Abstract
Simple Summary
The translocator protein 18 kDa (TSPO) gene is highly expressed in glioblastoma (GBM), the most common primary malignant brain tumor, which remains one of the most difficult tumors to treat. TSPO is located in the outer mitochondrial membrane and binds cholesterol through its C-terminal domain. One frequent single-nucleotide polymorphism (SNP) rs6971, which changes the alanine 147 into threonine (Ala147Thr), has been found in the C-terminal domain of the TSPO region and dramatically alters the affinity with which TSPO binds drug ligands. However, the potential association between the TSPO genetic variants and GBM clinical outcomes is not known. Here, we evaluated the effects of the Ala147Thr SNP localized in this TSPO region on biological, sex-specific, overall, and progression-free GBM survival. Our findings suggest an association between the TSPO rs6971 variant and adverse outcomes in male GBM patients but not in females. These findings also suggest that the TSPO rs6971 SNP could be used as a prognostic marker of survival in GBM patients.
Abstract
Glioblastoma (GBM) is the most common primary brain tumor in adults, with few available therapies and a five-year survival rate of 7.2%. Hence, strategies for improving GBM prognosis are urgently needed. The translocator protein 18kDa (TSPO) plays crucial roles in essential mitochondria-based physiological processes and is a validated biomarker of neuroinflammation, which is implicated in GBM progression. The TSPO gene has a germline single nucleotide polymorphism, rs6971, which is the most common SNP in the Caucasian population. High TSPO gene expression is associated with reduced survival in GBM patients; however, the relation between the most frequent TSPO genetic variant and GBM pathogenesis is not known. The present study retrospectively analyzed the correlation of the TSPO polymorphic variant rs6971 with overall and progression-free survival in GBM patients using three independent cohorts. TSPO rs6971 polymorphism was significantly associated with shorter overall survival and progression-free survival in male GBM patients but not in females in one large cohort of 441 patients. We observed similar trends in two other independent cohorts. These observations suggest that the TSPO rs6971 polymorphism could be a significant predictor of poor prognosis in GBM, with a potential for use as a prognosis biomarker in GBM patients. These results reveal for the first time a biological sex-specific relation between rs6971 TSPO polymorphism and GBM.
Keywords: TSPO, biomarker, glioblastoma, single nucleotide polymorphism, survival
1. Introduction
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults and accounts for 14.5% of all primary brain and central nervous system (CNS) neoplasms [1]. In the U.S., the incidence rate for GBM is 3.23 per 100,000 in the population, the highest for malignant tumors, and is 1.6 times higher in males than females [1]. Despite the aggressive and multimodal standard-of-care treatment, which includes maximal safe surgical resection followed by radiation in addition to concomitant and adjuvant chemotherapy, the prognosis remains extremely poor, with a five-year survival rate of 7.2% [1]. The median survival also shows a biological sex-based disproportion of 20.4 months for females and 17.5 months for males [2].
The unfavorable prognosis for GBM can be attributed to numerous features, including the intra- and inter-heterogeneity and intrinsic cell plasticity [3], a high rate of invasion to the brain parenchyma, a hypoxic intratumoral environment [4], the presence of cancer stem cells that contribute to the treatment resistance and recurrence [5], and an immunosuppressive tumor state [6]. Moreover, increasing evidence suggests that mitochondrial dysfunction plays a key role in the pathogenic events of GBM due to their role as central regulators of cell metabolism, cell death, oxidative stress, invasion, and inflammation [7].
The translocator protein 18 kDa (TSPO), previously known as the peripheral benzodiazepine receptor [8], has been extensively studied within the last two decades due to its location in the outer mitochondrial membrane (OMM). TSPO is known to play crucial roles in essential mitochondria-based physiological processes, such as mitochondrial respiration, metabolism, cellular bioenergetics, cholesterol transport and steroidogenesis, and heme biosynthesis, among others [8,9,10,11,12]. TSPO expression is most abundant in steroid-synthesizing tissues, whereas the heart and kidney express intermediate levels of TSPO. Interestingly, TSPO is expressed at low levels in the normal brain neuropil but becomes highly expressed following nervous system insults and neuroinflammation [13,14], highlighting the role of TSPO as a sensitive biomarker of brain injury and neuroinflammation. Given the clinical interest, TSPO positron-emission tomography (PET) imaging has been used in a wide variety of neuroinflammatory conditions [15,16,17,18,19,20], including high-grade gliomas [21,22,23,24].
The human TSPO gene located on chromosome 22q13.3 is made up of 4 exons with a large intron, containing repetitive sequences separating the first and second exons, a GC-rich promoter region, and multiple transcription initiation sites [25,26,27]. TSPO is a hydrophobic protein composed of 169 amino acids with a high degree of homology among species from bacteria to humans [9]. The structure of the protein has five putative transmembrane domains in which the C-terminus is exposed to the cytoplasm [28]. The cholesterol recognition amino acid consensus sequence has been identified on the interface between the fifth transmembrane domain and the cytosol-facing C-terminus domain (L144-S159) [29]. In addition, the TSPO gene has several polymorphisms, but only two variants, namely rs6971 and rs6972, are the most frequent in human populations and are located in the C-terminal domain [30]. The most studied variant, rs6971, is an A/G transition in exon 4 that leads to a non-conservative amino acid substitution, Ala147Thr, in the fifth transmembrane domain of the protein. This polymorphism destabilizes the protein, especially in the ligand-binding pocket, reducing the affinity of TSPO for cholesterol and its transport into mitochondria [31], consequently blunting the production of steroid precursors [29,32,33] and the neurosteroid pregnenolone in lymphomonocytes [34]. This single-nucleotide polymorphism (SNP) has been associated with psychiatric disorders, such as bipolar disorder, depression, and anxiety [35,36,37,38]. The structural alteration caused by the rs6971 polymorphism also decreases the distance between the second and fifth transmembrane domains [39], which dramatically reduces the binding affinity of second-generation TSPO-PET radiotracers [40,41]. The second most common though less studied polymorphism, rs6972, is a G/A transition also in exon 4 of the TSPO gene, leading to a single amino acid substitution, Arg162His, and has been reported to affect the conformation of the C-terminus through use of structural bioinformatics models [30].
Several studies have shown a positive correlation between TSPO expression and grade of malignancy and glioma cell proliferation and a negative correlation with survival in glioma patients [42,43,44]. Noteworthy, TSPO expression varies significantly depending on the GBM subtype and is significantly higher in isocitrate-dehydrogenase wild-type (IDHWT) compared to IDH mutant (IDHMUT) GBM [45]. However, the association between the most frequent TSPO genetic variant and GBM susceptibility, prognosis, and patient outcome is not known.
In the present study, we analyzed the association between rs6971 TSPO polymorphic variant and clinical outcomes for GBM patients. We used three independent cohorts of GBM patients to evaluate the correlation of TSPO genetic variant with the overall survival (OS) and progression-free survival (PFS) time, incorporating biological sex and treatment-specific analysis. Our results indicate allele-specific effects of TSPO rs6971 SNPs on the survival of GBM patients in a sex-specific manner.
2. Materials and Methods
2.1. Study Population
The Cleveland Clinic Foundation (CCF) cohort: Peripheral blood samples from 441 GBM patients were collected through the Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center (BBTC) at the Cleveland Clinic under IRB2559. White blood cells from each blood sample were isolated via Ficoll gradient and then snap-frozen and stored at −80 Celsius for research use. In this study, we selected all available GBM samples. Genomic DNA was extracted using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, MD, USA) following the manufacturer’s protocols. DNA purity and concentration were measured using a ThermoFisher NanoDrop spectrophotometer (ThermoFisher, Waltham, MA, USA).
The Case Western Reserve University (CWRU) cohort: Newly diagnosed, untreated brain tumor patients were identified for the Ohio Brain Tumor Study (OBTS) under approval from University Hospitals IRB CC296. Clinical and pathological data were gathered for each patient. Patient blood samples were collected at the time of consent. DNA extraction from whole blood samples was conducted using a Qiasymphony robotic platform (Qiagen) with Qiagen processing kits designed to maximize DNA (QIAsymphonyDSP DNA midi kit) yields and purity.
The Cancer Genome Atlas (TCGA) cohort: Raw BAM files from the TCGA-GBM cohort were utilized for the analysis of the TSPO rs6971 SNP using whole-exome sequencing data aligned by the TCGA. Alignment of the SNP rs6971 genotype was identified via the use of HaplotypeCaller, where samples with alternative counts at the reference position chr22:43162920 were identified. After classification of the samples by genotype, the phenotype data were downloaded via TCGA, and survival analysis was performed. Survival analysis was performed using a log-rank test via R version 4.1.0.
2.2. SNP Selection and Genotyping
Patient genotyping was performed using rhAmpTM SNP assays (IDT, custom design) for the human TSPO polymorphism: rs6971 (A→G). The genotyping reaction was carried out according to the manufacturer’s protocol. Briefly, 10 μL reaction volumes were prepared by combining 5.3 μL of combined master mix (IDT, 1076018) and reporter mix (IDT, 1076024), 0.5 μL rhAmpTM SNP assay, 2.2 μL nuclease-free water, and 2 μL diluted DNA sample. gBlocks gene fragments (IDT, custom design) for each SNP were used as positive controls for all genotyping. Reactions were performed in an Applied Biosystems QuantStudioTM 3 (Thermo Fisher, MA, USA) instrument with cycling parameters as specified by the assay manufacturer.
2.3. GlioVis Analysis
Gene expression and GBM patient survival data from the TCGA Project, the Chinese Glioma Genome Atlas (CGGA) database, REMBRANDT brain cancer dataset, and Gravendeel database, were analyzed using GlioVis (Version 0.20 (18 March 2016)) (http://gliovis.bioinfo.cnio.es/, accessed on 10 May 2021) [46]. We only included adult patients from all the databases and stratified patients by biological sex, when available. We used patients’ data from the TCGA-GBM database whose tumors had Affymetrix HG-U133A, Agilent-4502A, and RNA-seq RNA expression for our gene of interest. Pairwise comparisons using t-test (with Bonferroni correction) was performed. p-values of the pairwise comparisons are indicated in the graphs as *** p < 0.001; ** p < 0.01; * p < 0.05; and ns, not significant.
2.4. Statistical Analysis
Demographic and clinical characteristics were compared between rs6971 polymorphism genotype groups. Wilcoxon rank-sum test was used to assess differences in continuous data. Differences in categorical data were assessed through Pearson’s chi-square test and Fisher’s exact test where appropriate (cell counts less than 5 cases). Kaplan-Meier curves were generated and log-rank test performed to assess differences in OS and PFS between groups. These survival differences were also assessed for patients who had received standard of care and surgery (gross total or near total resection). Median survival and 95% confidence intervals for OS and PFS are reported. All statistical analyses were performed in R (version 4.0.5). R package gtsummary was used to generate descriptive tables, and packages survival and survminer were used to generate the survival curves. p-values under 0.05 are statistically significant.
3. Results
3.1. TSPO Expression Is Upregulated in GBM Patient Samples
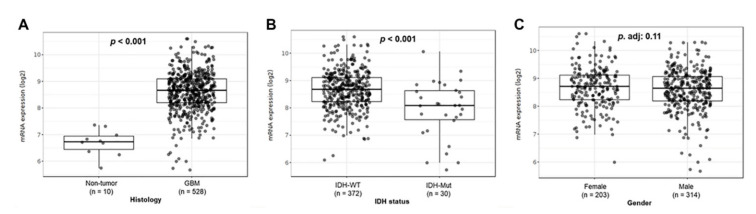
To evaluate TSPO expression in GBM, we conducted a TSPO mRNA expression analysis in TCGA-GBM dataset using GlioVis platform. We observed a significant increase in TSPO gene expression in GBM compared to that of non-tumor brain tissue (p < 0.001) (Figure 1A). A significant increase in the mRNA TSPO expression in GBM tumor samples was also observed comparing the three different RNA data platforms available from the TCGA-GBM database as well as in the REMBRANDT and Gravendeel independent datasets (p < 0.001) (Figure S1).
Figure 1.
TSPO gene-expression analysis in TCGA-GBM cohort. Patient GBM samples from the TCGA-GBM database, using the platform Affymetrix HG-U133A, were analyzed using GlioVis. (A) mRNA expression levels in GBM samples compared to tumor-free brain samples. (B) Expression levels of TSPO in GBM patients grouped by IDH status (IDH-WT vs. IDH-Mut). (C) TSPO expression levels between males and females with GBM. The number of patients (n) and p-value of the pairwise comparisons using t-test (with Bonferroni correction) are indicated in each graph.
Similarly, we also found higher gene expression of TSPO in IDH wild-type GBM subtype compared to IDH mutant GBM subtype in the TCGA-GBM dataset (p < 0.001) (Figure 1B). Interestingly, no significant difference was observed when comparing females versus males in the same GBM cohort (Figure 1C). In addition, we analyzed TSPO expression in the three different GBM subtypes as classified by Phillips and colleagues [47,48] using the TCGA, CGGA, Rembrandt, and Gravendeel independent datasets. We found higher TSPO expression that was statistically significant in the mesenchymal subtype as compared to classical and proneural subtypes in the analyzed datasets (Figure S2).
A recent study reported a new pathway-based classification of GBM identifying four different subtypes that included metabolic and developmental attributes, namely proliferative/progenitor (PPR), mitochondrial (MTC), neuronal (NEU), and glycolytic/plurimetabolic (GPM). As TSPO is a mitochondrial protein, we analyzed its expression in these four newly described subtypes and found higher gene expression of TSPO that was statistically significant in the MTC subtype as compared to PPR and NEU subtypes. We also found higher TSPO expression in the GPM subtype as compared to the PPR subtype (Figure S3).
Additionally, we analyzed the correlation of the TSPO gene expression with commonly mutated genes in GBM (PTEN, TP53, EGFR, PIK3R1, PIK3CA, NF1, and RB1) using the TCGA dataset. Out of all the genes, we only found higher TSPO gene expression that was statistically significant in TP53 wild-type GBM subtype compared to TP53 mutant (Figure S4).
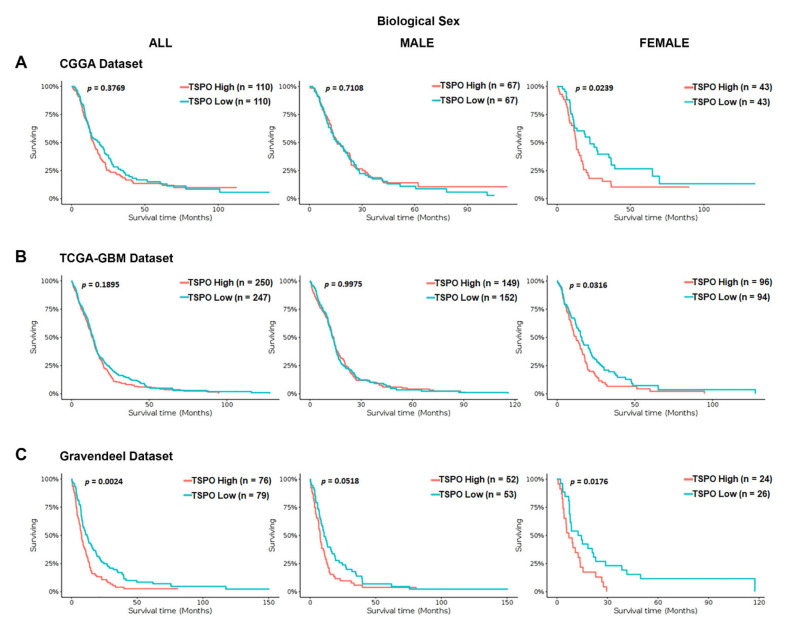
To investigate the association between TSPO mRNA expression and OS time in GBM patients, we interrogated a series of independent datasets (the Chinese glioma genome atlas (CGGA), TCGA-GBM using Affymetrix HG-U133A platform, and Gravendeel). We stratified the GBM patients in each database based on their biological sex. We found a statistically significant difference in the OS time for the entire population only in the Gravendeel dataset (p = 0.0024) (Figure 2). Interestingly, when GBM patients were stratified by biological sex in the three analyzed databases, high TSPO gene expression was associated with a significantly shorter median survival time only in the females and not in males (Figure 2).
Figure 2.
Kaplan–Meier survival curves of TSPO mRNA expression in GBM patients stratified by biological sex using the CGGA, TCGA-GBM, and Gravendeel datasets. Overall survival of GBM patients with a high or low expression of TSPO, using the median as the cutoff value, was analyzed using the (A) CGGA database, (B) the TCGA-GBM database (Affymetrix HG-U133A platform), and (C) Gravendeel database, available at GlioVis data portal. Survival curves analysis were stratified by biological sex, including the entire population (left panel), male population (middle panel), and the female population (right panel) for the three independent databases. Genotype patient numbers and p-values are indicated in each graph, and the median and statistical data are in Table S1.
3.2. Patients Characteristics in CCF and CWRU Datasets
Based on the observations of increased TSPO mRNA in GBM patients and an association to OS time in a sex-specific manner, we retrospectively analyzed a large dataset that included 441 GBM patients from CCF. The patient’s characteristics based on the TSPO rs6971 polymorphism genotype are detailed in Table 1. In total, we identified 34 patients with the homozygous wild-type A/A genotype, 175 patients with the heterozygous A/G genotype, and 232 patients with the homozygous variant G/G genotype. There was a borderline significant difference (p = 0.056) in the mean age at diagnosis between the wild-type A/A genotype (54 years old) and the variant G/G genotype (61 years old) or the heterozygous A/G genotype (61 years old). No significant difference was observed in the ratio of males to females between the three different genotypes, maintaining a higher prevalence in males (67%) than in females (33%) regardless of the genotype. Strikingly, the effect of the TSPO rs6971 polymorphism on OS time was statistically significant (p < 0.001), showing a median of 13 months for the variant G/G genotype patients or a median of 12 months for the heterozygous A/G genotype patients compared with a median of 25 months for wild-type A/A genotype patients. Furthermore, the TSPO rs6971 polymorphism was also significantly associated with the PFS time (p = 0.003). The variant G/G and the heterozygous A/G genotypes were associated with worse PFS times (seven and five months respectively) when compared with the wild-type A/A genotype (nine months). This indicates that the presence of only one mutated G allele is associated with a shorter OS and PFS time in GBM patients. Patients were also stratified into those receiving surgery and standard of care treatment, Karnofsky Performance Score (KPS), and tumor recurrence, but no significant association was found with any genotype.
Table 1.
Patient Characteristics by TSPO rs6971 polymorphism genotype from the CCF dataset.
| Characteristics | Overall N = 441 1 |
G/G Variant N = 232 1 |
A/G Heterozygous N = 175 1 |
A/A Wild Type N = 34 1 |
p-Value 2 |
|---|---|---|---|---|---|
| Age at Diagnosis | 61 (53, 70) | 61 (53, 70) | 61 (54, 70) | 54 (46, 67) | 0.056 |
| Sex | >0.9 | ||||
| Female | 145 (33%) | 76 (33%) | 57 (33%) | 12 (35%) | |
| Male | 296 (67%) | 156 (67%) | 118 (67%) | 22 (65%) | |
| Surgery | 0.9 | ||||
| Gross Total Resection | 152 (49%) | 79 (49%) | 59 (48%) | 14 (48%) | |
| Near Total Resection | 73 (23%) | 37 (23%) | 31 (25%) | 5 (17%) | |
| Subtotal Resection | 88 (28%) | 46 (28%) | 32 (26%) | 10 (34%) | |
| Unknown | 128 | 70 | 53 | 5 | |
| Standard of Care | 03 | ||||
| No | 66 (15%) | 36 (16%) | 28 (16%) | 2 (5.9%) | |
| Yes | 375 (85%) | 196 (84%) | 147 (84%) | 32 (94%) | |
| KPS | >0.9 | ||||
| <=70 | 62 (14%) | 32 (14%) | 25 (15%) | 5 (15%) | |
| 70+ | 370 (86%) | 195 (86%) | 147 (85%) | 28 (85%) | |
| Unknown | 9 | 5 | 3 | 1 | |
| Overall Survival (Months) | 14 (7, 26) | 13 (7, 27) | 12 (6, 20) | 25 (16, 52) | <0.001 |
| Vital Status | 0.009 | ||||
| Alive | 42 (9.6%) | 31 (13%) | 8 (4.6%) | 3 (8.8%) | |
| Deceased | 397 (90%) | 200 (87%) | 166 (95%) | 31 (91%) | |
| Unknown | 2 | 1 | 1 | 0 | |
| Progression Free Survival | 6 (3, 12) | 7 (3, 13) | 5 (3, 10) | 9 (4, 27) | 0.003 |
| Recurrence | 0.5 | ||||
| No | 114 (26%) | 60 (26%) | 48 (28%) | 6 (18%) | |
| Yes | 323 (74%) | 169 (74%) | 126 (72%) | 28 (82%) | |
| Unknown | 4 | 3 | 1 | 0 |
1 Median (IQR); n (%) 2 Kruskal–Wallis rank-sum test; Pearson’s chi-square test; Fisher’s exact test. Note: Unknown demographic and clinical characteristics are included in the table but are not assessed in statistical tests.
As a validation cohort, we retrospectively analyzed the CWRU dataset that included 131 GBM patients. Table 2 details the patient’s characteristics based on the TSPO rs6971 polymorphism genotype. Out of the 131 GBM patients included in CWRU database, we identified 10 patients with the homozygous wild-type A/A genotype, 67 patients with the heterozygous A/G genotype, and 54 patients with the homozygous variant G/G genotype. There was no difference in the mean age at diagnosis between any of the genotypes; however, there was a significant difference (p = 0.014) in the ratios of males to females between genotypes. The variant G/G genotype was identified in 22% of females and 78% of males as compared with the heterozygous A/G genotype, which was present in 43% of females and 57% of males, and the wild-type A/A genotype identified in 60% of females and 40% of males. No significant association was found between any genotype and the KPS score, tumor recurrence, vital status, and the treatment received.
Table 2.
Patient Characteristics by TSPO rs6971 polymorphism genotype from the CWRU dataset.
| Characteristic | Overall N = 131 1 |
G/G Variant N = 54 1 |
A/G Heterozygous N = 67 1 |
A/A Wild Type N = 10 1 |
p-Value 2 |
|---|---|---|---|---|---|
| Age at Diagnosis | 64 (55, 69) | 64 (55, 70) | 62 (56, 68) | 62 (50, 71) | 0.5 |
| Sex | 0.014 | ||||
| Female | 47 (36%) | 12 (22%) | 29 (43%) | 6 (60%) | |
| Male | 84 (64%) | 42 (78%) | 38 (57%) | 4 (40%) | |
| Surgery | 0.7 | ||||
| Gross Total Resection | 75 (59%) | 33 (62%) | 37 (57%) | 5 (50%) | |
| Subtotal Resection | 53 (41%) | 20 (38%) | 28 (43%) | 5 (50%) | |
| Unknown | 3 | 1 | 2 | 0 | |
| Standard of Care | 0.7 | ||||
| No | 45 (37%) | 20 (39%) | 21 (33%) | 4 (44%) | |
| Yes | 78 (63%) | 31 (61%) | 42 (67%) | 5 (56%) | |
| Unknown | 8 | 3 | 4 | 1 | |
| KPS | 0.7 | ||||
| ≤70 | 56 (60%) | 23 (61%) | 29 (62%) | 4 (44%) | |
| 70+ | 38 (40%) | 15 (39%) | 18 (38%) | 5 (56%) | |
| Unknown | 37 | 16 | 20 | 1 | |
| Overall Survival (Months) | 12 (5, 21) | 10 (5, 16) | 13 (6, 22) | 18 (9, 24) | 0.4 |
| Vital Status | 0.7 | ||||
| Alive | 3 (2.3%) | 2 (3.7%) | 1 (1.5%) | 0 (0%) | |
| Deceased | 128 (98%) | 52 (96%) | 66 (99%) | 10 (100%) | |
| Progression Free Survival | 8 (4, 12) | 6 (4, 10) | 10 (5, 13) | 9 (5, 17) | 0.11 |
| Unknown | 57 | 22 | 32 | 3 | |
| Recurrence | 0.6 | ||||
| No | 56 (43%) | 22 (41%) | 31 (47%) | 3 (30%) | |
| Yes | 74 (57%) | 32 (59%) | 35 (53%) | 7 (70%) | |
| Unknown | 1 | 0 | 1 | 0 |
1 Median (IQR); n (%) 2 Kruskal–Wallis rank-sum test; Fisher’s exact test Note: Unknown demographic and clinical characteristics are included in the table but are not assessed in statistical tests.
3.3. High Frequency of TSPO rs6971 Polymorphism in the CCF and CWRU Datasets
Out of the 441 GBM patients in the CCF dataset, 92% carry at least one mutated G allele for the TSPO rs6971 polymorphism, with a global minor allele frequency (MAF) of 0.725. This TSPO SNP polymorphism exhibit a higher frequency in the GBM population analyzed than the previously reported MAF of 0.3 in Caucasian populations [30]. We also observed that out of the 131 GBM patients in the CWRU dataset, 92% of the GBM patients carry at least one mutated G allele for the TSPO rs6971 polymorphism, with a MAF of 0.668. Notably, our cohorts are also mostly Caucasian (frequency >90%). Table 3 shows the comparison between both datasets.
Table 3.
Comparison between CCF and CWRU datasets for TSPO rs6971 polymorphism.
| Characteristic | Overall N = 572 1 |
CCF Dataset N = 411 1 |
CWRU Dataset N = 131 1 |
p-Value 2 |
|---|---|---|---|---|
| Age at Diagnosis | 62 (53, 70) | 61 (53, 70) | 64 (55, 69) | 0.2 |
| Sex | 0.6 | |||
| Female | 197 (34%) | 150 (33%) | 47 (36%) | |
| Male | 385 (66%) | 301 (67%) | 84 (64%) | |
| Race | 0.9 | |||
| Black/African-American | 12 (2.8%) | 4 (3.1%) | ||
| White | 408 (94%) | 127 (97%) | ||
| rs6971 SNP Status | 0.056 | |||
| G/G variant | 286 (50%) | 232 (53%) | 54 (41%) | |
| A/G heterozygous | 242 (42%) | 175 (40%) | 67 (51%) | |
| A/A wild type | 44 (7.7%) | 34 (7.7%) | 10 (7.6%) | |
| Unknown | 10 | 10 | 0 |
1 Median (IQR); n (%). 2 Wilcoxon rank-sum test; Pearson’s chi-square test; Fisher’s exact test.
3.4. The TSPO rs6971 Polymorphism Is Associated with Worse Overall Survival in GBM Patients Stratified by Biological Sex and Treatment
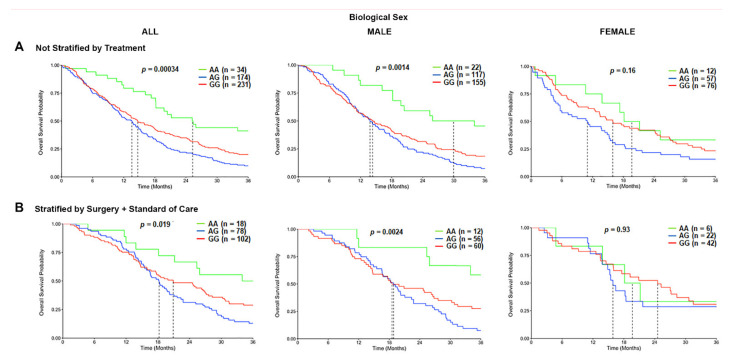
Based on the initial differences observed, we evaluated the association of the TSPO rs6971 polymorphism on the OS in GBM patients from the CCF cohort. In the overall population, a statistically significant difference was observed in the median survival time for the variant G/G genotype patients (14.7 months) and the heterozygous A/G genotype patients (13.6 months), as compared with wild-type A/A genotype patients (25.3 months) (p = 0.00034; log-rank test) (Figure 3A). When patients were stratified by the biological sex, the variant G/G genotype (14.3 months) and the heterozygous A/G genotype (13.8 months) were associated with a significantly worse OS time only in males as compared with the wild-type A/A genotype (29.9 months) (Figure 3A). This effect was not observed in females (Figure 3A). We further stratified the patients by treatment, comprising those who underwent surgical resection followed by the standard of care and biological sex. The OS time was significantly worse among those variant G/G genotype patients (20.9 months) and the heterozygous A/G patients (18.2 months) that received surgery and standard of care as compared with the wild-type A/A genotype patients (36.2 months) that also received the same treatment (p = 0.019; log-rank test) in the overall population. This association was only significantly different in the male subgroup that received surgery and standard of care treatment, where the variant G/G genotype patients (18.5 months) and the heterozygous A/G patients (18.8 months) exhibited a shorter OS time as compared with the wild-type A/A genotype patients (51.3 months) (p = 0.0024) (Figure 3B). Again, this effect was not observed in the females who also received the same therapeutic approach (Figure 3B). Similar trends were observed in the CWRU dataset (Figure S5A,B); however, it was not statistically significant due to the cohort being underpowered (only N = 10 of the wild type compared to N = 54 of the variant).
Figure 3.
Overall survival (OS) curves in the carriers of the TSPO rs6971 polymorphism stratified by genotype, biological sex, and treatment, using the CCF cohort. (A) Overall survival probability curves in the entire population (left panel), male subgroup (middle panel), and female subgroup (right panel) comparing the wild-type A/A, heterozygous A/G, and variant G/G genotypes for the TSPO rs6971 polymorphism using the patient data from the CCF cohort. (B) Overall survival curves stratifying by treatment (those who underwent surgical resection followed by standard of care treatment) in the entire population (left panel), the male subgroup (middle panel), and the female subgroup (right panel) comparing the wild-type A/A, heterozygous A/G, and variant G/G genotypes for the TSPO rs6971 polymorphism. Genotype patient numbers and p-values are indicated in each graph, and the median and statistical data are in Table S1.
In addition, we analyzed a third independent cohort using the TCGA-GBM database and found a borderline significant difference (p = 0.08; log-rank test) in the median survival time only in males for the variant G/G genotype patients (12.5 months) and the heterozygous A/G genotype patients (12 months) as compared with the wild-type A/A genotype patients (13.5 months). This effect was not observed in females (Figure S6). There was no significant association of the TSPO rs6971 polymorphism with the OS time in the overall population for any of the three genotypes (Figure S6). We could not stratify patients based on the treatment received due to a lack of clinical information in the database.
3.5. The TSPO rs6971 Polymorphism Is Associated with Worse Progression-Free Survival in GBM Patients Stratified by Biological Sex and Treatment
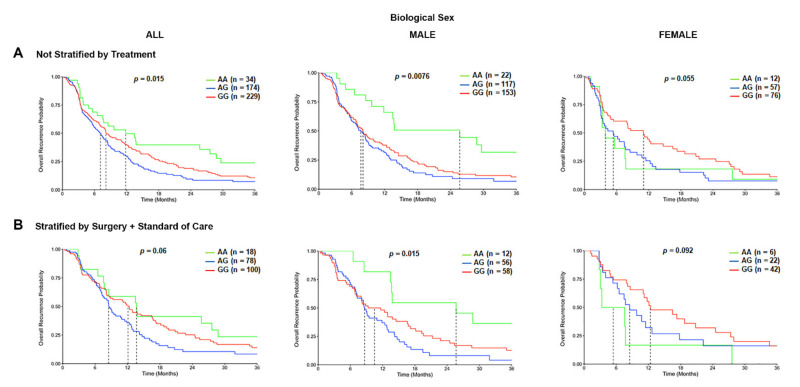
We further defined the association of TSPO rs6971 polymorphism on the overall recurrence probability or PFS of GBM patients in the CCF cohort. Patients with the variant G/G genotype or the heterozygous A/G genotype for the rs6971 SNP exhibited a significantly worse PFS time in the overall population (8.1 months and 7.1 months respectively) as compared with the wild-type A/A genotype (11.8 months) (p = 0.015) (Figure 4A). This difference was even more significant only in males (p = 0.0076) as compared with females (p = 0.055) (Figure 4A). When patients were also stratified by treatment received, the PFS time was significantly worse in males (p = 0.015) among those variant G/G genotype patients or the A/G genotype patients (10.5 months and 8.6 months, respectively) as compared with the wild-type A/A genotype patients (25.7 months) that also received the same treatment (Figure 4B). This association was not significant in the overall population nor in females who also received the same therapeutic approach (Figure 4B).
Figure 4.
Progression-free survival (PFS) curves in the carriers of TSPO rs6971 polymorphism stratified by genotype, biological sex, and treatment, using the CCF cohort. (A) Overall recurrence probability curves in the entire population (left panel), male subgroup (middle panel), and female subgroup (right panel) comparing the wild-type A/A, heterozygous A/G, and variant G/G genotypes for the TSPO rs6971 polymorphism using patient data from the CCF cohort. (B) Overall recurrence probability curves stratifying by treatment (those who underwent surgical resection followed by standard of care treatment) in the entire population (left panel), the male subgroup (middle panel), and the female subgroup (right panel) comparing the wild-type A/A, heterozygous A/G, and variant G/G genotypes for the TSPO rs6971 polymorphism. Genotype patient numbers and p-values are indicated in each graph, and the median and statistical data are in Table 1.
In the CWRU dataset, we also observed a similar trend to the CCF dataset (Figure S7A) that was not statistically significant due to underpower (only N = 10 of the wild type compared to N = 54 of the variant). When patients were further stratified by treatment received, the PFS time was worst only in the overall population (p = 0.018) among those variant G/G genotype patients that received surgery and standard of care (5.8 months) as compared with the wild-type A/A genotype patients (8.5 months) or the heterozygous A/G genotype patients (11.1 months) that also received the same treatment (Figure S7B). This significant difference was not observed when patients were stratified by biological sex. We could not analyze the PFS time in the TCGA database due to the lack of clinical information in the database.
4. Discussion
In this study, global TSPO gene expression was analyzed in publicly available, independent datasets where we observed a statistically significant worse survival time only in female GBM patients with high TSPO gene expression. These results showed an association of TSPO with biological sex-specific mechanisms in GBM clinical outcomes. This prompted us to perform genotyping of the most frequent TSPO polymorphism, rs6971, in GBM patient samples stratified by sex, using a large cohort from the Cleveland Clinic. We showed statistically significantly worse survival time in male carriers of at least one variant G allele in the TSPO rs6971 SNP as compared with the carriers of the A/A wild-type genotype. In addition, the rs6971 G/G variant genotype and A/G heterozygous genotype carriers also showed a significantly worse recurrence-free survival time among male GBM patients as compared with the carriers of A/A wild-type genotype. This indicates that the presence of only one mutated G allele in the TSPO rs6971 polymorphism could be sufficient to predict a shorter survival time in male GBM patients. We also used two independent GBM patient cohorts to validate our results, and although the OS and PFS analysis did not show statistically significant association with the TSPO SNP, we observed that the rs6971 wild-type A/A genotype carriers have a better survival time only in GBM male patients as compared with the carriers of at least one G allele, showing a similar trend to the CCF cohort.
The CCF dataset is a carefully curated cohort from a single institution, based on the clinical characteristics, where all the patients completed the cycles of the standard treatment. The CWRU dataset comprised fewer patients compared to the primary cohort (CCF dataset), and among these, only 10 patients exhibited the wild-type A/A genotype. Moreover, these numbers got even smaller when stratifying the patients by biological sex and treatment received, which drives the statistical power downward. The TCGA dataset is an older cohort where the data were collected from multiple institutions all over the world to reach their accrual targets (usually around 500 specimens per cancer type). This dataset is extremely heterogenous and was acquired as part of routine care and not as part of a controlled research study or clinical trial. The clinical survival endpoint analyses in the TCGA dataset are approximate because of the lack of absolute verification of the cause of death and relatively short and uniformly clinical follow-up records. All these differences among the different cohorts analyzed can explain the statistical limitations found in this study for the CWRU and the TCGA-GBM datasets [49]. To the best of our knowledge, this study is the first to evaluate the effect of TSPO genetic polymorphisms on mortality among patients with GBM in a sex-specific context.
In the CCF dataset, the TSPO rs6971 SNP reach a frequency of 0.725, which may indicate that in the GBM patient population, the most common allele for the TSPO rs6971 SNP is the mutated one and may have an important role in the pathogenesis of GBM. Because steroid molecules are involved in several biological functions, this TSPO polymorphism could be associated with the susceptibility to or protection against diseases that have been associated with a decreased or increased production of steroids.
Interestingly, in all the cohorts evaluated, the prevalence of GBM is higher in males than in females, corroborating the reported sex differences in the incidence of GBM [1]. Furthermore, our results in differential survival times among male and female carriers of distinct germline variants in the TSPO gene support the role of gender-specific molecular patterns in GBM. Current epidemiological data indicate that the male to female incidence ratio for GBM is 1.6:1 in the United States [1]. Several studies suggest that patient outcomes also differ between males and females in the adult GBM patient population [50,51]. It is evident that biological sex differences are inherent drivers of GBM incidence and survival; therefore, the elucidation of biological sex-specific mechanisms in GBM has the potential to improve patient outcomes and develop more personalized GBM therapies.
Elevated TSPO expression is a hallmark of gliosis (microglia and astrocytes), which has led to the use of TSPO as a robust marker for brain injury and neuroinflammation [13,14], using both ex-vivo and in-vivo imaging methods. TSPO-PET imaging is increasingly being used in neuro-oncology, especially in glioma patients, and has been shown to be a useful tool for assessing tumor progression at follow-up [21,52]. Moreover, several reports found a positive correlation between tumor grade and TSPO expression, with the highest expression observed in GBM [42,43]. However, little is known about the functional role of TSPO in GBM pathophysiology. Interestingly, we observed that TSPO has significantly different expression levels among the different GBM genetic subtypes, showing the highest expression in the mesenchymal subtype in the four independent datasets analyzed. Mesenchymal tumors are predominantly IDH wild-type [53,54], and this subtype is associated with poor radiation response and worse survival [55], which is in line with our finding that TSPO is highly expressed in IDH-wild type subtype and is correlated with worse overall survival. Furthermore, we found higher TSPO gene expression in the newly described mitochondrial and glycolytic/plurimetabolic GBM subtypes [56], which is consistent with the mitochondrial associated function of TSPO. While the mitochondrial GBM subtype was associated with the most favorable clinical outcome, with a marked vulnerability to inhibitors of oxidative phosphorylation, the glycolytic/plurimetabolic subtype exhibits a poor prognosis and is sustained by concurrent activation of multiple energy-producing pathways. Interestingly, this cluster is also enriched in mesenchymal and immune-related functions.
TSPO is overexpressed in activated microglia and macrophages in the surrounding areas of GBM [45] and could play an immunomodulatory role in the GBM microenvironment. However, more studies are required to assess the specific mechanisms of TSPO in the GBM pathophysiology and to specifically associate TSPO gene expression with any of these newly described GBM subgroups. Several reports indicate that TSPO impacts the cell bioenergetic profile by modulating ATP production, supporting a pro-proliferative role of TSPO in GBM [57,58,59,60]. In contrast, other studies support an anti-proliferative role of TSPO, showing that TSPO knockdown and TSPO ligands promote cell proliferation and migration due to a decrease in apoptosis [61,62,63]. Studies indicate that TSPO is involved in the regulation of cell death; however, the exact mechanism and to what extent TSPO regulates the resistance of GBM cells to apoptosis is not known. This contradictory evidence may be due to context-dependent factors that may influence the role of TSPO in GBM. This may include differences in cell lines, species, experimental conditions, and signaling pathways. Moreover, TSPO ligand effects are concentration- and tissue-specific [64,65]. It is important to note that these biological studies have lacked either analysis of or incorporation of TSPO polymorphic genotypes in the study design. Based on the results shown here, it may be plausible to conclude that the differences observed between studies may be due to varying influences/presence of TSPO polymorphisms in the study samples.
In addition, the large inter-individual variability found in TSPO-PET brain signal in several clinical trials may be explained by the presence of the genetic polymorphisms in the TSPO gene, which affects the binding affinity patterns among different TSPO radioligands [30,40]. While there is some evidence that TSPO genetic polymorphisms, specifically the rs6971 SNP, may be associated with certain psychiatric disorders, such as bipolar and panic disorders [37,66], it is not yet known whether TSPO polymorphisms may have clinical implications in the detection and treatment of GBM patients. Further studies will be required to examine these effects in greater detail.
Our findings provide evidence that the status of the TSPO polymorphic variant may be useful in predicting GBM patient outcomes in a sex-specific manner. This remains to be validated in a larger curated cohort. In an underpowered cohort, we observed the expected trends seen in this study. Unfortunately, due to the rarity of GBM, this cohort was unpowered to fully validate our initial findings in the CCF cohort. Additional in-vitro and in-vivo studies are warranted to fully assess the role of TSPO polymorphisms in GBM, with a specific focus on biological sex differences, but these studies are beyond the scope of this paper. Further studies are required to understand the exact mechanisms and signaling pathways involved in the regulation of GBM malignancy by the different TSPO genetic variants. There is evidence that steroid hormone receptors are expressed in GBM [67], and the inhibition of such receptors can promote GBM cell death in vivo and in vitro [68,69,70]. It is interesting to postulate that the association of TSPO genetic polymorphisms with steroid hormonal pathways may play a role in GBM development and/or progression, particularly in a sex-dependent manner.
5. Conclusions
Taken together, our data provide evidence suggesting that the TSPO rs6971 gene variant may be a useful indicator of survival time in GBM patients, with the presence suggesting a poorer survival outlook in male patients. This is the first study that reveals the potential for a biological sex-specific relation between rs6971 TSPO polymorphism and GBM clinical outcome. These results, taken together with other studies, provide evidence that TSPO may play a significant role in GBM pathogenesis. Moreover, our results suggest that Ala147Thr TSPO gene polymorphism has the potential to be useful as a prognostic marker of OS and PFS in GBM patients. Future studies will focus on independent validation of these sex-based results and the potential of TSPO rs6971 polymorphism to be used as a prognostic biomarker for GBM.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13184525/s1: Figure S1: TSPO Gene-Expression Analysis in Different GBM Cohort Datasets; Figure S2: TSPO Gene-Expression Analysis in GBM Subtypes Using Different GBM Cohort Datasets; Figure S3: TSPO Gene-Expression Levels in the Four Newly Described GBM Subtypes; Figure S4: Correlation of TSPO Gene-Expression Analysis with the Most Common Mutated Genes in GBM; Figure S5: Overall Survival Curves in the Carriers of the TSPO rs6971 Polymorphism Stratified by Genotype, Biological Sex, and Treatment using the CWRU Dataset; Figure S6: Overall Survival Curves in the Carriers of the TSPO rs6971 Polymorphism Stratified by Genotype and Biological Sex, Using the TCGA Dataset; Figure S7: Progression-Free Survival (PFS) Curves in the Carriers of TSPO rs6971 Polymorphisms Stratified by Genotype, Biological Sex, and Treatment using the CWRU dataset; Table S1: Statistical Analysis Data for Kaplan–Meier Survival Curves of Figure 2, Figure 3 and Figure 4.
Author Contributions
Original idea, T.R.G.; conceptualization, T.R.G., J.D.L., and D.J.A.; methodology, K.M.T., A.M.A.d.l.R., T.J.A., J.S.B.S., G.C., J.D.L., T.R.G., and D.J.A.; formal analysis, K.M.T., A.M.A.d.l.R., T.J.A., and G.C.; resources, J.S.B.S., J.D.L., T.R.G., and D.J.A.; data curation, K.A.W., J.S.B.S., J.D.L., K.M.T., and G.C.; writing—original draft preparation, A.M.A.d.l.R., D.J.A., and T.R.G.; writing—review and editing, K.M.T., A.M.A.d.l.R., T.J.A., M.M.G., B.O., G.C., K.A.W., J.S.B.S., J.D.L., T.R.G., and D.J.A.; funding acquisition, T.R.G. and D.J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIEHS, grant number R01ES007062-23 (Tomás R. Guilarte) and FIU-ORED, project ID: 800006850.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Cleveland Clinic (IRB2559) and University Hospitals Cleveland Medical Center (IRB CC296, approval 14 May 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Publicly available datasets were analyzed in this study. The TCGA dataset can be found here: (https://portal.gdc.cancer.gov/, accessed on 10 May 2021), the CGGA dataset can be found here: (http://www.cgga.org.cn/, accessed on 10 May 2021), The Gravendeel and Rembrandt datasets are available in the GEO database with the accession numbers (GSE16011) and (GSE108474), respectively. The CCF and CWRU datasets presented in this study are available upon request from the senior authors. The data are not publicly available as they are not deidentified.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ostrom Q.T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro. Oncol. 2020;22:iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gittleman H., Ostrom Q.T., Stetson L.C., Waite K., Hodges T.R., Wright C.H., Wright J., Rubin J.B., Berens M.E., Lathia J., et al. Sex is an important prognostic factor for glioblastoma but not for nonglioblastoma. Neurooncol. Pract. 2019;6:451–462. doi: 10.1093/nop/npz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suter R.K., Rodriguez-Blanco J., Ayad N.G. Epigenetic pathways and plasticity in brain tumors. Neurobiol. Dis. 2020;145:105060. doi: 10.1016/j.nbd.2020.105060. [DOI] [PubMed] [Google Scholar]
- 4.Colwell N., Larion M., Giles A.J., Seldomridge A.N., Sizdahkhani S., Gilbert M.R., Park D.M. Hypoxia in the glioblastoma microenvironment: Shaping the phenotype of cancer stem-like cells. Neuro. Oncol. 2017;19:887–896. doi: 10.1093/neuonc/now258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauko A., Lo A., Ahluwalia M.S., Lathia J.D. Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin. Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locarno C.V., Simonelli M., Carenza C., Capucetti A., Stanzani E., Lorenzi E., Persico P., Della Bella S., Passoni L., Mavilio D., et al. Role of myeloid cells in the immunosuppressive microenvironment in gliomas. Immunobiology. 2020;225:151853. doi: 10.1016/j.imbio.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Giampazolias E., Tait S.W. Mitochondria and the hallmarks of cancer. FEBS J. 2016;283:803–814. doi: 10.1111/febs.13603. [DOI] [PubMed] [Google Scholar]
- 8.Papadopoulos V., Baraldi M., Guilarte T.R., Knudsen T.B., Lacapere J.J., Lindemann P., Norenberg M.D., Nutt D., Weizman A., Zhang M.R., et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends. Pharmacol. Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Casellas P., Galiegue S., Basile A.S. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 2002;40:475–486. doi: 10.1016/S0197-0186(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch T., Decaudin D., Susin S.A., Marchetti P., Larochette N., Resche-Rigon M., Kroemer G. PK11195, a ligand of the mitochondrial benzodiazepine receptor, facilitates the induction of apoptosis and reverses Bcl-2-mediated cytoprotection. Exp. Cell. Res. 1998;241:426–434. doi: 10.1006/excr.1998.4084. [DOI] [PubMed] [Google Scholar]
- 11.Rupprecht R., Papadopoulos V., Rammes G., Baghai T.C., Fan J., Akula N., Groyer G., Adams D., Schumacher M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug. Discov. 2010;9:971–988. doi: 10.1038/nrd3295. [DOI] [PubMed] [Google Scholar]
- 12.Veenman L., Papadopoulos V., Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): Regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr. Pharm. Des. 2007;13:2385–2405. doi: 10.2174/138161207781368710. [DOI] [PubMed] [Google Scholar]
- 13.Chen M.K., Guilarte T.R. Translocator protein 18 kDa (TSPO): Molecular sensor of brain injury and repair. Pharmacol. Ther. 2008;118:1–17. doi: 10.1016/j.pharmthera.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilarte T.R. TSPO in diverse CNS pathologies and psychiatric disease: A critical review and a way forward. Pharmacol. Ther. 2019;194:44–58. doi: 10.1016/j.pharmthera.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banati R.B., Newcombe J., Gunn R.N., Cagnin A., Turkheimer F., Heppner F., Price G., Wegner F., Giovannoni G., Miller D.H., et al. The peripheral benzodiazepine binding site in the brain in multiple sclerosis: Quantitative in vivo imaging of microglia as a measure of disease activity. Brain. 2000;123:2321–2337. doi: 10.1093/brain/123.11.2321. [DOI] [PubMed] [Google Scholar]
- 16.Turner M.R., Cagnin A., Turkheimer F.E., Miller C.C., Shaw C.E., Brooks D.J., Leigh P.N., Banati R.B. Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: An [11C](R)-PK11195 positron emission tomography study. Neurobiol. Dis. 2004;15:601–609. doi: 10.1016/j.nbd.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Coughlin J.M., Wang Y., Munro C.A., Ma S., Yue C., Chen S., Airan R., Kim P.K., Adams A.V., Garcia C., et al. Neuroinflammation and brain atrophy in former NFL players: An in vivo multimodal imaging pilot study. Neurobiol. Dis. 2015;74:58–65. doi: 10.1016/j.nbd.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amhaoul H., Hamaide J., Bertoglio D., Reichel S.N., Verhaeghe J., Geerts E., Van Dam D., De Deyn P.P., Kumar-Singh S., Katsifis A., et al. Brain inflammation in a chronic epilepsy model: Evolving pattern of the translocator protein during epileptogenesis. Neurobiol. Dis. 2015;82:526–539. doi: 10.1016/j.nbd.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Pavese N., Gerhard A., Tai Y.F., Ho A.K., Turkheimer F., Barker R.A., Brooks D.J., Piccini P. Microglial activation correlates with severity in Huntington disease: A clinical and PET study. Neurology. 2006;66:1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 20.Werry E.L., Bright F.M., Piguet O., Ittner L.M., Halliday G.M., Hodges J.R., Kiernan M.C., Loy C.T., Kril J.J., Kassiou M. Recent Developments in TSPO PET Imaging as A Biomarker of Neuroinflammation in Neurodegenerative Disorders. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20133161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Z., Roncaroli F., Durrenberger P.F., Coope D.J., Karabatsou K., Hinz R., Thompson G., Turkheimer F.E., Janczar K., Du Plessis D., et al. The 18-kDa mitochondrial translocator protein in human gliomas: An 11C-(R)PK11195 PET imaging and neuropathology study. J. Nucl. Med. 2015;56:512–517. doi: 10.2967/jnumed.114.151621. [DOI] [PubMed] [Google Scholar]
- 22.Su Z., Herholz K., Gerhard A., Roncaroli F., Du Plessis D., Jackson A., Turkheimer F., Hinz R. [(1)(1)C]-(R)PK11195 tracer kinetics in the brain of glioma patients and a comparison of two referencing approaches. Eur. J. Nucl. Med. Mol. Imaging. 2013;40:1406–1419. doi: 10.1007/s00259-013-2447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasnain N., Mustafa R.M., Bakhshi S.K., Shamim M.S. Efficacy of Positron Emission Tomography in distinguishing brain tumours from inflammation. J. Pak. Med. Assoc. 2020;70:2291–2293. [PubMed] [Google Scholar]
- 24.Zinnhardt B., Muther M., Roll W., Backhaus P., Jeibmann A., Foray C., Barca C., Doring C., Tavitian B., Dolle F., et al. TSPO imaging-guided characterization of the immunosuppressive myeloid tumor microenvironment in patients with malignant glioma. Neuro. Oncol. 2020;22:1030–1043. doi: 10.1093/neuonc/noaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riond J., Mattei M.G., Kaghad M., Dumont X., Guillemot J.C., Le Fur G., Caput D., Ferrara P. Molecular cloning and chromosomal localization of a human peripheral-type benzodiazepine receptor. Eur. J. Biochem. 1991;195:305–311. doi: 10.1111/j.1432-1033.1991.tb15707.x. [DOI] [PubMed] [Google Scholar]
- 26.Chang Y.J., McCabe R.T., Rennert H., Budarf M.L., Sayegh R., Emanuel B.S., Skolnick P., Strauss J.F., 3rd The human "peripheral-type" benzodiazepine receptor: Regional mapping of the gene and characterization of the receptor expressed from cDNA. DNA Cell Biol. 1992;11:471–480. doi: 10.1089/dna.1992.11.471. [DOI] [PubMed] [Google Scholar]
- 27.Giatzakis C., Papadopoulos V. Differential utilization of the promoter of peripheral-type benzodiazepine receptor by steroidogenic versus nonsteroidogenic cell lines and the role of Sp1 and Sp3 in the regulation of basal activity. Endocrinology. 2004;145:1113–1123. doi: 10.1210/en.2003-1330. [DOI] [PubMed] [Google Scholar]
- 28.Bernassau J.M., Reversat J.L., Ferrara P., Caput D., Lefur G. A 3D model of the peripheral benzodiazepine receptor and its implication in intra mitochondrial cholesterol transport. J. Mol. Graph. 1993;11:236–244. doi: 10.1016/0263-7855(93)80003-A. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 30.Milenkovic V.M., Bader S., Sudria-Lopez D., Siebert R., Brandl C., Nothdurfter C., Weber B.H.F., Rupprecht R., Wetzel C.H. Effects of genetic variants in the TSPO gene on protein structure and stability. PLoS One. 2018;13:e0195627. doi: 10.1371/journal.pone.0195627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen D.R., Fan J., Campioli E., Venugopal S., Midzak A., Daly E., Harlay A., Issop L., Libri V., Kalogiannopoulou D., et al. TSPO mutations in rats and a human polymorphism impair the rate of steroid synthesis. Biochem J. 2017;474:3985–3999. doi: 10.1042/BCJ20170648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacapere J.J., Papadopoulos V. Peripheral-type benzodiazepine receptor: Structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids. 2003;68:569–585. doi: 10.1016/S0039-128X(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 33.Prossin A.R., Chandler M., Ryan K.A., Saunders E.F., Kamali M., Papadopoulos V., Zollner S., Dantzer R., McInnis M.G. Functional TSPO polymorphism predicts variance in the diurnal cortisol rhythm in bipolar disorder. Psychoneuroendocrinology. 2018;89:194–202. doi: 10.1016/j.psyneuen.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa B., Pini S., Gabelloni P., Da Pozzo E., Abelli M., Lari L., Preve M., Lucacchini A., Cassano G.B., Martini C. The spontaneous Ala147Thr amino acid substitution within the translocator protein influences pregnenolone production in lymphomonocytes of healthy individuals. Endocrinology. 2009;150:5438–5445. doi: 10.1210/en.2009-0752. [DOI] [PubMed] [Google Scholar]
- 35.Costa B., Pini S., Martini C., Abelli M., Gabelloni P., Landi S., Muti M., Gesi C., Lari L., Cardini A., et al. Ala147Thr substitution in translocator protein is associated with adult separation anxiety in patients with depression. Psychiatr. Genet. 2009;19:110–111. doi: 10.1097/YPG.0b013e32832080f6. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura K., Yamada K., Iwayama Y., Toyota T., Furukawa A., Takimoto T., Terayama H., Iwahashi K., Takei N., Minabe Y., et al. Evidence that variation in the peripheral benzodiazepine receptor (PBR) gene influences susceptibility to panic disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:222–226. doi: 10.1002/ajmg.b.30211. [DOI] [PubMed] [Google Scholar]
- 37.Colasanti A., Owen D.R., Grozeva D., Rabiner E.A., Matthews P.M., Craddock N., Young A.H. Bipolar Disorder is associated with the rs6971 polymorphism in the gene encoding 18 kDa Translocator Protein (TSPO) Psychoneuroendocrinology. 2013;38:2826–2829. doi: 10.1016/j.psyneuen.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da Pozzo E., Costa B., Martini C. Translocator protein (TSPO) and neurosteroids: Implications in psychiatric disorders. Curr. Mol. Med. 2012;12:426–442. doi: 10.2174/156652412800163451. [DOI] [PubMed] [Google Scholar]
- 39.Li F., Liu J., Zheng Y., Garavito R.M., Ferguson-Miller S. Protein structure. Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism. Science. 2015;347:555–558. doi: 10.1126/science.1260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen D.R., Yeo A.J., Gunn R.N., Song K., Wadsworth G., Lewis A., Rhodes C., Pulford D.J., Bennacef I., Parker C.A., et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J. Cereb. Blood Flow Metab. 2012;32:1–5. doi: 10.1038/jcbfm.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rojas C., Stathis M., Coughlin J.M., Pomper M., Slusher B.S. The Low-Affinity Binding of Second Generation Radiotracers Targeting TSPO is Associated with a Unique Allosteric Binding Site. J. Neuroimmune. Pharmacol. 2018;13:1–5. doi: 10.1007/s11481-017-9765-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miettinen H., Kononen J., Haapasalo H., Helen P., Sallinen P., Harjuntausta T., Helin H., Alho H. Expression of peripheral-type benzodiazepine receptor and diazepam binding inhibitor in human astrocytomas: Relationship to cell proliferation. Cancer Res. 1995;55:2691–2695. [PubMed] [Google Scholar]
- 43.Vlodavsky E., Soustiel J.F. Immunohistochemical expression of peripheral benzodiazepine receptors in human astrocytomas and its correlation with grade of malignancy, proliferation, apoptosis and survival. J. Neurooncol. 2007;81:1–7. doi: 10.1007/s11060-006-9199-9. [DOI] [PubMed] [Google Scholar]
- 44.Unterrainer M., Fleischmann D.F., Vettermann F., Ruf V., Kaiser L., Nelwan D., Lindner S., Brendel M., Wenter V., Stocklein S., et al. TSPO PET, tumour grading and molecular genetics in histologically verified glioma: A correlative (18)F-GE-180 PET study. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:1368–1380. doi: 10.1007/s00259-019-04491-5. [DOI] [PubMed] [Google Scholar]
- 45.Cai L., Kirchleitner S.V., Zhao D., Li M., Tonn J.C., Glass R., Kalin R.E. Glioblastoma Exhibits Inter-Individual Heterogeneity of TSPO and LAT1 Expression in Neoplastic and Parenchymal Cells. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21020612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowman R.L., Wang Q., Carro A., Verhaak R.G., Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro. Oncol. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., de Carvalho A.C., Lyu S., Li P., Li Y., et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2018;33:152. doi: 10.1016/j.ccell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 49.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V., et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrom Q.T., Rubin J.B., Lathia J.D., Berens M.E., Barnholtz-Sloan J.S. Females have the survival advantage in glioblastoma. Neuro. Oncol. 2018;20:576–577. doi: 10.1093/neuonc/noy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trifiletti D.M., Alonso C., Grover S., Fadul C.E., Sheehan J.P., Showalter T.N. Prognostic Implications of Extent of Resection in Glioblastoma: Analysis from a Large Database. World Neurosurg. 2017;103:330–340. doi: 10.1016/j.wneu.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 52.Roncaroli F., Su Z., Herholz K., Gerhard A., Turkheimer F.E. TSPO expression in brain tumours: Is TSPO a target for brain tumour imaging? Clin. Transl. Imaging. 2016;4:145–156. doi: 10.1007/s40336-016-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhat K.P.L., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J.D., Goodman L.D., et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garofano L., Migliozzi S., Oh Y.T., D’Angelo F., Najac R.D., Ko A., Frangaj B., Caruso F.P., Yu K., Yuan J., et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer. 2021;2:141–156. doi: 10.1038/s43018-020-00159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veenman L., Levin E., Weisinger G., Leschiner S., Spanier I., Snyder S.H., Weizman A., Gavish M. Peripheral-type benzodiazepine receptor density and in vitro tumorigenicity of glioma cell lines. Biochem. Pharmacol. 2004;68:689–698. doi: 10.1016/j.bcp.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Rechichi M., Salvetti A., Chelli B., Costa B., Da Pozzo E., Spinetti F., Lena A., Evangelista M., Rainaldi G., Martini C., et al. TSPO over-expression increases motility, transmigration and proliferation properties of C6 rat glioma cells. Biochim. Biophys. Acta. 2008;1782:118–125. doi: 10.1016/j.bbadis.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Levin E., Premkumar A., Veenman L., Kugler W., Leschiner S., Spanier I., Weisinger G., Lakomek M., Weizman A., Snyder S.H., et al. The peripheral-type benzodiazepine receptor and tumorigenicity: Isoquinoline binding protein (IBP) antisense knockdown in the C6 glioma cell line. Biochemistry. 2005;44:9924–9935. doi: 10.1021/bi050150s. [DOI] [PubMed] [Google Scholar]
- 60.Daniele S., Taliani S., Da Pozzo E., Giacomelli C., Costa B., Trincavelli M.L., Rossi L., La Pietra V., Barresi E., Carotenuto A., et al. Apoptosis therapy in cancer: The first single-molecule co-activating p53 and the translocator protein in glioblastoma. Sci. Rep. 2014;4:4749. doi: 10.1038/srep04749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bode J., Veenman L., Caballero B., Lakomek M., Kugler W., Gavish M. The 18 kDa translocator protein influences angiogenesis, as well as aggressiveness, adhesion, migration, and proliferation of glioblastoma cells. Pharmacogenet. Genomics. 2012;22:538–550. doi: 10.1097/FPC.0b013e3283539cdc. [DOI] [PubMed] [Google Scholar]
- 62.Fu Y., Wang D., Wang H., Cai M., Li C., Zhang X., Chen H., Hu Y., Zhang X., Ying M., et al. TSPO deficiency induces mitochondrial dysfunction, leading to hypoxia, angiogenesis, and a growth-promoting metabolic shift toward glycolysis in glioblastoma. Neuro Oncol. 2020;22:240–252. doi: 10.1093/neuonc/noz183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kugler W., Veenman L., Shandalov Y., Leschiner S., Spanier I., Lakomek M., Gavish M. Ligands of the mitochondrial 18 kDa translocator protein attenuate apoptosis of human glioblastoma cells exposed to erucylphosphohomocholine. Cell Oncol. 2008;30:435–450. doi: 10.3233/clo-2008-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bader S., Wolf L., Milenkovic V.M., Gruber M., Nothdurfter C., Rupprecht R., Wetzel C.H. Differential effects of TSPO ligands on mitochondrial function in mouse microglia cells. Psychoneuroendocrinology. 2019;106:65–76. doi: 10.1016/j.psyneuen.2019.03.029. [DOI] [PubMed] [Google Scholar]
- 65.Gavish M., Veenman L. Regulation of Mitochondrial, Cellular, and Organismal Functions by TSPO. Adv. Pharmacol. 2018;82:103–136. doi: 10.1016/bs.apha.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Vaht M. Variation rs6971 in the Translocator Protein Gene (TSPO) is Associated with Aggressiveness and Impulsivity but Not with Anxiety in a Population-Representative Sample of Young Adults. J. Genet. Psychol. 2021;182:149–162. doi: 10.1080/00221325.2021.1896470. [DOI] [PubMed] [Google Scholar]
- 67.Yague J.G., Lavaque E., Carretero J., Azcoitia I., Garcia-Segura L.M. Aromatase, the enzyme responsible for estrogen biosynthesis, is expressed by human and rat glioblastomas. Neurosci. Lett. 2004;368:279–284. doi: 10.1016/j.neulet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Yu X., Jiang Y., Wei W., Cong P., Ding Y., Xiang L., Wu K. Androgen receptor signaling regulates growth of glioblastoma multiforme in men. Tumour Biol. 2015;36:967–972. doi: 10.1007/s13277-014-2709-z. [DOI] [PubMed] [Google Scholar]
- 69.Bao D., Cheng C., Lan X., Xing R., Chen Z., Zhao H., Sun J., Wang Y., Niu C., Zhang B., et al. Regulation of p53wt glioma cell proliferation by androgen receptor-mediated inhibition of small VCP/p97-interacting protein expression. Oncotarget. 2017;8:23142–23154. doi: 10.18632/oncotarget.15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zalcman N., Canello T., Ovadia H., Charbit H., Zelikovitch B., Mordechai A., Fellig Y., Rabani S., Shahar T., Lossos A., et al. Androgen receptor: A potential therapeutic target for glioblastoma. Oncotarget. 2018;9:19980–19993. doi: 10.18632/oncotarget.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. The TCGA dataset can be found here: (https://portal.gdc.cancer.gov/, accessed on 10 May 2021), the CGGA dataset can be found here: (http://www.cgga.org.cn/, accessed on 10 May 2021), The Gravendeel and Rembrandt datasets are available in the GEO database with the accession numbers (GSE16011) and (GSE108474), respectively. The CCF and CWRU datasets presented in this study are available upon request from the senior authors. The data are not publicly available as they are not deidentified.