Abstract
Wild birds play an important role in the circulation and spread of pathogens that are potentially zoonotic or of high economic impact on zootechnical production. They include, for example, West Nile virus (WNV), Usutu virus (USUV), avian influenza virus (AIV), and Newcastle disease virus (NDV), which, despite having mostly an asymptomatic course in wild birds, have a strong impact on public health and zootechnical production. This study investigated the presence of these viruses in several wild bird species from North Italy during the biennium 2019–2020. Wild birds derived from 76 different species belonging to 20 orders. Out of 679 birds, 27 were positive for WNV (lineage 2) with a prevalence of 4%; all birds were negative for USUV; one gull was positive for H13N6 influenza virus, and 12 samples were positive for NDV with a prevalence of 2%. Despite the low prevalence observed, the analyses performed on these species provide further data, allowing a better understanding of the diffusion and evolution of diseases of both economic and zoonotic importance.
Keywords: wild birds, avian influenza, West Nile virus, Usutu virus, Newcastle disease, Italy
1. Introduction
Attention to wildlife monitoring has always been very high in relation to public health, especially considering that more than 70% of the emerging zoonotic diseases derive precisely from free ranging wildlife [1]. The recent identification of SARS-CoV-2 and its subsequent pandemic diffusion have sparked an increased interest in these issues and the close relations involving animals, the environment, and public health.
Among wild species, birds play a crucial role in the mechanisms of distribution, persistence, and evolution of certain pathogens, potentially zoonotic, with an economic impact on poultry farming. They are known to be reservoirs for several bacteria, such as Salmonella spp. [2], drug-resistant organisms, parasites, or viruses. In this regard, avian influenza virus (AIV), Newcastle disease virus (NDV), and vector-borne diseases such as West Nile (WNV) and Usutu virus (USUV), could represent important zoonotic agents and/or have a high economic impact on zootechnical production.
Avian influenza is caused by the influenza virus type A, belonging to the family Orthomyxoviridae. Wild waterbirds from the order Anseriformes (mainly ducks, swans, and geese) and Charadriiformes (gulls and terns) are natural reservoirs of low pathogenic influenza (LPAI) [3]. In these species, the virus LPAI usually circulates asymptomatically, whereas poultry can show mild to severe clinical forms. However, the possible evolution of the H5 and H7 subtypes into highly pathogenic influenza virus (HPAI) is the most relevant aspect, since they can cause high mortality in both wild species and domestic poultry, with severe economic consequences. In addition, reassortment between subtypes poses a risk for immune escape in these and in new hosts susceptible to infection, such as swine, equids, and humans [4], raising potential pandemic threats [5].
ND, which is of lesser zoonotic importance [6], is still a major problem in poultry farming. NDV (Orthoavulavirus-1), or avian paramyxovirus-1 (APMV-1) belongs to the family Paramyxoviridae and genus Avulavirus. It represents a major limiting factor for poultry production in many developing countries, and even in developed countries, despite increased biosecurity measures and vaccination, it causes some sporadic outbreaks. The clinical signs of infected birds vary depending on the host species, immune status, and age, as well as the virulence and dose of the virus; thus, it could be difficult to recognise the disease [7]. NDV has been isolated from numerous species of wild birds, considered natural reservoirs of lentogenic strains, and, occasionally, velogenic strains [8,9,10]. Most strains isolated from wild birds would appear to be non-pathogenic to poultry; however, outbreaks of virulent NDV in poultry originating from avirulent strains from natural reservoirs have been reported [11].
WNV and USUV are closely related arthropod-borne viruses (genus Flavivirus; family Flaviviridae). These agents are transmitted by mosquitoes, and the infections they cause represent important public and animal health concerns because of their wide geographical diffusion and the broad range of potentially affected hosts, particularly birds and mammals, including equids and humans. Mammals are considered accidental or dead-end hosts, rarely developing sufficient viremia to re-infect feeding mosquitoes [12]. Instead, wild birds are considered the main vertebrate reservoir hosts of WNV and USUV and the main amplifying hosts of the viruses in nature [13,14]. Indeed, they are able to develop a strong and long-term viremia and are capable of infecting bird-biting mosquitoes [15]. European epidemiological data show a severe increase in human cases that have occurred in the last ten years, raising the focus on surveillance of these arboviruses through combined monitoring of vectors, wild birds, humans, and the environment.
Considering the characteristics of the viruses described, wild birds may have a predominant role in the epidemiology of diseases. In particular, the ability to migrate over long distances allows them to play a role in the amplification and circulation of viruses [16], thus creating the potential for the establishment of new endemic foci of disease along migration routes from endemic areas. Indeed, infected migratory wild birds have been suspected of spreading HPAI virus subtype H5N1 from central Asia to Eastern Europe through migratory flyways in 2005 [17], and USUV is thought to have been introduced in Europe during bird migration, at different times starting in the 1950s [18]. In addition, for some agents, wild birds can also act as a mechanical vector passively carrying infected insects or parasites, even if birds are not a competent reservoir for that particular infection [19]. Wetlands along migratory routes allow the concentration of many birds of different species, thus promoting intra- and interspecific transmission and playing an important role in local and long-distance viral dispersal [20,21].
In this epidemiological context, monitoring of wild bird populations provides information about pathogen spread and virus circulation, allowing the early detection of possible new outbreaks. However, observation of clinical symptoms and signs is somewhat problematic in wild birds, considering the difficulty in capturing them individually for examination. Often, only the phenomena of high mortality allow the detection of the circulation of certain pathogens [22]. Therefore, these aspects, along with the severe impact of the described diseases, highlight the importance of systematic surveillance of the avifauna in the presence of viral agents.
In Italy, surveillance plans for wild target species for avian influenza [23] and arbovirosis [24] are active and regularly performed. Therefore, this study aimed to investigate the presence of the above-mentioned viruses in several species of wild birds, derived from passive surveillance within the Lombardy (Northern Italy) regional plan for wildlife monitoring.
2. Materials and Methods
The samples examined were gathered in the framework of the wild bird monitoring plan that took place in the Lombardy region (Northern Italy) during the years 2019–2020. Wild bird carcasses from the wildlife rehabilitation centres (CRAS) and regional veterinary units of Lombardy were committed to the Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna (IZSLER). In most cases, subjects required immediate euthanasia, sometimes they were already dead when conferred, and, in the remaining cases, the average time spent in the CRAS was 1–2 days before death.
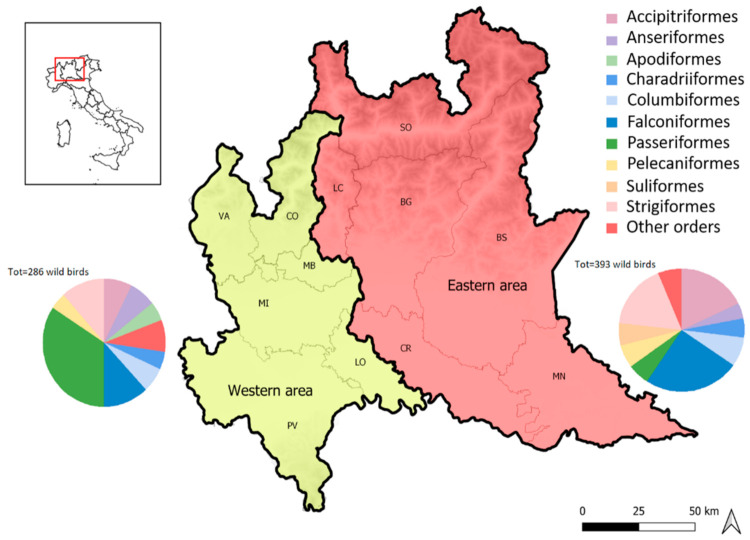
Two main areas of Lombardy, from which the largest number of samples have been sent, could be identified: (i) an eastern area that includes samples from Brescia, Bergamo, and Lecco provinces, in particular from Parco Adamello and Val Predina CRAS and (ii) a western area that includes Milano, Pavia, Monza, Varese, and Como provinces, referring to La Fagiana and Vanzago CRAS. From the eastern area, 393 samples were conferred, while 286 samples were from the western area, for a total of 679 wild birds analysed from 76 different species belonging to 20 orders (Figure 1).
Figure 1.
Representation of study areas (western and eastern). In the graphs, the most sampled orders for the two different areas are represented.
The greatest number of species belonged to the orders Strigiformes, Anseriformes, Accipitriformes, Passeriformes, Charadriiformes, and Falconiformes (for more details, and to consult the main results, see Table S1 in the Supplementary Materials).
For each wild bird, organs such as brain, lungs, heart, kidneys, spleen, and intestine were homogenised and subsequently centrifuged at 3750× g for 15 min to favour the precipitation of cells and proteins that could interfere in later stages. Total viral RNA was extracted from 100 µL of homogenised samples using Qiagen BioSprint 96 One-For-All Vet 100 (Qiagen, Hilden, Germany) and stored at −80 °C or immediately used. The internal control exogenous RNA template and carrier RNA were added to each sample prior to extraction. RNA from all samples was subjected to a panel of real-time RT-PCR to detect West Nile virus, Usutu virus, influenza A virus, and Newcastle virus. For any reaction, 5 μL of RNA was added to the master mix PCR, and positive and negative controls (no template) of amplification were included. The primers and probes used for the different PCRs are listed in Table 1.
Table 1.
Primer and probe sequences used for the PCRs and the corresponding target gene.
| Name | Primer-Probe Name | Sequence | Target Gene |
|---|---|---|---|
| WEST NILE IDENTIFICATION [25] | WN-10533-10522 | AAGTTGAGTAGACGGTGCTG | NS3 gene |
| WN-10625-10606 | AGACGGTTCTGAGGGCTTAC | ||
| WN-10560-10579-PROBE | fam-CTCAACCCCAGGAGGACTGG-bhq1 | ||
| WEST NILE TYPING [26,27] | WN-LCV-F1 | GTGATCCATGTAAGCCCTCAGAA | NS3 gene |
| WN-LCV-R1 | GTCTGACATTGGGCTTTGAAGTTA | ||
| WN-LCV-S1 PROBE | fam-AGGACCCCACATGTT-mgb | ||
| WN-LCV-S2 PROBE | vic-AGGACCCCACGTGCT-mgb | ||
| USUTU [28] | USUTU F | ACGGCCCAAGCGAACAGAC | NS5 gene |
| USUTU R | GGCTTGGGCCGCACCTAA | ||
| USUTU PROBE | CY5-CGAACTGTTCGTGGAAGG-BHQ3 | ||
| INFLUENZA A [29] | INFLU-124 MOD | TGC AAA GAC ACT TTC CAG TCT CTG | M gene |
| INFL-M124 | TGC AAA AAC ATC TTC AAG TCT CTG | ||
| INFLU M25 | AGA TGA GTC TTC TAA CCG AGG TCG | ||
| M+64 PROBE | fam TCAGGCCCCCTCAAAGCCGA tamra | ||
| INFLUENZA H5 [30] | H5LH1-F | ACATATGACTACCCACARTATTCAG | HA2 gene for emoagglutinin H 5 |
| H5RH1-R | AGACCAGCTAYCATGATTGC | ||
| H5PRO-probe | fam-TCWACAGTGGCGAGTTCCCTAGCA-tamra | ||
| INFLUENZA H7 [30] | H7-LH6H7-FOR | GGCCAGTATTAGAAACAACACCTATGA | HA2 gene for emoagglutinin H 7 |
| H7-RH4H7-REV | GCCCCGAAGCTAAACCAAAGTAT | ||
| H7PRO11-PROBE | fam-CCGCTGCTTAGTTTGACTGGGTCAATCT-bhq1 | ||
| NDV [31] | NDV-1M+4100 | AGTGATGTGCTCGGACCTTC | APMV-1 gene |
| NDV-M−4220 | CCTGAGGAGAGGCATTTGCTA | ||
| NDV-M+4169-PROBE | fam-TTCTCTAGCAGTGGGACAGCCTGC-tamra | ||
| NDV [32] | FOP 1 | TACACCTCATCCCAGACAGGGTC | F gene |
| NDV-FOP2 | AGGCAGGGGAAGTGATTTGTGGC |
Real-time RT-PCR for influenza, WNV, and USUV was performed using the QuantiFast Pathogen RT-PCR with IC Kit (Qiagen, Hilden, Germany), while for the detection of NDV and WNV Lineage 1–2, the QuantiTect Probe RT-PCR Kit (Qiagen, Hilden, Germany).
The amplification reaction for the detection of WNV [25] and USUV [28] was conducted with a thermal profile of 50 °C for 20 min, 95 °C for 5 min, and cycles 45 of 95 °C for 15 s and 60 °C for 30 s. West Nile positive samples were subjected to real-time RT-PCR to distinguish lineage 1 or 2 [26,27]. PCR was carried out using QuantiTect probe RT-PCR Kit (Qiagen, Hilden, Germany) in which, into each sample were added a pair of primers forward and reverse and 2 different probes: one specific for lineage 1 and the other specific for lineage 2. The thermal profile used was 50 °C for 30 min, 95 °C for 15 min, and 45 cycles at 95 °C for 15 s and 60 °C for 1 min.
For the detection of AIV [29], the amplification reaction was conducted with the same thermal profile used for WNV and USUV, but for 40 cycles instead of 45. Influenza-A-positive samples were subjected to two typing real-time RT-PCRs to define the subtype H5 or H7 [30]. Two reaction mixes were set up with the SuperScript III Platinum One-Step qRT-PCR kit (Invitrogen, Waltham, Mass., USA) in each of which specific H5 and H7 primers and probes were inserted, respectively, but they had the same thermal profile: 30 min for 50 °C, 2 min for 95 °C, and 40 of 10 s for 95 °C, 30 s for 54 °C, and 10 s for 72 °C. Influenza-A-positive samples that were negative for PCR subtypes H5 or H7 were subjected to complete sequencing by next-generation sequencing (NGS).
Finally, the thermal profile for the detection of NDV [31] was 50 °C for 30 min, 95 °C for 15 min, and 40 of 94 °C for 10 s, 52 °C for 30 s, and 72 °C for 10 s. Positive samples were subjected to reverse transcriptase polymerase chain reaction (RT-PCR) targeting the fusion (F) protein gene [32] with One-Step RT PCR Kit (Qiagen, Hilden, Germany) under the following amplification steps: 50 °C for 30 min, 95 °C for 15 min and 45 cycles of 94 °C for 30 s, 64 °C for 30 s, and 72 °C for 40 s and elongation at 72 °C for 5 min. The segments obtained at 532 bp were sequenced by the Sanger method. We performed phylogenetic and molecular analyses of the variable region of the F gene (42–421 nt), including the cleavage site, as standard tool for molecular characterization of NDV sequences and for determination of the potential virulence of isolates.
3. Results
Molecular analyses performed on wild bird samples in order to detect arboviruses showed a total prevalence of 4% (CI 95%: 3–5) for WNV, with 27 positive birds, mainly belonging to the order Strigiformes, Accipitriformes, and Passeriformes, and originating from different provinces of Lombardy. Real-time RT-PCR performed on positive samples to distinguish lineage 1 and 2 detected lineage 2 in 23 birds. Four samples from Phalacrocorax carbo, Larus michahellis, Pica pica, and Tachymarptis melba could not be identified. All birds were negative for the USUV PCR. Table 2 summarises the WNV-positive results in detail.
Table 2.
Orders and species of wild bird tested positive for WND.
| Order | Species | Number | Prevalence | Type |
|---|---|---|---|---|
| Accipitriformes | Accipiter nisus | 4 | 8% (4/50) | Lineage 2 |
| Buteo buteo | 1 | 4.1% (1/24) | Lineage 2 | |
| Pernis apivorus | 1 | 9% (1/11) | Lineage 2 | |
| Apodiformes | Tachymarptis melba | 1 | 33% (1/3) | not typed |
| Charadriiformes | Larus michahellis | 1 | 3.1% (1/32) | not typed |
| Columbiformes | Columba palumbus | 1 | 12.5.% 1/8) | Lineage 2 |
| Falconiformes | Falco tinnunculus | 4 | 3.4% (4/118) | Lineage 2 |
| Falco subbuteo | 1 | 16.7% (1/6) | Lineage 2 | |
| Passeriformes | Corvus cornix | 4 | 11.7% (4/34) | Lineage 2 |
| Pica pica | 1 | 7.1% (1/14) | not typed | |
| Turdus merula | 1 | 4.1% (1/24) | Lineage 2 | |
| Suliformes | Phalacrocorax carbo | 1 | 4.3% (1/23) | not typed |
| Strigiformes | Athene noctua | 3 | 4.9% (3/61) | Lineage 2 |
| Otus scops | 1 | 9.1% (1/11) | Lineage 2 | |
| Strix aluco | 2 | 10.5% (2/19) | Lineage 2 |
Regarding avian influenza, only one sample from Larus michahellis was positive, and the same gull was positive for WNV. NGS allows identification of the H13N6 virus. Phylogenetic analyses on virus segments highlighted its reassortant nature. Indeed, the segment PB2 belonged to the American lineage, while segments NA, HA, NP, PB1, M, PA, and NS clustered within the Eurasian lineage (see Table 3).
Table 3.
Analyses of virus segments belonging to Eurasian (Eu) and American (Am) lineages. Sequences with the highest % of identity are reported.
| Segment | NA | HA | NP | PB2 | PB1 | M | PA | NS |
|---|---|---|---|---|---|---|---|---|
| Lineage | Eu | Eu | Eu | Am | Eu | Eu | Eu | Eu |
| % Identity | 96% | 96% | 98% | 97% | 98% | 99% | 97% | 98% |
| Reference sequences |
KX978407.1 KX978025.1 |
MF682848.1 CY185497.1 |
MF461189.1 MF146988.1 |
MF461185.1 MH764127.1 |
MF145750.1 KX979824.1 |
MK192343.1 MF694084.1 |
MF148015.1 MF147897.1 |
MF694157.1 MF694139.1 |
Finally, 12 samples were positive for NDV PCR, in particular, eight Columba livia, three Streptopelia turtur, and one Cygnus olor, with a total prevalence of 1.7% (CI 95%: 1–3). Similarly to birds positive to WNV, subjects came from different provinces. Blast analysis of NDV sequences showed the highest % identity of the swan-origin sequence (99.38%) to NDV sequences detected in geese in Nigeria [33] belonging to the genotype I. All other Italian NDV sequences showed the highest % identity (from 99.58 to 98.53%) to the NDV strain SD18 of pigeon origin, which belongs to genotype VI.2.1.1.2.2. [34].
Molecular analysis of protein F revealed three different motifs at positions 112–117. The Italian swan-origin NDV sequence showed the 112GKQGRL117 motif typically found in low virulence NDVs [35]. All other sequences originating from pigeons and turtledoves showed two different motifs found in the velogenic NDV strains (112RRQKRF117 and 112RRRKKF117) characterized by multiple basic amino acids at positions 112–116 and a phenyl alanine at position 117.
The main results from the molecular analyses related to the different species are summarized in the Table S1 in the Supplementary Materials.
4. Discussion
This study highlights the presence and circulation of influenza viruses, even if limited to only a single H13N6 strain, West Nile, and NDVs in free living avian populations, although with a modest prevalence.
Most of the positive samples are attributable to the WNV, which involves a wide range of species. WNV represents a severe public health concern. In Italy, in recent years, there have been a large number of human cases, with the increasingly frequent manifestation of neuro-invasive forms. The peak was recorded in 2018 with more than 500 cases in humans and an increase in isolation in other susceptible species (equids), and in species involved in the virus life cycle: birds and mosquitoes. In wild birds, 321 specimens tested positive for the virus; of these, 215 were from surveillance of target species (crows, magpies, and jays), while 106 were from passive monitoring of wild animals found dead. The number of cases found in 2019 was significantly lower than that in the previous year, slightly rising by 2020 [36]. Indeed, the 2020 European bulletin reported in Italy 144 confirmed cases of WNV among wild birds; even in Germany and Bulgaria, several cases (28 and 2, respectively) in wild birds were reported [36].
The results of this study confirm positivity in orders characterised by species most susceptible to infection; in particular, Accipitriformes, Charadriiformes, Falconiformes, Passeriformes, and Strigiformes [20,37,38].
Of the 27 positive wild birds, four samples from magpie, cormorant, swift, and gull, respectively, could not be typed, probably because the positivity values were close to the threshold. In these cases, typing is difficult to obtain. Nevertheless, a technical issue making typing impossible cannot be excluded; in fact, samples derived from wildlife rehabilitation centres are usually stored in non-optimal conditions (−20 °C) for quite a long time, and they may have been subjected to several freeze and thawing steps during the preparation process, which may have compromised the virus and affected the quality of the analysis. Apart from the magpie, which is listed as a target species for WNV research in the national control plan, the other three species, whose lineage is unknown, are rarely involved in outbreaks of WNV. Currently, no other positive findings have been reported for the alpine swift, even though it is considered a species potentially able to introduce the virus from sub-Saharan Africa and amplify it, especially in dry areas [13].
Sporadic episodes of WNV have also been reported in cormorants. Recently, a serological survey carried out in Germany showed the presence of antibodies against West Nile virus in this species [39]; moreover, in the Volga delta area the virus was isolated from cormorants and ticks (Hyalomma marginatum) associated with them [40]. Of particular interest is the co-infection (West Nile and Influenza viruses) found in a gull in 2020. The presence of West Nile has already been documented in this species, but with an infrequent occurrence [41,42,43]. In Italy, only one other identification of WNV in a gull was reported in 2020; in particular, lineage 2 was detected [36]. Previous studies carried out in Italy in the same study areas, aimed to genetically characterize the WNV-2 isolated strains, by investigating the genetic diversity of sequences obtained in the period 2015–2018, from mosquitoes, birds, horses, and humans [44]. The study demonstrated the complete segregation of Italian sequences from those of other European countries, highlighting the presence of two different clades (A and B). The clade A became extinct in 2013–2014, while clade B gave rise to several sub-clades characterized by different spatial distribution. The segregation from other European sequences, and the continuous persistence of some sub-clades on the territory, suggest the presence of endemic clades, supporting the hypothesis of local overwintering in Italy [42]. It will therefore be interesting to deepen the study of these aspects by comparing present birds’ sequences with those of mosquitoes, horses, and humans from the same areas. These further analyses could confirm, as expected, the endemic nature of clades previously isolated.
With regard to the influenza virus, molecular analysis identified the type H13N6, which was isolated for the first time in a gull in 1977 [45]. This type of virus is classified as having low pathogenicity and seems to be typical of the order Charadriiformes. Indeed, gulls are widely recognised as reservoirs for LPAI viruses; among the several types of AIV, the hemagglutinin subtypes H13 and H16 are rarely detected in other avian groups, suggesting their maintenance almost exclusively within gull populations [46,47], but other avian species (i.e., turkeys and ducks) could be a spill-over host for the H13 type [46]. The present data and previous identification of H13N6 in gulls in Italy [23] suggest a low diffusion of this virus in the resident gulls’ population. However, only a small number of seagull species have been received, which does not provide a complete picture of the circulation of this flu type. The presence of segments belonging to different lineages has already been reported [48,49]. While it is relatively common to find European lineages in North America, not much data is available on the opposite phenomenon. Thus, the American origin of segment PB2 found in gulls is of particular interest. This could be due to the occasional migration of typically American species in Europe, or to those ubiquitous species (e.g., Larus ridibundus) that are widespread in both Europe and America, which could act as vector for European populations, favouring the virus exchange [50]. Molecular investigations showed the complete absence of other avian influenza strains, particularly H5 and H7 HPAI. The European Food Safety Authority (EFSA), every year, draws up a report about surveillance for avian influenza in poultry and wild birds. In the 2020 report, two cases of HPAI from wild birds, belonging to passive surveillance, were reported in Italy with a prevalence of 0.1%. The highest percentages of HPAI positive birds found by passive surveillance were in Denmark (32% of samples), Germany (14%), the Netherlands (12%), and Ireland (12%) [51]. Therefore, monitoring in wild species helped to expand surveillance data on influenza virus, even if the limited sampling in this study of reservoir species, particularly water birds, may have greatly reduced the overall amount of useful and available information.
With regard to NDV, molecular analyses identified almost all positives in the order of Columbiformes, particularly in pigeons and turtle doves. Pigeons are considered among the most important reservoirs of NDV [52], and strains of variable virulence have been isolated from wild pigeons and doves [35,53]. These bird species could play a very important role in the spread of NDV and other avian viruses because of their ethological characteristics and the overlap of their habitats with areas usually urbanised or used by human activity, particularly livestock and poultry farming. In recent decades, a great number of outbreaks of virulent NDV in chickens, originating from wild birds in European countries, have been reported [54]. Indeed, in the last decade, several cases of NDV in wild birds, above all from the Southern and Eastern Europe, were reported [55]. In most cases, positive birds were identified as collared and turtle doves, which are the most susceptible species. However other species, rarely reported, were infected (i.e., Accipitridae and Gaviidae) [54]. Interspecies transmission seems to be an important mechanism contributing to the maintenance, propagation, and spread of APMV-1 [56]. However, this aspect should be considered bi-directionally between domestic and wild bird populations, including the potential diffusion of live vaccine strains designed for domestic poultry into wild birds [57,58]. Most APMV-1 recognised in wild birds is avirulent in poultry; however, strains of low virulence are capable of naturally evolving into high virulence, although this has been documented only sporadically [33,54]. In addition to Columbiformes, several free ranging aquatic fowls are considered to be potential carriers of avulavirus-1 [59]. The present data highlight the presence of APMV-1 in a mute swan. In this species, isolation of NDV is mainly occasional [60]; however, it should not be underestimated considering the proven pathogenicity of the avulavirus from swans in chickens [61]. Although occasional serological studies conducted in Argentina reported a seroprevalence of 35% [59], sero-epidemiological surveys could certainly provide better indications of the true circulation of this virus in waterfowl populations. However, unfortunately, the use in this study of carcasses found dead or delivered from rehabilitation centres did not allow adequate blood samples to be obtained to conduct specific serological investigations.
Finally, all samples were negative for the presence of the USUV. This result seems strange compared to the positives reported in Italy in previous years and in the same biennium. In fact, since 2006, several outbreaks of USUV were recorded in Central–Northern Italy, with massive mortality in susceptible species [62], and, in 2009, the first human case was registered [63]. In recent years, an increase in the number of wild bird cases has been observed. Indeed, the national bulletins on arboviroses recorded 98, 26, and 87 USUV-positive wild birds, respectively, in 2018, 2019, and 2020 [36]. However, the great heterogeneity of the analysed species, in relation to the limited number of analyses on target species, could explain the negative results obtained during this survey.
In conclusion, the data presented in this study highlight the importance of passive monitoring of wildlife, particularly wild birds, for increasing the information derived from surveillance plans implemented at the regional and national levels, involving a higher number of wild species often less represented and considered in such control programs. From this perspective, wildlife rehabilitation centres may enhance and simplify surveillance efforts for avian-related viruses [64,65].
The analyses performed on these species, also supported by data collected from other neighbourhoods countries (see Table S2 in the Supplementary Materials), could provide a mass of information, which allows a better description of the epidemiological characteristics such as the diffusion and trends of diseases of economic and, especially, zoonotic importance.
The observed prevalence detected for three out of the four viral agents considered in this study was very low; however, it is possible that these were underestimated. Many viral diseases develop asymptomatically in wild birds, causing rare phenomena of intense mortality. Therefore, it is possible that the real reservoirs of infections were hardly contacted. Moreover, we must not underestimate the quality of the samples; although we constantly tried to keep them under optimal conditions, in field conditions this is not always completely applicable. Finally, in spite of the large number of species analysed (likely the most widespread), this sampling approach is not exhaustive of the real variety of species present in the territory, thus limiting the achievements of some and even more useful information. Therefore, although it provides important data on the spread of certain pathogens, it is essential that passive monitoring is always accompanied by active epidemiological surveillance, including sero-surveillance, which focuses on target species for the viruses under study.
Acknowledgments
The authors would like to thank the staff of rehabilitation centres “Associazione CRAS di Vanzago”, “CRAS La Fagiana”, “CRAS WWF Valpredina” and “CRAS Parco Adamello”; the staff of the diagnostic laboratories of Binago, Brescia, Cremona, Milano, Mantua, Pavia and Sondrio units; Marta Consoli, Elisa Bosio, Chiara Chiapponi and Ilaria Barbieri for their technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9091957/s1, Table S1: Orders and species of wild bird analysed during the years 2019–2020 in Lombardy and summary of the main molecular results; Table S2: Situation reported in European countries in recent years. Registered percentages refer to molecular analyses carried out on birds’ tissue. In some cases, values refer to serological surveys (in brackets). NF: data not found. In regard to the study period, data about NDV in wild birds are not available.
Author Contributions
Conceptualization, A.M. and D.L.; methodology, S.C.; formal analysis, T.T., S.C., S.S., C.T.; data collection, G.G., L.A., A.G., G.S., A.B., C.R., P.P., M.G.; resources, M.C., M.F.; data curation, T.T., S.C.; writing—original draft preparation, T.T., S.C., A.M.; writing—review and editing, A.L., A.M.; supervision, A.L., A.M., E.S., D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study that did not involve killing of animals. The samples did not originate from experimental trial but took advantage just from diagnostic activity with Regional Surveillance Plans for wildlife diseases (DDG, 5 December 2012, no. 11358). Therefore, since the sampling was not specifically programmed as experimental study, but originating from diagnostic activity, we believed that it does not fall in the provisions of the National Law (e.g., DLSG 4/3 2014, n. 26. Application at national level of the EU Directive 2010/63/UE) and no ethical approval or permit for animal experimentation was required.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data generated or analysed during this study are included in the published article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beleza A.J.F., Maciel W.C., Lopes E.D.S., Albuquerque Á.H.D., Carreira A.S., Nogueira C.H.G., de Melo Bandeira J., Vasconcelos R.H., Teixeira R.S.D.C. Evidence of the role of free-living birds as disseminators of Salmonella spp. Arq. Inst. Biol. 2002;87 [Google Scholar]
- 3.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joseph U., Su Y.C., Vijaykrishna D., Smith G.J. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir. Viruses. 2017;11:74–84. doi: 10.1111/irv.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutton T.C. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses. 2018;10:461. doi: 10.3390/v10090461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman A.U., Ishaq H.M., Raza M.A., Shabbir M.Z. Zoonotic potential of Newcastle disease virus: Old and novel perspectives related to public health. Rev. Med. Virol. 2021:1–12. doi: 10.1002/rmv.2246. [DOI] [PubMed] [Google Scholar]
- 7.Suarez D.L., Miller P.J., Koch G., Mundt E., Rautenschlein S. Newcastle disease, other avian paramyxoviruses, and avian metapneumovirus infections. Dis. Poult. 2020:109–166. [Google Scholar]
- 8.Liu H., Zhang P., Wu P., Chen S., Mu G., Duan X., Hao H., Du E., Wang X., Yang Z. Phylogenetic characterization and virulence of two Newcastle disease viruses isolated from wild birds in China. Infect. Genet. Evol. 2013;20:215–224. doi: 10.1016/j.meegid.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Hao H., Liu Q., Wang R., Zhang P., Wang X., Du E., Yang Z. Phylogenetic and pathogenic analyses of two virulent Newcastle disease viruses isolated from Crested Ibis (Nipponia nippon) in China. Virus Genes. 2013;46:447–453. doi: 10.1007/s11262-013-0881-7. [DOI] [PubMed] [Google Scholar]
- 10.Vidanović D., Šekler M., Ašanin R., Milić N., Nišavić J., Petrović T., Savić V. Characterization of velogenic Newcastle disease viruses isolated from dead wild birds in Serbia during 2007. J. Wildl. Dis. 2011;47:433–441. doi: 10.7589/0090-3558-47.2.433. [DOI] [PubMed] [Google Scholar]
- 11.Zanetti F., Berinstein A., Carrillo E. Effect of host selective pressure on Newcastle disease virus virulence. Microb. Pathog. 2008;44:135–140. doi: 10.1016/j.micpath.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kramer L.D., Styer L.M., Ebel G.D. A global perspective on the epidemiology of West Nile virus. Annu. Rev. Entomol. 2008;53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- 13.Jourdain E., Toussaint Y., Leblond A., Bicout D.J., Sabatier P., Gauthier-Clerc M. Bird species potentially involved in introduction, amplification, and spread of West Nile virus in a Mediterranean wetland, the Camargue (Southern France) Vector-Borne Zoonotic Dis. 2007;7:15–33. doi: 10.1089/vbz.2006.0543. [DOI] [PubMed] [Google Scholar]
- 14.Roesch F., Fajardo A., Moratorio G., Vignuzzi M. Usutu virus: An arbovirus on the rise. Viruses. 2019;11:640. doi: 10.3390/v11070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komar N., Langevin S., Hinten S., Nemeth N., Edwards E., Hettler D., Davis B., Bowen R., Bunning M. Experimental Infection of North American Birds with the New York 1999 Strain of West Nile Virus. Emerg. Infect. Dis. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissenböck H., Kolodziejek J., Url A., Lussy H., Rebel-Bauder B., Nowotny N. Emergence of Usutu virus, an African mosquito-borne Flavivirus of the Japanese encephalitis virus group, central Europe. Emerg. Infect. Dis. 2002;8:652. doi: 10.3201/eid0807.020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert M., Xiao X.M., Domenech J., Lubroth J., Martin V., Slingenbergh J. Anatidae migration in the Western Palearctic and spread of highly pathogenic avian influenza H5N1 virus. Emerg. Infect. Dis. 2006;12:1650–1656. doi: 10.3201/eid1211.060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel D., Jost H., Wink M., Borstler J., Bosch S., Garigliany M.M., Jost A., Czajka C., Luhken R., Ziegler U., et al. Reconstruction of the Evolutionary History and Dispersal of Usutu Virus, a Neglected Emerging Arbovirus in Europe and Africa. MBio. 2016;7:e01938-15. doi: 10.1128/mBio.01938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurt D.R., Meece J.K., Henkel J.S., Shukla S.K. Birds, migration and emerging zoonoses: West Nile virus, lyme disease, influenza A and enteropathogens. Clin. Med. Res. 2002;1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jourdain E., Gauthier-Clerc M., Bicout D.J., Sabatier P. Bird migration routes and risk for pathogen dispersion into western Mediterranean wetlands. Emerg. Infect. Dis. 2007;13:365–372. doi: 10.3201/eid1303.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malkinson M., Banet C. The role of birds in the ecology of West Nile virus in Europe and Africa. Curr. Top. Microbiol. Immunol. 2002;267:309–322. doi: 10.1007/978-3-642-59403-8_15. [DOI] [PubMed] [Google Scholar]
- 22.Chvala S., Kolodziejek J., Nowotny N., Weissenböck H. Pathology and viral distribution in fatal Usutu virus infections of birds from the 2001 and 2002 outbreaks in Austria. J. Comp. Pathol. 2004;131:176–185. doi: 10.1016/j.jcpa.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 23.National Surveillance Plan for Avian Influenza–2021. [(accessed on 25 July 2021)]. Available online: https://www.izsvenezie.it/documenti/temi/influenza-aviaria//piani-sorveglianza/piano-nazionale-influenza-aviaria-2021.pdf.
- 24.National Arbovirosis Plan-2020–2025. [(accessed on 25 July 2021)]; Available online: https://www.salute.gov.it/imgs/C_17_pubblicazioni_2947_allegato.pdf.
- 25.Tang Y., Hapip C.A., Liu B., Fang C.T. Highly sensitive TaqMan RT-PCR assay for detection and quantification of both lineages of West Nile virus RNA. J. Clin. Virol. 2006;36:177–182. doi: 10.1016/j.jcv.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Del Amo J., Sotelo E., Fernández-Pinero J., Gallardo C., Llorente F., Agüero M., Jiménez-Clavero M.A. A novel quantitative multiplex real-time RT-PCR for the simultaneous detection and differentiation of West Nile virus lineages 1 and 2, and of Usutu virus. J. Virol. Methods. 2013;189:321–327. doi: 10.1016/j.jviromet.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez-Clavero M.A., Agüero M., Rojo G., Gómez-Tejedor C. A new fluorogenic real-time RT-PCR assay for detection of lineage 1 and lineage 2 West Nile viruses. J. Vet. Diagn Investig. 2006;18:459–462. doi: 10.1177/104063870601800505. [DOI] [PubMed] [Google Scholar]
- 28.Cavrini F., Della Pepa M.E., Gaibani P., Pierro A.M., Rossini G., Landini M.P., Sambri V. A rapid and specific real-time RT-PCR assay to identify Usutu virus in human plasma, serum, and cerebrospinal fluid. J. Clin. Virol. 2011;50:221–223. doi: 10.1016/j.jcv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monne I., Ormelli S., Salviato A., De Battisti C., Bettini F., Salomoni A., Drago A., Zecchin B., Capua I., Cattoli G. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J. Clin. Microbiol. 2008;46:1769–1773. doi: 10.1128/JCM.02204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise M.G., Suarez D.L., Seal B.S., Pedersen J.C., Senne D.A., King D.J., Kapczynski D.R., Spackman E. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004;42:329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kho C.L., Mohd-Azmi M.L., Arshad S.S., Yusoff K. Performance of an RT-nested PCR ELISA for detection of Newcastle disease virus. J. Virol. Methods. 2000;86:71–83. doi: 10.1016/S0166-0934(99)00185-8. [DOI] [PubMed] [Google Scholar]
- 33.Snoeck C.J., Marinelli M., Charpentier E., Sausy A., Conzemius T., Losch S., Muller C.P. Characterization of newcastle disease viruses in wild and domestic birds in Luxembourg from 2006 to 2008. Appl. Environ. Microbiol. 2013;79:639–645. doi: 10.1128/AEM.02437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y., Xue R., Yang W., Li Y., Xue J., Zhang G. Characterization of ten paramyxovirus type 1 viruses isolated from pigeons in China during 1996–2019. Vet. Microbiol. 2020;244:108661. doi: 10.1016/j.vetmic.2020.108661. [DOI] [PubMed] [Google Scholar]
- 35.Kim L.M., King D.J., Guzman H., Tesh R.B., Travassos da Rosa A.P.A., Bueno Jr. R., Dennett J.A., Afonso C.L. Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. J. Clin. Microbiol. 2008;46:3303–3310. doi: 10.1128/JCM.00644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epidemiological Bulletins of WND. [(accessed on 25 July 2021)]. Available online: https://westnile.izs.it/j6_wnd/home.
- 37.Pérez-Ramírez E., Llorente F., Jiménez-Clavero M.Á. Experimental infections of wild birds with West Nile virus. Viruses. 2014;6:752–781. doi: 10.3390/v6020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taieb L., Ludwig A., Ogden N.H., Lindsay R.L., Iranpour M., Gagnon C.A., Bicout D.J. Bird Species Involved in West Nile Virus Epidemiological Cycle in Southern Québec. Int. J. Environ. Res. Public Health. 2020;17:4517. doi: 10.3390/ijerph17124517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel F., Sieg M., Fischer D., Keller M., Eiden M., Reuschel M., Schmidt V., Schwehn R., Rinder M., Urbaniak S., et al. Evidence for West Nile virus and Usutu virus infections in wild and resident birds in Germany, 2017 and 2018. Viruses. 2019;11:674. doi: 10.3390/v11070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.L’vov D.K., Dzharkenov A.F., L’vov D.N., Aristova V.A., Kovtunov A.I., Gromashevskiĭ V.L., Vyshemirskiĭ O.I., Galkina I.V., Al’khovskiĭ S.V., Samokhvalov E.I., et al. Isolation of the West Nile fever virus from the great cormorant Phalacrocorax carbo, the crow Corvus corone, and Hyalomma marginatum ticks associated with them in natural and synanthroic biocenosis in the Volga delta (Astrakhan region, 2001) Vopr. Virusol. 2002;47:7–12. [PubMed] [Google Scholar]
- 41.Ternovoĭ V.A., Protopopova E.V., Surmach S.G., Gazetdinov M.V., Zolotykh S.I., Shestopalov A.M., Pavlenko E.V., Leonova G.N., Loktev V.B. The genotyping of the West Nile virus in birds in the far eastern region of Russia in 2002–2004. Mol. Gen. Mikrobiol. Virusol. 2006;4:30–35. [PubMed] [Google Scholar]
- 42.Monaco F., Savini G., Calistri P., Polci A., Pinoni C., Bruno R., Lelli R. 2009 West Nile disease epidemic in Italy: First evidence of overwintering in Western Europe? Res. Vet. Sci. 2011;91:321–326. doi: 10.1016/j.rvsc.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Petrović T., Blázquez A.B., Lupulović D., Lazić G., Escribano-Romero E., Fabijan D., Kapetanov M., Lazić S., Saiz J.C. Monitoring West Nile virus (WNV) infection in wild birds in Serbia during 2012: First isolation and characterisation of WNV strains from Serbia. Eurosurveillance. 2013;18:20622. doi: 10.2807/1560-7917.ES2013.18.44.20622. [DOI] [PubMed] [Google Scholar]
- 44.Veo C., Della Ventura C., Moreno A., Rovida F., Percivalle E., Canziani S., Torri D., Calzolari M., Baldanti F., Galli M., et al. Evolutionary dynamics of the lineage 2 West Nile virus that caused the largest European epidemic: Italy 2011–2018. Viruses. 2019;11:814. doi: 10.3390/v11090814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinshaw V.S., Air G.M., Gibbs A.J., Graves L., Prescott B., Karunakaran D. Antigenic and genetic characterization of a novel hemagglutinin subtype of influenza A viruses from gulls. J. Virol. 1982;42:865–872. doi: 10.1128/jvi.42.3.865-872.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown J., Poulson R., Carter D., Lebarbenchon C., Pantin-Jackwood M., Spackman E., Shepherd E., Killian M., Stallknecht D. Susceptibility of avian species to North American H13 low pathogenic avian influenza viruses. Avian Dis. 2012;56:969–975. doi: 10.1637/10158-040912-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z.J., Kikutani Y., Nguyen L.T., Hiono T., Matsuno K., Okamatsu M., Krauss S., Webby R., Lee Y.J., Kida H., et al. H13 influenza viruses in wild birds have undergone genetic and antigenic diversification in nature. Virus Genes. 2018;54:543–549. doi: 10.1007/s11262-018-1573-0. [DOI] [PubMed] [Google Scholar]
- 48.Benkaroun J., Shoham D., Kroyer A.N., Whitney H., Lang A.S. Analysis of influenza A viruses from gulls: An evaluation of inter-regional movements and interactions with other avian and mammalian influenza A viruses. Cog Biol. 2016;2:1234957. doi: 10.1080/23312025.2016.1234957. [DOI] [Google Scholar]
- 49.Wille M., Robertson G.J., Whitney H., Bishop M.A., Runstadler J.A., Lang A.S. Extensive geographic mosaicism in avian influenza viruses from gulls in the northern hemisphere. PLoS ONE. 2011;6:e20664. doi: 10.1371/journal.pone.0020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebarbenchon C., Chang C.M., Gauthier-Clerc M., Thomas F., Renaud F., van der Werf S. H9N2 avian influenza virus in a Mediterranean gull. J. Mol. Gen. Med. Int. J. Biol. Res. 2009;3:121. doi: 10.4172/1747-0862.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Annual Report on Surveillance for Avian Influenza in Poultry and Wild Birds in Member States of the European Union in 2020. [(accessed on 30 August 2021)]. Available online: https://www.efsa.europa.eu/sites/default/files/2021-07/9985.pdf. [DOI] [PMC free article] [PubMed]
- 52.Teske L., Ryll M., Rautenschlein S. Epidemiological investigations on the role of clinically healthy racing pigeons as a reservoir for avian paramyxovirus-1 and avian influenza virus. Avian Pathol. 2013;42:557–565. doi: 10.1080/03079457.2013.852157. [DOI] [PubMed] [Google Scholar]
- 53.Schuler K.L., Green D.E., Justice-Allen A.E., Jaffe R., Cunningham M., Thomas N.J., Spalding M.G., Ip H.S. Expansion of an exotic species and concomitant disease outbreaks: Pigeon paramyxovirus in free-ranging Eurasian collared doves. EcoHealth. 2012;9:163–170. doi: 10.1007/s10393-012-0758-6. [DOI] [PubMed] [Google Scholar]
- 54.Alexander D.J. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 2011;40:547–558. doi: 10.1080/03079457.2011.618823. [DOI] [PubMed] [Google Scholar]
- 55.OIE-WHAIS Disease Situation. [(accessed on 31 August 2021)]. Available online: https://wahis.oie.int/#/dashboards/country-or-disease-dashboard.
- 56.Hicks J.T., Dimitrov K.M., Afonso C.L., Ramey A.M., Bahl J. Global phylodynamic analysis of avian paramyxovirus-1 provides evidence of inter-host transmission and intercontinental spatial diffusion. BMC Evol. Biol. 2019;19:1–15. doi: 10.1186/s12862-019-1431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cardenas G.S., Navarro L.R., Morales R., Olvera M.A., Marquez M.A., Merino R., Miller P.J., Afonso C.L. Molecular epidemiology of Newcastle disease in Mexico and the potential spillover of viruses from poultry into wild bird species. Appl Environ. Microbiol. 2013;79:4985–4992. doi: 10.1128/AEM.00993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayala A.J., Dimitrov K.M., Becker C.R., Goraichuk I.V., Arns C.W., Bolotin V.I., Ferreira H.L., Gerilovych A.P., Goujgoulova G.V., Martini M.C., et al. Presence of vaccine-derived Newcastle disease viruses in wild birds. PLoS ONE. 2016;11:e0162484. doi: 10.1371/journal.pone.0162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanetti F., Berinstein A., Pereda A., Taboga O., Carrillo E. Molecular characterization and phylogenetic analysis of Newcastle disease virus isolates from healthy wild birds. Avian Dis. 2005;49:546–550. doi: 10.1637/7381-051605R.1. [DOI] [PubMed] [Google Scholar]
- 60.Dodovski A., Popova Z., Savić V. Characterization of a Novel Avian Avulavirus 1 of Class I isolated from a Mute Swan (Cygnus Olor) in Macedonia in 2012. Maced. Vet. Rev. 2019;42:115–122. doi: 10.2478/macvetrev-2019-0015. [DOI] [Google Scholar]
- 61.Zhao S., Han Z., Shao Y., Kong X., Liu S. Pathogenicity of an avian paramyxovirus serotype 1 isolate from swan to chickens. Zhongguo Yufang Shouyi Xuebao/Chin. J. Prev. Vet. Med. 2015;37:140–142. [Google Scholar]
- 62.Manarolla G., Bakonyi T., Gallazzi D., Crosta L., Weissenböck H., Dorrestein G.M., Nowotny N. Usutu virus in wild birds in northern Italy. Vet. Microbiol. 2010;141:159–163. doi: 10.1016/j.vetmic.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 63.Pecorari M., Longo G., Gennari W., Grottola A., Sabbatini A.M., Tagliazucchi S., Savini G., Monaco F., Simone M.L., Lelli R., et al. First human case of Usutu virus neuroinvasive infection, Italy, August-September 2009. Eurosurveillance. 2009;14:19446. doi: 10.2807/ese.14.50.19446-en. [DOI] [PubMed] [Google Scholar]
- 64.Nemeth N.M., Beckett S., Edwards E., Klenk K., Komar N. Avian mortality surveillance for West Nile virus in Colorado. Am. J. Trop. Med. Hyg. 2007;76:431–437. doi: 10.4269/ajtmh.2007.76.431. [DOI] [PubMed] [Google Scholar]
- 65.Llopis I.V., Rossi L., Di Gennaro A., Mosca A., Teodori L., Tomassone L., Grego E., Monaco F., Lorusso A., Savini G. Further circulation of West Nile and Usutu viruses in wild birds in Italy. Infect. Genet. Evol. 2015;32:292–297. doi: 10.1016/j.meegid.2015.03.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analysed during this study are included in the published article.