Abstract
Vesicular stomatitis (VS) is a reportable viral disease which affects horses, cattle, and pigs in the Americas. Outbreaks of vesicular stomatitis virus New Jersey serotype (VSV-NJ) in the United States typically occur on a 5–10-year cycle, usually affecting western and southwestern states. In 2019–2020, an outbreak of VSV Indiana serotype (VSV-IN) extended eastward into the states of Kansas and Missouri for the first time in several decades, leading to 101 confirmed premises in Kansas and 37 confirmed premises in Missouri. In order to investigate which vector species contributed to the outbreak in Kansas, we conducted insect surveillance at two farms that experienced confirmed VSV-positive cases, one each in Riley County and Franklin County. Centers for Disease Control and Prevention miniature light traps were used to collect biting flies on the premises. Two genera of known VSV vectors, Culicoides biting midges and Simulium black flies, were identified to species, pooled by species, sex, reproductive status, and collection site, and tested for the presence of VSV-IN RNA by RT-qPCR. In total, eight positive pools were detected from Culicoides sonorensis (1), Culicoides stellifer (3), Culicoides variipennis (1), and Simulium meridionale (3). The C. sonorensis- and C. variipennis-positive pools were from nulliparous individuals, possibly indicating transovarial or venereal transmission as the source of virus. This is the first report of VSV-IN in field caught C. stellifer and the first report of either serotype in S. meridionale near outbreak premises. These results improve our understanding of the role midges and black flies play in VSV epidemiology in the United States and broadens the scope of vector species for targeted surveillance and control.
Keywords: vesicular stomatitis virus, Indiana, VSV-IN, Rhabdoviridae, Vesiculovirus, Culicoides, Simulium
1. Introduction
Vesicular stomatitis virus (VSV; family Rhabdoviridae, genus Vesiculovirus) is the causative agent of vesicular stomatitis (VS) disease in horses, cattle, and pigs in the Americas. Two serotypes have been described, VSV New Jersey (VSV-NJ) and Indiana (VSV-IN). In the United States (U.S.), the disease primarily affects horses and cattle, producing lesions and ulceration of the mouth, teats, and coronary bands of affected animals. While infection does not typically result in mortality in any of the species, clinical disease in cattle and swine closely resembles foot and mouth disease. Therefore, VSV is a reportable disease that results in quarantines and animal movement restrictions on premises with positive animals [1,2]. Due to these restrictions and effects of the virus itself, outbreaks of VSV on dairy and beef operations have resulted in significant economic impacts. These include estimated average financial losses of USD 15,565/beef cattle ranch in Colorado [3], approximately USD 50,000 in losses to a single dairy farm in Idaho [4], from USD 200 to USD 250/dairy cow affected with oral lesions in California [5] and Colorado [6], and up to USD 570/dairy cow for those with teat lesions [6].
VSV is maintained in an endemic transmission cycle in South and Central America. Sporadic outbreaks with the VSV-NJ serotype in the U.S. occur on a 5–10-year cycle, often with viral incursions into the southwestern states of Texas, Arizona, and New Mexico, and spreading northward into Colorado and Wyoming [7]. Outbreaks with the VSV-IN serotype in the U.S. are much less frequent with prior outbreaks only reported in 1966 and 1997–1998 [7,8]. Transmission can occur animal to animal by direct contact, through fomites contaminated with virus from shedding animals, and through mechanical and biological vector transmission [9]. The suspected biological vector range for this pathogen is extensive, encompassing black flies (Simuliidae), biting midges (Ceratopogonidae), mosquitoes (Culicidae), and sand flies (Psychodidae subfamily Phlebotiminae) [9]. Information on specific vector species and regional variation in vector communities is sparse. To date, confirmed competent North American vectors are from three genera of these biting Diptera: Lutzomyia shannoni [10], Culicoides sonorensis [11,12], Simulium vittatum, and S. notatum [13].
To estimate vector-borne VSV transmission risk to animals in any given region, it is critical to identify all competent vector species, and determine which species are present. These incriminated Dipteran vectors differ substantially in their individual ecologies, especially as it pertains to larval habitat requirements, which vary from fully aquatic to fully terrestrial. Black fly larvae are fully aquatic and require fast-flowing fresh water to complete their development, making them common in rivers and streams proximal to bloodmeal sources [14]. Biting midge larvae inhabit semi-aquatic habitats, typically at the water–soil interface of waterbodies such as ponds, streams, puddles, and springs [15]. Sand fly larvae require moist, organic terrestrial soil conditions without standing water. Therefore, the primary vector taxa present in an area, and the relative risk of VSV transmission, is often a byproduct of larval habitat availability. Adult dispersal, which also differs significantly between these taxa, can also assist with the movement of vectors and associated pathogens. Sand flies have characteristically weak flight ability and short dispersal of just a few hundred meters [16]. Biting midges are slightly stronger fliers with average dispersal distances of 1–2.5 km [17]. Finally, black flies are the strongest fliers of these three, with some species dispersing more than 100 km [18] and single recorded continuous flight distances of up to 5.25 km [19]. Due to their presence in running water, black fly larvae can also disperse both passively and actively by water flow [20] and both biting midges and black flies are suspected to move on air currents over long distances [21,22,23,24,25,26], potentially contributing to the movement of VSV over large geographic areas.
The latest U.S. VSV-IN outbreak started in 2019 affecting eight states (Colorado, Kansas, Nebraska, New Mexico, Oklahoma, Texas, Utah, and Wyoming) with a total of 1144 affected premises [27]. In 2020, a second year of the outbreak affected eight states (Arizona, Arkansas, Kansas, Missouri, Nebraska, New Mexico, Oklahoma, and Texas) with a total of 326 affected premises [28]. The 2020 outbreak expanded further eastward than in typical years, reaching farms as far east as Kansas and Missouri for the first time in decades. In total, 26 counties in Kansas reported VSV-positive premises, mainly within the southeastern portion of the state [29]. This expansion into an area representing unique vector communities provided an opportunity to further investigate the potential vector breadth of VSV-IN outside the southwest. We report on virus detection efforts conducted on field-caught Culicoides biting midges and Simulium black flies at two VSV-positive premises in eastern Kansas in 2020.
2. Results
2.1. Field Collections
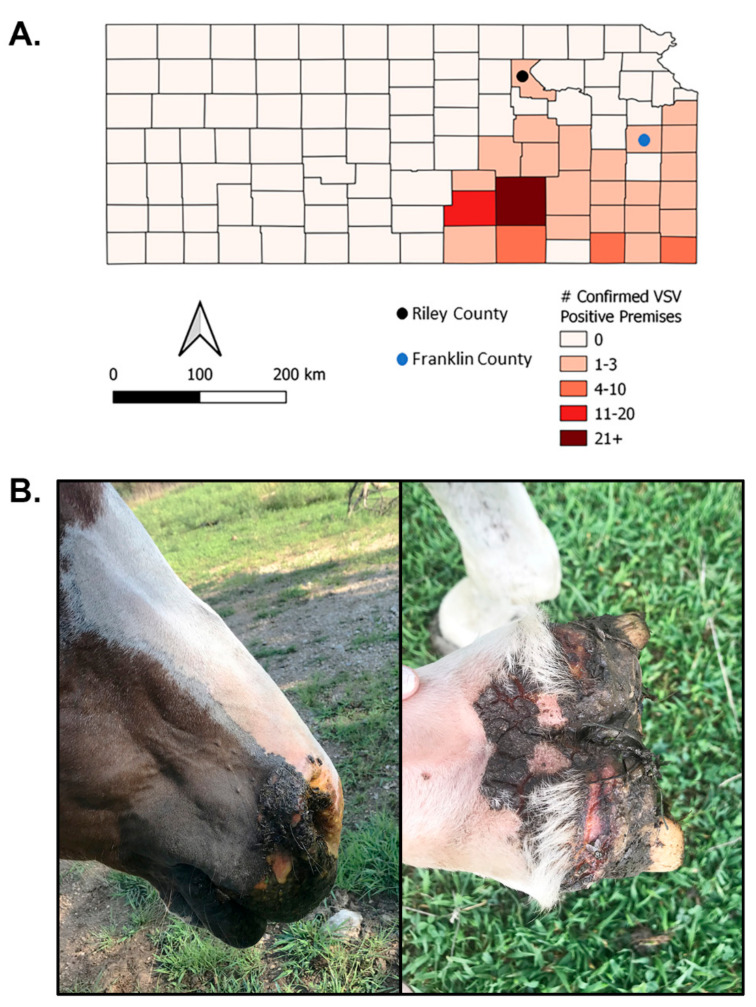
Collections were conducted on two farms in Kansas: one in Riley County and one in Franklin County (Figure 1). In total, 1396 biting insects were collected and sorted into 154 pools for VSV testing including 40 Simulium and 1356 Culicoides individuals. All Simulium were collected from the Franklin County site and identified as female S. meridionale. Eight species of Culicoides were identified and pooled separately by sex, reproductive status, and collection site (Table 1 and Table 2). Of the Culicoides collected, 469 were from Franklin County and 927 were from Riley County.
Figure 1.
Insect collections on vesicular stomatitis virus-Indiana (VSV-IN)-positive premises in Kansas, USA. (A) Map depicting the number of USDA-APHIS confirmed VSV-IN cases per county. Insects were collected in Riley and Franklin counties (solid circles). (B) Clinically affected horse on Franklin County premises. Map generated using QGis with publicly available shapefiles and data. (Photographs used with permission from owner, 2021).
Table 1.
Culicoides and Simulium individuals collected at the Franklin and Riley County sites.
| Species | Franklin N |
Franklin Pools Total (+) |
Riley N |
Riley Pools Total (+) |
|---|---|---|---|---|
| Culicoides crepuscularis | 16 | 6 (0) | 160 | 16 (0) |
| Culicoides guttipennis | 1 | 1 (0) | 0 | 0 (0) |
| Culicoides haematopotus | 289 | 29 (0) | 86 | 14 (0) |
| Culicoides sonorensis | 1 | 1 (0) | 148 | 14 (1) |
| Culicoides stellifer | 120 | 15 (0) | 266 | 19 (3) |
| Culicoides variipennis | 0 | 0 (0) | 266 | 27 (1) |
| Culicoides venustus | 1 | 1 (0) | 1 | 1 (0) |
| Culicoides villosipennis | 1 | 1 (0) | 0 | 0 (0) |
| Simulium meridionale | 40 | 9 (3) | 0 | 0 (0) |
| Total | 469 | 63 (3) | 927 | 91 (5) |
Total number (N) is shown as well as the number of pools for each species and the total positive pools in parenthesis.
Table 2.
Sex and parity status of Culicoides biting midge pools tested for VSV-IN by RT-qPCR. Parity status was not determined for nine pools of female Simulium meridionale.
| Female | ||||
|---|---|---|---|---|
| Culicoides Species | Nulliparous | Parous/Gravid | Blood Fed | Male |
| C. crepuscularis | 6 (0) | 13 (0) | 1 (0) | 2 (0) |
| C. guttipennis | 0 (0) | 0 (0) | 0 (0) | 1 (0) |
| C. haematopotus | 8 (0) | 21 (0) | 8 (0) | 6 (0) |
| C. sonorensis | 2 (1) | 5 (0) | 3 (0) | 5 (0) |
| C. stellifer | 5 (0) | 24 (3) | 2 (0) | 3 (0) |
| C. variipennis | 4 (1) | 11 (0) | 3 (0) | 9 (0) |
| C. venustus | 1 (0) | 1 (0) | 0 (0) | 0 (0) |
| C. villosipennis | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
Positive pools indicated in parentheses.
2.2. VSV Detection by RT-qPCR
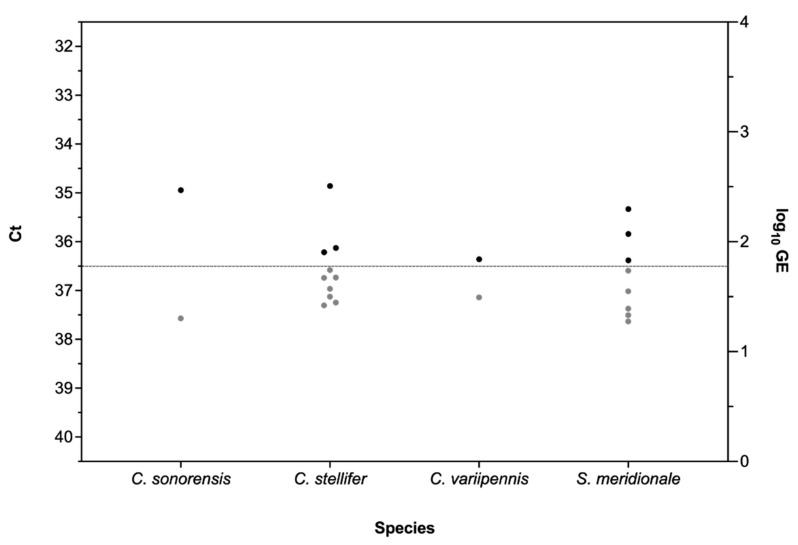
Of the 154 pools tested, RT-qPCR-positive results (Ct ≤ 36.5) were identified for eight pools (Figure 2), including three from Franklin County and five from Riley County. The positive Franklin County pools were all female Simulium meridionale (Ct = 35.84, 36.38, 35.33). The positive Riley County pools included three pools of gravid C. stellifer (Ct = 35.84, 36.38, 35.33), one pool of nulliparous C. sonorensis (Ct = 34.94), and one pool of nulliparous C. variipennis (Ct = 36.36).
Figure 2.
Detection of VSV-IN in field-collected insects from two counties in Kansas during the 2020 outbreak. Positive insect species (black dots) above RT-qPCR cycle threshold (Ct) cutoff point (≤36.5). Ct values (left Y-axis) and calculated log10 viral genome equivalents (right Y axis).
2.3. Sequencing
Partial sequencing of the VSV hypervariable phosphoprotein (P) gene was successfully conducted on one pool of nulliparous C. sonorensis, producing a 337 nucleotide (nt) sequence (GenBank accession: MZ703188). As of July 12, 2021, only seven VSV-IN 2019–2020 outbreak sequences were publicly available (BLASTN 2.12.0; [30]). The C. sonorensis sequence most closely matched four near-complete genomes from the 2020 outbreak in Kansas (GenBank accessions: MW373776 to MW373779) and three genome sequences from the 2019 outbreak in the southwestern U.S. (GenBank accessions: MT437283 to MT437285) (92.6% identity, 100% query cover, 4e-140 E-value for all matches) (Supplemental Table S1). Additionally, 140 nt sequences were obtained for pooled samples of the Riley County C. stellifer (GenBank accession: MZ703186) and the Franklin County S. meridionale (GenBank accession: MZ703187). These sequences had 100% identity to each other and 95.68% identity to the available 2019/2020 sequences (99% query cover, 1e-61 E-value) (Supplemental Table S1). Unfortunately, whole genome sequencing attempts from positive pools were unsuccessful.
2.4. Field Infection Rates
Two field infection rate metrics were calculated to provide a measure of the comparative epidemiological contribution of the implicated vector species in this study. The metrics calculated include the minimum infection rate (MIR), a conservative estimate, and the maximum likelihood estimate (MLE), a more robust estimate of field infection rates. Both MIR and MLE were calculated for positive biting midge species at the Riley County site and expressed as total positive insects per 1000 in the population. The MIR for C. stellifer was 11.24 with an MLE of 12.25 (95% CI: 2.61–46.39). For C. sonorensis, the calculated MIR was 6.76 and MLE was 6.81 (95% CI: 0.40–33.61). The MIR for C. variipennis was 3.76 with an MLE of 3.66 (95% CI: 0.22–17.47). At the Franklin County site, the MIR for S. meridionale was 75. MLE could not be calculated for S. meridionale since the few pools tested did not allow the generation of robust estimates.
3. Discussion
This is the first report with data documenting putative vector species for VSV transmission during an active outbreak in eastern Kansas. While C. sonorensis has been confirmed as a competent vector [11,12], our finding of positive nulliparous C. sonorensis is novel. Nulliparous midges have never taken a blood meal, indicating that this positive pool is the result of an alternative infection pathway such as venereal or transovarial transmission. Venereal transmission has been documented in experimentally infected C. sonorensis [31] and the presence of VSV in developing oocytes and the ovarial epithelium of experimentally infected females suggest that transovarial transmission may also be possible [12]. Although few sequences are currently available in public databases for the 2019–2020 VSV-IN outbreak, analysis of sequencing data for this pool showed it was divergent from, but most closely associated with, 2019 bovine and 2020 horse isolates. Several factors may have contributed to the nulliparous pool having lower percent identity with the available mammalian isolate sequences than did parous pools that would have fed on infected animals. VSV is a non-segmented negative-stranded RNA virus with extremely wide host tropism and a relatively constant mutation rate of approximately 6 Log10 substitutions per nucleotide per round of replication in mammalian cells [32]. An insect-to-insect transmitted virus, with the genetic bottlenecks and selective pressures inherent to transovarial or venereal transmission, may be expected to show greater divergence from mammalian-derived (blood meal) virus populations. The paucity of publicly available 2019–2020 isolate sequences and our inability to obtain sequence from more than a single fragment of the hypervariable region of the P gene limited the robustness of our comparative analysis. However, the VSV RT-qPCR and confirmatory sequence results support our hypothesis of vertical or venereal VSV transmission occurring in the field-collected nulliparous C. sonorensis pool.
In addition to the C. sonorensis nulliparous pool, one pool of nulliparous C. variipennis was also RT-qPCR-positive. C. variipennis is closely related to C. sonorensis, having only been recognized as distinct species after a reassessment in 2000 [33]. Despite their close phylogenetic association, it is generally accepted that C. sonorensis is the vastly more competent vector species for most Culicoides-borne viruses [34,35] and the majority of vector competence studies focus on C. sonorensis due to availability of laboratory colonies. Our results indicate that at least some degree of vector competence for VSV is possible for C. variipennis, although infection rates for this species were low (3.66 MLE for C. variipennis compared to 6.81 for C. sonorensis) suggesting a low likelihood that this species is contributing substantially to transmission. Despite this, our finding of VSV-positive nulliparous C. variipennis emphasizes the need for additional vector competence studies involving Culicoides species other than C. sonorensis.
Culicoides stellifer has never formally been implicated as a vector for VSV, although VSV-NJ was isolated from a single pool of this species during an outbreak in Colorado [36]. This species is also a suspected vector for the orbiviruses bluetongue virus and epizootic hemorrhagic disease virus [37,38]. Moreover, C. stellifer has a broad geographic distribution throughout the U.S. [39], is often found in areas inhabited by grazing herbivores, and has documented blood-feeding events on horses [40,41,42,43]. In some parts of the U.S., C. stellifer often dominates collections of biting midges [44,45], leading to further speculation that even at low levels of competence for arboviruses, higher abundance could lead to an increase in overall vectorial capacity for this species [38,46]. Calculated infection rates for C. stellifer were high compared with the other positive Culicoides species in this study at 11.24 MIR and 12.25 MLE, suggesting that this species may play a significant role in VSV-IN transmission. However, because insects were collected in ethanol, virus isolations to confirm potential transmission were not possible.
Two black fly species, S. vittatum and S. notatum, have been implicated as competent vectors of VSV-IN in the U.S. [13]. For VSV-NJ, only S. notatum has been found to be competent in laboratory assays [47], while S. bivittatum has been implicated due to virus detections in field-collected specimens [48]. While S. vittatum is known to occur within the sampled region of Kansas, no individuals of this species were collected during this effort at either of the outbreak premises sampled. Simulium meridionale has not previously been implicated as a vector of either VSV serotype and historically was believed to feed primarily on birds [49]. This species is known to transmit the turkey parasite Leucocytozoon smithi [50], and large swarms have resulted in avian simuliotoxicosis [51]. However, additional evidence has shown that this species will feed on mammals, including humans and cattle [52,53]. This black fly species was the third most common biting insect sampled on the Franklin County farm at which the primary available hosts were horses. An exceptionally high MIR value (75) was calculated for this species, which could indicate a substantial potential for VSV-IN transmission. However, comparably low numbers of individuals of this species were available for testing, so this infection rate may be artificially high. Further studies to elucidate the vector competence of this Simulium species are warranted to better understand its potential role in the transmission of VSV-IN in the U.S. Because neither C. stellifer nor S. meridionale have been previously implicated as vectors, sequencing was attempted to confirm the positive RT-qPCR result. A hypervariable P gene sequence was obtained; however, the minimal length (140 nt) made sequence analysis possible but less robust than desired. In addition to alignments and comparisons with all available sequences, we sequenced our positive control strain to rule out potential plate contamination. Sequences from the two species pools, from two different counties over 100 miles apart, were 100% identical to each other and divergent from the plate control.
The vectors collected on these two farms reflect the availability of larval habitats for biting midges and black flies. The Franklin County farm has a flowing stream moving through the center of the property that is conducive to the development of black fly larvae. In addition to the stream, numerous low-lying areas and a pond were present on this property, creating conditions favorable for the breeding of several biting midge species. While black flies have been collected around Riley County [54], the lack of adequate flowing water resources near the VSV-positive premises and proliferation of low lying, semi-aquatic habitats led to great abundance of biting midges and the absence of black flies in our collections from this site. Black flies are capable fliers that can disperse over great distances. Thus, while the lack of great abundance of black flies was expected, the lack of any individuals in our collections was surprising. It is likely that the availability of prime larval habitat and hosts closer to their emergence sites within the county likely precluded black flies from dispersing to the VSV-positive premises. This result highlights the importance of considering the ecology of vector species when assessing which VSV vectors to target and predicting vector-borne disease transmission risk.
For both C. stellifer and S. meridionale, these first reports of VSV-IN positives fulfill two of the four criteria necessary for the implication of vector species [55]. Virus was detected in field-collected specimens of both species that lacked any visible blood during an active VSV-IN outbreak, which fulfills the first criterium. Our collections also fulfilled the fourth criterium by showing an association of the infected arthropods with affected vertebrate populations, although additional studies on host associations, especially for S. meridionale, would strengthen this criterium. The remaining two criteria require completion of laboratory vector competence studies showing the ability for these species to become infected and then biologically transmit the virus. Completion of these laboratory assays with these two species is greatly needed considering the high calculated infection rates for both during this study. At this time, these species can be categorized as suspected, although not confirmed, vectors of VSV-IN until the completion of all criteria.
No positive samples were recovered from other midge species collected, including C. haematopotus and C. crepuscularis, both species that have been implicated as suspected vectors of other arboviruses [56]. Despite testing 65 pools from these two species, the lack of positives gives us confidence that these species were not likely contributing to the transmission of VSV-IN on the study premises. However, it is also possible that conducting collections solely with blacklight LED traps could have biased against the collection of some species that are attracted to other light wavelengths [57,58], or even against infected vectors, which has been proposed for C. sonorensis infected with BTV [59].
In this study, insect collections were performed within two weeks of VSV lesion onset dates. No additional cases were reported for the locations and no additional collections were made. Therefore, it is unclear how long infected insects persisted on these premises and whether persistent infection in midge populations could have the potential for overwintering. A study evaluating the phylogenetics of isolates associated with VSV-NJ outbreaks in the U.S. in 2005 and 2006 found that the 2006 outbreak was the result of overwintering rather than a novel introduction event [60] with S. bivittatum involved at the start of the outbreak [48]. An overwintering event is especially concerning considering the potential transovarial or venereal transmission suspected due to VSV-IN detection in nulliparous C. sonorensis and C. variipennis. Based on laboratory studies, both transovarial or venereal transmission are speculated maintenance mechanisms for VSV in Phlebotomine sandflies [61] and C. sonorensis [31]. As of August 24, 2021, no VSV-IN cases have been documented within the U.S. in 2021. Although Culicoides are well-adapted to survive cold temperatures, a particularly hard freeze from a polar vortex in February 2021 may have impacted the survival of overwintering biting midges. It is unclear how venereally or vertically transmitted viral infections could be affected by this particularly extreme weather event.
In conclusion, we have identified four species of biting Diptera that were positive for VSV-IN during the 2020 expansion into Kansas: C. sonorensis, C. stellifer, C. variipennis, and S. meridionale. Calculated infection rates for these species were variable, with the greatest rates calculated in S. meridionale and C. stellifer and the lowest rates in C. variipennis. Each of these species represents unique biology and ecology that must be considered when optimizing control and surveillance strategies moving forward. This is also the first field detection of VSV-IN-positive nulliparous individuals indicating transovarial or venereal transmission outside of a laboratory environment. These data contribute substantially to our knowledge of the epidemiology of VSV-IN in the U.S. and will improve our ability to conduct targeted surveillance and control of vector species in the event of continued outbreaks.
4. Materials and Methods
4.1. Field Collections
Insects were collected during the 2020 outbreak at two sites in Kansas where confirmed VSV-IN-positive horses were present. The premises in Riley County reported one equine case (of 14 horses on site) and the Franklin County premises had three equine cases (of 7 horses on site) (Figure 1). At both sites, Centers for Disease Control and Prevention (CDC) miniature light traps baited with black light LED arrays (BioQuip Inc., Rancho Dominguez, CA, USA) were used to collect insects. When available, CO2 (dry ice) was used to improve trap efficacy. Collections took place overnight on July 30 and August 5–6 on the Franklin County site (lesion onset date was July 23) [62,63] using five traps per night and July 31 on the Riley County site (estimated lesion onset date July 20) [63,64] using seven traps. Insects were collected directly into 95% ethanol. Traps were recovered the following morning and collection tubes were immediately returned to the lab and placed into a freezer at −80 °C prior to processing.
4.2. Species Identification and Pooling
All insect sorting and identification was conducted on a chill table (BioQuip Inc.). Midges were identified to species and sex using morphological keys [15,33] and further identified as to physiological status (parous, nulliparous, gravid) using abdominal cuticular pigmentation [65,66]. Black flies were also identified to species and sex [14]. Insects were pooled in 2 mL microcentrifuge tubes containing 500 µL TRIzol reagent according to species, physiological status (for Culicoides only), site, and date. Culicoides were pooled in groups of ≤20 individuals and Simulium in pools of ≤5 individuals. Nulliparous, blood fed, and male midges were all pooled separately. Gravid and parous midges were combined in some pools.
4.3. RNA Extraction and RT-qPCR for VSV-IN Detection
Frozen pools of midges and black flies stored in 500 μL of TRIzol were thawed on ice, two 2.4 mm stainless steel beads (Omni Inc., Kennesaw, GA, USA) were added, and tubes were homogenized by shaking at 3.1 m/s with a Bead Mill Homogenizer (Omni Inc.). Additional 500 μL of TRIzol were added for a final volume of 1000 μL. Samples were centrifuged at 12,000 × g for 6 min to pellet debris. Total RNA was extracted using Trizol-BCP (1-Bromo-3-chloropropane; ThermoFisher Life Technologies, Inc. Waltham, MA, USA). RNA was precipitated using isopropanol, washed in 75% ethanol, and eluted in 40 μL of nuclease-free water. RNA extracts were analyzed using TaqMan Fast Virus 1-Step MasterMix (Applied Biosystems; Thermo Fisher Scientific, Inc.) in a RT-qPCR targeting the L segment [67]: forward primer VSVIN+7230: 5′-TGATACAGTACAATTATTTTTGGGAC-3; reverse primer VSVIN-7456: 5′-GAGACTTTCTGTTACGGGATCTGG-3′; probe: 5′-FAM- ATGATGCATGATCCAGC BHQ-1 plus-3′. For amplification, the following temperature profile was used: Reverse-transcription 1 cycle at 50 °C for 5 min, denaturing and polymerase activation at 95 °C for 20 s, and amplification: 45 cycles of 95 °C for 15 s and 60 °C for 60 s. VSV-IN VR-158 (ATCC, Manassas, VA, USA) was used as an internal control for each PCR reaction and plate control for inter-run variation. Water was used for non-template negative controls. Samples were tested in triplicate and average Ct values used for analysis. Based on positive and negative controls, samples with a minimum of two RT-qPCR reactions with Ct ≤ 36.5 were considered positive for VSV-IN RNA. This cutoff value was based on our previous work with Culicoides midges and VSV-NJ [31,68]. Standard curves and calculation of Ct values were carried out with the 7500 Fast Dx software (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The linear regression (y = −3.3239x + 18.815) was used to determine the amount of viral genomic ssRNA per midge. Genome equivalents were calculated with the published VSV genome molecular weight [69] and the NEBioCalulator (https://nebiocalculator.neb.com/#!/ssrnaamt, access date: 13 March 2021).
4.4. Sequencing and Sequence Analysis
The remaining RNA from positive samples was pooled by species and submitted to the Kansas State Veterinary Diagnostic Laboratory for Illumina next generation deep sequencing of the hypervariable region of the phosphoprotein (P) gene. Full-length sequences obtained were compared to each other and to published sequences in GenBank using BLASTN 2.12.0 [30]. This included the seven sequences currently available from the 2019–2020 VSV-IN outbreak (Table S1).
4.5. Infection Rate Calculations
Two field infection rate metrics were calculated at each site including the MIR, a conservative estimate, and the MLE, a more robust estimate of field infection rates. MIR was calculated by dividing the total number of positive pools by the total number of insects sampled and multiplying by 1000 such that the resulting number is the number of infected insects on the landscape out of 1000 individuals. The MLE was calculated using the Excel PooledInfRate program (available through the CDC) adapted for variable pool sizes, which uses a binomial distribution and a probabilistic model to find a more likely and less conservative estimate of field infection rates [70,71].
Acknowledgments
We thank the owners and managers of the VSV-positive properties from which insects were collected for this study. We thank Dustin Swanson for assistance with identification of the Simuliidae specimens, Lindsey Reister-Hendricks for assistance in the field, and Ashton Harlan for laboratory technical assistance. We thank Rachel Palinski at the Kansas State University Veterinary Diagnostic Laboratory for sequencing our samples. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The conclusions in this report are those of the authors and do not necessarily represent the views of the USDA. USDA is an equal opportunity provider and employer.
Supplementary Materials
The following is available online at https://www.mdpi.com/article/10.3390/pathogens10091126/s1, Table S1: Vesicular stomatitis virus Indiana sequences downloaded from GenBank.
Author Contributions
Conceptualization, B.L.M. and B.S.D.; data curation, B.L.M., P.R.-L. and T.M.D.; formal analysis, B.L.M. and P.R.-L.; funding acquisition, B.L.M. and B.S.D.; investigation, B.L.M., P.R.-L. and T.M.D.; methodology, B.L.M., P.R.-L., T.M.D. and B.S.D.; project administration, B.L.M. and B.S.D.; resources, B.L.M. and B.S.D.; software, B.L.M. and P.R.-L.; supervision, B.S.D. and B.L.M.; validation, B.L.M. and P.R.-L.; visualization, B.L.M., P.R.-L., B.S.D.; writing—original draft preparation, B.L.M., P.R.-L. and B.S.D.; writing—review and editing, B.L.M., P.R.-L., T.M.D. and B.S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the U.S. Department of Agriculture, Agricultural Research Service, NP-103 Animal Health National Program Project #3020-32000-013-00D and NP-104 Medical, Veterinary, and Urban Entomology National Program Project #3020-32000-018-00D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding authors and through the USDA, Agricultural Research Information System.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kitching R.P. OIE List A disease as a constraint to international trade. Ann. N. Y. Acad. Sci. 2000;916:50–54. doi: 10.1111/j.1749-6632.2000.tb05273.x. [DOI] [PubMed] [Google Scholar]
- 2.Peck D.E., Reeves W.K., Pelzel-McCluskey A.M., Derner J.D., Drolet B., Cohnstaedt L.W., Swanson D., McVey D.S., Rodriguez L.L., Peters D.P.C. Management strategies for reducing the risk of equines contracting vesicular stomatitis virus (VSV) in the western United States. J. Equine Vet. Sci. 2020;90:103026. doi: 10.1016/j.jevs.2020.103026. [DOI] [PubMed] [Google Scholar]
- 3.Hayek A.M., McCluskey B.J., Chavez G.T., Salman M.D. Financial impact of the 1995 outbreak of vesicular stomatitis on 16 beef ranches in Colorado. J. Am. Vet. Med. Assoc. 1998;212:820–823. [PubMed] [Google Scholar]
- 4.Leder R.R., Maas J., Lane V.M., Evermann J.F. Epidemiologic investigation of vesicular stomatitis virus in a dairy and its economic impact. Bov. Practitioner. 1983;18:45–49. [Google Scholar]
- 5.Goodger W.J., Thurmond M., Nehay J., Mitchell J., Smith P. Economic impact of an epizootic of bovine vesicular stomatitis in California. J. Am. Vet. Med. Assoc. 1985;186:370–373. [PubMed] [Google Scholar]
- 6.Alderink F.J. Vesicular stomatitis epidemic in Colorado: Clinical observations and financial losses reported by dairymen. Prev. Vet. Med. 1984;3:29–44. doi: 10.1016/0167-5877(84)90022-9. [DOI] [Google Scholar]
- 7.Rodriguez L.L., Bunch T.A., Fraire M., Llewellyn Z.N. Re-emergence of vesicular stomatitis in the western United States is associated with distinct viral genetic lineages. Virology. 2000;271:171–181. doi: 10.1006/viro.2000.0289. [DOI] [PubMed] [Google Scholar]
- 8.McCluskey B.J., Hurd H.S., Mumford E.L. Review of the 1997 outbreak of vesicular stomatitis in the western United States. J. Am. Vet. Med. Assoc. 1999;215:1259–1262. [PubMed] [Google Scholar]
- 9.Rozo-Lopez P., Drolet B.S., Londono-Renteria B. Vesicular stomatitis virus transmission: A comparison of incriminated vectors. Insects. 2018;9:190. doi: 10.3390/insects9040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer J.A., Tesh R.B., Modi G.B., Corn J.L., Nettles V.F. Vesicular stomatitis virus, New Jersey serotype: Replication in and transmission by Lutzomyia shannoni (Diptera: Psychodidae) Am. J. Trop. Med. Hyg. 1990;42:483–490. doi: 10.4269/ajtmh.1990.42.483. [DOI] [PubMed] [Google Scholar]
- 11.Nunamaker R.A., Perez de Leon A.A., Campbell C.C., Lonning S.M. Oral infection of Culicoides sonorensis (Diptera: Ceratopogonidae) by vesicular stomatitis virus. J. Med. Entomol. 2000;37:784–786. doi: 10.1603/0022-2585-37.5.784. [DOI] [PubMed] [Google Scholar]
- 12.Drolet B.S., Campbell C.L., Stuart M.A., Wilson W.C. Vector competence of Culicoides sonorensis (Diptera: Ceratopogonidae) for vesicular stomatitis virus. J. Med. Entomol. 2005;42:409–418. doi: 10.1093/jmedent/42.3.409. [DOI] [PubMed] [Google Scholar]
- 13.Mead D.G., Ramberg F.B., Mare C.J. Laboratory vector competence of black flies (Diptera: Simuliidae) for the Indiana serotype of vesicular stomatitis virus. Ann. N. Y. Acad. Sci. 2000;916:437–443. doi: 10.1111/j.1749-6632.2000.tb05323.x. [DOI] [PubMed] [Google Scholar]
- 14.Adler P.H., Currie D.C., Woods D.M., Idema R.M., Zettler L.W. The Black Flies (Simuliidae) of North America. Cornell University Press; Ithaca, NY, USA: 2004. [Google Scholar]
- 15.Blanton F.S., Wirth W.W. The sand flies (Culicoides) of Florida (Diptera: Ceratopogonidae) Division of Plant Industry, Florida Department of Agriculture and Consumer Services; Gainesville, FL, USA: 1979. pp. 1–205. [Google Scholar]
- 16.Alexander J.B. Dispersal of phlebotomine sand flies (Diptera: Psychodidae) in a Colombian coffee plantation. J. Med. Entomol. 1987;24:552–558. doi: 10.1093/jmedent/24.5.552. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman R.H., Turner E.C., Jr. Dispersal and gonotrophic age of Culicoides variipennis (Diptera: Ceratopogonidae) at an isolated site in southwestern Virginia, USA. J. Med. Entomol. 1984;21:527–535. doi: 10.1093/jmedent/21.5.527. [DOI] [PubMed] [Google Scholar]
- 18.Noamesi G.K. Dry season survival and associated longevity and flight range of Simulium damnosum Theobald in Northern Ghana. Ghana Med. J. 1966;5:95–102. [Google Scholar]
- 19.Cooter R.J. Studies on the flight of black-flies (Diptera: Simuliidae). II. Flight performance of three cytospecies in the complex of Simulium damnosum Theobald. Bull. Entomol. Res. 1983;73:275–288. doi: 10.1017/S0007485300008865. [DOI] [Google Scholar]
- 20.Fonseca D.M., Hart D.D. Density-dependent dispersal of black fly neonates is mediated by flow. Oikos. 1996;75:49–58. doi: 10.2307/3546320. [DOI] [Google Scholar]
- 21.Service M.W. Effects of wind on the behavior and distribution of mosquitoes and blackflies. Int. J. Biometeorol. 1980;24:347–353. doi: 10.1007/BF02250577. [DOI] [Google Scholar]
- 22.Ducheyne E., De Deken R., Becu S., Codina B., Nomikou K., Mangana-Vougiaki O., Georgiev G., Purse B.V., Hendickx G. Quantifying the wind dispersal of Culicoides species in Greece and Bulgaria. Geospat Health. 2007;1:177–189. doi: 10.4081/gh.2007.266. [DOI] [PubMed] [Google Scholar]
- 23.Braverman Y., Chechik F. Air streams and the introduction of animal diseases borne on Culicoides (Diptera, Ceratopogonidae) into Israel. Rev. Sci. Tech. 1996;15:1037–1052. doi: 10.20506/rst.15.3.968. [DOI] [PubMed] [Google Scholar]
- 24.Burgin L.E., Gloster J., Sanders C., Mellor P.S., Gubbins S., Carpenter S. Investigating incursions of bluetongue virus using a model of long-distance Culicoides biting midge dispersal. Transbound. Emerg. Dis. 2013;60:263–272. doi: 10.1111/j.1865-1682.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 25.Sedda L., Brown H.E., Purse B.V., Burgin L., Gloster J., Rogers D.J. A new algorithm quantifies the roles of wind and midge flight activity in the bluetongue epizootic in northwest Europe. Proc. Biol. Sci. 2012;279:2354–2362. doi: 10.1098/rspb.2011.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellers R.F., Maarouf A.R. Trajectory analysis of winds and vesicular stomatitis in North America, 1982-5. Epidemiol. Infect. 1990;104:313–328. doi: 10.1017/S0950268800059495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.USDA-APHIS . 2019 Vesicular Stomatitis Virus (VSV) Situation Report—January 6, 2020. USDA-APHIS; Riverdale Park, MD, USA: 2020. pp. 1–10. [Google Scholar]
- 28.USDA-APHIS . 2020 Vesicular Stomatitis Virus (VSV) Situation Report—November 13, 2020. USDA-APHIS; Riverdale Park, MD, USA: 2020. pp. 1–8. [Google Scholar]
- 29.USDA-APHIS . 2020 Vesicular Stomatitis Virus (VSV) Situation Report—August 6, 2020. USDA-APHIS; Riverdale Park, MD, USA: 2020. pp. 1–8. [Google Scholar]
- 30.Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schaffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozo-Lopez P., Londono-Renteria B., Drolet B.S. Venereal transmission of vesicular stomatitis virus by Culicoides sonorensis midges. Pathogens. 2020;9:316. doi: 10.3390/pathogens9040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combe M., Sanjuan R. Variation in RNA virus mutation rates across host cells. PLoS Pathog. 2014;10:e1003855. doi: 10.1371/journal.ppat.1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holbrook F.R., Tabachnick W.J., Schmidtmann E.T., McKinnon C.N., Bobian R.J., Grogan W.L. Sympatry in the Culicoides variipennis complex (Diptera: Ceratopogonidae): A taxonomic reassessment. J. Med. Entomol. 2000;37:65–76. doi: 10.1603/0022-2585-37.1.65. [DOI] [PubMed] [Google Scholar]
- 34.Tabachnick W.J. Culicoides and the global epidemiology of bluetongue virus infection. Veterinaria Italiana. 2004;40:145–150. [PubMed] [Google Scholar]
- 35.Becker M.E., Roberts J., Schroeder M.E., Gentry G., Foil L.D. Prospective Study of Epizootic Hemorrhagic Disease Virus and Bluetongue Virus Transmission in Captive Ruminants. J. Med. Entomol. 2020;57:1277–1285. doi: 10.1093/jme/tjaa027. [DOI] [PubMed] [Google Scholar]
- 36.Kramer W.L., Jones R.H., Holbrook F.R., Walton T.E., Calisher C.H. Isolation of arboviruses from Culicoides midges (Diptera: Ceratopogonidae) in Colorado during an epizootic of vesicular stomatitis New Jersey. J. Med. Entomol. 1990;27:487–493. doi: 10.1093/jmedent/27.4.487. [DOI] [PubMed] [Google Scholar]
- 37.Mullen G.R., Jones R.H., Braverman Y., Nusbaum K.E. Laboratory infections of Culicoides debilipalpis and C. stellifer (Diptera: Ceratopogonidae) with bluetongue virus. Prog. Clin. Biol. Res. 1985;178:239–243. [PubMed] [Google Scholar]
- 38.McGregor B.L., Sloyer K.E., Sayler K.A., Goodfriend O., Krauer J.M.C., Acevedo C., Zhang X., Mathias D., Wisely S.M., Burkett-Cadena N.D. Field data implicating Culicoides stellifer and Culicoides venustus (Diptera: Ceratopogonidae) as vectors of epizootic hemorrhagic disease virus. Parasit Vectors. 2019;12:258. doi: 10.1186/s13071-019-3514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borkent A., Grogan W.L., Jr. Catalog of the New World Biting Midges North of Mexico (Diptera: Ceratopogonidae) Zootaxa. 2009;2273:1–48. doi: 10.11646/zootaxa.2273.1.1. [DOI] [Google Scholar]
- 40.Schmidtmann E.T., Jones C.J., Gollands B. Comparative host-seeking activity of Culicoides (Diptera: Ceratopogonidae) attracted to pastured livestock in central New York state, USA. J. Med. Entomol. 1980;17:221–231. doi: 10.1093/jmedent/17.3.221. [DOI] [Google Scholar]
- 41.Zimmerman R.H., Turner E.C., Jr. Host-feeding patterns of Culicoides (Diptera: Ceratopogonidae) collected from livestock in Virginia, USA. J. Med. Entomol. 1983;20:514–519. doi: 10.1093/jmedent/20.5.514. [DOI] [PubMed] [Google Scholar]
- 42.Greiner E.C., Fadok V.A., Rabin E.B. Equine Culicoides hypersensitivity in Florida: Biting midges collected in light traps near horses. Med. Vet. Entomol. 1988;2:129–135. doi: 10.1111/j.1365-2915.1988.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 43.Greiner E.C., Fadok V.A., Rabin E.B. Equine Culicoides hypersensitivity in Florida: Biting midges aspirated from horses. Med. Vet. Entomol. 1990;4:375–381. doi: 10.1111/j.1365-2915.1990.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman R.H., Turner E.C., Jr. Seasonal abundance and parity of common Culicoides collected in blacklight traps in Virginia pastures. Mosquito News. 1983;43:63–69. [Google Scholar]
- 45.Smith K.E., Stallknecht D.E. Culicoides (Diptera: Ceratopogonidae) collected during epizootics of hemorrhagic disease among captive white-tailed deer. J. Med. Entomol. 1996;33:507–510. doi: 10.1093/jmedent/33.3.507. [DOI] [PubMed] [Google Scholar]
- 46.Kramer L.D., Ciota A.T. Dissecting vectorial capacity for mosquito-borne viruses. Curr. Opin. Virol. 2015;15:112–118. doi: 10.1016/j.coviro.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mead D.G., Mare C.J., Cupp E.W. Vector competence of select black fly species for vesicular stomatitis virus (New Jersey serotype) Am. J. Trop. Med. Hyg. 1997;57:42–48. doi: 10.4269/ajtmh.1997.57.42. [DOI] [PubMed] [Google Scholar]
- 48.Drolet B.S., Reeves W.K., Bennett K.E., Pauszek S.J., Bertram M.R., Rodriguez L.L. Identical viral genetic sequence found in black flies (Simulium bivittatum) and the equine index case of the 2006 U.S. vesicular stomatitis outbreak. Pathogens. 2021;10:929. doi: 10.3390/pathogens10080929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson J.R., DeFoliart G.R. Feeding behavior and host preferences of some black flies (Diptera: Simuliidae) in Wisconsin. Annu. Meet. Entomol. Soc. Am. 1961;54:716–729. doi: 10.1093/aesa/54.5.716. [DOI] [Google Scholar]
- 50.Pinkovsky D.D., Forrester D.J., Butler J.F. Investigations on black fly vectors (Diptera: Simuliidae) of Leucocytozoon smithi (Sporozoa: Leucocytozoidae) in Florida. J. Med. Entomol. 1981;18:153–157. doi: 10.1093/jmedent/18.2.153. [DOI] [PubMed] [Google Scholar]
- 51.Schnellbacher R.W., Holder K., Morgan T., Foil L., Beaufrere H., Nevarez J., Tully T.N., Jr. Avian simuliotoxicosis: Outbreak in Louisiana. Avian Dis. 2012;56:616–620. doi: 10.1637/9647-070711-CASE.1. [DOI] [PubMed] [Google Scholar]
- 52.Defoliart G.R., Rao M.R. The ornithophilic black fly Simulium meridionale Riley (Diptera: Simuliidae) feeding in man during autumn. J. Med. Entomol. 1965;2:84–85. doi: 10.1093/jmedent/2.1.84. [DOI] [PubMed] [Google Scholar]
- 53.Westwood A.R., Brust R.A. Ecology of black flies (Dipetera: Simuliidae) of the Souris River, Manitoba as a basis for control strategy. Can. Entomol. 1981;113:223–234. doi: 10.4039/Ent113223-3. [DOI] [Google Scholar]
- 54.Swanson D.A., Kapaldo N.O., Maki E., Carpenter J.W., Cohnstaedt L.W. Diversity and abundance of nonculicid biting flies (Diptera) in a zoo environment. J. Am. Mosq. Control. Assoc. 2018;34:265–271. doi: 10.2987/18-6761.1. [DOI] [PubMed] [Google Scholar]
- 55.WHO . Arboviruses and Human Disease: Report of a WHO Scientific Group. WHO; Geneva, Switzerland: 1967. p. 22. [PubMed] [Google Scholar]
- 56.Becker M.E., Reeves W.K., Dejean S.K., Emery M.P., Ostlund E.N., Foil L.D. Detection of bluetongue virus RNA in field-collected Culicoides spp. (Diptera: Ceratopogonidae) following the discovery of bluetongue virus serotype 1 in white-tailed deer and cattle in Louisiana. J. Med. Entomol. 2010;47:269–273. doi: 10.1603/ME09211. [DOI] [PubMed] [Google Scholar]
- 57.Bishop A.L., Bellis G.A., McKenzie H.J., Spohr L.J., Worrall R.J., Harris A.M., Melville L. Light trappingof biting midges Culicoides spp. (Diptera: Ceratopogonidae) with green light-emitting diodes. Aust. J. Entomol. 2006;45:202–205. doi: 10.1111/j.1440-6055.2006.00538.x. [DOI] [Google Scholar]
- 58.Hope A., Gubbins S., Sanders C., Denison E., Barber J., Stubbins F., Baylis M., Carpenter S. A comparison of commercial light-emitting diode baited suction traps for surveillance of Culicoides in northern Europe. Parasit Vectors. 2015;8:239. doi: 10.1186/s13071-015-0846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDermott E.G., Mayo C.E., Gerry A.C., Laudier D., MacLachlan N.J., Mullens B.A. Bluetongue virus infection creates light averse Culicoides vectors and serious errors in transmission risk estimates. Parasit Vectors. 2015;8:460. doi: 10.1186/s13071-015-1062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez A.M., Pauszek S.J., Jimenez D., Kelley W.N., Whedbee Z., Rodriguez L.L. Spatial and phylogenetic analysis of vesicular stomatitis virus over-wintering in the United States. Prev. Vet. Med. 2010;93:258–264. doi: 10.1016/j.prevetmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Tesh R.B., Chaniotis B.N., Johnson K.M. Vesicular stomatitis virus (Indiana serotype): Transovarial transmission by phlebotomine sandflies. Science. 1972;175:1477–1479. doi: 10.1126/science.175.4029.1477. [DOI] [PubMed] [Google Scholar]
- 62.USDA-APHIS . 2020 Vesicular Stomatitis Virus (VSV) Situation Report—July 30, 2020. USDA-APHIS; Riverdale Park, MD, USA: 2020. pp. 1–8. [Google Scholar]
- 63.Pelzel-McCluskey A.M. ((USDA APHIS Veterinary Services, Fort Collins, CO 80526, USA)). Personal communication. 2021.
- 64.USDA-APHIS . 2020 Vesicular Stomatitis Virus (VSV) Situation Report—July 27, 2020. USDA-APHIS; Riverdale Park, MD, USA: 2020. pp. 1–8. [Google Scholar]
- 65.Dyce A.L. The recognition of nulliparous and parous Culicoides (Diptera: Ceratopogonidae) without dissection. Aust. J. Entomol. 1969;8:11–15. doi: 10.1111/j.1440-6055.1969.tb00727.x. [DOI] [Google Scholar]
- 66.Akey D.H., Potter H.W. Pigmentation associated with oogenesis in the biting fly Culicoides variipennis (Diptera: Ceratopogonidae): Determination of parity. J. Med. Entomol. 1979;16:67–70. doi: 10.1093/jmedent/16.1.67. [DOI] [PubMed] [Google Scholar]
- 67.Hole K., Velazquez-Salinas L., Clavijo A. Improvement and optimization of a multiplex real-time reverse transcription polymerase chain reaction assay for the detection and typing of Vesicular stomatitis virus. J. Vet. Diagn. Investig. 2010;22:428–433. doi: 10.1177/104063871002200315. [DOI] [PubMed] [Google Scholar]
- 68.Rozo-Lopez P., Londono-Renteria B., Drolet B.S. Impacts of infectious dose, feeding behavior, and age of Culicoides sonorensis biting midges on infection dynamics of vesicular stomatitis virus. Pathogens. 2021;10:816. doi: 10.3390/pathogens10070816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ball L.A., White C.N. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biggerstaff B.J., Petersen L.R. Estimated risk of transmission of the West Nile virus through blood transfusion in the US, 2002. Transfusion. 2003;43:1007–1017. doi: 10.1046/j.1537-2995.2003.00480.x. [DOI] [PubMed] [Google Scholar]
- 71.Gu W., Lampman R., Novak R.J. Problems in estimating mosquito infection rates using minimum infection rate. J. Med. Entomol. 2003;40:595–596. doi: 10.1603/0022-2585-40.5.595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available upon request from the corresponding authors and through the USDA, Agricultural Research Information System.