Abstract
Antibiotics are important disruptors of the intestinal microbiota establishment, linked to immune and metabolic alterations. The intrapartum antibiotics prophylaxis (IAP) is a common clinical practice that is present in more than 30% of labours, and is known to negatively affect the gut microbiota composition. However, little is known about how it affects to Bifidobacterium (sub)species level, which is one of the most important intestinal microbial genera early in life. This study presents qualitative and quantitative analyses of the bifidobacterial (sub)species populations in faecal samples, collected at 2, 10, 30 and 90 days of life, from 43 healthy full-term babies, sixteen of them delivered after IAP use. This study uses both 16S rRNA–23S rRNA internal transcribed spacer (ITS) region sequencing and q-PCR techniques for the analyses of the relative proportions and absolute levels, respectively, of the bifidobacterial populations. Our results show that the bifidobacterial populations establishment is affected by the IAP at both quantitative and qualitative levels. This practice can promote higher bifidobacterial diversity and several changes at a compositional level. This study underlines specific targets for developing gut microbiota-based products for favouring a proper bifidobacterial microbiota development when IAP is required.
Keywords: Bifidobacterium, ITS, q-PCR, gut microbiota, antibiotics, IAP, GBS, C-section
1. Introduction
Antibiotics are lifesaving drugs, due to their powerful ability to battle infections. Since the discovery of penicillin in 1928, numerous antibiotics have been described [1]. Among them, beta-lactams are the most-commonly administered compounds and make up 65% of the antibiotic market [2]. Antibiotics are also extensively used for prophylactic purposes, and they are by far the most common prescription drugs given in the perinatal and neonatal environment, being present in more than 30% of labours [3,4,5]. In this context, the administration of intrapartum antibiotic prophylaxis (IAP) is commonly used for the prevention of early-onset Group-B-streptococci (GBS) infection, in pre-labour rupture of membranes or C-section deliveries to avoid surgical infections [3,6]. However, the use of antibiotics also presents disadvantages, such as promoting antimicrobial resistance, adverse drug events or alterations of the gut microbiota [7,8,9].
The gut microbiota is an ever-changing and complex microbial ecosystem harboured by the gastrointestinal tract (GIT). The microbiota is a key factor in a range of biological processes in the host [10,11,12,13]. After birth, the GIT is rapidly colonised by a wide diversity of microorganisms. Accumulating evidence shows that a correct establishment of this microbiota in the early days of life plays an important role in the reduction of the later development of chronic diseases, such as obesity, allergies, infections, inflammatory or brain disorders, and is an overall determinant for the health of the individual [14,15,16]. Thereby, the period of the gut microbiota colonisation and establishment constitutes a critical window-of-opportunity for its modulation towards a healthy status [17,18,19]. The gut microbiota establishment begins with the settlement of facultative anaerobes and aerotolerant microorganisms, such as enterobacteria or lactobacilli, which pave the way by reducing the oxygen in the gut to the colonisation by other strict anaerobic bacteria, such as bifidobacteria or bacteroides [20,21]. Several factors have an impact in shaping the gut microbiota colonisation process [6,14,19,22,23]. Among them, antibiotics are one of the most pivotal factors promoting alterations on the intestinal microbiota establishment at the beginning of life, linked to immune and metabolic alterations [24,25,26].
Animal studies have shown that when the early-life gut microbiota is altered by antibiotics, enduring physiological effects are observed, even though there is a later microbiota restoration [27,28,29,30,31]. On the other hand, different epidemiological studies have demonstrated the association between early exposure to antibiotics and different diseases, such as allergy [32,33], asthma [31,34], celiac disease [35,36], overweight [37,38,39,40] or inflammatory bowel disease [41]. In this regard, it is proven that early-life antibiotic treatment disrupts the proper and natural development of gut microbiota with potential negative influence on later health [42,43,44]. During the last years, several studies have seen the advent reporting an impact on the gut microbiota after IAP treatment. Lower relative proportions of Actinobacteria and Bacteroidetes and increased of Proteobacteria and Firmicutes were observed during the first weeks of life, with Bifidobacterium being one of the most affected genera [45,46,47,48]. Nogacka et al., by using 16S rRNA gene profiling of faeces, observed reduced proportions of Bifidobacteriaceae family in full-term babies during the first days of life after IAP treatment [45], which is in good agreement with the results obtained by Corvaglia et al. by using q-PCR for Bifidobacterium genus [47]. Moreover, Mazzola et al. [46] also reported lower levels of Bifidobacterium at seven days of life. These differences were also observed at a later age, even at 6 and 12 months [49], with a higher impact of IAP even than that of later administration of oral antibiotics in infants [50,51].
Bifidobacterium is a genus belonging to the Actinobacteria phylum and is regarded as a keystone taxon in the gut microbiota early in life, with a strong eco-physiological impact on microbiota composition and function [52,53,54]. Some species can dominate the gut of breast-fed infants [55,56,57]. Convincing evidence has accumulated showing that the presence of bifidobacteria in the gut is associated with health improvement [53,58]. This is particularly true in the case of infants. In the early gut microbiota, bifidobacteria drive the intestinal microbiome development and their removal or failure to colonise may lead to the development of chronic diseases [52]. Several studies have shown lower levels or reduced relative abundances of bifidobacteria on infant populations in different scenarios, such as prematurity, obesity, later sepsis, colics [19,59,60,61,62]. This occurs because the levels and diversity of bifidobacteria depend on different perinatal factors, as was reported in a recent publication [63]. Among them, antibiotics deeply affected the Bifidobacterium genus, which could disrupt the crosstalk with the immune system [44]. However, the impact of IAP on gut-specific bifidobacterial populations is still unexplored.
To date, the studies exploring this field have focused their investigations on using the 16S rRNA gene sequencing approach, quantification of the Bifidobacterium genus by q-PCR, or hybridisation-based technique. However, this has provided limited knowledge on the impact of IAP at lower taxonomical levels. To overcome this limitation, developing next-generation techniques, such as the sequencing of the Internally Transcribed Spacer (ITS) within the rRNA locus [64], was useful as marker of Bifidobacterium species, as it has been demonstrated in studies focused on the vertical transmission of bifidobacteria [65], the effect of using donor and own-mother’s milk on the premature bifidobacterial community [66] or the impact of several perinatal factors on the establishment of Bifidobacterium species in premature and full-term babies [63].
In this context, by using the ITS sequencing technique as a marker of bifidobacterial (sub)species and q-PCR, we aimed at exploring the effect of the IAP on the establishment and development of the bifidobacterial populations in full-term babies during the first three months of life.
2. Materials and Methods
2.1. Volunteers and Faecal Samples Collection
The study included 43 Caucasian full-term healthy neonates (22 females/21 males) born after an uncomplicated pregnancy at a gestational age ranging between 38 and 41 weeks (mean 39.2) and recruited at the Neonatology Unit of University Central Hospital of Asturias (Oviedo, Spain). Thirty-four neonates were vaginally delivered and nine were born through C-section. Twenty-seven, twenty-six, twenty-three and twenty neonates were exclusively breast-fed at 2, 10, 30 and 90 days of life, respectively. None received antibiotics, pro- or prebiotics during the sampling time considered for the study. Sixteen mothers received antibiotics during the labour period. Seven of them followed a vaginal labour and were administered with an initial dose of five million units of penicillin followed by 2.5 million units every 4 h until delivery (in most cases, the mothers received three or fewer doses) as prophylaxis, due to confirmed or suspected vaginal colonisation by GBS. Whereas the other nine mothers received a single dose of 2 g of intravenous cefazolin, due to C-Section delivery. The rest of the mothers were culture-negative for GBS, and following a vaginal birth, were not exposed to antibiotics (control group). None of the mothers received antibiotics during pregnancy or the postnatal period, other than the above mentioned (Table S1).
Fresh faecal samples were collected at 2 (between 24 and 48 h after birth), 10, 30 and 90 days of age in a sterile container and immediately frozen at −20 °C.
The study was approved by the Regional Ethical Committee of Asturias Public Health Service (SESPA) (Ref. 51/18), and an informed written consent was obtained from each infant’s parents.
2.2. Faecal DNA Isolation
Faecal samples were homogenised (1:10 w/v) in a sterile phosphate-buffered-saline (PBS) solution in a LabBlender 400 stomacher (Seward Medical, London, UK) at full speed for 3 min. Then they were centrifuged for 15 min at full speed to the separate cell pellet and supernatant, which were kept at −20° for further analysis. DNA was isolated from faecal pellets following Qiagen manufacturer’s instructions (QIAmp DNA stool kit, Qiagen GmbH, Hilden, Germany), and kept at −20 °C until use for intergenic ribosomal transcriber spacer (ITS) and q-PCR analyses.
2.3. Analyses of Faecal Bifidobacterial Populations by ITS Region Profiling
The isolated DNA was used as a target to amplify and further sequencing the ITS region (the 16S–23S internal transcriber spacer of the ribosomal DNA) as described elsewhere [64]. Briefly, primers Probio_bif_uni and Probio_bif_rev and Illumina technology were used for bifidobacterial ITS region sequencing. Sequences obtained were assembled, filtered and annotated by following an in-house protocol, and an improved bifidobacterial ITS database [66]. The number of reads and relative abundances were determined for each bifidobacterial species in each sample analysed.
2.4. Analysis of the Faecal Bifidobacterial Levels by Specific Quantitative PCR
Absolute levels of the most relevant gut bifidobacterial species, B. bifidum, B. breve, B. catenulatum, B. dentium, B. longum, B. angulatum, B. animalis ssp. lactis and B. adolescentis were determined by q-PCR using primers and methodology described elsewhere [66]. Standard curves were created with pure cultures of each strain, which were grown overnight in MRS medium (Difco, BectoneDickinson and Company, Le Pont de Claix, France) supplemented with 0.25% L-cysteine (Sigma Chemical Co, St. Louis, MO, USA) under anaerobic conditions. Cultures were plate-counted, and DNA isolation was completed following the same protocol used for faecal samples (QIAmp DNA stool kit, Qiagen GmbH, Hilden, Germany). Samples were analysed in duplicates in at least two independent PCR runs.
2.5. Statistical Analysis
Results were analysed using the SPSS software version 26 (SPSS Inc., Chicago, IL, USA) and Calypso software (version 8.84) [67] with sum normalisation (TSS) and cumulative-sum scaling (CSS) to account for the non-normal distribution of taxonomic count data [68]. Multivariate redundancy analysis (RDA) was conducted. Species number and alpha diversity indices (Chao1 and Shannon) were calculated and analysed by t-test. To assess the differences in gut bifidobacterial relative abundance and levels related to the use of intrapartum antibiotics, and to avoid bias derived from delivery mode, an ANCOVA test with delivery mode as covariable was conducted. LefSe test (linear discriminant analysis effect size) was also used to detect species features between groups (LDA scores > 2 and significance of p < 0.05 as determined by Wilcoxon’s signed-rank test) [69]. Data were considered statistically significant at p < 0.05.
2.6. Nucleotide Sequence Accession Numbers
The raw sequences from the samples have been deposited in the National Centre for Biotechnology Information (NCBI)—Short Read Archive (SRA) under the BioProject ID code PRJNA750917.
3. Result
3.1. Bifidobacterium Populations on Full-Term Infants
B. longum ssp. longum was the most abundant species detected along the three first months of life, followed by B. breve, B. dentium, B. adolescentis, B. bifidum and B. pseudocatenulatum (Table S2). Although bifidobacterial ITS profiling revealed 35 different (sub)species in the infant population analysed, at the age of two days, there was a large inter-individual variability with babies harbouring a single species (B. breve, n = 1) and babies with even 30 different ones (n = 1). On average, at this age, the babies harboured 11 different bifidobacterial species, with a range between 9 ± 4 and 11 ± 6 (mean ± SD) (sub)species along with the study. B. longum ssp. longum, B. breve and B. adolescentis were the species with the highest average relative proportions (29.18%, 22.14% and 10.18%, respectively) and highest occurrence: B. longum ssp. longum was present in 98% of babies, B. breve in 95% and B. adolescentis in 81% at two days of life. At the age of 10 days, B. dentium became the third most abundant species and was present in 69% of babies, following the same trend at one month of age. At the three months’ time, we observed that more than 70% of the sequences were assigned to B. longum ssp. longum, B. breve, B. bifidum and B. pseudocatenulatum in descending order (Table S2). Absolute determinations corroborated those observations. B. longum and B. breve showed the highest counts, along with B. catenulatum during the first month of life. At three months of age, the count levels of B. longum and B. breve were followed by B. bifidum and B. catenulatum, in this order. B. dentium and B. adolescentis remained at relatively low levels by q-PCR (Table S2), despite showing high relative abundances by ITS-sequencing at some points.
Other than these general observations, clear differences in the bifidobacterial community composition and levels were observed because of the perinatal antibiotic administration, as is depicted below.
3.2. Gut Bifidobacterial Diversity Is Affected by Intrapartum Antibiotics
Neonates from the antibiotic group presented an increased number of coexisting bifidobacterial (sub)species at the different time points (11 ± 5 at 2 days, 11 ± 3 at 10 days, 12 ± 4 at 30 days, 10 ± 5 at 90 days, as average mean ± sd) in contrast to the average number of different species observed in the non-antibiotic control group (10 ± 6 at 2 days, 9 ± 5 at 10 days, 8 ± 3 at 30 days, 9 ± 5 at 90 days, mean ± sd). Those differences also reached statistical significance (p < 0.01) at one month of life.
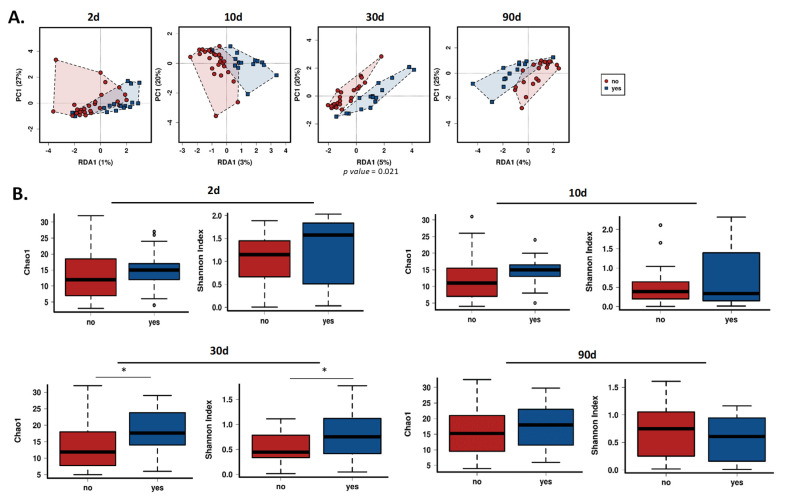
With the objective to assess how the intrapartum antibiotic treatment to mothers affects the structure of the gut bifidobacterial community, a multivariate redundant discriminant analysis (RDA) and two indices of alpha-diversity metrics were performed. Statistically significant differences were neither observed during the first days of life, nor at three months (Figure 1). However, at one month of life, both infants’ groups differed between them (RDA p < 0.05) (Figure 1A). Moreover, Chao1 and Shannon indices showed higher bifidobacterial alpha diversity in the group of babies whose mothers received antibiotics (p < 0.05) (Figure 1B).
Figure 1.
Gut bifidobacterial diversity. (A) Redundancy analysis (RDA) and (B) Chao1 and Shannon indexes between the control and antibiotic group of babies at 2, 10, 30 and 90 days of life. No: Control group (non-antibiotic); Yes: Antibiotic group. * Indicates statistically significant differences (p < 0.05).
3.3. Antibiotics Impact on the Bifidobacterium Species Establishment
It was recently described [63] that the delivery mode affects the bifidobacterial establishment. To investigate the impact of the mother’s antibiotic administration on the gut bifidobacterial composition independently of the mode of labour, we used an ANVOCA method controlling the delivery mode as covariable.
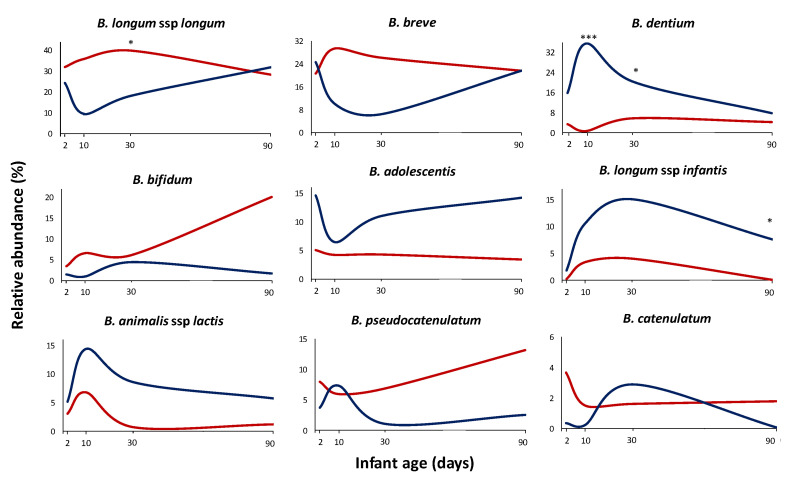
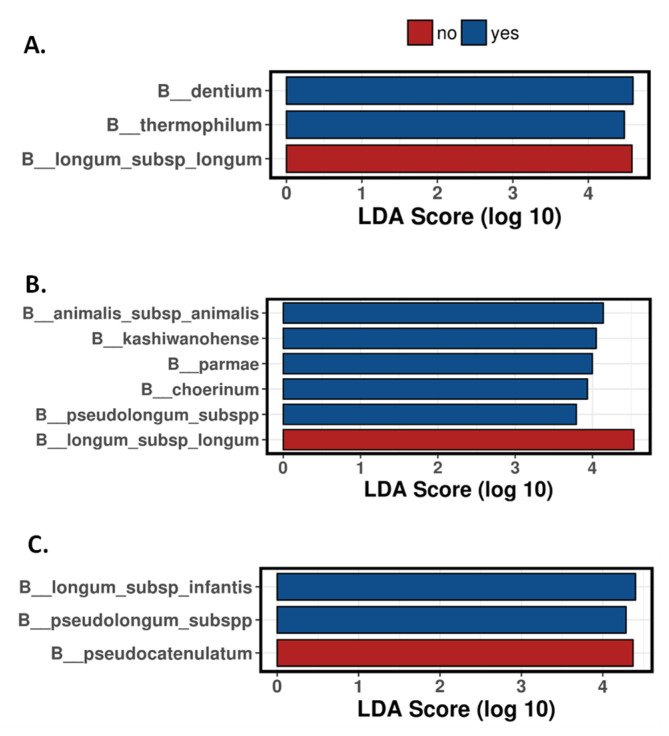
We could observe that the antibiotic administration to mothers in the perinatal time had an impact, at both qualitative and quantitative levels, on developing the bifidobacterial population in the neonate during the first months of life. Overall, the relative and absolute levels of the most abundant species, such as B. longum, B. breve, B. bifidum, or B. pseudocatenulatum, were negatively affected by the treatment, whilst B. dentium or B. adolescentis were increased, this getting statistical significance at some time points (Figure 2, Figure S1). Moreover, other (sub)species with low relative abundance also showed differences among the groups of babies whose mothers received antibiotics with respect to the control group of babies (no antibiotics administered to mothers) (Table S3). For example, B. pseudolongum ssp. pseudolongum was significantly increased (p < 0.05) in the antibiotic group (1.97% of relative abundance; 44% of occurrence) compared to the control group (0.23% of relative abundance; 30% of occurrence) on the second day of life. As with regard to the majority (sub)species (Figure 2) at 2 days of age, B. adolescentis showed increased proportions in antibiotic group infants with respect to the control group (relative abundances of 14.58% vs. 5.05% respectively; p = 0.068). At 10 days of age, B. dentium was significantly higher (p < 0.001) in the antibiotic group with respect to the control group (36% vs. 0.63% relative abundance, respectively) (Figure 2). A linear discriminant analysis effect size (LEfSe) (Figure 3) indicated that this bifidobacterial species was also the most discriminant (4.59 LDA score) between the two groups of babies established based on mother’s antibiotic treatment at ten days of age. We also observed that, although not reaching statistically significant differences (p = 0.093), B. breve decreased in the group of babies whose mothers received antibiotics (10% of relative average abundance) in comparison with the control group (29.48%) (Figure 2). The former observations were further confirmed when absolute levels of these species were determined by q-PCR (p < 0.05 for B. dentium; p = 0.085 for B. breve) (Figure S1).
Figure 2.
Bifidobacterial relative abundance. Average relative proportions of the dominant (sub)species during the first three months of life in the control group (red line) and antibiotic group (blue line). * Indicates statistically significant differences (* p < 0.05; *** p < 0.001) at the corresponding sampling times (2, 10, 30, or 90 days of age).
Figure 3.
Bifidobacterial composition. Linear discriminant analysis (LDA) scores of bifidobacterial species with differentially abundance between antibiotic and control groups at 10 (A), 30 (B) and 90 (C) days of age. No: Control group (non-antibiotic); Yes: Antibiotic group.
One month of life was the age where more differences between the two groups of babies were observed. B. longum ssp. longum showed significantly lower relative proportions (p < 0.05) in the IAP group (18% vs. 40% in the control group) (Figure 2), which also was confirmed by a reduction of absolute levels analysed by q-PCR (7.21 vs. 8.16 log10 cfu/g; p < 0.01) (Figure S1). This species also remained at this age as the species was more discriminant (4.53 LDA score) between the two groups of babies (Figure 3). In the same way, the absolute levels of B. breve were reduced in the antibiotic group of babies (5.87 vs. 6.96 log10 cfu/g; p < 0.05) (Figure S1), which was also reflected in its relative abundances (6.40 vs. 26.25% respectively; p = 0.097) (Figure 2). In the faecal samples of one-month old babies, the mother’s antibiotic treatment was found to promote higher presence and relative proportions of other species. Thus, B. dentium, with an 87.50% of occurrence in the antibiotic group and 66.67% in the control group, showed a significantly (p < 0.05) higher proportion in the antibiotic group (20.39 vs. 5.71%, respectively); B. animalis ssp. animalis was found to be in the 56.25% of the babies whose mothers received antibiotics (12.50% control group) and showed higher relative proportions in these babies (0.68 vs. 0.03%; p < 0.05); B. kashiwanogense, present in 56.25% of the antibiotic-babies (20.83% control group), also displayed higher proportions in this group (1.81 vs. 0.04%; p < 0.05), as well as B. mongoliense (0.13 vs. 0.00% relative abundance; p < 0.05), B. parmae (0.05 vs. 0.00% relative abundance; p < 0.01) and B. reuteri (0.04 vs. 0.00% relative abundance; p < 0.05) (Table S3).
After three months, some of these alterations seem to be reduced, although slight differences persisted (Figure 2, Table S3). B. pseudocatenulatum for the non-antibiotic group and B. longum ssp infantis for the antibiotic group remained as the most discriminant species between groups (Figure 3). Moreover, while ITS results do not reveal differences in B. bifidum between the two groups of babies, probably due to the intrinsic variability among individuals, q-PCR analyses still unveiled significantly lower levels (p < 0.05) of this species in antibiotic-treated babies at three months of age (Figure S1).
4. Discussion
This study analysed the effect of the IAP on the Bifidobacterium (sub)species establishment in full-term babies until three months of age. The ITS sequencing technique, which has been previously employed to assess other perinatal factors, such as gestational age, delivery and feeding mode [63], the effect of donated milk on the premature bifidobacterial community [66] or to study the vertical transmission of bifidobacteria [65], allowed us to thoroughly unveil the effect of the IAP—going a step beyond conventional studies which only used 16S sequencing information at the genus level. This technique can be used for tracking bifidobacterial (sub)species in a more accurate way than the use of other techniques. Moreover, the use of specific q-PCR methods for the most abundant bifidobacterial species allowed us to complete the study at a truly quantitative level. Thus, to the best of our knowledge, this is the first study encompassing qualitative and quantitative information about Bifidobacterium (sub)species establishment after the use of one of the most common clinical perinatal practices, the IAP.
The study of the microbiome during the last years laid the foundation for considering it as a vital organ or key factor for maintaining a healthy status. Moreover, it is known that its correct establishment at the beginning of life will entail consequences for later life [14,70]. Bifidobacterium is one of the dominant genera in the early microbiota and was recurrently linked to a healthy status [53,58]. Alterations on gut microbiota after IAP treatment were repeatedly observed by several authors. At the family and genus level, this antibiotic exposure was linked to the higher relative abundance of members of Clostridiaceae, Enterococcaceae, Campylobacteriaceae families and lower proportions of Bacteriodes and Bifidobacterium [45,46,47,48]. Moreover, it was observed that the duration of IAP administration exerts a major persistent negative impact on the bifidobacterial genus after 12 weeks of life [71]. Bifidobacterium genus is one of the microbial groups severely and negatively impacted by this prophylactic practice; however, its effect at the species level had not been previously explored.
In general, the dominant bifidobacterial species identified in this work, B. longum ssp. longum, B. breve, B. dentium, B. adolescentis, are in good agreement with those showed as dominant in other studies [63,72,73]. Our results demonstrated that IAP impacts the bifidobacterial community during the first months of life. A multivariate redundant discriminant analysis showed a clear separation between babies whose mothers were administered IAP with respect to the control group, which was evident and significant at one month of life. Moreover, we also found significantly higher alpha diversity in the bifidobacterial populations of the IAP-babies group at the age of one month. Our results are in contrast with that observed when the global gut microbiota community was studied, and the differences with respect to the IAP practice appear in the early time-points analysed, the microbiota becoming more uniform from the first month of life [45,46]. Our data, although interesting, are not surprising, since an increased bifidobacteria diversity when the scenario is not optimum for the correct gut microbiota establishment was previously reported [63,66]. Despite being an every-changing process, early-life gut microbiome development is characterised by low bacterial diversity, unlike that of adults [74,75], and dominance by certain Bifidobacterium species reflects a healthy status. However, our results show that when the correct gut microbiota establishment is affected by antibiotics, IAP in this case, the dominance of a few bifidobacterial (sub)species is altered, and the diversity is increased, favouring a non-natural gut environment. We are starting to understand that unfavourable situations early in life could entail changes in the gut microbiome from the whole structure to even at the species level.
IAP treatment also affects the bifidobacterial community at a compositional level. Although in the first days of life, slight differences could be observed among the two groups of infants, at 10 days, a clear differential pattern was evidenced. Thus, B. dentium was increased in the antibiotic group—this species was also discriminant for those babies, while B. breve showed lower levels in the antibiotic group. These differences become even larger at one month of age. Moreover, at this time, B. animalis spp. animalis was also significantly increased in babies whose mothers received antibiotics, whereas B. longum spp. longum was negatively affected by antibiotics. These results are slightly different to those reported by Aloisio et al., the sole work currently available focusing on IAP influence on bifidobacterial species to date [76]. These authors observed a lower diversity and lower abundance of B. breve, B. bifidum and B. dentium in babies at seven days of life by using the DGGE technique. These differences among both studies may be mainly due to the different techniques used in each case. However, our data suggest a pattern of Bifidobacterium species in concordance with that observed by other authors in other undesirable or unideal circumstances. Saturio et al. found lower relative proportions of B. longum spp. longum and B. breve in C-section babies and lower proportions of the former species in preterm neonates [63]. Nagpal et al., using a q-PCR method, also observed lower levels of B. longum in C-Section infants [77]. Moreover, when the gut bifidobacterial population were compared between premature babies fed with their own mother’s milk or donated milk, B. dentium appears to be significantly higher in babies fed with donated milk [66]. B. longum is the species that predominantly inhabit the human intestines [78]. It was shown in our study, and in concordance with other previously published, that when unideal perinatal conditions exist, this species is affected, and its abundance and levels are decreased. On the other hand, it seems that B. dentium has an advantage in those negative circumstances and increases its presence. At three months of age, despite the larger differences that appear to be recovered, specific species remain as discriminant signatures in each group of babies. It discloses that the effect of the IAP treatment on Bifidobacterium species persists over time.
Our results indicate that IAP treatment impacts intestinal bifidobacteria and that, in good agreement with previous studies, the bifidobacterial community is highly sensitive to suffer aberrancies by different factors commonly present early in life.
5. Conclusions
In summary, this study is among the first ones assessing the influence of the IAP in developing the bifidobacterial microbiota in the newborn during the first months of life by both qualitative and quantitative techniques. The data reveals the negative effect of the IAP in some of the most important communities in the early microbiota, such as bifidobacteria. Our results underline specific targets for developing gut microbiota-based products for favouring proper healthy microbial infant development when this clinical practice is required.
Acknowledgments
We would like to extend a warm thank you to all the “small” volunteers participating in the study and their families.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9091867/s1, Figure S1: Bifidobacterial levels. Average levels (log10 cfu/g) of the dominant species during the first three months of life in the control group (red line) and antibiotic group (blue line). * Indicates statistically significant differences (* p < 0.05, ** p < 0.01) at the corresponding sampling times (2, 10, 30, or 90 days of age). Table S1: Basal characteristics of the infant groups included in this study. Table S2: Relative abundance (%) and levels (log10 cfu/g) of bifidobacterial (sub)species during the first three months of life in the general population. Table S3: Bifidobacterial relative abundance. Average relative proportions of the minority (sub)species during the first three months of life.
Author Contributions
Conceptualisation, M.G., S.A., G.S.; volunteers’ recruitment, M.S., N.F., L.M. (Laura Mantecón) and G.S.; methodology, S.A., S.S. and L.M. (Leonardo Mancabelli); formal analysis, S.A.; writing—original draft preparation, S.A. and M.G.; reviewing, S.S., M.S., L.M. (Leonardo Mancabelli), N.F., L.M. (Laura Mantecón), C.G.d.l.R.-G., M.V., M.G., S.A. and G.S. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the Project AGL2017-83653R funded by the Spanish “Ministerio de Ciencia, Innovación y Universidades (MCIU), Agencia Estatal de Investigación (AEI) and FEDER” and by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 749255. S.A. was the recipient of a postdoctoral Juan de la Cierva Contract (Ministry of Science, Innovation and Universities, Ref. IJCI-2017-32156).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Regional Ethical Committee of Asturias Public Health Service (SESPA) (Ref. 51/18).
Informed Consent Statement
Informed written consent was obtained from each infant’s parents involved in the study.
Data Availability Statement
The raw sequences reported in this article were deposited in the NCBI-SRA under the accession number PRJNA750917. Other additional data presented in this study is available upon reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Simeis D., Serra S. Actinomycetes: A never-ending source of bioactive compounds-an overview on antibiotics production. Antibiotics. 2021;10:483. doi: 10.3390/antibiotics10050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey N., Cascella M. StatPearls. StatPearls Publishing LLC.; Treasure Island, FL, USA: 2021. Beta lactam antibiotics. StatPearls Publishing Copyright© 2021. [PubMed] [Google Scholar]
- 3.Van Dyke M.K., Phares C.R., Lynfield R., Thomas A.R., Arnold K.E., Craig A.S., Mohle-Boetani J., Gershman K., Schaffner W., Petit S., et al. Evaluation of universal antenatal screening for group B streptococcus. N. Engl. J. Med. 2009;360:2626–2636. doi: 10.1056/NEJMoa0806820. [DOI] [PubMed] [Google Scholar]
- 4.Nogacka A.M., Salazar N., Arboleya S., Suárez M., Fernández N., Solís G., de Los Reyes-Gavilán C.G., Gueimonde M. Early microbiota, antibiotics and health. Cell Mol. Life Sci. 2018;75:83–91. doi: 10.1007/s00018-017-2670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chai G., Governale L., McMahon A.W., Trinidad J.P., Staffa J., Murphy D. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics. 2012;130:23–31. doi: 10.1542/peds.2011-2879. [DOI] [PubMed] [Google Scholar]
- 6.Arboleya S., Suárez M., Fernández N., Mantecón L., Solís G., Gueimonde M., de Los Reyes-Gavilán C.G. C-section and the neonatal gut microbiome acquisition: Consequences for future health. Ann. Nutr. Metab. 2018;73:17–23. doi: 10.1159/000490843. [DOI] [PubMed] [Google Scholar]
- 7.Hagiya H., Kokado R., Ueda A., Okuno H., Morii D., Hamaguchi S., Yamamoto N., Yoshida H., Tomono K. Association of adverse drug events with broad-spectrum antibiotic use in hospitalized patients: A single-center study. Intern. Med. 2019;58:2621–2625. doi: 10.2169/internalmedicine.2603-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ianiro G., Tilg H., Gasbarrini A. Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 9.Gibson M.K., Crofts T.S., Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 2015;27:51–56. doi: 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wopereis H., Oozeer R., Knipping K., Belzer C., Knol J. The first thousand days—intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014;25:428–438. doi: 10.1111/pai.12232. [DOI] [PubMed] [Google Scholar]
- 11.O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morais L.H., Golubeva A.V., Moloney G.M., Moya-Pérez A., Ventura-Silva A.P., Arboleya S., Bastiaanssen T.F.S., O’Sullivan O., Rea K., Borre Y., et al. Enduring behavioral effects induced by birth by caesarean section in the mouse. Curr. Biol. 2020;30:3761–3774.e3766. doi: 10.1016/j.cub.2020.07.044. [DOI] [PubMed] [Google Scholar]
- 13.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 14.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., Belzer C., Delgado Palacio S., Arboleya Montes S., Mancabelli L., et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017;81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien M., Alvarez A.S., de Vos W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27:997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang L., Chen H., Zhang S., Zhuang J., Li Q., Feng Z. Intestinal microbiota in early life and its implications on childhood health. Genom. Proteom. Bioinform. 2019;17:13–25. doi: 10.1016/j.gpb.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Villares J.M., Collado M.C., Larqué E., Leis Trabazo R., Saenz De Pipaón M., Moreno Aznar L.A. The first 1000 days: An opportunity to reduce the burden of noncommunicable diseases. Nutr. Hosp. 2019;36:218–232. doi: 10.20960/nh.02453. [DOI] [PubMed] [Google Scholar]
- 19.Arboleya S., Binetti A., Salazar N., Fernández N., Solís G., Barranco A.H., Margolles A., Reyes-Gavilan C.D.L., Gueimonde M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2011;79:763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 20.Matamoros S., Guen C.G.-L., Le Vacon F., Potel G., de La Cochetiere M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Houghteling P.D., Walker W.A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 2015;60:294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arboleya S., Sanchez B., Milani C., Duranti S., Solis G., Fernandez N., de los Reyes-Gavilan C.G., Ventura M., Margolles A., Gueimonde M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 2015;166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 23.Isolauri E., Rautava S., Salminen S., Collado M.C. Early-life nutrition and microbiome development. Nestle. Nutr. Inst. Workshop Ser. 2019;90:151–162. doi: 10.1159/000490302. [DOI] [PubMed] [Google Scholar]
- 24.Zeissig S., Blumberg R.S. Life at the beginning: Perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 2014;15:307–310. doi: 10.1038/ni.2847. [DOI] [PubMed] [Google Scholar]
- 25.Nobel Y.R., Cox L.M., Kirigin F.F., Bokulich N.A., Yamanishi S., Teitler I., Chung J., Sohn J., Barber C.M., Goldfarb D.S., et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaser M.J. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D., et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lach G., Fülling C., Bastiaanssen T.F.S., Fouhy F., Donovan A.N.O., Ventura-Silva A.P., Stanton C., Dinan T.G., Cryan J.F. Enduring neurobehavioral effects induced by microbiota depletion during the adolescent period. Transl. Psychiatry. 2020;10:1–16. doi: 10.1038/s41398-020-01073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Mahony S.M., Felice V.D., Nally K., Savignac H.M., Claesson M.J., Scully P., Woznicki J., Hyland N.P., Shanahan F., Quigley E.M., et al. Disturbance of the gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience. 2014;277:885–901. doi: 10.1016/j.neuroscience.2014.07.054. [DOI] [PubMed] [Google Scholar]
- 30.Leclercq S., Mian F.M., Stanisz A.M., Bindels L.B., Cambier E., Ben-Amram H., Koren O., Forsythe P., Bienenstock J. Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nat. Commun. 2017;8:15062. doi: 10.1038/ncomms15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell S.L., Gold M.J., Willing B.P., Thorson L., McNagny K.M., Finlay B.B. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pascal M., Perez-Gordo M., Caballero T., Escribese M.M., Lopez Longo M.N., Luengo O., Manso L., Matheu V., Seoane E., Zamorano M., et al. Microbiome and allergic diseases. Front. Immunol. 2018;9:1584. doi: 10.3389/fimmu.2018.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vercelli D. Microbiota and human allergic diseases: The company we keep. Curr. Opin. Immunol. 2021;72:215–220. doi: 10.1016/j.coi.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murk W., Risnes K.R., Bracken M.B. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: A systematic review. Pediatrics. 2011;127:1125–1138. doi: 10.1542/peds.2010-2092. [DOI] [PubMed] [Google Scholar]
- 35.Sander S.D., Andersen A.-M.N., Murray J.A., Karlstad Ø., Husby S., Størdal K. Association between antibiotics in the first year of life and celiac disease. Gastroenterology. 2019;156:2217–2229. doi: 10.1053/j.gastro.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 36.Leonard M.M., Valitutti F., Karathia H., Pujolassos M., Kenyon V., Fanelli B., Troisi J., Subramanian P., Camhi S., Colucci A., et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc. Natl. Acad. Sci. USA. 2021;118:e2020322118. doi: 10.1073/pnas.2020322118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelimo C., Camargo C.A., Jr., Morton S.M.B., Grant C.C. Association of repeated antibiotic exposure up to age 4 years with body mass at age 4.5 years. JAMA Netw. Open. 2020;3:e1917577. doi: 10.1001/jamanetworkopen.2019.17577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox L.M., Blaser M.J. Antibiotics in early life and obesity. Nat. Rev. Endocrinol. 2015;11:182–190. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saari A., Virta L.J., Sankilampi U., Dunkel L., Saxen H. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics. 2015;135:617–626. doi: 10.1542/peds.2014-3407. [DOI] [PubMed] [Google Scholar]
- 40.Trasande L., Blustein J., Liu M., Corwin E., Cox L.M., Blaser M.J. Infant antibiotic exposures and early-life body mass. Int. J. Obes. 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kronman M.P., Zaoutis T.E., Haynes K., Feng R., Coffin S.E. Antibiotic exposure and IBD development among children: A population-based cohort study. Pediatrics. 2012;130:e794–e803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bokulich N.A., Chung J., Battaglia T., Henderson N., Jay M., Li H., Lieber A.D., Wu F., Perez-Perez G.I., Chen Y., et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstantinidis T., Tsigalou C., Karvelas A., Stavropoulou E., Voidarou C., Bezirtzoglou E. Effects of antibiotics upon the gut microbiome: A review of the literature. Biomedicines. 2020;8:502. doi: 10.3390/biomedicines8110502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vangay P., Ward T., Gerber J.S., Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;17:553–564. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nogacka A., Salazar N., Suarez M., Milani C., Arboleya S., Solis G., Fernandez N., Alaez L., Hernandez-Barranco A.M., de Los Reyes-Gavilan C.G., et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome. 2017;5:1–10. doi: 10.1186/s40168-017-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazzola G., Murphy K., Ross R.P., Di Gioia D., Biavati B., Corvaglia L.T., Faldella G., Stanton C. Early gut microbiota perturbations following intrapartum antibiotic prophylaxis to prevent group B Streptococcal Disease. PLoS ONE. 2016;11:e0157527. doi: 10.1371/journal.pone.0157527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corvaglia L., Tonti G., Martini S., Aceti A., Mazzola G., Aloisio I., Di Gioia D., Faldella G. Influence of intrapartum antibiotic prophylaxis for group b streptococcus on gut microbiota in the first month of life. J. Pediatr. Gastroenterol. Nutr. 2016;62:304–308. doi: 10.1097/MPG.0000000000000928. [DOI] [PubMed] [Google Scholar]
- 48.Tapiainen T., Koivusaari P., Brinkac L., Lorenzi H.A., Salo J., Renko M., Pruikkonen H., Pokka T., Li W., Nelson K., et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-46964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azad M.B., Konya T., Persaud R.R., Guttman D.S., Chari R.S., Field C.J., Sears M.R., Mandhane P.J., Turvey S.E., Subbarao P., et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG. 2016;123:983–993. doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 50.Coker M.O., Hoen A.G., Dade E., Lundgren S., Li Z., Wong A.D., Zens M.S., Palys T.J., Morrison H.G., Sogin M.L., et al. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: A prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2020;127:217–227. doi: 10.1111/1471-0528.15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ainonen S., Tejesvi M.V., Mahmud M.R., Paalanne N., Pokka T., Li W., Nelson K.E., Salo J., Renko M., Vänni P., et al. Antibiotics at birth and later antibiotic courses: Effects on gut microbiota. Pediatric Res. 2021:1–9. doi: 10.1038/s41390-021-01494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar H., Collado M.C., Wopereis H., Salminen S., Knol J., Roeselers G. The bifidogenic effect revisited-ecology and health perspectives of bifidobacterial colonization in early life. Microorganisms. 2020;8:1855. doi: 10.3390/microorganisms8121855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arboleya S., Watkins C., Stanton C., Ross R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alessandri G., van Sinderen D., Ventura M. The genus bifidobacterium: From genomics to functionality of an important component of the mammalian gut microbiota running title: Bifidobacterial adaptation to and interaction with the host. Comput. Struct. Biotechnol. J. 2021;19:1472–1487. doi: 10.1016/j.csbj.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada C., Gotoh A., Sakanaka M., Hattie M., Stubbs K.A., Katayama-Ikegami A., Hirose J., Kurihara S., Arakawa T., Kitaoka M., et al. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome bifidobacterium longum. Cell Chem. Biol. 2017;24:515–524.e515. doi: 10.1016/j.chembiol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill C.J., Lynch D.B., Murphy K., Ulaszewska M., Jeffery I.B., O’Shea C.A., Watkins C., Dempsey E., Mattivi F., Tuohy K., et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017;5:1–18. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henrick B.M., Rodriguez L., Lakshmikanth T., Pou C., Henckel E., Arzoomand A., Olin A., Wang J., Mikes J., Tan Z., et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 59.Gao X., Jia R., Xie L., Kuang L., Feng L., Wan C. Obesity in school-aged children and its correlation with gut E.coli and Bifidobacteria: A case-control study. BMC Pediatr. 2015;15:1–4. doi: 10.1186/s12887-015-0384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madan J.C., Salari R.C., Saxena D., Davidson L., Toole G.A., Moore J.H., Sogin M.L., Foster J.A., Edwards W.H., Palumbo P., et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child.-Fetal Neonatal Ed. 2012;97:F456. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mai V., Torrazza R.M., Ukhanova M., Wang X., Sun Y., Li N., Shuster J., Sharma R., Hudak M.L., Neu J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE. 2013;8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Weerth C., Fuentes S., Puylaert P., de Vos W.M. Intestinal microbiota of infants with colic: Development and specific signatures. Pediatrics. 2013;131:e550–e558. doi: 10.1542/peds.2012-1449. [DOI] [PubMed] [Google Scholar]
- 63.Saturio S., Nogacka A.M., Suárez M., Fernández N., Mantecón L., Mancabelli L., Milani C., Ventura M., de los Reyes-Gavilán C.G., Solís G., et al. Early-life development of the bifidobacterial community in the infant gut. Int. J. Mol. Sci. 2021;22:3382. doi: 10.3390/ijms22073382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milani C., Lugli G.A., Turroni F., Mancabelli L., Duranti S., Viappiani A., Mangifesta M., Segata N., van Sinderen D., Ventura M. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol. Ecol. 2014;90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 65.Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K., Ferretti P., Gorfer V., Ferrario C., Milani C., et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:1–13. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arboleya S., Saturio S., Suárez M., Fernández N., Mancabelli L., de Los Reyes-Gavilán C.G., Ventura M., Solís G., Gueimonde M. Donated human milk as a determinant factor for the gut bifidobacterial ecology in premature babies. Microorganisms. 2020;8:760. doi: 10.3390/microorganisms8050760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zakrzewski M., Proietti C., Ellis J.J., Hasan S., Brion M.-J., Berger B., Krause L. Calypso: A user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerperien J., Schouten B., Boehm G., Willemsen L., Garssen J., Knippels L., Land B. Development of the immune system—early nutrition and consequences for later life. In: Kanwar J.R., editor. Recent Advances in Immunology to Target Cancer, Inflammation and Infections. InTech; London, UK: 2012. pp. 315–334. [Google Scholar]
- 71.Stearns J.C., Simioni J., Gunn E., McDonald H., Holloway A.C., Thabane L., Mousseau A., Schertzer J.D., Ratcliffe E.M., Rossi L., et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-16606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy K., O’Shea C.A., Ryan C.A., Dempsey E.M., O’ Toole P.W., Stanton C., Ross R.P. The gut microbiota composition in dichorionic triplet sets suggests a role for host genetic factors. PLoS ONE. 2015;10:e0122561. doi: 10.1371/journal.pone.0122561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C., Mangifesta M., Viappiani A., Ferretti P., Gorfer V., et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsen O.F.A., Claassen E. The mechanistic link between health and gut microbiota diversity. Sci. Rep. 2018;8:1–5. doi: 10.1038/s41598-018-20141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kriss M., Hazleton K.Z., Nusbacher N.M., Martin C.G., Lozupone C.A. Low diversity gut microbiota dysbiosis: Drivers, functional implications and recovery. Curr. Opin. Microbiol. 2018;44:34–40. doi: 10.1016/j.mib.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aloisio I., Mazzola G., Corvaglia L.T., Tonti G., Faldella G., Biavati B., Di Gioia D. Influence of intrapartum antibiotic prophylaxis against group B Streptococcus on the early newborn gut composition and evaluation of the anti-Streptococcus activity of Bifidobacterium strains. Appl. Microbiol. Biotechnol. 2014;98:6051–6060. doi: 10.1007/s00253-014-5712-9. [DOI] [PubMed] [Google Scholar]
- 77.Nagpal R., Kurakawa T., Tsuji H., Takahashi T., Kawashima K., Nagata S., Nomoto K., Yamashiro Y. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-10711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Odamaki T., Bottacini F., Kato K., Mitsuyama E., Yoshida K., Horigome A., Xiao J.Z., van Sinderen D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequences reported in this article were deposited in the NCBI-SRA under the accession number PRJNA750917. Other additional data presented in this study is available upon reasonable request from the corresponding author.