Med. Assoc. 2007;230:244–250. doi: 10.2460/javma.230.2.244. [DOI] [PubMed] [Google Scholar]
- 36.Segalés J., Calsamiglia M., Olvera A., Sibila M., Badiella L., Domingo M. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, trachea-bronchial, urinary and fecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS) Vet. Microbiol. 2005;111:223–229. doi: 10.1016/j.vetmic.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Saif L.J., Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J. Clin. Microbiol. 2020;58:e01355-20. doi: 10.1128/JCM.01355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Reeth K., Pensaert M. Prevalence of infection with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Vet. Rec. 1994;135:594–597. [PubMed] [Google Scholar]
- 39.Van Reeth K., Nauwynck H., Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: A clinical and virological study. Vet. Microbiol. 1996;48:325–335. doi: 10.1016/0378-1135(95)00145-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettenleiter T.C., Ehlers B., Müller T., Yoon K.J., Teifke J.P. Herpesviruses: Porcine cytomegalovirus. In: Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. Wiley-Blackwell; Chichester, UK: 2012. pp. 421–446. [Google Scholar]
- 41.Gottschalk M. Actinobacillosis. In: Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. Wiley-Blackwell; Chichester, UK: 2012. pp. 653–669. [Google Scholar]
- 42.Calsamiglia M., Pijoan C. Colonisation state and colostral immunity to Mycoplasma hyopneumoniae of different parity sows. Vet. Rec. 2000;146:530–532. doi: 10.1136/vr.146.18.530. [DOI] [PubMed] [Google Scholar]
- 43.Fablet C., Marois C., Kobisch M., Madec F., Rose N. Estimation of the sensitivity of four sampling methods for Mycoplasma hyopneumoniae detection in live pigs using a Bayesian approach. Vet. Microbiol. 2010;143:238–245. doi: 10.1016/j.vetmic.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Marois C., Le Carrou J., Kobisch M., Gautier-Bouchardon A.V. Isolation of Mycoplasma hyopneumoniae from different sampling sites in experimentally infected and contact SPF pigs. Vet. Microbiol. 2007;120:96–104. doi: 10.1016/j.vetmic.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Vangroenweghe F., Karriker L., Main R., Christianson E., Marsteller T., Hammen K., Bates J., Thomas P., Ellingson J., Harmon K., et al. Assessment of litter prevalence of Mycoplasma hyopneumoniae in preweaned piglets utilizing an antemorten tracheobronchial mucus collection technique and real-time polymerase chain reaction assay. J. Vet. Diagn. Investig. 2015;27:606–610. doi: 10.1177/1040638715595062. [DOI] [PubMed] [Google Scholar]
- 46.Vangroenweghe F.A.C.J., Labarque G.L., Piepers S., Strutzberg-Minder K., Maes D. Mycoplasma hyopneumoniae infections in peri-weaned and post-weaned pigs in Belgium and The Netherlands: Prevalence and associations with climatic conditions. Vet. J. 2015;205:93–97. doi: 10.1016/j.tvjl.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 47.Vangroenweghe F. Ph.D. Thesis. Applied Biological Sciences, Ghent University; Ghent, Belgium: 2018. Early Detection of Mycoplasma hyopneumoniae under Field Conditions; pp. 1–205. [Google Scholar]
- 48.Lung O., Ohene-Adjei S., Buchanan C., Joseph T., King R., Erickson A., Detmer S., Ambagala A. Multiplex PCR and microarray for detection of swine respiratory pathogens. Transbound. Emerg. Dis. 2017;64:834–848. doi: 10.1111/tbed.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraile L., Alegre A., López-Jiménez R., Nofrarías M., Segalés J. Risk factors associated with pleuritic and cranio-ventral pulmonary consolidation in slaughter-age pigs. Vet. J. 2010;184:326–333. doi: 10.1016/j.tvjl.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 50.Meyns T., Van Steelant J., Rolly E., Dewulf J., Haesebrouck F., Maes D. A cross-sectional study of risk factors associated with pulmonary lesions in pigs at slaughter. Vet. J. 2011;187:388–392. doi: 10.1016/j.tvjl.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 51.Calsamiglia M., Pijoan C., Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J. Vet. Diagn. Investig. 1999;11:246–251. doi: 10.1177/104063879901100307. [DOI] [PubMed] [Google Scholar]
- 52.Marois C., Dory D., Fablet C., Madec F., Kobish M. Development of a quantitative Real-Time TaqMan PCR assay for determination of the minimal dose of Mycoplasma hyopneumoniae strain 116 required to induce pneumonia in SPF pigs. J. Appl. Microbiol. 2010;108:1523–1533. doi: 10.1111/j.1365-2672.2009.04556.x. [DOI] [PubMed] [Google Scholar]
- 53.Vangroenweghe F., Willems E., Malásek J., Thas O., Maes D. Use of trachea-bronchial swab qPCR testing to confirm Mycoplasma hyopneumoniae seropositivity in an SPF breeding herd. Porc. Health Manag. 2018;4:12–17. doi: 10.1186/s40813-018-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vangroenweghe F., Willems E., Thas O., Maes D. Short communication: Confirmation of Mycoplasma hyopneumoniae in a breeding herd through tracheo-bronchial swab sampling and PCR. Vet. Rec. 2018;183:325. doi: 10.1136/vr.104712. [DOI] [PubMed] [Google Scholar]
- 55.Pieters M., Pijoan C., Fano E., Dee S. An assessment of the duration of Mycoplasma hyopneumoniae infection in an experimentally infected population of pigs. Vet. Microbiol. 2009;134:261–266. doi: 10.1016/j.vetmic.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 56.Giacomini E., Ferrari N., Pitozzi A., Remistani M., Giardiello D., Maes D., Loris Alborali G. Dynamics of Mycoplasma hyopneumoniae seroconversion and infection in pigs in the three main production systems. Vet. Res. Commun. 2016;40:81–88. doi: 10.1007/s11259-016-9657-6. [DOI] [PubMed] [Google Scholar]
- 57.Desrosiers R. Survival and transmission of swine influenza A virus within and between farms. J. Swine Health Prod. 2021;29:133–138. [Google Scholar]
- 58.Kothalawala H., Toussaint M.J.M., Gruys E. An overview of swine influenza. Vet. Q. 2006;28:45–53. doi: 10.1080/01652176.2006.9695207. [DOI] [PubMed] [Google Scholar]
- 59.Lin C.-N., Ke N.-J., Chiou M.-T. Cross-sectional study on the sero- and viral dynamics of porcine circovirus type 2 in the field. Vaccines. 2020;8:339. doi: 10.3390/vaccines8020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segalés J., Valero O., Espinal A., Lopez-Soria S., Nofrarias M., Calsamiglia M., Sibila M. Exploratory study on the influence of climatological parameters on Mycoplasma hyopneumoniae infection dynamics. Int. J. Biometeorol. 2012;56:1167–1171. doi: 10.1007/s00484-011-0487-5. [DOI] [PubMed] [Google Scholar]
- 61.Dee S., Otake S., Deen J. Use of a production region model to assess the efficacy of various air filtration systems for preventing airborne transmission of porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae: Results from a 2-year study. Virus Res. 2010;154:177–184. doi: 10.1016/j.virusres.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 62.Strait E.L., Madsen M., Minion F., Christopher-Hennings J., Dammen M., Jones K., Thacker E. Real-time PCR assays to address genetic diversity among strains of Mycoplasma hyopneumoniae. J. Clin. Microbiol. 2008;46:2491–2498. doi: 10.1128/JCM.02366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hothorn T., Bretz F., Westfall P. Simulaneous inference in general parametric models. Biometr. J. J. Mathematic. Meth. Biosci. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 64.Storey J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 65.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2021. [(accessed on 26 August 2021)]. Available online: https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.