Abstract
Yin Yang 1 (YY1) is a zinc finger-containing transcription factor and a target of viral oncoproteins. To determine the biological role of YY1 in mammalian development, we generated mice deficient for YY1 by gene targeting. Homozygosity for the mutated YY1 allele results in embryonic lethality in the mouse. YY1 mutants undergo implantation and induce uterine decidualization but rapidly degenerate around the time of implantation. A subset of YY1 heterozygote embryos are developmentally retarded and exhibit neurulation defects, suggesting that YY1 may have additional roles during later stages of mouse embryogenesis. Our studies demonstrate an essential function for YY1 in the development of the mouse embryo.
Yin Yang 1 (YY1) is a multifunctional transcription factor that can act as a transcriptional repressor, an activator, or an initiator element-binding protein that directs and initiates transcription in vitro (8, 12, 22, 25, 27). Recent studies have focused on mechanisms by which YY1 regulates transcription and have identified repression and activation domains in YY1 (2, 9, 17, 27) as well as interactions of YY1 with coactivators and corepressors (16, 34). These findings have suggested potential molecular mechanisms that may underlie the ability of YY1 to regulate transcription but have not elucidated how these molecular events contribute to the biological activities of YY1.
Previous studies have shown that YY1 is a target of the adenovirus E1A oncoprotein (27). Mutations that abrogate the ability of E1A to induce oncogenic transformation also disrupt the ability of E1A to regulate YY1 (16), suggesting that YY1 is likely to play a role in cell proliferation. Studies performed with cell culture systems suggest that YY1 might also play a role in differentiation in multiple cell types (reviewed in references 26 and 28). In addition, although YY1 appears to regulate many genes that encode proteins with diverse biological activities, the genes that have been shown to be repressed by YY1 are largely associated with differentiation (reviewed in references 26 and 28). Taken together, these in vitro studies suggest a global role of YY1 in the regulation of differentiation and cell proliferation, possibly in a variety of cell types. These studies further predict that YY1 might play a crucial and exciting role in the development of higher organisms such as the mouse. However, the in vivo function of mammalian YY1 remains unclear to date.
To address the role of YY1 in vivo, we disrupted one YY1 allele in mouse embryonic stem (ES) cells by homologous recombination and generated mice harboring the mutant YY1 allele. Homozygosity for the mutant YY1 allele results in embryonic lethality in the mouse. By genotyping embryos at different gestational times, we identified YY1−/− embryos at the blastocyst stage. The YY1-deficient embryos were implanted in the uterine tissue but failed to develop to the gastrulation stage, resulting in embryonic death around the time of implantation. These findings suggest that YY1 plays an indispensable role in early mouse embryogenesis. In addition, a subset of YY1 heterozygotes display growth retardation and neurulation defects. Although YY1 is ubiquitously expressed, significantly higher levels of YY1 mRNA are detected in the somites, limb bud, and tail tip. The phenotype of these YY1 heterozygotes and the enriched YY1 expression in selective tissues together suggest that YY1 is likely to play a role in later stages of mouse embryonic development, such as organogenesis. Our findings demonstrate for the first time a critical role for YY1 at multiple stages during mouse embryogenesis.
MATERIALS AND METHODS
Disruption of the YY1 gene.
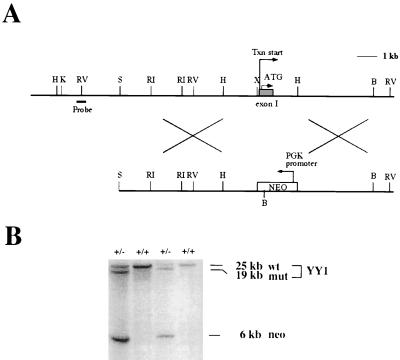
An 18-kb mouse YY1 genomic DNA was isolated by screening a 129/Sv mouse genomic library (Stratagene) with the murine YY1 cDNA (12). This fragment contains the entire YY1 exon I as well as the sequences 11 and 6 kb 5′ and 3′ to exon I, respectively. The targeting vector was constructed by replacing the XhoI/HindIII fragment containing the entire exon I and the promoter-proximal region important for YY1 transcription (24) with the bacterial neomycin gene driven by the phosphoglycerate kinase 1 promoter pgk-neo expression cassette. To disrupt the YY1 gene, the J1 ES cell line was electroporated with the linearized targeting vector DNA and neomycin-resistant colonies were selected in G418-containing medium (18). These colonies were expanded and screened for homologous recombination at the YY1 locus by Southern blot hybridization with an external YY1 probe shown in Fig. 1A. Four positive clones were identified from a total of 96 colonies and confirmed by hybridization with a 600-bp PstI neo fragment isolated from the pgk-neo vector. One such YY1+/− ES clone was expanded and microinjected into C57BL/6 blastocysts to obtain chimeric mice as determined by agouti coat color. Male chimeras with germ line transmission were subsequently bred either to 129/Sv females to establish an inbred strain or to C57BL/6 females to establish hybrid F1 progeny. Tail biopsy specimens were obtained from pups 3 to 4 weeks of age for genotyping.
FIG. 1.
Generation of a targeted mutation at the mouse YY1 locus and identification of the YY1 mutant allele. (A) Genomic organization of the mouse YY1 locus and design of the targeting construct. An 18-kb YY1 fragment was obtained from a genomic library of the mouse strain 129/Sv. The YY1 fragment targeted for gene replacement contains exon I and the sequences 11 kb 5′ and 6 kb 3′ to exon I. A XhoI/HindIII fragment (approximately 2 kb) containing the entire exon I including both the transcription and the translational start sites was replaced with the neo cassette. The arrow marked “Txn start” denotes the transcriptional start site. The arrow labeled “ATG” denotes the translational start site. The probe used in Southern hybridization to detect homologous recombination is indicated by a thick bar and labeled as such. The size of a 1-kb sequence is also indicated. B, BamHI; H, HindIII; K, KpnI; RV, EcoRV; S, SacI; X, XhoI; RI, EcoRI; PGK, phosphoglycerate kinase. (B) Identification of ES cells containing a mutant YY1 allele. The YY1 targeting vector was introduced into ES cells and selected with G418. Southern blot hybridization was performed on genomic DNA extracted from ES cells, digested with BamHI, and hybridized to a radioactively labeled neo and external YY1 probe as shown in panel A. Wild-type (wt) ES cells contain a single 25-kb YY1 fragment, while YY1+/− ES cells contain the 25-kb and the mutant (mut) 19-kb YY1 fragment. The 6-kb fragment containing the neo gene is detected for the YY1+/− ES cells only. The genotypes of the ES cells are indicated above the lanes. The size of the hybridized bands is indicated on the right.
As an alternative to genotyping by Southern hybridization, PCR was performed on DNA isolated from early-stage embryos, blastocyst outgrowths, or tail biopsy specimens. For instance, to genotype blastocysts, single, flushed blastocysts were suspended in 20 μl of lysis buffer (50 mM Tris [pH 8.0], 0.5% Triton X-100, and 200 μg of proteinase K per ml). DNA samples were incubated overnight at 50°C and heated for 5 min at 95°C. PCR amplification with the neo primers yielded a 500-bp fragment, and the YY1 primers yielded a 211-bp PCR product. In some cases, visualization of the PCR products was aided by Southern blot hybridization with [γ-32P]ATP-labeled neo (neo-forward) or YY1 probes. Primers used for PCRs were the following: neo-forward, 5′-ATGAACTGCAGGACGAGGCAGCG-3′; neo-reverse, 5′-GGCGATAGAAGGCGATGCGCTG-3′; YY1 forward, 5′-TCGCGCTGCAGCCGCTGGTGAC-3′; YY1 reverse, 5′-CGCCACGGTGACCAGCGTCTGC-3′; YY1 probe, 5′-CACCAGGATCACCTCCTGGTGGTGGTGCAC-3′.
Histological and immunohistochemical analysis of embryos.
Uteri were obtained from YY1 heterozygous intercrosses, resuspended in phosphate-buffered saline, fixed overnight in 4% paraformaldehyde at 4°C, dehydrated, and embedded in paraffin as described by Kaufman (14). Serial sections were cut at a 7-μm thickness and stained with hematoxylin and eosin.
Confocal analysis of preimplantation embryos.
Preimplantation embryos were obtained from wild-type, superovulated females crossed with wild-type males as described below at 12, 36, and 93 h postcoitum to obtain 1-cell, 2-cell, and blastocyst (60-cell)-stage embryos, respectively. A whole-mount immunofluorescence assay was performed as described by Palmieri et al. (21). Embryos were fixed in freshly prepared 4% paraformaldehyde, permeabilized, blocked for nonspecific antibody staining, and incubated with primary antibody. Primary antibodies used were rabbit polyclonal anti-TATA-binding protein (TBP) (Sl-1, 1:25 dilution; Santa Cruz), rabbit anti-Sp1 (PEP 2, 1:25 dilution; Santa Cruz), monoclonal anti-YY1 1A12 (1:5 dilution, a gift from Anny Usheva), and affinity-purified rabbit anti-YY1 (1:150 dilution) (16). Secondary antibodies used were fluorescein 5-isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) (Cappel; 1:150 dilution), Texas red-conjugated goat anti-rabbit IgG (1:100 dilution; Cappel), and FITC-conjugated goat anti-rabbit IgG (1:500 dilution; Cappel). DNA was visualized by propidium iodide-DAPCO (1,4-diazabicyclo-[2,2,2]-octane). The embryos were analyzed on a Zeiss laser scanning confocal microscope with an excitation wavelength of 568 nm from an argon-ion laser for Texas red-conjugated antibodies and 488 nm from a helium-neon laser for FITC-conjugated secondary antibodies.
Whole-mount in situ analysis of YY1 mRNA expression.
The YY1 riboprobe was constructed by digesting the murine YY1/δ cDNA with SacI (12) and religating this plasmid to generate YY1delSacI. This construct spans YY1 amino acids 1 to 337. The antisense RNA probe was transcribed with the T7 polymerase from the YY1delSacI plasmid linearized with EcoRI, while the sense RNA probe was transcribed with the SP6 polymerase from the same template DNA linearized with HindIII. The above riboprobes were generated with digoxigenin-UTP (Boehringer Mannheim). To obtain embryos of various developmental stages, 129/B6 mice were mated overnight and the morning of vaginal plug detection was defined as 0.5 days of gestation. Embryos were dissected, fixed in 4% paraformaldehyde plus 0.1% Tween 20 in phosphate-buffered saline, and stored overnight at 4°C. Whole-mount in situ hybridization with the digoxigenin-labeled riboprobes (Boehringer Mannheim) was performed as described elsewhere (32).
RESULTS
Generation of mice lacking a functional YY1 gene.
We isolated a YY1 genomic DNA clone from a mouse strain 129/Sv genomic library. This YY1 genomic fragment contains the entire exon I, which represents more than 50% of the YY1 coding region (227 of 414 amino acids). As shown in Fig. 1A, exon I as well as the proximal promoter region of YY1 essential for YY1 gene transcription (24, 35) was replaced by the bacterial neomycin resistance (Neor) gene. The promoter and the translational start site of YY1 were removed to minimize the possibility of residual transcription or translation that might yield a truncated YY1 protein. The linearized targeting vector was electroporated into J1 ES cells (18) and subjected to G418 selection. Of 96 G418-resistant ES colonies analyzed for homologous recombination, 4 displayed YY1 gene replacement. A representative Southern blot with both the external YY1 and neo probes that detected YY1+/− ES cells is shown in Fig. 1B. One of these YY1+/− ES cells was expanded and microinjected into C57BL/6 female blastocysts. Chimeric mice obtained were then bred to C57BL/6 and 129/Sv females to produce heterozygous mice. Male and female heterozygous YY1+/− mice were fertile and appeared phenotypically normal as observed over a year, with the exception of a small subset (see below).
Disruption of YY1 results in early embryonic lethality.
Heterozygous YY1 mice were mated, and genotypes of newborn offspring were determined by Southern blot analysis or PCR (detailed in Materials and Methods) of biopsied tail samples. Of 58 newborn animals genotyped from 12 independent litters, 21 were YY1+/+, 37 were YY1+/−, and none were YY1−/− (Table 1), indicating a Mendelian ratio typical for an embryonic lethal phenotype (1:2:0). To determine the time of death during embryogenesis, embryos obtained from heterozygous intercrosses were dissected at various gestational times, and DNA was isolated and analyzed by Southern blot hybridization (Fig. 1B) or PCR (data not shown). As shown in Table 1, embryos at various developmental stages that could be genotyped were either wild type or heterozygous for the YY1 allele. The resorbed embryos in the empty decidual sacs had little embryonic material for genotyping and therefore were assumed to be YY1−/− based on the Mendelian distribution. For instance, at embryonic day 8.5 (E8.5, mid-somite stage), all decidua obtained from YY1 heterozygous matings were externally indistinguishable from those of wild-type littermates, but dissection revealed that 23 of 107 (22%) implantation sites lacked discernible embryos, yolk sacs, or ectoplacental cones, suggesting that these defective embryos were likely to be YY1−/−. Embryos from E7.5 to E5.0 were studied by histological analysis. These results show that embryonic defects can be detected as early as E5.0 (see below).
TABLE 1.
Genotyped offspring from YY1 heterozygous intercrosses
| Age | No. of litters | No. of normal embryos | No. of resorbed embryos | No. with YY1 genotype:
|
||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | ||||
| Postnatal | 12 | 58 | NAa | 21 | 37 | 0 |
| E12.5 | 9 | 60 | 5 | 12 | 38 | 0 |
| E11.5 | 5 | 28 | 4 | 8 | 20 | 0 |
| E10.5 | 6 | 32 | 11 | 12 | 20 | 0 |
| E9.5 | 7 | 42 | 18 | 10 | 32 | 0 |
| E8.5 | 14 | 84 | 23 | 22 | 62 | 0 |
| E3.5 | 12 | 116 | 23 | 72 | 21 | |
NA, not available.
To determine whether we could identify YY1−/− embryos prior to E5.0, we isolated E3.5 blastocysts directly from heterozygous intercrosses by flushing uteri. These blastocysts were cultured in vitro for 5 to 7 days, and DNAs were isolated for genotyping. As shown in Table 1, of a total of 116 blastocyst outgrowths genotyped, 23 were YY1+/+, 72 were YY1+/−, and 21 were YY1−/−. We also identified YY1−/− blastocysts generated from cultured, fertilized one-cell embryos in KSOMAA medium supplemented with amino acids (15), a result consistent with the examination of the blastocysts directly flushed from the uteri (data not shown). Our ability to detect YY1−/− blastocysts isolated in utero suggests that zygotic YY1 is not essential for preimplantation cell viability. The majority of the cultured blastocysts were phenotypically normal with the formation of inner cell mass and trophoblast giant cells (data not shown). Taken together, these findings suggest that YY1−/− blastocysts are capable of implantation but die shortly thereafter.
Developmental deficiencies in YY1−/− embryos occur at peri-implantation.
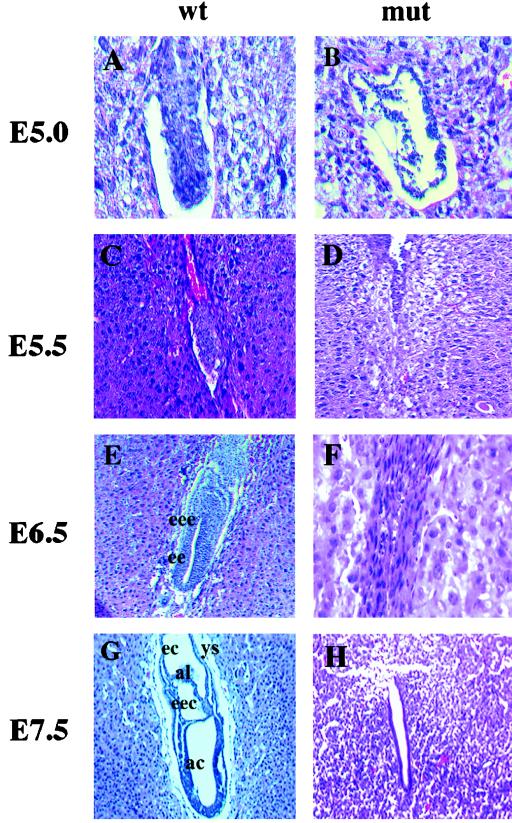
To examine the tissue defects and the appearance of the mutant embryos, in utero histological sections were obtained from YY1 heterozygous crosses at E5.0, 5.5, 6.5, and 7.5. As shown in Fig. 2, normal embryonic development at E5.0 (Fig. 2A) and E5.5 (Fig. 2C) is demonstrated by the proliferative expansion of the embryo. During the egg cylinder stage at E6.5, growth and elongation of the embryo mark the differentiation of the visceral and parietal endoderm and yolk sac formation (Fig. 2E). Further embryonic differentiation is observed at E7.5 as the formation of the primitive streak and mesoderm (Fig. 2G). As shown in Fig. 2, in contrast to the normal morphogenic events seen for YY1+/+ and YY1+/− embryos, the presumptive YY1−/− embryos (12 of 46 embryos examined) exhibit a decidual reaction (demonstrated by a swollen and edematous endometrium as a result of implantation) but show a decreased proliferation, as evidenced by a decreased number of cells and disorganized embryos (compare Fig. 2B with A and 2D with C). These mutants fail to form a distinctive egg cylinder (Fig. 2F; compare with 2E) and reach nearly complete resorption by the time of gastrulation at E7.5 (Fig. 2H) and complete resorption at E8.5 and onwards (data not shown). These results corroborate our above findings (Table 1) that approximately 25% of dissected decidual swellings have resorbed embryos. Therefore, YY1−/− embryos elicit a decidual response and invade the uterine epithelium by attachment to the basement membrane but fail to differentiate to form egg cylinders prior to resorption.
FIG. 2.
Histological examination of in utero embryos obtained from YY1 heterozygous matings. The uteri of female YY1+/− mice were dissected between 5.0 and 7.5 days after intercross mating as described in Materials and Methods. All uterine decidua (wild-type and mutant) were sectioned transversely at a 7-μm thickness and stained with hematoxylin and eosin. Wild-type or heterozygous embryos are shown in the column labeled wt, and presumptive YY1−/− mutants are shown in the column labeled mut. (A and B) E5.0, early egg cylinder stage; (C and D) E5.5, egg cylinder stage; (E and F) E6.5, late egg cylinder stage; (G and H) E7.5, late primitive streak stage. The wild-type embryos show a differentiating early egg cylinder stage embryo at E5.0 (A) and E5.5 (C), whereas the mutant embryos show decreased proliferation and a lack of organized embryos (B and D) at these embryonic stages. Note the appearance of a proamniotic cavity in the E6.5 elongated late-egg-cylinder wild-type embryo (E) including the formation of the embryonic ectoderm (ee) and extraembryonic ectoderm (eee) and the lack of these clearly differentiated tissues in the mutant embryos (F). Wild-type embryos at the late primitive streak stage (G) show clearly defined yolk sac (ys), ectoplacental cavity (ec), extraembryonic coelomic cavity (eec), amniotic cavities (ac), and allantois (al). In contrast, the presumptive YY1−/− mutant at E7.5 is nearly completely resorbed as shown by the appearance of an empty uterine crypt (H).
YY1 expression in early mouse development.
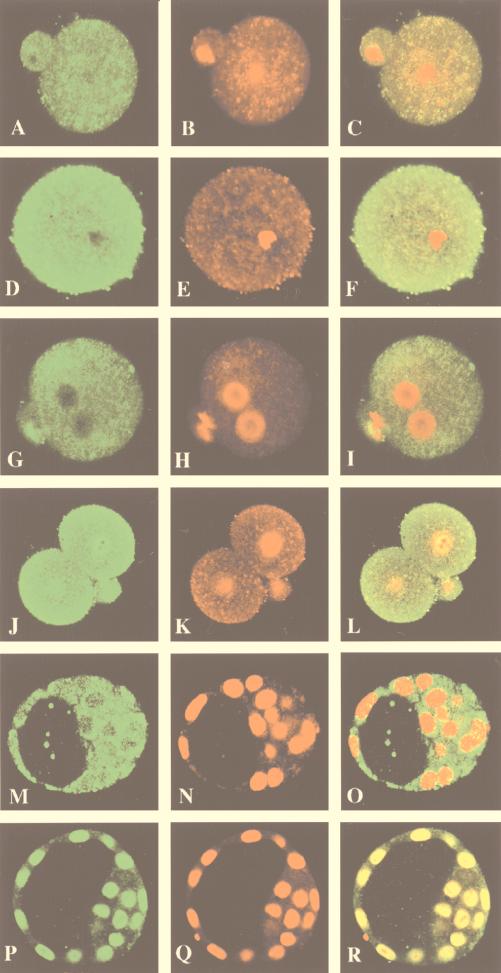
The peri-implantation defects of the YY1−/− embryos prompted us to examine YY1 protein expression in early mouse embryos by confocal laser microscopy. Immunostaining was done with either affinity-purified polyclonal YY1 antibodies (16) or a monoclonal YY1 antibody (a gift from Anny Usheva), and identical results were obtained. As negative controls, we applied either primary or secondary antibodies alone to the embryos and found virtually no staining (data not shown). As shown in Fig. 3A, we found YY1 expression in the one-cell unfertilized oocyte, suggesting that YY1 may be a maternally derived protein. Figure 3C shows a superimposed image of YY1 signal (green) and DNA staining by propidium iodide (red). Note the DNA staining in the pronucleus and polar body and the lack of YY1 signal in the nucleus. As a positive control, we show that TBP is highly expressed in the one-cell, unfertilized oocyte (Fig. 3D) as previously described (33). YY1 protein is also prominently expressed in the one-cell, fertilized (Fig. 3G), two-cell (Fig. 3J), and blastocyst (Fig. 3M) embryos. YY1 signal becomes detectable in the nucleus of the two-cell embryos as shown by the yellow signal (Fig. 3L), which represents the coincidence of the YY1 signal (green) and the DNA (red). Interestingly, the appearance of the YY1 signal in the nucleus at the two-cell stage coincides with the onset of zygotic transcription (reviewed in reference 20) (compare Fig. 3I and L). In E3.5 blastocysts, YY1 protein is detected in both the inner cell mass and the trophectoderm (Fig. 3M), which later give rise to the embryo itself and contribute to the placental tissue, respectively. YY1 protein shows nuclear (see superimposition in Fig. 3L and O) as well as cytoplasmic expression in both the inner cell mass and the trophectodermal cells similar to that described for TBP (33). This is in contrast to cultured cells in which YY1 shows a predominant nuclear staining pattern. The significance of the cytoplasmic localization of YY1, if any, is currently unclear. Sp1 expression is mainly restricted to the nucleus, commencing at the two-cell embryonic stage and in the blastocyst stage (Fig. 3P) as previously described (33). Interestingly, while Sp1 seems uniformly distributed in the nuclei, YY1 staining in the nuclei of the blastocysts appears punctate. These results show that YY1 is expressed at the earliest stage of mouse development, in both the inner cell mass and the trophectoderm of the blastocyst.
FIG. 3.
Expression of YY1 protein in wild-type murine preimplantation embryos. Immunofluorescent detection of YY1 protein was performed on wild-type preimplantation embryos by confocal microscopy. Embryos were stained with monoclonal and polyclonal (not shown) anti-YY1 antibodies and polyclonal anti-TBP and anti-Sp1 antibodies. The leftmost column represents embryos stained with these primary antibodies and visualized with FITC-conjugated secondary antibodies. The middle column represents the same embryos stained with propidium iodide that visualizes DNA. The rightmost column shows superimposed images of the antibody and DNA staining. (A to C) YY1 expression in a one-cell unfertilized oocyte; (D to F) TBP expression in a one-cell unfertilized oocyte; (G to I) YY1 expression in a one-cell fertilized embryo; (J to L) YY1 expression in a two-cell embryo. Note the pronounced YY1 expression in the nucleus as shown by the intense yellow signal as a result of the colocalization of the YY1 signal (green) and the DNA signal (red) (L). (M to O) YY1 expression in the inner cell mass and trophoblast of an E3.5 blastocyst; (P to R) Sp1 expression in the nucleus of a blastocyst. Note the intense yellow signal as a result of colocalization of the Sp1 signal (green) with DNA signal (red) (R).
We analyzed YY1 expression in later embryogenesis between E7.5 and E12.5 by whole-mount in situ hybridization. Wild-type mouse embryos were obtained at different embryonic days. With a YY1 antisense riboprobe, we find YY1 mRNA ubiquitously expressed at gestational stages E7.5, E8.5, E9.5, and E12.5 with a relatively elevated expression in the ectoplacental cone (Fig. 4A), somites, limb bud, and tail tip (Fig. 4B, C, and D). As a negative control, a YY1 sense riboprobe did not hybridize to the embryos. The results of a representative negative control experiment are shown in Fig. 4E, where an E12.5 embryo was probed with a YY1 sense riboprobe. The ubiquitous expression of YY1 in early mouse embryos is consistent with a critical role for YY1 at the time around implantation.
FIG. 4.
YY1 has a widespread expression pattern in the developing embryo. Whole-mount in situ hybridization of E7.5 (A), E8.5 (B), E9.5 (C), and E12.5 (D) wild-type embryos with a YY1 antisense riboprobe. Hybridization of an E12.5 embryo with a YY1 sense riboprobe was included as a negative control (E). The Brachyury (T) riboprobe was used as a positive control (5a).
A subset of YY1 heterozygous embryos demonstrate growth defects and exencephaly.
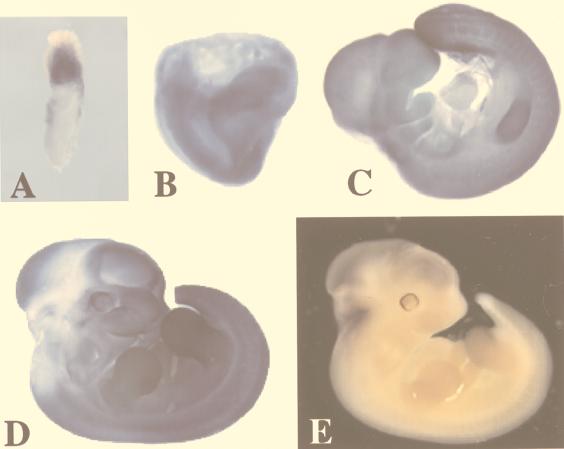
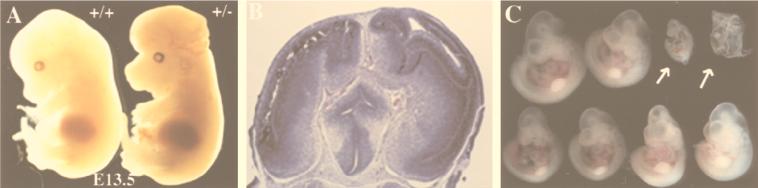
Although the majority of YY1 heterozygous embryos are phenotypically normal, a small subset of heterozygous embryos are developmentally retarded in growth and exhibit neurulation defects. In a mixed-strain background (129/Sv × C57BL/6), we observed growth retardation or a phenotype resembling exencephaly (3, 4) in 6 of 63 total embryos (9.5%). These six embryos are genotypically YY1 heterozygous and represent 16.7% of the total YY1 heterozygotes analyzed (6 of 36). Examples of an E13.5 exencephalic YY1+/− embryo and its phenotypically normal YY1+/+ littermate are shown in Fig. 5A. The YY1+/− embryo is on the right (Fig. 5A) and is exencenphalic with an open brain. A coronal histological section of the head region of this abnormal embryo is shown in Fig. 5B, demonstrating asymmetry and the presence of pseudoventricles consistent with exencephaly. Midgestational embryo sections immunostained with anti-YY1 antibodies exhibit increased YY1 protein expression in the developing midbrain, hindbrain, and cerebellar primordia consistent with a role for YY1 in neural development (5a). When backcrossed onto the inbred C57BL/6 genetic background, 7 of 29 YY1 heterozygotes (approximately 24%) displayed retarded development and neurulation defects. Figure 5C shows a representative litter of E10.5 embryos obtained from a YY1+/− heterozygote crossed to the YY1+/+ wild type on an inbred C57BL/6 genetic background. The two YY1+/− embryos in the upper right-hand corner of Fig. 5C (indicated by arrows) clearly show delayed development and alteration in morphology compared with the other six embryos, which were genotyped as either YY1+/− or YY1+/+. These observations suggest that YY1 may play a role in later mouse embryogenesis that is not revealed in the homozygotes due to the early embryonic lethality. This possibility is consistent with our YY1 expression studies, which show a relatively high level of YY1 expression in the somites, limb bud, and tail tip of the mouse embryo. These results being taken together, YY1 is likely to play a critical role before as well as after gastrulation (i.e., organogenesis) and there may be a dosage effect of YY1 in the developing mouse embryo.
FIG. 5.
Exencephaly and growth retardation in a subset of YY1+/− mice. (A) E13.5 littermates obtained from mixed-strain heterozygous intercrosses (129/Sv × C57BL/6) genotyped as YY1+/+ on the left and YY1+/− on the right. The YY1+/− embryo that is exencephalic with an open brain is shown on the right. (B) A coronal histological section (4-μm thickness) of the exencephalic YY1+/− embryo observed in panel A stained with hematoxylin and eosin as described in Materials and Methods shows pseudoventricles and asymmetry. (C) An entire litter of E10.5 mice obtained from a YY1+/− male crossed to a YY1+/+ wild-type female on the inbred C57BL/6 genetic background. The two smaller embryos in the right-hand corner indicated by arrows were genotyped as YY1+/− whereas the other embryos were either YY1+/− or YY1+/+.
DISCUSSION
In the present study, we have demonstrated a critical role for YY1 in mouse embryonic development. Mouse embryos lacking YY1 develop to the blastocyst stage and are implanted but die shortly thereafter. These embryos show a severe defect in the development of the embryonic and extraembryonic tissues at a developmental stage that coincides with rapid cell proliferation and differentiation. Our findings provide the first demonstration of a pivotal role for YY1 in vertebrate development.
During mouse development, cleavage of the fertilized embryo to the blastocyst-stage embryo (E3.5) marks the first differentiation event. This event is the first morphological asymmetry observed in the embryo highlighted by the formation of the blastocyst inner cell mass and the trophectoderm which give rise to the embryo proper and contribute to the placental tissue, respectively (reviewed in references 13 and 31). The blastocyst-stage embryo prepares for uterine implantation by generating two specialized tissues, the trophoblast and primitive endoderm, which later contribute to the formation of the placenta. At the time of implantation, proteolytic degradation of the uterine epithelium and attachment of the hatched blastocyst occur. Following blastocyst implantation and prior to gastrulation, mouse embryos must achieve a rapid proliferative expansion of the inner cell mass and its associated trophectoderm, resulting in the formation of the egg cylinder (E6.5), which consists of the inner epiblast and the outer visceral endoderm. This event marks the differentiation of the three earliest cell types, the endoderm, the mesoderm, and the ectoderm. Embryos that lack a threshold number of epiblast cells due to either proliferative defects or cell losses fail to gastrulate and are arrested prior to the formation of the primitive streak (reviewed in reference 31). The YY1 mutants fail prior to the formation of the primitive streak, suggesting that YY1 is likely to be necessary for epiblast proliferation or differentiation events prior to gastrulation. Taken together, these findings suggest that YY1 is a key molecule that is involved in regulating genes whose products are pivotal for differentiation and/or proliferation during early embryogenesis. The fact that YY1 is detected in both the inner cell mass and the trophectoderm of the preimplantation blastocyst (Fig. 3M) is consistent with such a role for YY1 in regulating genes for both embryonic and extraembryonic tissues.
What might be the downstream target genes for YY1 during early embryogenesis whose misregulation may account for the observed defects in the YY1−/− embryos? The phenotypes of the YY1 mutant embryos are reminiscent of those described for the evx-1 (29), Fgf-4 (7), fug-1 (5), rad51 (19), and β1 integrin (6, 30) mutant mice. These mice share a feature with the YY1−/− embryos in which proliferative expansion of the embryo does not allow the morphogenesis of the mutant embryos to the pregastrulation stage. All of these genes are crucial for the implanting embryo. Therefore, it is possible that YY1 may be essential for regulating the expression of these or other genes important for the formation of the pregastrulation embryo (reviewed in reference 4).
Recently, a putative Drosophila YY1 homolog, pleiohomeotic (Pho), has been described (1). Mutations in Pho result in Drosophila embryonic lethality (10, 11). Pho and YY1 have extensive amino acid identity in the zinc finger region (95% in the entire zinc finger region and 100% in zinc fingers 2 and 3), suggesting that they are likely to recognize similar (if not identical) DNA sequences. Outside the zinc finger region, there is very little sequence conservation except for a 22-amino-acid region located in the central portion of the proteins (1). The transcription activation domain situated at the N terminus of YY1 (2, 17) appears to be absent in Pho. Therefore, it is not clear whether the same molecular mechanism underlies the biological functions of both proteins. Regardless, our results indicate that murine YY1 may have a crucial role similar to that of Drosophila Pho in early embryonic development, suggesting an evolutionarily conserved function for YY1 prior to the separation of arthropods and vertebrates.
In summary, we have shown that mice lacking YY1 exhibit early embryonic lethality at the time around implantation, revealing a crucial role for YY1 in early mouse development. We postulate that YY1 may regulate genes whose products are essential for the rapid proliferation and differentiation of mouse embryos around the time of implantation. The expression pattern of YY1 and the phenotypes displayed by a subset of the YY1 heterozygotes raise the additional possibility that YY1 may also be required for later-stage embryogenesis. The observation that a subset of YY1 heterozygotes is growth retarded and has neurulation defects suggests that both alleles of YY1 are necessary for normal embryonic growth and development. Future experiments will focus on delineating YY1 function and the mechanism of action in later embryonic development by selective inhibition of YY1 expression with conditional knockout technology. These results being taken together, YY1 appears indispensable for mouse embryonic development and is one of the few transcription factors characterized to date that has such an early role in mouse embryogenesis. Given the fact that YY1 is highly conserved among human (22, 27), mouse (8, 12), avian (5a), and Xenopus (23) species, it is likely that YY1 plays a crucial developmental role in these organisms as well.
ACKNOWLEDGMENTS
We thank Grace Gill, Keith Blackwell, Malcolm Logan, Zhenyu Gu, and Andrew Lassar for insightful discussions and for critical comments on the manuscript. We are grateful to Andrew McMahon for the T (Brachyury) probe and Anny Usheva for the monoclonal YY1 antibody. We thank L. Yu, H. Lei, M. Raffin, and S. Witte for excellent technical assistance and Keith Ketterer and Tim Lis for computer graphic assistance.
This work is supported by a grant from the NIH (GM 53874) to Y.S. M.E.D. was supported by the training grant T32CA09031-22.
REFERENCES
- 1.Brown J L, Lesley D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 2.Bushmeyer S, Park K-S, Atchison M L. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1996;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 3.Campbell L R, Datton D H, Sohal G S. Neural tube defects: a review of human and animal studies on the etiology of neural tube defects. Teratology. 1986;34:171–187. doi: 10.1002/tera.1420340206. [DOI] [PubMed] [Google Scholar]
- 4.Copp A J. Deaths before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 5.DeGregori J, Russ A, von Melchner H, Rayburn H, Priyaranjan P, Jenkins N A, Copeland N G, Ruley H E. A murine homolog of the yeast RNA1 gene is required for postimplantation development. Genes Dev. 1994;8:265–276. doi: 10.1101/gad.8.3.265. [DOI] [PubMed] [Google Scholar]
- 5a.Donohoe, M. E., and Y. Shi. Unpublished observations.
- 6.Fassler R, Meyer M. Consequences of lack of β1 integrin gene expression in mice. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 7.Feldman B, Poueymirou W, Papaioannou V E, Dechiara T M, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 8.Flanagan J R, Becker K, Ennist D, Gleason S L, Driggers P H, Levi B-Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvin K M, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehring W J. A recessive lethal (1(4)29) with a homeotic effect in D. melanogaster. Drosophila Information Service. 1970;45:103. [Google Scholar]
- 11.Girton J R, Jeon S H. Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev Biol. 1994;161:393–407. doi: 10.1006/dbio.1994.1040. [DOI] [PubMed] [Google Scholar]
- 12.Hariharan N, Kelley D, Perry R P. δ, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 14.Kaufman M H. The atlas of mouse development. San Diego, Calif: Academic Press; 1992. [Google Scholar]
- 15.Lawitts J A, Biggers J D. Culture of preimplantation embryos. In: Wassarman P M, DePamphilis M L, editors. Guide to techniques in mouse development. Vol. 294. New York, N.Y: Academic Press; 1993. pp. 153–164. [Google Scholar]
- 16.Lee J-S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 17.Lee J S, See R H, Galvin K M, Wang J, Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 19.Lim D-S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethality that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nothias J Y, Majumder S, Kaneko K J, DePamphilis M L. Regulation of the gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 21.Palmieri S, Payne J, Stiles C D, Biggers J, Mercola M. Expression of the mouse PDGF-A and PDGF-α-receptor genes during pre- and post-implantation development: evidence for a developmental shift from an autocrine to a paracrine mode of action. Mech Dev. 1992;39:181–191. doi: 10.1016/0925-4773(92)90045-l. [DOI] [PubMed] [Google Scholar]
- 22.Park K, Atchison M. Isolation of a candidate repressor/activator, NF-E1 (YY1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain μE1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pisaneschi G, Ceccotti S, Falchetti M L, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- 24.Safrany G, Perry R P. Characterization of the mouse gene that encodes the δ/YY1/EF-E1/UCRBP transcription factor. Proc Natl Acad Sci USA. 1993;90:5559–5563. doi: 10.1073/pnas.90.12.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-specific binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Lee J S, Galvin K. Everything you have ever wanted to know about Yin Yang 1. BBA Rev Cancer. 1997;1332(2):F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 28.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spyropoulos D D, Capecchi M R. Targeted disruption of the even-skipped gene, evx 1, causes early postimplantation lethality of the mouse conceptus. Genes Dev. 1994;8:1949–1961. doi: 10.1101/gad.8.16.1949. [DOI] [PubMed] [Google Scholar]
- 30.Stephens L E, Sutherland A E, Klimanskaya I V, Andrieux A, Meneses J, Pederson R A, Damsky C H. Deletion of β1 integrin in mice results in inner cell mass failure and peri-implantation lethality. Genes Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- 31.Tam P P L, Behringer R R. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson D G. In situ hybridization: a practical approach. London, United Kingdom: Oxford University Press; 1992. [Google Scholar]
- 33.Worrad D M, Ram P T, Schultz R M. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120:2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 34.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Y L, Dupont B R, Ghosh S, Fang Y, Leach R J, Seto E. Cloning, chromosomal localization and promoter analysis of the human transcription factor YY1. Nucleic Acids Res. 1998;26:3776–3783. doi: 10.1093/nar/26.16.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]