Abstract
The 26S proteasome is a eukaryotic ATP-dependent protease, but the molecular basis of its energy requirement is largely unknown. Ornithine decarboxylase (ODC) is the only known enzyme to be degraded by the 26S proteasome without ubiquitinylation. We report here that the 26S proteasome is responsible for the irreversible inactivation coupled to sequestration of ODC, a process requiring ATP and antizyme (AZ) but not proteolytic activity. Neither the 20S proteasome (catalytic core) nor PA700 (the regulatory complex) by itself contributed to this ODC inactivation. Analysis with a C-terminal mutant ODC revealed that the 26S proteasome recognizes the C-terminal degradation signal of ODC exposed by attachment of AZ, and subsequent ATP-dependent sequestration of ODC in the 26S proteasome causes irreversible inactivation, possibly unfolding, of ODC and dissociation of AZ. These processes may be linked to the translocation of ODC into the 20S proteasomal inner cavity, centralized within the 26S proteasome, for degradation.
Polyamines are indispensable for cell growth, but they become harmful, displaying severe cytotoxic effects, if they accumulate to excess in cells (2, 35, 43). To prevent abnormal accumulation of polyamines in cells, a unique feedback regulatory system controlling the biosynthesis and uptake of polyamines has developed during the course of evolution. Since ornithine decarboxylase (ODC) is a rate-limiting enzyme catalyzing the first reaction in the multiple concerted pathways of polyamine biosynthesis, one strategy for preventing overproduction of cellular polyamines is precise control of ODC activity in response to alterations in cellular polyamine levels. Antizyme (AZ), an ODC inhibitory protein, is a key player in this scenario, since it is induced by polyamines, end products of the metabolic pathway, through programmed ribosomal frameshifting (30, 45) of the AZ mRNA that is abundant in cells (29).
The regulation of ODC by AZ is of interest. Active ODC consists of two identical monomer subunits with two active sites formed at their interfaces. The enzymatically active dimer form of ODC is in rapid equilibrium with the inactive monomer form (7). AZ preferentially binds with the inactive ODC monomer to form an ODC-AZ complex (31) and thus inhibits ODC by preventing reassociation of its inactive subunits. However, the maximum level of ODC-AZ complex in cells is much less than one-tenth of that of total ODC, suggesting that the AZ-induced inhibition of ODC activity does not have much significance in ODC regulation. An important role of AZ could be to cause conformational change in the ODC subunit, resulting in exposure of the carboxy-terminal region to attack by the 26S proteasome. ODC is broken down by the proteasome, whereas most of the AZ molecules are recycled to destabilize more ODC monomers. Another function of AZ is the suppression of polyamine uptake at the cell membrane (32, 46). Thus, AZ plays a pivotal role in an autoregulatory loop maintaining normal polyamine levels in cells. The characteristics of the ODC degradation directed by AZ were recently reviewed by Hayashi et al. (18) and Coffino (6).
Antizyme inhibitor (AIn), another regulatory protein, is also present in cells. It is highly homologous with ODC but is clearly distinct and has no enzymatic activity. It binds to AZ with higher affinity than that of ODC to AZ, releasing active ODC from the inactive ODC-AZ complex (37). Accordingly, AIn can be defined as a negative regulatory factor canceling the feedback control mechanism mediated by AZ. Taken together, these two regulators, AZ and AIn, presumably contribute to fine control of the cellular polyamine concentration.
The 26S proteasome, a eukaryotic ATP-dependent protease, is a 2,000-kDa multisubunit proteolytic complex consisting of a central catalytic machine (called the 20S proteasome or simply 20S) and two terminal regulatory subcomplexes, termed PA700 (also known as the 19S regulatory complexes), which are attached to both ends of the central portion in opposite orientations. The 20S proteasome is a protease complex with a molecular mass of 700 to 750 kDa and is composed of 28 subunits. It is a barrel-like particle formed by the axial stacking of four rings made up of two outer α-rings and two inner β-rings, associated in the order αββα (each ring is composed of seven homologous subunits). PA700 is a 700-kDa protein complex composed of about 20 subunits with display sizes of 25 to 110 kDa. These subunits can be divided into two subgroups: 6 homologous ATPases and approximately 14 non-ATPase subunits that are structurally unrelated. The functions of many of these subunits are still unknown (reviewed in references 3, 8, and 44). There is no experimental evidence to explain how or why the energy is consumed. One possible explanation is that the energy released in ATP hydrolysis is necessary for formation of the 26S proteasome by the association of the 20S proteasome with PA700 or other factors. Indeed, the energy-dependent formation of a high-molecular-weight complex containing proteasome/PA700 has been reported (1, 5, 22). Another possible function of the ATPases is to unfold substrate proteins to allow their passage through the narrow entry ports of the proteasome and their translocation into the interior of the 20S proteasome core, which harbors proteolytically active sites (3, 17). However, evidence that the 26S proteasome indeed unfolds the substrate proteins in an ATP-dependent manner before their degradation has not yet been obtained (for reviews, see references 3, 8, 44, and 47). Most cellular proteins are targeted for degradation by the 26S proteasome after they have been covalently attached to ubiquitin (Ub) in the form of a poly-Ub chain which functions as a degradation signal (19, 20). The 26S proteasome has been shown to act as protein-destroying machinery responsible for the selective degradation of numerous ubiquitinylated cellular proteins. However, ODC is degraded in an ATP-dependent manner by the 26S proteasome without ubiquitinylation (4, 11, 38).
It is plausible that the 26S proteasome degrades both ODC and polyubiquitinylated proteins by the same mechanism requiring ATP, except for the manner of substrate recognition. It was found recently that one subunit of the mammalian 26S proteasome, named S5a (equivalent to yeast Rpn10/Sun1/Mcb1), can bind specifically to proteins conjugated to poly-Ub chains and thus functions as a poly-Ub receptor (abbreviated as pUb-R) of the 26S proteasome (14, 44, 47). Intriguingly, the pUb-R protein is present not only as a component associated with the PA700 complex but also in a free form. The latter presumably is capable of recruiting target polyubiquitinylated proteins to the 26S proteasome for their breakdown. On the other hand, it would be interesting to know how the 26S proteasome recognizes a nonubiquitinylated protein, such as ODC, prior to its degradation. Recent studies suggested that the “C-terminal degradation domain” of ODC may be exposed in collaboration with the N-terminal region of AZ and that the exposure is needed for proteolytic attack by the 26S proteasome (23, 24). However, the mechanism by which the 26S proteasome can interact with the ODC molecule remains obscure.
In the present study, we investigated the mechanism of ODC degradation mediated by the 26S proteasome and propose that degradation of ODC by the 26S proteasome involves multiple sequential processes: first, exposure of the C-terminal degradation signal of ODC by attachment of the AZ molecule; second, a process involving ATP-dependent sequestration of ODC into the 26S proteasome associated with irreversible ODC inactivation and dissociation of AZ; and finally, degradation of ODC in the 26S supercomplex after its translocation into the 20S inner cavity, the proteolytic center of the 26S proteasome. ATP may be required to induce a conformational change of ODC and/or to translocate the unfolded ODC. Both processes are thought to be prerequisites for degradation. This is the first report of how the 26S proteasome degrades a target protein in an experimental model.
MATERIALS AND METHODS
Materials.
A plasmid, p9T7ODC71, containing the rat ODC cDNA sequence was kindly provided by H. Van Steeg of the National Institute of Public Health and Environmental Protection, Bilthoven, The Netherlands. N-Benzyloxycarbonyl-Leu-Leu-Leu-aldehyde (Z-LLL-CHO) was purchased from the Peptide Institute, Inc., Osaka, Japan. Clasto-lactacystin β-lactone was kindly supplied by MBL (Nagoya, Japan). Proteinase K (PK) was purchased from Merck (Germany).
ODC, AZ, and AIn.
Rat AZ cDNA Z1 (30) was expressed in Escherichia coli, an extract (800-μg protein) was applied to a monoclonal anti-AZ antibody (HZ-2E9) (28) AffiGel 10 column (1 ml), and the column was washed with buffer A (25 mM Tris buffer, pH 7.5, containing 1 mM EDTA, 1 mM dithiothreitol [DTT], and 0.01% Tween 80) containing 4 M NaCl. AZ was eluted with 4 ml of 3 M MgCl2, and the eluate was dialyzed against buffer A. Purified glutathione S-transferase GST-AIn was prepared as described previously (37). ODC was purified by DEAE-cellulose chromatography and immunoaffinity chromatography (40) from rat liver. 35S-labeled wild-type or mutant ODC was synthesized with an in vitro transcription/translation system (34) and purified by immunoaffinity chromatography (40). Where indicated, ODC was metabolically labeled and purified similarly; ODC was induced in FM3A or hepatoma tissue culture (HTC) cells by replacing the growth medium with fresh medium. After incubation for 2.5 h, a mixture of l-[35S]methionine and l-[35S]cysteine (Du Pont NEN) was added at 40 to 100 μCi/ml and the cells were incubated further for 1 h.
20S proteasome, PA700, and 26S proteasome.
The 20S proteasome and the 26S proteasome were purified from rat liver as described previously (48). PA700 was purified from bovine erythrocytes by a modification of the method of Chu-Ping et al. (5). Briefly, the supernatant of bovine erythrocyte lysate was applied to a column of DE 52. The proteins were eluted with a linear gradient of KCl from 0 to 0.5 M. The fractions containing high activity were centrifuged at 100,000 × g for 10 h. The pellets were dissolved and applied to a hydroxyapatite column. The flow-through fractions were applied to a heparin-Sepharose column that had been equilibrated with 100 mM potassium phosphate buffer (pH 6.8) containing 1 mM DTT. The proteins were eluted with a linear gradient of NaCl from 0 to 0.4 M. The fractions with high activity were subjected to velocity sedimentation centrifugation with glycerol gradients from 10 to 30% in 25 mM Tris buffer containing 1 mM DTT and 2 mM ATP. Proteasome activity was measured as hydrolysis of the synthetic peptide succinyl-Leu-Leu-Val-Tyr-4-methyl-coumaryl-7-amide (Suc-LLVY-AMC), as described previously (48). One unit of proteasome activity is defined as the amount degrading 1 nmol of Suc-LLVY-AMC per min.
ODC inactivation and degradation assays of cell extract.
HTC cells grown to 80 to 100% confluency in 10-cm dishes were washed twice with cold phosphate-buffered saline and lysed by three cycles of freezing-thawing. Then, 0.1 ml of 50 mM Tris-HCl, pH 7.5, containing 1 mM DTT was added. Cell extract was obtained by centrifugation of the homogenate at 12,000 rpm (rav, 6 cm) for 20 min. The reaction mixture (50 μl) for the inactivation assay contained cell extract (15 to 30 μg of protein), 1.5 units of purified rat liver ODC, 1.0 unit of AZ (Z1 antizyme) (30), an ATP-regenerating system (2 mM ATP, 20 mM phosphocreatine, 10 μg of creatine kinase, and 10 mM MgCl2), 2.5 mM DTT, and 30 mM Tris-HCl, pH 7.4. When degradation alone or degradation together with inactivation was assayed, 35S-ODC (2,000 to 3,000 cpm) was added to the assay mixture. The inactivation/degradation mixture was incubated at 37°C for 60 min, and the amount of trichloroacetic acid (TCA)-soluble radioactivity of a part of the mixture was measured for determination of ODC degradation, which was expressed as the percentage of total ODC added. To determine ODC inactivation, pyridoxal phosphate, [14C]ornithine, and about 1.5 units of AIn were added to another part of the mixture, and the remaining ODC activity was determined (37). The zero time control was determined similarly but without incubation for the first inactivation/degradation reaction. The decrease in ODC activity on incubation should be caused by degradation and/or irreversible inactivation of ODC and is expressed as a percent decrease in ODC activity.
Antibodies and immunological analysis.
Antibodies against ODC, HO101 (27), and the 20S proteasome (48) were prepared as previously described. For immunodepletion of the proteasome, HTC cell extract (300 μg of protein) was incubated in a total volume of 200 μl containing 1 mM DTT, 2 mM ATP, and 25 mM Tris-HCl, pH 7.5, at 4°C for 60 min with the indicated amounts of anti-proteasome immunoglobulin G (IgG) or control IgG and then centrifuged at 14,000 × g for 30 min. Aliquots of the supernatants were used for proteolytic assay. For immunoprecipitation, antibody was added and incubated at 4°C for 60 min. Formalin-fixed Staphylococcus aureus Cowan I cells (ZYSORBIN; Zymed) or protein A-Sepharose (Pharmacia) was then added and shaken gently at 4°C for 60 min. The pellet was washed four times with 25 mM Tris buffer, pH 7.5, containing 0.1% sodium dodecyl sulfate (SDS), 0.1% Triton X-100, 2 mM EDTA, and 1 mM DTT (for pellet with polyclonal antibody) or with 20 mM Tris buffer, pH 7.5, containing 0.15 M NaCl and 0.05% Tween 20 (for monoclonal antibody).
Assay for PK susceptibility of inactivated ODC (ODC bound to the proteasome).
ODC was preincubated in the inactivation assay mixture (50 μl) containing extracts of HTC cells (56 μg of protein) that had been treated with clasto-lactacystin β-lactone and then treated with PK at a final concentration of 80 μg/ml for the indicated times at 0 or 37°C. Digestion was halted by addition of phenylmethylsulfonyl fluoride (PMSF) (2 mM). Anti-20S proteasome antibody (2 μg) and 20 μl of formalin-fixed S. aureus Cowan I cells (10% suspension) were added to the reaction mixture, and bound ODC in the immunoprecipitates was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by radioautography. Unbound ODC in the supernatant was also analyzed for comparison. In addition, as another control, ODC without preincubation was treated similarly with PK, except that bovine serum albumin (BSA) instead of crude cell extracts was added, and then analyzed by SDS-PAGE.
RESULTS
Irreversible inactivation of ODC in HTC cell extracts.
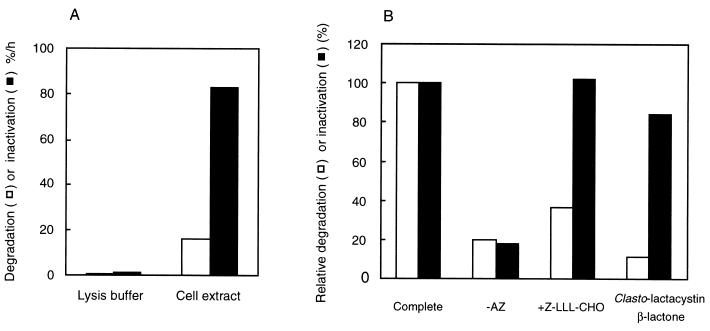
ODC was induced in HTC cells in the presence of [35S]methionine and purified. The labeled ODC was incubated with a fresh extract of HTC cells in a degradation/inactivation assay mixture containing AZ and ATP. After incubation, the amount of TCA-soluble radioactivity of a part of the mixture was measured for determination of ODC degradation. Pyridoxal phosphate, [14C]ornithine, and AIn were added to another part of the mixture, and the remaining ODC activity was determined. AIn was added to liberate ODC from the inactive ODC-AZ complex, resulting in reactivation of ODC that had been reversibly inhibited with AZ. Surprisingly, we found that the extent of decrease in ODC activity (irreversible inactivation) was much greater than that of ODC protein (degradation) during incubation with the cell extract, indicating that inactivated ODC protein was formed and remained at least in part without degradation in the reaction mixture (Fig. 1A). Similar results were obtained when in vitro-translated 35S-labeled rat ODC or metabolically labeled 35S-ODC from mouse FM3A cells, both mixed with purified rat liver ODC, was used as an ODC substrate (data not shown). Thereafter, the mixture of in vitro-translated 35S-labeled rat ODC and purified rat liver ODC was used as the ODC substrate unless otherwise noted. The ratio of inactivation to degradation, however, varied considerably (from about 1 to 5) in different cell extracts for some unknown reason, indicating that the finding was not necessarily reproducible. Later experiments showed that reproducibility of overinactivation required the presence of proteasome inhibitor (see below).
FIG. 1.
Irreversible inactivation of ODC by cell extracts. (A) ODC metabolically labeled with [35S]methionine in HTC cells was incubated with or without cell extracts that had been supplemented with AZ and ATP. Irreversible inactivation and degradation of ODC were determined as described in Materials and Methods. (B) ODC (a mixture of in vitro-translated labeled rat ODC and cold ODC purified from rat liver) was incubated with cell extracts in an inactivation/degradation reaction mixture. The assay was the same as for Fig. 1A, except that AZ was withdrawn or Z-LLL-CHO (100 μM) was added. When the effect of clasto-lactacystin β-lactone was examined, cell extracts were preincubated with the inhibitor (200 μM) at 37°C for 15 min in an inactivation/degradation reaction mixture without ODC, and then a one-sixth volume of ODC was added. The inactivation and degradation of ODC are represented by filled and open columns, respectively. Results are shown as percentages of the values obtained in the complete reaction mixture without inhibitors.
In these experiments, a decrease in ODC activity measured after the addition of AIn to the postincubation mixture of ODC, AZ, and cell extract was defined as “irreversible inactivation of ODC” (or simply “inactivation of ODC”). That is, inactivation of ODC corresponded to a loss of ODC activity that could not be recovered with AIn. It should be noted that this definition differs from “inhibition of ODC activity” by AZ, which can be reactivated fully by AIn, and that the value for irreversible inactivation of ODC measured by the present method includes loss of activity caused by ODC degradation (see below).
Energy and AZ requirement for irreversible inactivation of ODC.
Previously, we reported that degradation of ODC by an HTC cell extract was energy dependent (38). Therefore, we examined the energy requirement for irreversible inactivation of ODC on incubation with HTC cell extract. First, we tested the requirement for Mg2+. In the absence of exogenous Mg2+ and with the addition of 5 mM EDTA to remove endogenous Mg2+ in the extract, no significant inactivation of ODC was observed, suggesting that energy from ATP is necessary for irreversible inactivation of ODC (Table 1). Moreover, nonhydrolyzable ATP analogues, such as α,β-methylene-ATP and β,γ-methylene-ATP, had no appreciable effects on the inactivation of ODC, as observed for ODC degradation (Table 1) (38). Thus, irreversible inactivation of ODC is energy dependent. Next, we examined the effects of various ribonucleotide triphosphates on the inactivation of ODC. As shown in Table 1, all of the nucleotide triphosphates tested stimulated ODC inactivation. Similar nucleotide dependence was observed for the degradation of ODC (Table 1) (38) and ubiquitinylated proteins by the 26S proteasome (1) and nucleotidase activities of the 26S proteasome or PA700 (21).
TABLE 1.
Both degradation and inactivation of ODC require a hydrolyzable nucleotide triphosphate
| Nucleotide (5 mM) | Degradation
|
Inactivation
|
||
|---|---|---|---|---|
| %/h | % ATP | %/h | % ATP | |
| None | 1.9 | 0 | 15.8 | 18 |
| ATP | 49.5 | 100 | 87.4 | 100 |
| CTP | 47.5 | 89 | 91.4 | 105 |
| UTP | 34.7 | 42 | 73.4 | 84 |
| GTP | 41.0 | 69 | 83.3 | 95 |
| α,β-Methylene-ATP | 3.4 | 7 | 25.1 | 29 |
| β,γ-Methylene-ATP | 0.9 | 2 | 3.8 | 4 |
| ATP with no Mg2+ and 5 mM EDTA | 0 | 0 | 3.0 | 3 |
Previously, AZ has been shown to be involved in in vitro ODC degradation by cell extracts (39) and by the purified 26S proteasome (38). Therefore, we examined whether or not the irreversible inactivation of ODC requires AZ. As shown in Fig. 1B, inactivation of ODC was greatly reduced in the absence of AZ, as observed for ODC degradation, clearly indicating that the irreversible inactivation of ODC is AZ dependent. The requirements of a nucleotide and AZ for both irreversible inactivation and degradation of ODC suggest that there is a functional relationship between the two processes.
Proteolytic activity of the proteasome is not required for its irreversible inactivation of ODC.
We then tested the requirement of proteolytic activity of the proteasome for its irreversible inactivation of ODC with proteasome-selective inhibitors. When the substrate-related peptidyl aldehyde inhibitor Z-LLL-CHO, also named MG132, was added at a final concentration of 100 μM, degradation of ODC was markedly inhibited; intriguingly, however, the irreversible inactivation was not decreased appreciably, resulting in an increase in the ratio of inactivation to degradation of ODC (Fig. 1B). A further increase in the ratio was observed in assays using extracts prepared from HTC cells that had been pretreated with Z-LLL-CHO (50 μM) for 2 h (data not shown). Z-LLL-CHO is known to affect the activities of both the proteasome and other proteases, such as calpains and lysosomal thiol proteases (48). Therefore, we studied the effect of clasto-lactacystin β-lactone, spontaneously formed from lactacystin, which is a more potent and specific inhibitor of the proteasome (10, 13). Clasto-lactacystin β-lactone strongly inhibited ODC degradation in a concentration-dependent manner (data not shown) but had only a weak effect on the irreversible inactivation of ODC even at the high concentration of 200 μM, resulting in the accumulation of inactivated ODC, as observed in the treatment with Z-LLL-CHO (Fig. 1B). These results clearly indicate that, unlike ODC degradation, the proteolytic function of the proteasome is not required for irreversible inactivation of ODC. Thereafter, to measure the irreversible inactivation of ODC accurately and reproducibly, we assayed ODC inactivation routinely in the presence of Z-LLL-CHO or clasto-lactacystin β-lactone to repress degradation activity. Unless otherwise mentioned, cell extracts or 26S proteasome preparations were preincubated with clasto-lactacystin β-lactone (200 μM) for 15 min at 37°C and then added to the inactivation assay mixture.
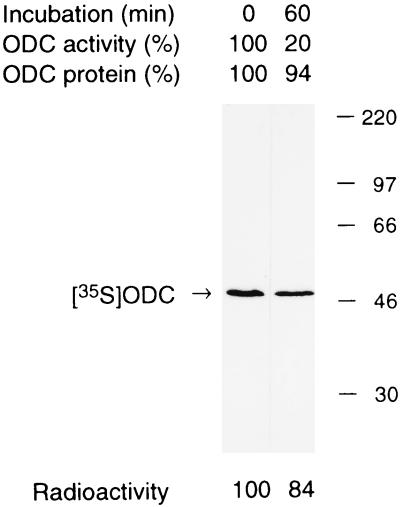
We analyzed the electrophoretic mobility of the inactivated ODC. 35S-ODC was incubated with a fresh extract of HTC cells in an inactivation assay mixture containing 100 μM Z-LLL-CHO for 1 h. Under this condition, approximately 80% of the ODC was inactivated irreversibly, but only about 6% was degraded (Fig. 2). We then analyzed the 35S-ODC by SDS-PAGE, followed by autoradiography. As shown in Fig. 2, the inactivated 35S-ODC had a size similar to the original 35S-ODC, estimated to be approximately 50 kDa. The decrease in the radioactive intensity of 35S-ODC estimated with an image analyzer (Fujix BAS2000) was calculated to be only 16%, which was comparable to the value obtained as TCA-soluble radioactivity, indicating that the irreversibly inactivated ODC retained its native size without any digestion. The inactivated 35S-ODC was also analyzed by nondenaturing PAGE (4.5%). Its electrophoretic mobility was much smaller than that of the native 35S-ODC (data not shown).
FIG. 2.
Molecular size of irreversibly inactivated ODC. ODC (mixture of purified rat ODC and metabolically labeled 35S-ODC from FM3A mouse cells) was incubated for 60 min at 37°C in an inactivation/degradation reaction mixture containing an extract of HTC cells that had been treated with Z-LLL-CHO. The decreases in ODC activity and ODC protein during incubation were 80 and 6%, respectively. Aliquots of the reaction mixture were subjected to SDS-PAGE and analyzed by autoradiography. Relative intensities of ODC bands determined with an image analyzer (Fujix BAS2000) are shown below each lane.
Involvement of the proteasome in irreversible inactivation of ODC.
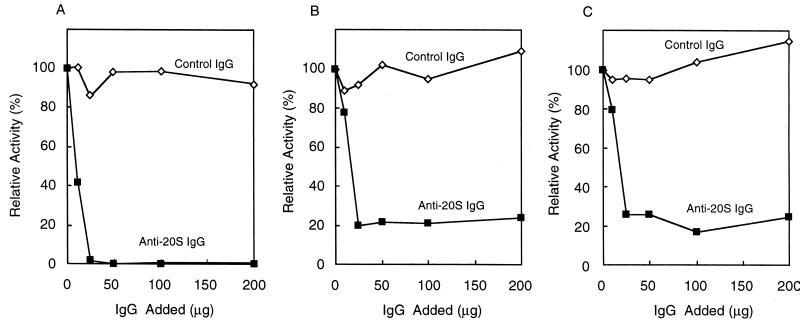
As mentioned above, the proteolytic activity of the proteasome is not necessary for the irreversible inactivation of ODC observed in cell extracts, but whether the proteasome itself was involved in the event remained unknown. Therefore, we examined the effect of immunodepletion of the proteasome from cell extracts on ODC inactivation. As shown in Fig. 3A, the addition of polyclonal antibody against the 20S proteasome resulted in almost complete loss of proteasome activity in a concentration-dependent fashion, judging from the hydrolysis of a fluorogenic peptide, Suc-LLVY-AMC, a typical substrate for the 20S proteasome (48). The disappearance of both 20S and 26S proteasomes was confirmed by Western blotting analysis (data not shown). The polyclonal antibody was also found to cause marked and dose-dependent suppression of ODC inactivation (Fig. 3C), as well as its degradation (Fig. 3B), whereas control IgG had no significant effect in parallel experiments. These results indicate clearly that the irreversible inactivation of ODC is catalyzed by the proteasome, but that its proteolytic activity is not required for the process.
FIG. 3.
Effects of immunodepletion of the proteasome on inactivation of ODC by an HTC cell extract. Samples (300 μg) of HTC cell extracts were treated with anti-proteasome IgG (■) or control IgG (◊), and aliquots of the supernatants were assayed for Suc-LLVY-AMC degradation (A), ODC degradation (B), and ODC inactivation (C). Values are percentages of the activities measured with no added IgG, namely, 600 nmol/h for peptide hydrolysis, 25%/h for ODC inactivation, and 10%/h for ODC degradation.
The 26S proteasome, not PA700 or the 20S proteasome alone, catalyzes ATP- and AZ-dependent ODC inactivation.
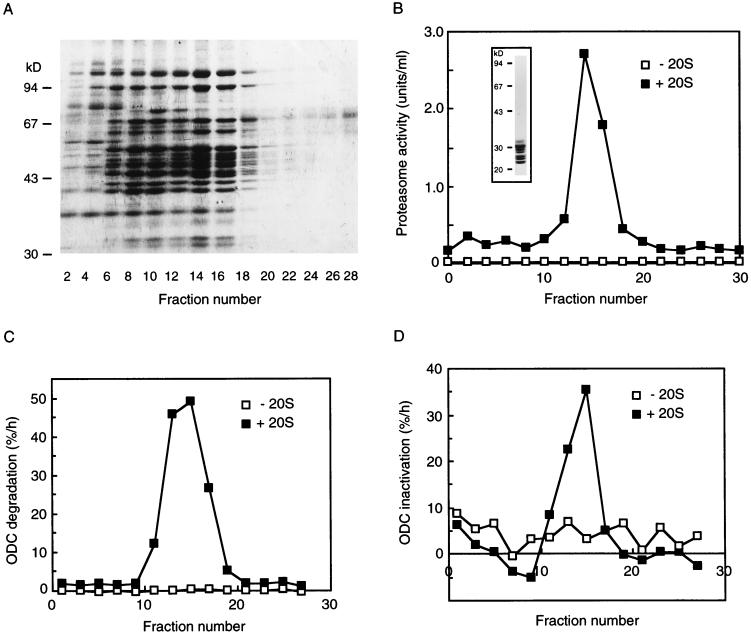
Previously we reported that the 26S proteasome, but not the 20S proteasome, catalyzes ATP- and AZ-dependent ODC degradation in vitro (38). In the present work we have demonstrated involvement of the proteasome, but not its proteolytic activity, in the ATP-dependent irreversible inactivation of ODC. Judging from the energy dependency, it seemed likely that the 26S proteasome was the enzyme responsible for the inactivation of ODC. However, it was still considered possible that PA700 alone had the ability to inactivate ODC and then recruited it to the 20S proteasome for degradation. Therefore, we examined whether ODC was inactivated by free PA700. To test this possibility, we isolated PA700 in a highly purified state from bovine erythrocytes as described in Materials and Methods. Figure 4A shows the protein-staining profile analyzed by SDS-PAGE of PA700 preparations obtained by fractionation after glycerol density gradient centrifugation. Components of 25 to 110 kDa were observed, in excellent accord with the broad distribution of PA700 components reported previously (5). We detected no proteasome activity or immunoreactivity on Western analysis with the anti-20S proteasomal antibody in these PA700 preparations (data not shown), indicating that the preparations were free from the 20S proteasome. Curiously, however, many 25- to 110-kDa components were detected in a broad range of fractions from lightly to heavily sedimenting fractions, as judged by the protein-staining pattern (Fig. 4A). Since these protein components were broadly distributed and appeared to be similar, but not identical, we assumed that the complexes recovered in the lightly sedimenting fractions probably lack some components or have altered conformations. To clarify the functional relationship between PA700 and the 20S proteasome, we assayed the chymotryptic activity of the 20S proteasome by using Suc-LLVY-AMC as a substrate after addition of purified latent 20S proteasome. In contrast to the broad distribution of the PA700 component on the SDS-polyacrylamide gel, a sharp peak of chymotryptic activity was recovered only in the heavily sedimenting fractions around fraction 16 (Fig. 4B). This activation required incubation with the 20S proteasome for at least 15 min at 37°C in the presence of ATP-Mg2+, like that required for formation of the 26S proteasome by association of PA700 with the 20S proteasome (5). Subsequently, we examined the inactivation and degradation of ODC by using this reconstituted system. No fraction of PA700 had significant activity for ODC degradation without the 20S proteasome, but incubation of the same fractions with the 20S proteasome for over 15 min at 37°C in the presence of ATP-Mg2+ generated significant activity for degradation of ODC (Fig. 4C). Similarly, irreversible inactivation of ODC was observed in the same fractions possessing ODC degradation activity under entirely the same assay conditions except that clasto-lactacystin β-lactone was added to inhibit ODC degradation (Fig. 4D). These results indicate that PA700 itself cannot irreversibly inactivate ODC. Since the 20S proteasome was added to all fractions examined, it is clear that the 20S proteasome itself also had no effect on the inactivation or degradation of ODC. From these findings, we concluded that the 26S proteasome reconstituted from PA700 and the 20S proteasome was the main enzyme responsible for ATP- and AZ-dependent ODC inactivation. We also confirmed that the highly purified 26S proteasome irreversibly inactivated ODC in an ATP- and AZ-dependent manner (data not shown).
FIG. 4.
PA700 inactivates ODC in the presence of the 20S proteasome but not in the absence of the proteasome. Purified PA700 was separated by glycerol density gradient centrifugation. The gradient was separated into 30 fractions of 1 ml each. Samples of 150 μl of the gradient fractions were precipitated with 750 μl of acetone, and the precipitates were subjected to SDS-PAGE and stained with Coomassie blue (A). Twenty microliters of the samples were assayed for proteasome activity (B) and ODC degradation (C) and inactivation (D) in the presence (■) and absence (□) of purified 20S proteasome. For the ODC inactivation assay, samples were preincubated with clasto-lactacystin β-lactone (200 μM) at 37°C for 15 min. The inset in panel B shows SDS-PAGE of the 20S proteasome used.
Binding of inactivated ODC to the 26S proteasome.
To examine immunoreactivity of inactivated ODC, immunoprecipitates with the HO101 ODC monoclonal antibody were analyzed by SDS-PAGE separation followed by autoradiography. The HO101 antibody did not react with the ODC protein, which had been denatured by a high concentration of MgCl2 (27) or by SDS (data not shown). After incubation for inactivation (Fig. 5A, right lane), 35S-ODC had decreased greatly compared to that in the nonincubated control (Fig. 5A, left lane). The intensity of the radioactivity measured with the image analyzer was determined to be 37%, which was comparable to the value of the remaining activity of ODC that escaped inactivation, indicating that the irreversibly inactivated 35S-ODC resisted immunoprecipitation with the antibody used. These results suggest that the irreversible inactivation of ODC was associated with a drastic change in its protein structure and/or that, owing to its sequestration in some macromolecules such as the proteasome, inactivated ODC lost its accessibility to the antibody. Furthermore, we found that inactivated ODC also lost immunoreactivity against polyclonal anti-ODC antibody (data not shown).
FIG. 5.
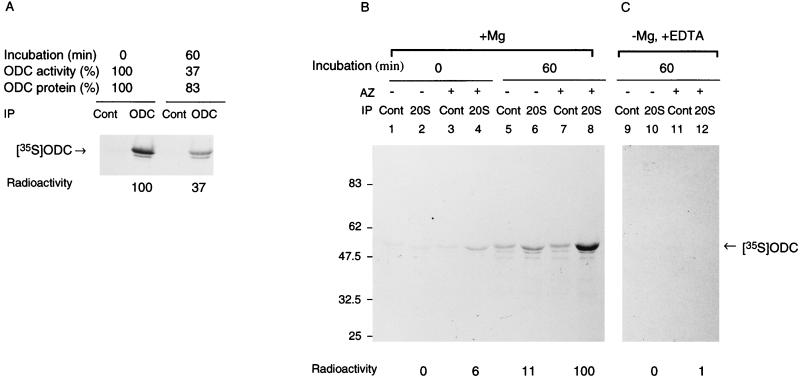
Energy- and AZ-dependent binding of ODC to the 26S proteasome. (A) ODC was incubated with a crude cell extract of HTC cells in the inactivation assay mixture for 60 min at 37°C. Aliquots of the reaction mixture before and after incubation were examined for ODC activity and ODC protein, and aliquots were immunoprecipitated with monoclonal antibody to ODC (HO101) or control IgG followed by SDS-PAGE and autoradiography. Intensities of ODC bands were determined with an image analyzer (Fujix BAS2000), and specific bindings were obtained by subtracting the value with control IgG and are expressed relative to the amount of time zero control. (B and C) ODC was incubated with partially purified 26S proteasome in the inactivation assay mixture. Where indicated, AZ was removed from the reaction mixture or EDTA was added to it instead of MgCl2. The incubated mixture was immunoprecipitated with anti-20S proteasome antibody (20S) or control IgG (Cont). The mixture without incubation was similarly treated with antibodies. The immunoprecipitates were washed extensively with buffer containing 0.1% SDS and 0.1% Triton X-100 and subjected to SDS-PAGE and then to autoradiography. Panels B and C represent separate experiments. The control (with MgCl2, without EDTA) is not shown in panel C but was almost the same as in panel B. Intensities of ODC bands were determined by an image analyzer, and the specific precipitation of ODC with anti-20S proteasome antibody was obtained by subtracting the value with control IgG and is expressed relative to the amount immunoprecipitated when ODC was incubated in the presence of AZ and MgCl2.
We examined whether or not the inactivated ODC was trapped in the proteasome. ODC was preincubated with partially purified 26S proteasome in an inactivation assay mixture, and then the proteasome was immunoprecipitated with specific antibody against the 20S proteasome. The immunoprecipitates were washed extensively and analyzed by SDS-PAGE. As shown in Fig. 5B (lane 8), anti-20S antibody effectively coimmunoprecipitated 35S-ODC which had been preincubated with the 26S proteasome in the presence of AZ. For this coimmunoprecipitation, AZ was essential during preincubation, because only small amounts of 35S-ODC were immunoprecipitated after preincubation without AZ (Fig. 5B, lane 6). However, after preincubation, AZ was not necessary for coimmunoprecipitation (data not shown). ATP appears to be necessary for this coimmunoprecipitation, because the addition of EDTA, instead of Mg2+, during preincubation inhibited coimmunoprecipitation almost completely (Fig. 5C, lane 12). In the presence of AZ, a very small amount of 35S-ODC was detected in the immunoprecipitates of the reaction mixture without incubation (Fig. 5B, lane 4). Control IgG did not precipitate 35S-ODC, irrespective of the incubation and/or addition of AZ (lanes 1, 3, 5, and 7). These findings indicate that the proteasome effectively entraps AZ-bound ODC but not free ODC, as for its AZ-dependent inactivation by the 26S proteasome. Thus, the inactivated ODC was likely to be entrapped by the 26S proteasome.
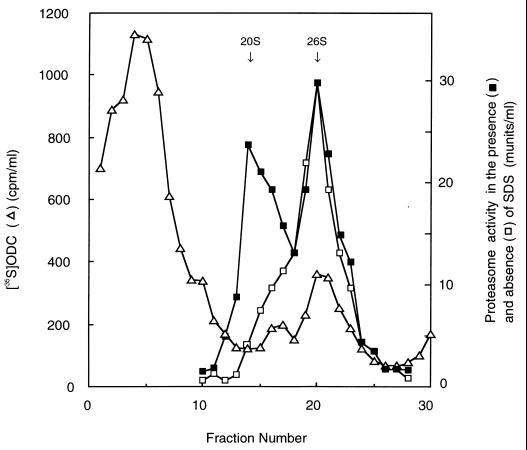
To further confirm the binding of ODC to the proteasome, we determined whether ODC cosedimented with the proteasome in a glycerol gradient. ODC was preincubated for 1 h with a crude extract of HTC cells in the inactivation assay mixture, and then the mixture was centrifuged at 128,000 × g for 5 h to enrich the proteasome and roughly separate ODC bound to the proteasome from large amounts of unbound ODC and its degradation products. The precipitates were suspended and subjected to glycerol density gradient centrifugation (Fig. 6). The 20S and 26S proteasomes were sedimented at fractions with Suc-LLVY-AMC degrading activity observed in the presence and absence of SDS, respectively (see arrows). The radioactivity derived from free 35S-ODC appeared as light fractions 2 through 6. Moreover, two additional unequal peaks were detected in more heavily sedimenting fractions 15 through 24. The first small peak had a slightly greater Mr than that of the 20S proteasome and was suggested to be consistent with that of PA700 detected by Western blotting in similar experiments (data not shown). The second peak, with greater radioactivity, was sedimented at a position corresponding to the 26S proteasome. This result confirmed the binding of ODC to the proteasome and at the same time indicated that most of the inactivated ODC was bound preferentially to the 26S form but not to free PA700. However, it was not shown whether 35S-ODC was incorporated into the interior of the core or trapped by PA700 of the 26S proteasome.
FIG. 6.
Cosedimentation of inactivated ODC with the 26S proteasome in glycerol density gradient centrifugation. 35S-ODC was incubated for 1 h with a crude extract of HTC cells (640 μg of protein) in the inactivation assay mixture (500 μl), and the mixture was centrifuged at 128,000 × g for 5 h. The precipitates were resuspended and subjected to glycerol density gradient centrifugation as for Fig. 4. Fraction 1 represents the top of the gradient (10% glycerol), and fraction 30 represents the bottom of the gradient (40% glycerol). Samples (0.8 ml) of the gradient fractions were used for determinations of ODC radioactivity, and proteasome activities in the presence and absence of SDS were determined for each 20 μl of the fractions. Arrows indicate the positions of elution of purified 20S and 26S proteasomes.
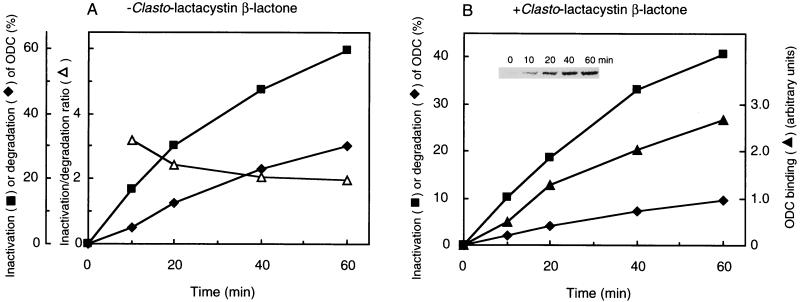
To address the relationship between ODC binding, inactivation, and degradation by the 26S proteasome, we examined the time courses of these reactions with or without clasto-lactacystin β-lactone. In the absence of the proteasome inhibitor, both ODC inactivation and degradation were time dependent, and the inactivation/degradation ratio decreased slightly but clearly (Fig. 7A). In the presence of proteasome inhibitor, the time courses of ODC inactivation and degradation were essentially the same as those in the absence of the inhibitor except for a reduced degradation rate, and binding of ODC to the proteasome was observed almost in parallel with the inactivation of ODC (Fig. 7B). These results indicated that ODC binding and inactivation were almost simultaneous events and preceded ODC degradation.
FIG. 7.
Time courses of ODC inactivation, degradation, and binding. ODC was incubated for the indicated times at 37°C in an inactivation/degradation assay mixture containing partially purified 26S proteasome that had been treated (B) or not treated (A) with clasto-lactacystin β-lactone (200 μM). ODC inactivation, degradation, and binding were determined as described in Materials and Methods. The inset (B) shows SDS-PAGE of ODC coimmunoprecipitated with anti-20S proteasome antibody after the inactivation reaction at the indicated times. The relative amounts of coimmunoprecipitated ODC (ODC binding) were quantitated with an image analyzer.
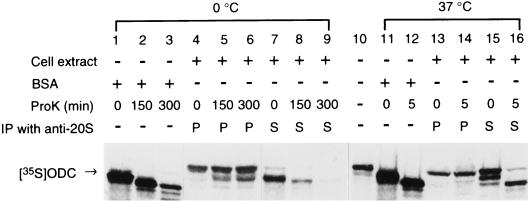
Next, we examined whether or not the inactivated ODC was sequestrated in the 26S proteasome by a proteinase protection assay. 35S-ODC was preincubated for 1 h with a crude extract of HTC cells in the inactivation assay mixture. The preincubated reaction mixtures were then treated with PK for varying times at 0 or 37°C. After digestion was halted by the addition of PMSF, the proteasome was precipitated with anti-20S antibody. The supernatant and washed immunoprecipitates were analyzed for unbound and proteasome-bound ODCs, respectively, by SDS-PAGE. As a control, unpreincubated 35S-ODC was treated with PK under the same conditions except that BSA was used instead of a crude extract of HTC cells. As shown in Fig. 8, control ODC was found to be rapidly digested at both 0°C (lanes 1 through 3) and 37°C (lanes 11 and 12). No protease resistance was observed with the ODC unbound to the proteasome contained in the preincubated mixture (ODC in the supernatant fraction after immunoprecipitation with anti-20S proteasome antibody) (lanes 7 through 9, 15, and 16). By contrast, ODCs bound to the proteasome were clearly resistant to proteinase K digestion (lanes 4 through 6, 13, and 14). The apparent increase of ODC in the proteasome (lanes 4 through 6) may be due to incorporation of full-length or truncated ODC into the 26S proteasome during incubation at 0°C. These results indicated that inactivated ODC was sequestrated into the proteasome.
FIG. 8.
Resistance of inactivated ODC to PK digestion. 35S-ODC was preincubated for 1 h with a crude extract of HTC cells in the inactivation assay mixture. The mixtures were treated with PK (80 μg/ml) for varying times at the indicated temperatures (lanes 4 to 9 and 13 to 16). Digestion was halted by the addition of PMSF. ODCs bound to the proteasome and unbound (shown by P and S, respectively) were separated with anti-20S proteasome antibody, and both ODCs were analyzed by SDS-PAGE followed by autoradiography. As another control, unpreincubated ODC was similarly treated with PK in the presence of BSA instead of a crude cell extract and then analyzed by SDS-PAGE (shown in lanes 1 to 3, 11, and 12). Purified substrate 35S-ODC is shown in lane 10.
Role of the carboxyl-terminal region of ODC in its irreversible inactivation.
The C-terminal domain of ODC is required for its rapid degradation (6, 15). A single amino acid replacement of Cys-441 by Trp-441 in this domain also has a strongly stabilizing effect (33, 34). Notably, this mutant ODC, ODC(C441W), has the same enzymatic activity as normal ODC and can bind to AZ (42) but resists rapid degradation, resulting in abnormal accumulation of its complex with AZ in the cells (36). Thus, it has become clear that AZ prompts the degradation of ODC mediated by the 26S proteasome perhaps by exposing the C-terminal domain of ODC as a degradation signal (23). Therefore, it was of particular interest to examine whether the mutant ODC shows irreversible inactivation similar to that of normal ODC, as found in this study, and whether it binds to the 26S proteasome.
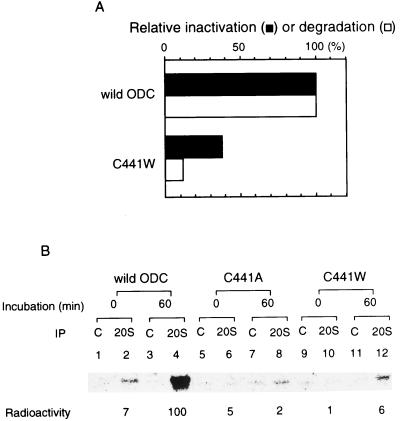
We incubated ODC(C441W) with the 26S proteasome in the presence of AZ and ATP-Mg2+. Unlike wild-type ODC, the mutant ODC considerably resisted inactivation (Fig. 9A). Moreover, after preincubation, as shown in Fig. 7A, 35S-ODC(C441W) was not significantly immunoprecipitated with anti-20S proteasome antibodies (Fig. 9B). Similar resistance to 26S proteasome entrapment was observed with another mutant, ODC(C441A), in which Cys at position 441 was replaced by Ala (Fig. 9B). Like ODC(C441W), mutant ODC(C441A) was strongly stabilized (34). These findings show that the C-terminal region of ODC exposed by AZ is recognized by the 26S proteasome for binding, followed by its irreversible inactivation. Hence, the binding signal governs ODC stability.
FIG. 9.
The C-terminal region of ODC is necessary for binding to and inactivation by the 26S proteasome. (A) Recombinant ODC with replacement of cysteine at position 441 by tryptophan (C441W) was obtained by using a pET vector expression system, purified by immunoaffinity chromatography, and tested for inactivation by the 26S proteasome. mRNA encoding ODC (C441W) was translated in a rabbit reticulocyte lysate in the presence of [35S]methionine. The protein product was purified by immunoaffinity chromatography and tested for degradation by the 26S proteasome. Wild-type ODC was similarly treated for comparison. (B) Purified 35S-labeled stable ODCs were prepared as described above, and binding to the 26S proteasome was examined as in Fig. 5B. Results with wild-type ODC treated similarly are shown for comparison.
DISCUSSION
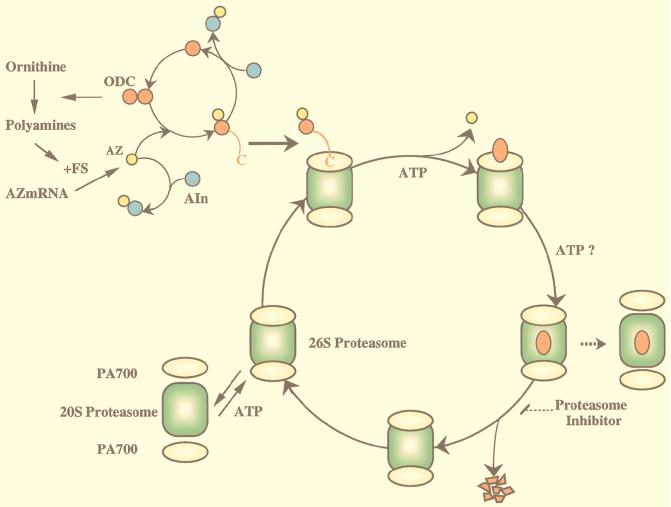
In the present study, we found that the 26S proteasome, but not the 20S proteasome or PA700, irreversibly inactivated ODC prior to its degradation. The inactivation was coupled to sequestration of ODC within the 26S proteasome. These processes required AZ, ATP hydrolysis, and the presence of a native ODC carboxy-terminal region but not proteolytic activity of the 26S proteasome. Based on the present and previous observations, we propose a new model (Fig. 10) for the energy- and AZ-dependent pathway for ODC degradation mediated by the 26S proteasome which involves two distinct steps: irreversible inactivation and ODC degradation. Here, we discuss several steps involved in the process.
FIG. 10.
Model of ODC degradation by the 26S proteasome. ODC is active as a homodimer complex. AZ, which is induced by translational frameshifting, binds to inactive monomeric ODC to form an ODC-AZ complex. Two terminal regulatory subcomplexes, termed PA700, are attached in an ATP-dependent manner to both ends of the 20S proteasome (central catalytic machinery) in opposite orientations to form enzymatically active proteasomes. The 26S proteasome may attack the exposed ODC C-terminal region and pull the ODC molecule, but not AZ, into the interior, which is associated with ATP-dependent substrate unfolding. The continuous translocation of unfolded ODC (inactivated ODC) may be required for further continuous unfolding of ODC (ODC inactivation), and the two processes may proceed in concert with each other. The unfolded ODC translocated into the cavity of the 20S proteasome, which harbors proteolytically active sites, is degraded, but it is unknown whether ATP consumption is needed for the degradative process itself. After degradation, the 26S proteasome traps another substrate(s) or may be in part dissociated into its constituents, the 20S proteasome and PA700. See the text for details.
How does the 26S proteasome recognize ODC for degradation?
It is of particular interest that two mutant ODCs, ODC(C441W) and ODC(C441A), which carry a single amino acid exchange in their C-terminal regions, show high resistance to ODC degradation as well as to binding and inactivation. As these mutated ODCs both retained normal enzymatic activity and AZ-binding ability that led to loss of their activity, the mutant ODCs should maintain an almost normal tertiary conformation and resemble the wild-type ODC in many respects, except for one critical difference in their defective stable phenotype. Therefore, it is likely that the C-terminal region of ODC plays an essential role in its rapid turnover: the in vitro findings obtained in the present study are consistent with previous reports showing that rapid ODC degradation depends on the presence of a cis-acting element in its carboxyl terminus in addition to a trans-acting factor, AZ (23), and that a single amino acid replacement in the carboxyl-terminal region (C441W) results in ODC stabilization in living HMOA cells (34). The most likely interpretation of these observations is that “the C-terminal degradation signal” of ODC exposed by attachment of AZ is somehow recognized by the 26S proteasome.
The alternative possibility, that AZ bound to ODC provides a recognition site for the 26S proteasome, cannot be ruled out completely. However, it seems unlikely that AZ is directly recognized by the 26S proteasome. We presume that AZ is dissociated from ODC before sequestration of ODC into the 26S proteasome (Fig. 10), since a few AZ molecules could promote the degradation of a large number of ODC molecules (26, 39), and ODC, but not AZ, was degraded to oligopeptides of similar size in an in vitro ODC degradation assay by the 26S proteasome (49). The recycling of AZ seems to be similar to that of Ub, but their action mechanisms may be different. Ub is directly recognized by pUb-R (see the introduction) of the 26S proteasome, and Ub or the polyubiquitin chain is removed from the protein substrate by isopeptidase associated with the 26S proteasome. Furthermore, the degradation of polyubiquitinylated protein is inhibited by excessive amounts of both Ub-R (9) and poly-Ub (41), whereas ODC degradation is inhibited by C-terminal peptides of ODC (unpublished results) but not excessive AZ (39). Finally, ODC can be degraded slowly by the purified 26S proteasome in the absence of AZ. These findings suggest that ODC, rather than AZ, is directly recognized by the 26S proteasome.
The question that arises is how can the 26S proteasome recognize the C-terminal degradation signal of ODC, and which molecule(s) or subunit(s) of the 26S proteasome recognizes it? At present, we do not have any clear evidence to explain the direct relationship between the degradation signal of ODC and its ability to be recognized by the 26S proteasome. Very recently, Glickman et al. (16) reported that yeast PA700 can be separated into two subcomplexes: a lid and a base complex. The base consists of six ATPases and two other subunits of Rpn1/p97/S2 and Rpn2/p112/S1, whereas the lid complex is composed of the other 10 subunits. Interestingly, association of the base with the 20S proteasome results in marked activation of peptidase activity but not ATP-dependent degradation of ubiquitinylated proteins. We observed sometimes that purified PA700 lost activities to inactivate and degrade ODC even when incubated with the 20S proteasome, but the activation of peptidase activity for Suc-LLVY-AMC degradation remained, suggesting that the attachment of the base to the α-ring outer complex of the 20S proteasome may be a stable form retaining activity for peptide degradation. From this, we speculate that a certain component(s) present in the lid complex is capable of recognizing the C-terminal degradation signal of ODC. It will be important to determine the physical features of ODC attached to AZ and to identify the subunit(s) of the 26S proteasome that interacts directly with ODC molecules.
How is ODC sequestrated in the 26S proteasome?
Recent structural analyses of 20S and 26S proteasomes have shown that the catalytically active sites of the 20S proteasome face the interior of the cylinder and reside in a chamber formed by the centers of the abutting β-rings. Substrates gain access to the active sites only after passing through a narrow opening corresponding to the center of the rings, and the amino termini of the subunits form an additional physical barrier for substrates to reach the active sites (see the introduction). The latency of the 20S proteasome is supported by the above-described observation that the center of the α-ring is almost closed, preventing penetration of proteins into the inner surface of the β-ring, on which the proteolytically active sites are located (17). The PA700 complex contains six ATPases that may attach directly to the α-ring of the 20S proteasome and open the channel for translocation of the protein (16). However, there is no evidence that substrate proteins are unfolded by the 26S proteasome before their penetration of the channel into the α- and β-rings of the 20S proteasome, and no ATP-dependent unfolding of a substrate has been shown experimentally. Here, we demonstrated a pathway for energy- and AZ-dependent binding of ODC to the 26S proteasome and inactivation of ODC. This clearly demonstrated that ATP-energy is required for the process before proteolysis itself. The finding that ODC bound to the proteasome was almost completely protected from PK attack indicates that the inactivated ODC was sequestrated in the 26S proteasome, although it was not shown whether the ODC was sequestrated into the interior of the 20S proteasome or retained in PA700. For the reason that ATP hydrolysis is needed for proteasome-dependent inactivation of ODC, liberated energy may be required to induce a conformational change of ODC (unfolding) and/or to translocate the unfolded ODC. We assume that sequestration and unfolding (irreversible inactivation of ODC) may be coupled to substrate translocation into the 20S proteasomal inner cavity centralized within the 26S proteasome complex. However, it is difficult to separate the processes of sequestration and translocation of ODC as distinct biochemical steps, because they appear to occur simultaneously in the 26S proteasome. Further studies are required to answer this fundamental question.
It would be interesting to know the structural features of the ODC that enters the 26S proteasome. It was not shown whether 35S-ODC trapped by the 26S proteasome is enzymatically active or is inactivated by unfolding. We initially found that ODC was irreversibly inactivated after its incubation with cell extracts or the 26S proteasome and ATP and AZ, judging from its insensitivity to reactivation with AIn. From additional data showing the loss of immunoreactivity of the inactivated ODC with anti-ODC antibodies, we now assume that the tertiary structure of the inactivated ODC is altered dramatically, perhaps due to unfolding. However, it is also possible that the insensitivities to AIn and the antibody used may be because they have no access to the ODC molecule trapped in the 26S proteasome complex, even if the sequestrated ODC has the native structure without being unfolded or denatured. We favor the possibility that the inactivated ODC formed by incubation with the 26S proteasome was unfolded, since the inactivated ODC had the same molecular size as native ODC and was free from AZ, as discussed above, but no ODC activity was detected, suggesting that the entrapped ODC lost the functional tertiary structure responsible for enzymatic activity. This finding is in marked contrast to findings with α2-macroglobulin a large protease inhibitor complex that can entrap a broad spectrum of proteases, all of which have enzymatic activities for the small substrates tested, indicating that when entrapped by the large α2-macroglobulin complex they retain their native tertiary structures (12). However, we cannot exclude the possibility that the ODC monomer with a folded structure cannot form an active enzyme in the inner cavity of the 26S proteasome. Whether or not ODC entrapped in the 26S proteasome is actually unfolded remains unclear and requires further study.
Inactivation of ODC was dependent on energy but not on the proteolytic activity of the proteasome. However, isolated PA700 alone could not inactivate ODC: its association with the 20S proteasome was required for its inactivating function. These findings suggest that the functions of both the 20S proteasome and PA700 are required for efficient function, hence the 20S proteasome is necessary for the function of PA700 and vice versa. Recently, Rechsteiner (44) proposed two possible mechanisms of proteolysis catalyzed by the 26S proteasome: the “ribosome model” and the “solid-state model.” According to the ribosome model, free PA700 can trap target proteins, mostly ubiquitinylated, and recruit them to the 20S catalytic proteasome, and then the 26S proteasome formed degrades the substrate. In the solid-state model, the 26S proteasome can directly trap target proteins for degradation. These two models were proposed without any experimentation, but considering the action mechanism of the 26S proteasome, such speculations are interesting. Our present findings that PA700 itself could not inactivate ODC and that the 26S proteasome, but not the PA700 peak, contained most of inactivated 35S-ODC (Fig. 6) appear to support the second solid-state model. However, the ribosome model cannot be excluded completely, because of the possibility that PA700 can trap ODC without conformational change, and hence without irreversible inactivation of ODC, or that the small number of PA700 molecules is sufficient to recruit the target protein to the 20S catalytic proteasome. In fact, the possible protein-binding ability of PA700, the chaperone activity of isolated PA700 towards nonubiquitinylated misfolded proteins, has been pointed out recently (25).
ACKNOWLEDGMENTS
This work was supported in part by grants-in-aid for scientific research on priority areas (intracellular proteolysis) from the Ministry of Education, Science, Sports, and Culture of Japan, by a grant from the Uehara Memorial Foundation, and by a grant from the Human Frontier Science Promotion Organization.
REFERENCES
- 1.Armon T, Ganoth D, Hershko A. Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990;265:20723–20726. [PubMed] [Google Scholar]
- 2.Auvinen M, Paasinen A, Andersson L C, Hölttä E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–359. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 4.Bercovich Z, Kahana C. Involvement of the 20S proteasome in the degradation of ornithine decarboxylase. Eur J Biochem. 1993;213:205–210. doi: 10.1111/j.1432-1033.1993.tb17749.x. [DOI] [PubMed] [Google Scholar]
- 5.Chu-Ping M, Vu J H, Proske R J, Slaughter C A, DeMartino G N. Identification, purification, and characterization of a high molecular weight ATP-dependent activator (PA700) of the 20S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 6.Coffino P. Degradation of ornithine decarboxylase. In: Peters J-M, Harris J D, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 411–428. [Google Scholar]
- 7.Coleman C S, Stanley B A, Viswanath R, Pegg A E. Rapid exchange of subunits of mammalian ornithine decarboxylase. J Biol Chem. 1994;269:3155–3158. [PubMed] [Google Scholar]
- 8.Coux O, Tanaka K, Goldberg A L. Structure and function of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 9.Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M. Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J Biol Chem. 1995;270:29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- 10.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V T, Perent L A, Plamondon L, Stein R. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 11.Elias S, Bercovich B, Kahana C, Coffino P, Fischer M, Hilt W, Wolf D H, Ciechanover A. Degradation of ornithine decarboxylase by the mammalian and yeast 26S proteasome complexes requires all the components of the protease. Eur J Biochem. 1995;229:276–283. [PubMed] [Google Scholar]
- 12.Feldman S R, Gonias S L, Pizzo S V. Model of α2-macroglobulin structure and function. Proc Natl Acad Sci USA. 1985;82:5700–5704. doi: 10.1073/pnas.82.17.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 14.Finley D, Tanaka K, Mann C, Feldmann H, Hochstrasser M, Vierstra R, Johnston S, Hampton R, Haber J, Silver P, et al. Unified nomenclature for subunits of the S. cerevisiae proteasome regulatory particle. Trends Biochem Sci. 1998;271:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- 15.Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxy-terminal truncation. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- 16.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9 signalosome and elF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 17.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartnik H D, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci. 1996;21:27–30. [PubMed] [Google Scholar]
- 19.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman L, Rechsteiner M. Nucleotidase activities of the 26S proteasome and its regulatory complex. J Biol Chem. 1996;271:32538–32545. doi: 10.1074/jbc.271.51.32538. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman L, Pratt G, Rechsteiner M. Multiple forms of 20S multicatalytic and the 26S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J Biol Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 23.Li X, Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-indelible inhibitory protein. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Stubbiness B, Hoffman L, Pratt G, Rechsteiner M, Coffino P. The N-terminus of antizyme promotes degradation of heterologous proteins. J Biol Chem. 1996;271:4441–4446. doi: 10.1074/jbc.271.8.4441. [DOI] [PubMed] [Google Scholar]
- 25.Lin L, DeMartino G N, Greene W C. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 26.Mamroud-Kidron E, Omer-Itsicovich M, Bercovich Z, Tobias K, Rom E, Kahana C. A unified pathway for degradation of ornithine decarboxylase in reticulocytes requires interaction with the polyamine-induced protein, ornithine decarboxylase antizyme. Eur J Biochem. 1994;226:547–555. doi: 10.1111/j.1432-1033.1994.tb20079.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsufuji S, Fujita K, Kameji T, Kanamoto R, Murakami Y, Hayashi S. Monoclonal antibody to rat liver ornithine decarboxylase. J Biochem. 1984;96:1525–1530. doi: 10.1093/oxfordjournals.jbchem.a134981. [DOI] [PubMed] [Google Scholar]
- 28.Matsufuji S, Kanamoto R, Murakamia Y, Hayashi S. Monoclonal antibody studies on the properties and regulation of murine ornithine decarboxylase antizyme. J Biochem. 1990;107:87–91. doi: 10.1093/oxfordjournals.jbchem.a123018. [DOI] [PubMed] [Google Scholar]
- 29.Matsufuji S, Miyazaki Y, Kanamoto R, Kameji T, Murakami Y, Baby T G, Fujita K, Ohno T, Hayashi S. Analyses of ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J Biochem. 1990;108:365–371. doi: 10.1093/oxfordjournals.jbchem.a123207. [DOI] [PubMed] [Google Scholar]
- 30.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell J L, Chen H J. Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim Biophys Acta. 1990;1037:115–121. doi: 10.1016/0167-4838(90)90109-s. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell J L, Judd G G, Bareyal-Leyser A. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem J. 1994;299:19–22. doi: 10.1042/bj2990019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell J L A, Choe C-Y, Judd G G, Daghfal D J, Kurzeja R J, Leyser A. Overproduction of stable ornithine decarboxylase and antizyme in the difluoromethylornithine-resistant cell line DH23b. Biochem J. 1996;317:811–816. doi: 10.1042/bj3170811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki Y, Matsufuji S, Murakami Y, Hayashi S. Single amino-acid replacement is responsible for the stabilization of ornithine decarboxylase in HMOA cells. Eur J Biochem. 1993;214:837–844. doi: 10.1111/j.1432-1033.1993.tb17987.x. [DOI] [PubMed] [Google Scholar]
- 35.Moshier J A, Dosescu J, Skunca M, Luk G D. Transformation of NIH/3T3 cells by ornithine decarboxylase overexpression. Cancer Res. 1993;53:2618–2622. [PubMed] [Google Scholar]
- 36.Murakami Y, Fujita K, Kameji T, Hayashi S. Accumulation of ornithine decarboxylase-antizyme complex in HMOA cells. Biochem J. 1985;225:689–697. doi: 10.1042/bj2250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami Y, Ichiba T, Matsufuji S, Hayashi S. Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J Biol Chem. 1996;271:3340–3342. doi: 10.1074/jbc.271.7.3340. [DOI] [PubMed] [Google Scholar]
- 38.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 39.Murakami Y, Tanaka K, Matsufuji S, Miyazaki Y, Hayashi S. Antizyme, a protein induced by polyamines, accelerates the degradation of ornithine decarboxylase in Chinese hamster ovary-cell extracts. Biochem J. 1992;283:661–664. doi: 10.1042/bj2830661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama M, Matsufuji S, Kanamoto R, Takano M, Murakami Y, Hayashi S. Two-step purification of mouse kidney ornithine decarboxylase. Prep Biochem. 1988;18:227–238. doi: 10.1080/00327488808062524. [DOI] [PubMed] [Google Scholar]
- 41.Piotrowski J, Beal R, Hoffman L, Wilkinson K D, Cohen R, Pickart C C. Inhibition of the 26S proteasome by polyubiquitin chains synthesized to have defined lengths. J Biol Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 42.Pritchard M L, Pegg A E, Jefferson L S. Ornithine decarboxylase from hepatoma cells and a variant cell line in which the enzyme is more stable. J Biol Chem. 1982;257:5892–5899. [PubMed] [Google Scholar]
- 43.Poulin R, Pelletier G, Pegg A E. Induction of apoptosis by excessive polyamine accumulation in ornithine decarboxylase-overproducing L1210 cells. Biochem J. 1995;311:723–727. doi: 10.1042/bj3110723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rechsteiner M. The 26S proteasomes. In: Peters J-M, Harris J D, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 147–189. [Google Scholar]
- 45.Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci USA. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T, He Y, Kashiwagi K, Murakami Y, Hayashi S, Igarashi K. Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc Natl Acad Sci USA. 1994;91:8930–8934. doi: 10.1073/pnas.91.19.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K. Molecular biology of the proteasome. Biochem Biophys Res Commun. 1998;247:537–541. doi: 10.1006/bbrc.1998.8617. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka K, Tanahashi N. Preparation of proteasomes. In: Celis J E, editor. Cell biology: a laboratory handbook. 2nd ed. New York, N.Y: Academic Press; 1997. pp. 129–134. [Google Scholar]
- 49.Tokunaga F, Goto T, Koide T, Murakami Y, Hayashi S, Tamura T, Tanaka K, Ichihara A. ATP- and antizyme-dependent endoproteolysis of ornithine decarboxylase to oligopeptides by the 26S proteasome. J Biol Chem. 1994;269:17382–17385. [PubMed] [Google Scholar]