Abstract
Trypanosomatids are easy to cultivate and they are (in many cases) amenable to genetic manipulation. Genome sequencing has become a standard tool routinely used in the study of these flagellates. In this review, we summarize the current state of the field and our vision of what needs to be done in order to achieve a more comprehensive picture of trypanosomatid evolution. This will also help to illuminate the lineage-specific proteins and pathways, which can be used as potential targets in treating diseases caused by these parasites.
Keywords: trypanosomatids, next-generation sequencing, genomics
1. Introduction
The flagellates of the family Trypanosomatidae represent one of the most evolutionarily successful groups of parasitic protists, adapted to an extremely wide range of hosts—from various animals (mainly insects and vertebrates) to flowering plants and even ciliates. Depending on whether their life cycle includes a single host or there is an obligate alternation between two different hosts, trypanosomatids are subdivided into monoxenous (predominantly insect parasites) and dixenous (typically insect-transmitted parasites of vertebrates or plants) [1]. Most research efforts have been focused on studying dixenous trypanosomatids of the genera Trypanosoma and Leishmania, which cause severe (often fatal) diseases in humans and domestic animals. Therefore, sequencing of trypanosomatid genomes started from the three important human pathogens: Trypanosoma brucei, T. cruzi, and Leishmania major [2,3,4]. A comparative study has shown that despite differences in genome size and gene content, these species share a relatively high level of gene order conservation (synteny) and overall genomic organization: most protein-coding genes are intron-less and form conserved polycistronic gene clusters, whereas species-specific genes predominate sub-telomeric or internal non-syntenic chromosomal regions [5]. The subsequent genomic studies expectedly focused on other species of these two genera with the clear preference for Leishmania, since it contains more species infective to humans. At the time of writing this review, the assembled genome sequences for multiple isolates of 24 species of Leishmania and about a dozen species and subspecies of the genus Trypanosoma are available in public databases (Table S1).
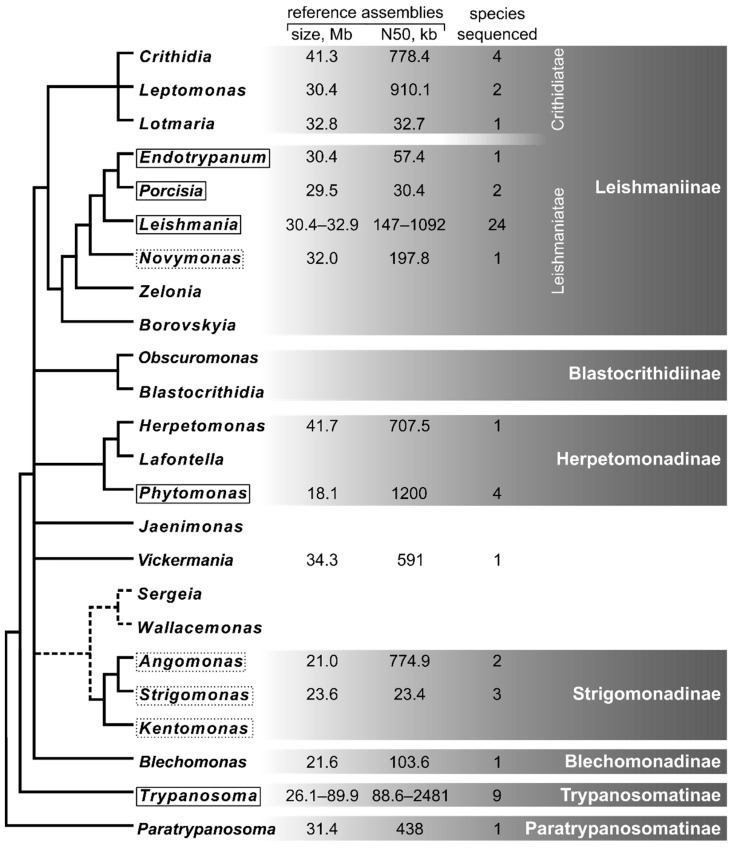
However, the diversity of trypanosomatids is predominantly represented by monoxenous parasites, from which their dixenous kin have originated at least three times independently [1]. These cases are Leishmania (along with Endotrypanum and Porcisia) spp. within subfamily Leishmaniinae, Phytomonas spp. in the subfamily Herpetomonadinae, and Trypanosoma spp. constituting a separate early-diverging lineage (Figure 1). The research interest in the monoxenous trypanosomatids has significantly increased in the last decade; of note, 12 out of the 19 currently recognized genera of these flagellates have been described within this short period [1].
Figure 1.
Schematic phylogenetic tree of Trypanosomatidae with a summary on sequenced genomes. Solid and dashed boxes mark dixenous and endosymbiont-bearing genera, respectively.
The studies of insect-dwelling flagellates are important for better understanding not only the biology of their dixenous relatives, but also eukaryotic evolution in general [6]. For example, the members of the genus Blastocrithidia evolved an idiosyncratic genetic code with all three stop codons used for coding amino acids [7]. Some trypanosomatids, namely Novymonas and the three genera of the subfamily Strigomonadinae (Angomonas, Strigomonas, and Kentomonas) harbor intracellular bacterial symbionts [8,9,10]. These endosymbionts complement the metabolic requirements of their flagellate hosts with pathways responsible for the synthesis of amino acids, vitamins, and heme [11,12,13,14]. The unusual genus Vickermania became biflagellate by disrupting the processes of cell division and flagellum duplication to resist the fly midgut peristaltic flow in the absence of an opportunity to attach to the intestinal wall [15]. Various monoxenous trypanosomatids independently acquired thermotolerance, a prerequisite of the transition to dixeny, and some of them have even been documented in vertebrates [16,17,18]. Below, we review the current state of genomic research in trypanosomatids with a focus on monoxenous species. The taxonomy is presented in accordance with [1].
2. Trypanosoma spp.
The first trypanosome, whose genome had been sequenced and analyzed, was the agent of African animal trypanosomiasis—T. brucei brucei [3,19] (Table S1). The studies of human-infective T. b. gambiense and T. b. rhodesiense demonstrated extremely high similarity of the genomes in all three subspecies, conservation of the variant surface glycoprotein (VSG) repertoire, and only rare segmental duplications [20,21]. In T. b. evansi, mechanically transmitted by insects and lacking kinetoplast, the procyclin-associated genes needed for the development in the vector have been lost or disrupted, and the γ-subunit of ATP synthase, which is involved in generation of the mitochondrial membrane potential in the absence of kDNA, has mutated [22,23]. The comparison of the genomes of all the above subspecies did not allow identification of factors leading to pathogenicity in humans. Two draft genome assemblies of T. b. equiperdum, which is dyskinetoplastic (lacks part of its kDNA) due to the loss of the vector part of its life cycle, have been published with no accompanied analysis [24,25]. Several studies of the genome of the tsetse-transmitted T. congolense focused on the analysis of its VSG repertoire and its comparison to that of T. brucei [26,27,28,29]. They revealed several important differences in the organization and functioning of the VSG expression sites, including the absence of conserved repeats flanking the VSG loci and the scarcity of expression site associated genes in T. congolense, and the scale of recombination. Trypanosoma vivax genome encodes the most diverse VSG repertoire among all investigated trypanosomes [26,30].
The studies of the T. cruzi genome involved numerous strains of this species, allowing to improve the quality of the existing assemblies and providing a deeper insight into its population structure [31,32,33,34,35,36,37,38,39,40,41]. A recent genome analysis of two T. cruzi strains revealed that the rapid evolution of gene families involved in immune evasion is one of the major contributors to the intraspecific genome variation in this species [42]. Interestingly, despite the shorter overall length, multiple genes were acquired by lateral gene transfer and some gene families underwent expansions in the genome of a bat-infecting species T. marinkellei, which is closely related to T. cruzi [43]. Genomes of human non-pathogenic T. rangeli and the bat parasite T. conorhini, representing a clade related to that of T. cruzi, have less retrotransposons and multigene family copies, but more genes involved in the biosynthesis of carbohydrates [44,45]. The crocodile-infecting species T. grayi was shown to lack surface proteins (mucins and VSGs), which are characteristic for other trypanosomes investigated thus far [46]. The genome analysis of ruminant-parasitizing T. theileri revealed several new families of surface proteins, as well as a general conservation of core cellular metabolic pathways [47].
What needs to be done: The genus Trypanosoma corresponds rather to a subfamily than to a single genus—it is very speciose (over 500 described species) and diverse. According to the latest taxonomical revision, it includes sixteen subgenera and several undescribed lineages of the same level [1]. Only a few of these have been analyzed to date, and this significantly limits our understanding of the evolution of parasitism in this group (Table S1). Surprisingly, the genome of one of the most common trypanosome species, flea-transmitted T. lewisi, which typically inhabits rats [48], but occasionally infects humans [49], has not been analyzed yet. Of special interest would be the genomic analyses of anuran trypanosomes (subgenus Trypanosoma), which gave rise to the parasites of fish and may represent the ancestral group for all terrestrial subgenera [50]. The representatives of this subgenus are expected to keep archaic traits of genomic organization, inherent to the common ancestor of trypanosomes, and their study using NGS might shed light on the origin and evolution of some important gene families, such as VSGs, procyclins, mucins, etc.
3. Dixenous Leishmaniinae
Out of the four Leishmania subgenera, i.e., Leishmania, Mundinia, Sauroleishmania, and Viannia, early genomic studies have focused on the first one (in particular, L. major, L. donovani, L. infantum, L. mexicana), and only L. (V.) braziliensis was used for comparison. Those studies revealed extremely high synteny levels, interspecific differences in the gene content, and associations of some genes with drug resistance phenotype [2,51,52] (Table S1). More L. (Leishmania) species and strains were analyzed later [53,54,55,56,57,58,59,60,61,62,63].
Later on, the subgenus Viannia started to receive more attention. Comparative genomic analysis of L. braziliensis and L. peruviana demonstrated substantial differences in gene content, chromosome copy number, as well as numerous SNPs and indels [64,65,66]. Sequencing of L. panamensis genome uncovered several mobile elements absent from the genomes of L. (Leishmania), along with a higher number of pseudogenes compared to the latter [67]. The study of L. naiffi and L. guyanensis genomes identified common features of the subgenus Viannia, such as aneuploidy, the presence of about 20 subgenus-specific gene families, and a high content of TATE transposons [68,69].
The early genomic study of a lizard parasite L. (Sauroleishmania) tarentolae demonstrated the loss of genes involved in oxidative stress protection and vesicular-mediated protein transport, as well as those expressed in L. (Leishmania) amastigotes. Meanwhile, the surface glycoprotein GP63 and promastigote surface antigen PSA31C gene families are expanded in this species [70,71]. Other studies of a species from this subgenus—L. adleri infecting rodents and lizards—has identified gene amplification, changes in chromosome copy number, and chromosome fission events [72,73].
The genome assemblies of L. (Mundinia) spp. were found to be similar in size to those of Sauroleishmania, but smaller than those of Leishmania and Viannia, due to multiple gene losses and gene family contractions [74]. The absence or reduction in the number of lipophosphoglycan-modifying side chain galactosyltransferases and arabinosyltransferases, as well as β-amastins has confirmed previous reports on the differences in cell surface architecture in L. (Mundinia) and other Leishmania spp. [75,76,77].
Endotrypanum monterogeii and Porcisia spp., being dixenous parasites of sloths and porcupines, respectively, represent the closest known relatives of Leishmania. The recently published analysis of their genomic sequences shed light on the evolution of pathogenicity in dixenous Leishmaniinae, which appears to be shaped mainly by changes in the amastin repertoire [78].
L. donovani and L. braziliensis, are the only trypanosomatids, to which single-cell genome sequencing approach has been applied thus far [79]. While the respective methods are widely used in human and cancer research, their application is restricted to just a handful of pathogenic species, including some apicomplexans and Leishmania [80]. Single-cell genome sequencing is instrumental in investigation of the haplotype diversity and de novo mutations in populations of pathogens. It allowed to characterize the karyotypes of L. braziliensis cells demonstrating mosaic aneuploidy [79]. A combination of multiple types of omics data originating from single trypanosomatid cells will provide a holistic view on the interactions of these pathogens with their hosts.
What needs to be done: The genus Leishmania is not as speciose as Trypanosoma, and the genomes for most of its representatives have been already sequenced with the exception of the poorly studied subgenus Sauroleishmania, for which 19 species have been described [81]. The peculiarities of the life cycles of these lizard-dwelling flagellates, such as their presence in the host gut and ability to infect a wide range of the mononuclear cells, erythrocytes, and thrombocytes [82,83], warrant further studies. Meanwhile, only one species has been analyzed for the genus Endotrypanum—E. monterogeii, and adding at least E. colombiensis (previously classified into Leishmania [84]), which can infect humans, would be important for understanding the pathogenesis of these flagellates. In addition, several genome assemblies of Leishmania spp. are available in public databases waiting to be analyzed and put into the context of comparative studies [85,86,87,88].
4. Monoxenous Leishmaniinae
Genomes for several monoxenous representatives of the subfamily Leishmaniinae have been sequenced and analyzed. The study of Lotmaria passim, Crithidia bombi, and C. expoeki, parasitizing agriculturally important Hymenoptera (honeybees and bumblebees), demonstrated numerous examples of horizontal gene transfer [89,90]. Genomic analysis of the latter two species at the population level has revealed that different strains vary considerably in terms of single nucleotide polymorphisms and gene copy number with a pattern fitting a scenario of rapid host-parasite coevolution, where the selective advantage of a given parasite strain is only temporary [91]. The genome and transcriptome sequencing of Leptomonas seymouri, the species repeatedly found in clinical samples along with Leishmania donovani [92], has allowed identifying its pre-adaptations to dixeny [17]. The genomic data of Leptomonas pyrrhocoris, an omnipresent parasite of firebugs, which has been proposed as a new model trypanosomatid species, were used to find new virulence factors of Leishmania [93]. The transcriptomic study of Crithidia thermophila showed a clear distinction in the mechanisms of thermotolerance in this species and L. seymouri [16]. The C. fasciculata RNA-seq data were used to elucidate potential mechanisms for insect-specific adhesion in trypanosomatids [94]. The available genomic data of C. acanthocephali made possible the comparative analysis of the endosymbiont-bearing and aposymbiotic species [14]. Two species closely related to C. fasciculata have been recently reported from human infections and their genomes have been sequenced [18,95]. The genome of the endosymbiont-bearing Novymonas esmeraldas, the closest known relative of dixenous Leishmaniinae, revealed a very similar gene content to the latter with the large number of GP63 proteases and pteridin/biopterin transporters, recognized virulence factors of Leishmania spp. Owing to the presence of the endosymbiont, this flagellate became prototrophic for all amino acids, heme, and most vitamins, i.e., even more independent of the presence of essential nutrients in the host than Strigomonadinae [12,96].
What needs to be done: Sequencing of additional species belonging to the non-monophyletic genera Crithidia and Leptomonas will help to delineate the entangled taxonomy of the infrafamily Crithidiatae (Figure 1). In addition, this lineage presents good examples of species with narrow and broad host specificity, which would be interesting to compare from the genomic point of view (e.g., L. pyrrhocoris is restricted to firebugs [97], while various species of true bugs and flies are documented for C. brevicula [98,99]). Although Novymonas is the closest relative of dixenous Leishmaniinae, the acquisition of endosymbionts resulted in very specific adaptations. Therefore, sequencing the genomes of other monoxenous trypanosomatids of the infrafamily Leishmaniatae (genera Zelonia [84] and Borovskyia [100]) is needed to illuminate the evolutionary origin and molecular signatures of dixenous Leishmaniinae.
5. Herpetomonadinae
The less studied lineage Herpetomonadinae is another subfamily containing dixenous parasites (plant-dwelling Phytomonas spp.) along with their monoxenous relatives (Figure 1). Some of the latter appear to be on the way to dixeny, as judged by their detection in plants [101,102] or vertebrates [103]. The analysis of four available genomes of Phytomonas spp. (those are Phytomonas spp. (isolates EM1 and Hart1) [104], P. serpens (isolate 9T) [105], and P. françai [106]) revealed additional peculiarities of these plant-inhabiting flagellates, such as significant genome streamlining at the expense of intergenic regions, mobile elements and narrowed gene repertoires, as well as the absence of some electron transport chain proteins. The only Herpetomonas species whose genome has been sequenced to date is H. muscarum [14,107]. It was used as a reference for the comparative analyses either with endosymbiont-bearing or dixenous trypanosomatids, therefore it is not clear what are its own peculiarities.
What needs to be done: Of special interest would be genomic studies of the speciose genus Herpetomonas, which actively explores various ecological niches. Ancestrally, these flagellates are parasites of (brachyceran) flies, but some of them switched to parasitism in true bugs, cockroaches, mosquitoes, or biting midges, while one species, H. samuelpessoai, demonstrates an astonishing ecological plasticity and has been isolated also from plants and even a human patient [1]. These should shed light on the adaptation of trypanosomatids to different hosts and environments. Although the genomes of Phytomonas spp. have been already investigated, the analysis was restricted to only four species inhabiting the phloem, latex or fruit and representing the “crown” of this lineage. Thus, the genomic features observed in these flagellates represent a derived state and it is still not clear what has allowed these flagellates to become dixenous. Therefore, the genomes of some early-branching species, such as P. lipae and P. oxycareni infecting seeds [108,109] need to be analyzed and compared with those of the two closely related monoxenous genera Herpetomonas and Lafontella [110], which would serve as outgroups. Of special interest would be to study the genomes of the secondarily monoxenous P. nordicus [111] (to identify genomic features associated with dixeny in this genus) and its closely related species—P. borealis, possessing a bacterial endosymbiont, the relationship with which is likely distinct from those in Novymonas and Strigomonadinae [112].
6. Strigomonadinae
The genomes of endosymbiont-bearing Strigomonadinae and their intracellular bacteria (Ca. Kinetoplastibacterium spp.) have been studied quite intensively. A series of papers characterized the genomes of Angomonas deanei, A. desouzai, Strigomonas oncopelti, S. galati, and S. culicis, as well as the metabolic interactions with their symbiotic partners [13,14,113]. It was demonstrated that the amino acid biosynthetic pathways are interlaced between the endosymbionts and their flagellate hosts and that many genes had been acquired by Strigomonadinae from various groups of bacteria. The importance of Strigomonadinae led to the establishment of the first genetically-trackable system in the model species, A. deanei [114]. A recent study using genomic data of two A. ambiguus strains, A. deanei, and their endosymbionts demonstrated that bacteria from the latter species repeatedly replaced bacteria in the former [115].
What needs to be done: The genus Kentomonas represent the earliest branch within the subfamily [8] and, therefore, may keep in its genome some archaic traits inherent to the common ancestor of the subfamily. In addition, it has been shown to differ from its cousins in the dependence of external source of heme (or its precursors) [11] and may also diverge in other aspects of its metabolism. Hence, a genomic analysis of this trypanosomatid is warranted.
7. Other Monoxenous Lineages
There are three more monoxenous species, whose genomes have been sequenced and analyzed. Genome sequencing of the early-diverging Paratrypanosoma confusum and a representative of the flea-parasitizing genus Blechomonas ayalai has allowed to draw preliminary conclusions concerning the evolution of metabolic pathways in the family Trypanosomatidae [116,117]. The most recent addition to the collection of trypanosomatid genomes was that of Vickermania ingenoplastis, a species lacking mitochondrial respiratory complexes III and IV and, thus, mainly relying on glycolysis, similarly to Phytomonas spp. However, in contrast to the plant trypanosomatids, the genome of this flagellate did not shrink, but experienced a substantial expansion of some protein families, in particular, the glycolytic enzymes [118].
What needs to be done: Representatives of numerous trypanosomatid genera have not been sequenced and some of them have not even been studied since their original description. (1) Blastocrithidia and Obscuromonas of the subfamily Blastocrithidiinae (Figure 1) share a unique resistant developmental stage—the cyst-like amastigote [119]. Moreover, some of them demonstrate quite a complex development in insects, comparable to that in dixenous parasites [120,121]. It would be interesting to find the genomic basis of these peculiarities. (2) Jaenimonas drosophilae inhabits fruit flies and have been proposed as a model to study the insect immune response to trypanosomatid parasites [122]. Sequencing the genome of this parasite would ease using it as such and understanding its intimate relationships with the host. (3) The genus Sergeia parasitizes biting midges and sandflies [123] and thus represents a good model to study the challenges faced and solutions used by trypanosomatids in blood-sucking nematoceran Diptera. Importantly, the same host groups are used by medically relevant Leishmania spp. and, therefore, finding parallels in the genome evolution between them and Sergeia might provide additional information on the biology of the former [124]. (4) The symbiont-free genus Wallacemonas is closely related to Strigomonadinae (Figure 1) and is similar to them in morphology and lifestyle [119]. Thus, it represents a promising reference to reconstruct the metabolism of the ancestors of these endosymbiont-bearing flagellates and answers the question of why some trypanosomatids need endosymbionts, while others successfully live without them in the same hosts.
8. Other Applications of the Trypanosomatid Genomic Data
The availability of multiple representative genome sequences from various Trypanosomatidae enabled a robust analysis of the evolution of different gene families in this group. Some examples are provided below. The analysis of amastins, a large family of surface glycoproteins expressed primarily in amastigotes, revealed that δ-amastin subfamily is restricted to the dixenous Leishmaniinae and its expansion has likely happened in the ancestor of the genus Leishmania [78,125]. The repertoire of adenylate cyclases has expanded in dixenous trypanosomatids and many genes encoding these proteins pseudogenized in those subspecies of T. brucei, which lost the ability to develop in insects [126]. The analysis of myosin gene family suggested that these proteins were already diversified in the kinetoplastid common ancestor and secondarily, lost multiple times afterwards [127]. Genomic studies revealed that at least three trypanosomatid lineages—Leishmaniinae, Blastocrithidiinae, and Vickermania—independently acquired catalase from different groups of bacteria, whereas dixenous Leishmaniinae secondarily lost it [118,128,129]. The study of tubulin gene arrays demonstrated that while in the majority of trypanosomatid lineages and in the free-living bodonids that the α- and β-tubulin genes are alternated, in Leishmaniinae, these multicopy genes are organized in homogeneous (α-only and β-only) stretches [130]. The analysis of the evolution of trypanosomatid UDP-glycosyltransferases, the superfamily of enzymes participating in the modification of various surface macromolecules, showed their independent diversification in distinct groups of these parasites. Interestingly, one of the ancient lineages of these enzymes present in the free-living Bodo saltans has been lost from all trypanosomatids except stercorarian trypanosomes [131]. Side chain galactosyl and arabinosyltransferases of that large superfamily ensure lipophosphoglycan modifications needed for Leishmania attachment and detachment inside insects [132,133,134]. The analysis demonstrated differences in the repertoires of these enzymes between the subgenera Leishmania and Viannia correlating with the affinity of the flagellates to different intestinal sections of their different insect hosts [135,136]. In Leptomonas pyrrhocoris, which does not attach to the intestinal wall of its firebug host, the orthologs of these genes showed early divergence and expansion, suggesting distinct functions [137].
The analysis of gene families and comparative genomic studies discussed above can be hampered by the absence of contiguous assemblies with well-resolved repetitive regions. Although trypanosomatid genomes are relatively small (typically around 20–30 Mb), they contain many repeats and, therefore, it is challenging to obtain a chromosome-level assembly based on short sequencing reads [138]. For several trypanosomatid genera, more contiguous hybrid assemblies based on the combination of short and long sequencing reads have become available (Table S1). Application of a combination of long read sequencing and genome-wide chromosome conformation capture (Hi-C) enabled haplotype-specific assembly of T. brucei 427 Lister genome and revealed that antigen-encoding sub-telomeric regions are folded into distinct compact structures [139]. For T. cruzi, the trypanosomatid having the largest genome sequenced so far, a newer assembly obtained using Nanopore data, led to a significant increase of the number of identified single-copy orthologs and repetitive transposable elements as well as overall estimated genome size [37]. By far, the most contiguous genome assembly, which we suggest to use as a new reference for this species, was obtained recently using a combination of PacBio Single-Molecule Real-Time sequencing and proximity ligation methods [42]. One more example of a substantial quality improvement is the recently published Nanopore-based genome assembly for Angomonas deanei, which identified new chromosome-level features such as a supernumerary chromosome, a long inversion and a translocation [140]. After careful annotation of such genome assemblies based on multiple types of evidence (including transcriptomic and proteomic data), the trypanosomatid research community should consider using these new assemblies instead of the old references based solely on short reads and sometimes erroneous annotations.
Although no DNA viruses have been reported in trypanosomatids so far, the available genomic data for several Leptomonas pyrrhocoris strains has allowed identification of an endogenous viral element related to LeppyrTLV1 (a tombus-like single-stranded positive sense RNA virus), which was apparently captured via reverse transcription and integrated into the trypanosomatid genome [141].
Finally, next-generation sequencing data can be used for analyzing the composition and (to some extent) function of the kinetoplast. In this respect, the kinetoplast genomes of the two model species, dixenous T. brucei and monoxenous Leptomonas pyrrhocoris, have been scrutinized. Their analyses revealed novel non-canonical mechanisms, as well as species-specific differences in RNA editing [142,143]. Such studies can delineate not only the structure of maxicircles and minicircles [144,145,146,147], but also predict the guide RNA repertoire in a given species [143,148,149]. As judged from pre-genomic studies carried out on single genes, different lineages of trypanosomatids possess distinct kDNA editing patterns [150]. This, along with the abovementioned degradation of kDNA in two T. brucei subspecies, demonstrates the underestimated importance of the kinetoplast genome in the trypanosomatid development. Performing comparative studies on the editing using whole kinetoplast genomes with a wide range of trypanosomatid phylogroups should shed light on their particular life strategies and allow better understanding of the evolution of this fascinating group of parasites.
9. Conclusions
A fair number of trypanosomatid genomes have been sequenced and there is a significant progress in understanding their evolution, structure, and function. Nevertheless, many questions still remain unanswered and more of them arise, as new representatives of this group of flagellates are discovered and/or analyzed in broadscale biodiversity assays.
The relatively small size of trypanosomatid genomes makes these parasites an attractive model to study how the evolution of traits and genomes are correlated. This is further facilitated by the possibility to cultivate and genetically modify many trypanosomatids, combined with a knowledge of their diversity. However, as judged from the environmental screens (for example, refs. [151,152] and many others), there are still taxa of the generic level and above to be described. Meanwhile, the lack of data on the biology of many trypanosomatid groups still represents an important obstacle in interpreting the observed genomic differences, therefore, more data on trypanosomatid development, strategies of transmission, host-parasite interactions, etc., are needed.
Acknowledgments
We thank Evgeny Gerasimov (Moscow State University) for the discussion on kDNA analyses.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091124/s1, Table S1: Available genomic data for Trypanosomatidae.
Author Contributions
Conceptualization, V.Y. and A.Y.K.; formal analysis, A.B. and A.Y.K.; data curation, all authors; writing—original draft preparation, all authors; writing—review and editing, all authors; funding acquisition, V.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Regional Funds (CZ.02.1.01/16_019/0000759 to V.Y. and A.Y.K.), the Grant Agency of Czech Republic (20-07186S to V.Y. and A.Y.K. and 21-09283S to V.Y.), and the State Assignment AAAA-A19-119031390116-9 for ZIN RAS to A.Y.K.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kostygov A.Y., Karnkowska A., Votýpka J., Tashyreva D., Maciszewski K., Yurchenko V., Lukeš J. Euglenozoa: Taxonomy, diversity and ecology, symbioses and viruses. Open Biol. 2021;11:200407. doi: 10.1098/rsob.200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 4.El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 5.El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 6.Lukeš J., Butenko A., Hashimi H., Maslov D.A., Votýpka J., Yurchenko V. Trypanosomatids are much more than just trypanosomes: Clues from the expanded family tree. Trends Parasitol. 2018;34:466–480. doi: 10.1016/j.pt.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Záhonová K., Kostygov A., Ševčíková T., Yurchenko V., Eliáš M. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr. Biol. 2016;26:2364–2369. doi: 10.1016/j.cub.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 8.Votýpka J., Kostygov A.Y., Kraeva N., Grybchuk-Ieremenko A., Tesařová M., Grybchuk D., Lukeš J., Yurchenko V. Kentomonas gen. n.; a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist. 2014;165:825–838. doi: 10.1016/j.protis.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Kostygov A., Dobáková E., Grybchuk-Ieremenko A., Váhala D., Maslov D.A., Votýpka J., Lukeš J., Yurchenko V. Novel trypanosomatid—Bacterium association: Evolution of endosymbiosis in action. mBio. 2016;7:e01985-15. doi: 10.1128/mBio.01985-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira M.M., Borghesan T.C., Ferreira R.C., Santos M.A., Takata C.S., Campaner M., Nunes V.L., Milder R.V., de Souza W., Camargo E.P. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Silva F.M., Kostygov A.Y., Spodareva V.V., Butenko A., Tossou R., Lukes J., Yurchenko V., Alves J.M.P. The reduced genome of Candidatus Kinetoplastibacterium sorsogonicusi, the endosymbiont of Kentomonas sorsogonicus (Trypanosomatidae): Loss of the haem-synthesis pathway. Parasitology. 2018;145:1287–1293. doi: 10.1017/S003118201800046X. [DOI] [PubMed] [Google Scholar]
- 12.Kostygov A.Y., Butenko A., Nenarokova A., Tashyreva D., Flegontov P., Lukeš J., Yurchenko V. Genome of Ca. Pandoraea novymonadis, an endosymbiotic bacterium of the trypanosomatid Novymonas esmeraldas. Front. Microbiol. 2017;8:1940. doi: 10.3389/fmicb.2017.01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alves J.M., Serrano M.G., Maia da Silva F., Voegtly L.J., Matveyev A.V., Teixeira M.M., Camargo E.P., Buck G.A. Genome evolution and phylogenomic analysis of Candidatus Kinetoplastibacterium, the beta-proteobacterial endosymbionts of Strigomonas and Angomonas. Genome Biol. Evol. 2013;5:338–350. doi: 10.1093/gbe/evt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves J.M., Klein C.C., da Silva F.M., Costa-Martins A.G., Serrano M.G., Buck G.A., Vasconcelos A.T., Sagot M.F., Teixeira M.M., Motta M.C., et al. Endosymbiosis in trypanosomatids: The genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol. Biol. 2013;13:190. doi: 10.1186/1471-2148-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostygov A., Frolov A.O., Malysheva M.N., Ganyukova A.I., Chistyakova L.V., Tashyreva D., Tesařová M., Spodareva V.V., Režnarová J., Macedo D.H., et al. Vickermania gen. nov.; trypanosomatids that use two joined flagella to resist midgut peristaltic flow within the fly host. BMC Biol. 2020;18:187. doi: 10.1186/s12915-020-00916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishemgulova A., Butenko A., Kortišová L., Boucinha C., Grybchuk-Ieremenko A., Morelli K.A., Tesařová M., Kraeva N., Grybchuk D., Pánek T., et al. Molecular mechanisms of thermal resistance of the insect trypanosomatid Crithidia thermophila. PLoS ONE. 2017;12:e0174165. doi: 10.1371/journal.pone.0174165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraeva N., Butenko A., Hlaváčová J., Kostygov A., Myškova J., Grybchuk D., Leštinová T., Votýpka J., Volf P., Opperdoes F., et al. Leptomonas seymouri: Adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 2015;11:e1005127. doi: 10.1371/journal.ppat.1005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama S.R., de Santana A.K.M., Takamiya N.T., Takahashi T.Y., Rogerio L.A., Oliveira C.A.B., Milanezi C.M., Trombela V.A., Cruz A.K., Jesus A.R., et al. Non-Leishmania parasite in fatal visceral leishmaniasis–like disease, Brazil. Emerg. Infect. Dis. 2019;25:2088–2092. doi: 10.3201/eid2511.181548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brems S., Guilbride D.L., Gundlesdodjir-Planck D., Busold C., Luu V.D., Schanne M., Hoheisel J., Clayton C. The transcriptomes of Trypanosoma brucei Lister 427 and TREU927 bloodstream and procyclic trypomastigotes. Mol. Biochem. Parasitol. 2005;139:163–172. doi: 10.1016/j.molbiopara.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Jackson A.P., Sanders M., Berry A., McQuillan J., Aslett M.A., Quail M.A., Chukualim B., Capewell P., MacLeod A., Melville S.E., et al. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl. Trop. Dis. 2010;4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sistrom M., Evans B., Benoit J., Balmer O., Aksoy S., Caccone A. De novo genome assembly shows genome wide similarity between Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS ONE. 2016;11:e0147660. doi: 10.1371/journal.pone.0147660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carnes J., Anupama A., Balmer O., Jackson A., Lewis M., Brown R., Cestari I., Desquesnes M., Gendrin C., Hertz-Fowler C., et al. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl. Trop. Dis. 2015;9:e3404. doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Jiang N., Sang X., Zhang N., Zhang K., Chen H., Yang N., Feng Y., Chen R., Suo X., et al. In-depth analysis of the genome of Trypanosoma evansi, an etiologic agent of surra. Sci. China Life Sci. 2019;62:406–419. doi: 10.1007/s11427-018-9473-8. [DOI] [PubMed] [Google Scholar]
- 24.Davaasuren B., Yamagishi J., Mizushima D., Narantsatsral S., Otgonsuren D., Myagmarsuren P., Battsetseg B., Battur B., Inoue N., Suganuma K. Draft genome sequence of Trypanosoma equiperdum strain IVM-t1. Microbiol. Resour. Announc. 2019;8:e01119-18. doi: 10.1128/MRA.01119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hébert L., Moumen B., Madeline A., Steinbiss S., Lakhdar L., Van Reet N., Buscher P., Laugier C., Cauchard J., Petry S. First draft genome sequence of the dourine causative agent: Trypanosoma Equiperdum strain OVI. J. Genom. 2017;5:1–3. doi: 10.7150/jgen.17904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson A.P., Berry A., Aslett M., Allison H.C., Burton P., Vavrova-Anderson J., Brown R., Browne H., Corton N., Hauser H., et al. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc. Natl. Acad. Sci. USA. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbas A.H., Silva Pereira S., D’Archivio S., Wickstead B., Morrison L.J., Hall N., Hertz-Fowler C., Darby A.C., Jackson A.P. The structure of a conserved telomeric region associated with variant antigen loci in the blood parasite Trypanosoma congolense. Genome Biol. Evol. 2018;10:2458–2473. doi: 10.1093/gbe/evy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silvester E., Ivens A., Matthews K.R. A gene expression comparison of Trypanosoma brucei and Trypanosoma congolense in the bloodstream of the mammalian host reveals species-specific adaptations to density-dependent development. PLoS Negl. Trop. Dis. 2018;12:e0006863. doi: 10.1371/journal.pntd.0006863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awuoche E.O., Weiss B.L., Mireji P.O., Vigneron A., Nyambega B., Murilla G., Aksoy S. Expression profiling of Trypanosoma congolense genes during development in the tsetse fly vector Glossina morsitans morsitans. Parasit. Vectors. 2018;11:380. doi: 10.1186/s13071-018-2964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greif G., Ponce de Leon M., Lamolle G., Rodriguez M., Pineyro D., Tavares-Marques L.M., Reyna-Bello A., Robello C., Alvarez-Valin F. Transcriptome analysis of the bloodstream stage from the parasite Trypanosoma vivax. BMC Genom. 2013;14:149. doi: 10.1186/1471-2164-14-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callejas-Hernández F., Rastrojo A., Poveda C., Girones N., Fresno M. Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep. 2018;8:14631. doi: 10.1038/s41598-018-32877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callejas-Hernández F., Gironès N., Fresno M. Genome sequence of Trypanosoma cruzi strain Bug2148. Genome Announc. 2018;6:e01497-17. doi: 10.1128/genomeA.01497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez I., Rastrojo A., Sanchez-Luque F.J., Lorenzo-Diaz F., Macias F., Valladares B., Aguado B., Requena J.M., Lopez M.C., Thomas M.C. Draft genome sequence of the Trypanosoma cruzi B. M. Lopez strain (TcIa), isolated from a Colombian patient. Microbiol. Resour. Announc. 2020;9:e00031-20. doi: 10.1128/MRA.00031-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez I., Rastrojo A., Lorenzo-Diaz F., Sanchez-Luque F.J., Macias F., Aguado B., Valladares B., Requena J.M., Lopez M.C., Thomas M.C. Trypanosoma cruzi Ikiakarora (TcIII) draft genome sequence. Microbiol. Resour. Announc. 2020;9:e00453-20. doi: 10.1128/MRA.00453-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berná L., Rodriguez M., Chiribao M.L., Parodi-Talice A., Pita S., Rijo G., Alvarez-Valin F., Robello C. Expanding an expanded genome: Long-read sequencing of Trypanosoma cruzi. Microb. Genom. 2018;4:e000177. doi: 10.1099/mgen.0.000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeCuir J., Tu W., Dumonteil E., Herrera C. Sequence of Trypanosoma cruzi reference strain SC43 nuclear genome and kinetoplast maxicircle confirms a strong genetic structure among closely related parasite discrete typing units. Genome. 2021;64:525–531. doi: 10.1139/gen-2020-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Díaz-Viraqué F., Pita S., Greif G., de Souza R.C.M., Iraola G., Robello C. Nanopore sequencing significantly improves genome assembly of the protozoan parasite Trypanosoma cruzi. Genome Biol. Evol. 2019;11:1952–1957. doi: 10.1093/gbe/evz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franzén O., Ochaya S., Sherwood E., Lewis M.D., Llewellyn M.S., Miles M.A., Andersson B. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl. Trop. Dis. 2011;5:e984. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grisard E.C., Teixeira S.M., de Almeida L.G., Stoco P.H., Gerber A.L., Talavera-Lopez C., Lima O.C., Andersson B., de Vasconcelos A.T. Trypanosoma cruzi clone Dm28c draft genome sequence. Genome Announc. 2014;2:e01114-13. doi: 10.1128/genomeA.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reis-Cunha J.L., Rodrigues-Luiz G.F., Valdivia H.O., Baptista R.P., Mendes T.A., de Morais G.L., Guedes R., Macedo A.M., Bern C., Gilman R.H., et al. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genom. 2015;16:499. doi: 10.1186/s12864-015-1680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baptista R.P., Reis-Cunha J.L., DeBarry J.D., Chiari E., Kissinger J.C., Bartholomeu D.C., Macedo A.M. Assembly of highly repetitive genomes using short reads: The genome of discrete typing unit III Trypanosoma cruzi strain 231. Microb. Genom. 2018;4:e000156. doi: 10.1099/mgen.0.000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Peng D., Baptista R.P., Li Y., Kissinger J.C., Tarleton R.L. Strain-specific genome evolution in Trypanosoma cruzi, the agent of Chagas disease. PLoS Pathog. 2021;17:e1009254. doi: 10.1371/journal.ppat.1009254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franzén O., Talavera-Lopez C., Ochaya S., Butler C.E., Messenger L.A., Lewis M.D., Llewellyn M.S., Marinkelle C.J., Tyler K.M., Miles M.A., et al. Comparative genomic analysis of human infective Trypanosoma cruzi lineages with the bat-restricted subspecies T. cruzi marinkellei. BMC Genom. 2012;13:531. doi: 10.1186/1471-2164-13-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradwell K.R., Koparde V.N., Matveyev A.V., Serrano M.G., Alves J.M.P., Parikh H., Huang B., Lee V., Espinosa-Alvarez O., Ortiz P.A., et al. Genomic comparison of Trypanosoma conorhini and Trypanosoma rangeli to Trypanosoma cruzi strains of high and low virulence. BMC Genom. 2018;19:770. doi: 10.1186/s12864-018-5112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoco P.H., Wagner G., Talavera-Lopez C., Gerber A., Zaha A., Thompson C.E., Bartholomeu D.C., Luckemeyer D.D., Bahia D., Loreto E., et al. Genome of the avirulent human-infective trypanosome Trypanosoma rangeli. PLoS Negl. Trop. Dis. 2014;8:e3176. doi: 10.1371/journal.pntd.0003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly S., Ivens A., Manna P.T., Gibson W., Field M.C. A draft genome for the African crocodilian trypanosome Trypanosoma grayi. Sci. Data. 2014;1:140024. doi: 10.1038/sdata.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly S., Ivens A., Mott G.A., O’Neill E., Emms D., Macleod O., Voorheis P., Tyler K., Clark M., Matthews J., et al. An alternative strategy for trypanosome survival in the mammalian bloodstream revealed through genome and transcriptome analysis of the ubiquitous bovine parasite Trypanosoma (Megatrypanum) theileri. Genome Biol. Evol. 2017;9:2093–2109. doi: 10.1093/gbe/evx152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoare C.A. The Trypanosomes of Mammals. Blackwell Scientific Publications; Oxford, UK: 1972. p. 768. [Google Scholar]
- 49.Truc P., Buscher P., Cuny G., Gonzatti M.I., Jannin J., Joshi P., Juyal P., Lun Z.R., Mattioli R., Pays E., et al. Atypical human infections by animal trypanosomes. PLoS Negl. Trop. Dis. 2013;7:e2256. doi: 10.1371/journal.pntd.0002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spodareva V.V., Grybchuk-Ieremenko A., Losev A., Votýpka J., Lukeš J., Yurchenko V., Kostygov A.Y. Diversity and evolution of anuran trypanosomes: Insights from the study of European species. Parasit. Vectors. 2018;11:447. doi: 10.1186/s13071-018-3023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Real F., Vidal R.O., Carazzolle M.F., Mondego J.M., Costa G.G., Herai R.H., Wurtele M., de Carvalho L.M., Carmona e Ferreira R., Mortara R.A., et al. The genome sequence of Leishmania (Leishmania) amazonensis: Functional annotation and extended analysis of gene models. DNA Res. 2013;20:567–581. doi: 10.1093/dnares/dst031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tschoeke D.A., Nunes G.L., Jardim R., Lima J., Dumaresq A.S., Gomes M.R., de Mattos Pereira L., Loureiro D.R., Stoco P.H., de Matos Guedes H.L., et al. The comparative genomics and phylogenomics of Leishmania amazonensis parasite. Evol. Bioinform. Online. 2014;10:131–153. doi: 10.4137/EBO.S13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valdivia H.O., Almeida L.V., Roatt B.M., Reis-Cunha J.L., Pereira A.A., Gontijo C., Fujiwara R.T., Reis A.B., Sanders M.J., Cotton J.A., et al. Comparative genomics of canine-isolated Leishmania (Leishmania) amazonensis from an endemic focus of visceral leishmaniasis in Governador Valadares, southeastern Brazil. Sci. Rep. 2017;7:40804. doi: 10.1038/srep40804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aoki J.I., Muxel S.M., Zampieri R.A., Laranjeira-Silva M.F., Muller K.E., Nerland A.H., Floeter-Winter L.M. RNA-seq transcriptional profiling of Leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl. Trop. Dis. 2017;11:e0006026. doi: 10.1371/journal.pntd.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patino L.H., Muskus C., Ramirez J.D. Transcriptional responses of Leishmania (Leishmania) amazonensis in the presence of trivalent sodium stibogluconate. Parasit. Vectors. 2019;12:348. doi: 10.1186/s13071-019-3603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Downing T., Imamura H., Decuypere S., Clark T.G., Coombs G.H., Cotton J.A., Hilley J.D., de Doncker S., Maes I., Mottram J.C., et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh N., Chikara S., Sundar S. SOLiD sequencing of genomes of clinical isolates of Leishmania donovani from India confirm Leptomonas co-infection and raise some key questions. PLoS ONE. 2013;8:e55738. doi: 10.1371/journal.pone.0055738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franssen S.U., Durrant C., Stark O., Moser B., Downing T., Imamura H., Dujardin J.C., Sanders M.J., Mauricio I., Miles M.A., et al. Global genome diversity of the Leishmania donovani complex. eLife. 2020;9:e51243. doi: 10.7554/eLife.51243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.González-de la Fuente S., Peiro-Pastor R., Rastrojo A., Moreno J., Carrasco-Ramiro F., Requena J.M., Aguado B. Resequencing of the Leishmania infantum (strain JPCM5) genome and de novo assembly into 36 contigs. Sci. Rep. 2017;7:18050. doi: 10.1038/s41598-017-18374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishemgulova A., Hlaváčová J., Majerová K., Butenko A., Lukeš J., Votýpka J., Volf P., Yurchenko V. CRISPR/Cas9 in Leishmania mexicana: A case study of LmxBTN1. PLoS ONE. 2018;13:e0192723. doi: 10.1371/journal.pone.0192723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iantorno S.A., Durrant C., Khan A., Sanders M.J., Beverley S.M., Warren W.C., Berriman M., Sacks D.L., Cotton J.A., Grigg M.E. Gene expression in Leishmania is regulated predominantly by gene dosage. mBio. 2017;8:e01393-17. doi: 10.1128/mBio.01393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valdivia H.O., Reis-Cunha J.L., Rodrigues-Luiz G.F., Baptista R.P., Baldeviano G.C., Gerbasi R.V., Dobson D.E., Pratlong F., Bastien P., Lescano A.G., et al. Comparative genomic analysis of Leishmania (Viannia) peruviana and Leishmania (Viannia) braziliensis. BMC Genom. 2015;16:715. doi: 10.1186/s12864-015-1928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.González-de la Fuente S., Camacho E., Peiro-Pastor R., Rastrojo A., Carrasco-Ramiro F., Aguado B., Requena J.M. Complete and de novo assembly of the Leishmania braziliensis (M2904) genome. Mem. Inst. Oswaldo Cruz. 2018;114:e180438. doi: 10.1590/0074-02760180438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruy P.C., Monteiro-Teles N.M., Miserani Magalhaes R.D., Freitas-Castro F., Dias L., Aquino Defina T.P., Rosas De Vasconcelos E.J., Myler P.J., Kaysel Cruz A. Comparative transcriptomics in Leishmania braziliensis: Disclosing differential gene expression of coding and putative noncoding RNAs across developmental stages. RNA Biol. 2019;16:639–660. doi: 10.1080/15476286.2019.1574161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llanes A., Restrepo C.M., Del Vecchio G., Anguizola F.J., Lleonart R. The genome of Leishmania panamensis: Insights into genomics of the L. (Viannia) subgenus. Sci. Rep. 2015;5:8550. doi: 10.1038/srep08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coughlan S., Taylor A.S., Feane E., Sanders M., Schonian G., Cotton J.A., Downing T. Leishmania naiffi and Leishmania guyanensis reference genomes highlight genome structure and gene evolution in the Viannia subgenus. R. Soc. Open Sci. 2018;5:172212. doi: 10.1098/rsos.172212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batra D., Lin W., Rowe L.A., Sheth M., Zheng Y., Loparev V., de Almeida M. Draft genome sequence of French Guiana Leishmania (Viannia) guyanensis strain 204–365, assembled using long reads. Microbiol. Resour. Announc. 2018;7:e01421-18. doi: 10.1128/MRA.01421-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raymond F., Boisvert S., Roy G., Ritt J.F., Legare D., Isnard A., Stanke M., Olivier M., Tremblay M.J., Papadopoulou B., et al. Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res. 2012;40:1131–1147. doi: 10.1093/nar/gkr834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goto Y., Kuroki A., Suzuki K., Yamagishi J. Draft genome sequence of Leishmania tarentolae Parrot Tar II, obtained by single-molecule real-time sequencing. Microbiol. Resour. Announc. 2020;9:e00050-20. doi: 10.1128/MRA.00050-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coughlan S., Mulhair P., Sanders M., Schonian G., Cotton J.A., Downing T. The genome of Leishmania adleri from a mammalian host highlights chromosome fission in Sauroleishmania. Sci. Rep. 2017;7:43747. doi: 10.1038/srep43747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harkins K.M., Schwartz R.S., Cartwright R.A., Stone A.C. Phylogenomic reconstruction supports supercontinent origins for Leishmania. Infect. Genet. Evol. 2016;38:101–109. doi: 10.1016/j.meegid.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 74.Butenko A., Kostygov A.Y., Sádlová J., Kleschenko Y., Bečvář T., Podešvová L., Macedo D.H., Žihala D., Lukeš J., Bates P.A., et al. Comparative genomics of Leishmania (Mundinia) BMC Genom. 2019;20:726. doi: 10.1186/s12864-019-6126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paranaiba L.F., Pinheiro L.J., Torrecilhas A.C., Macedo D.H., Menezes-Neto A., Tafuri W.L., Soares R.P. Leishmania enriettii (Muniz & Medina, 1948): A highly diverse parasite is here to stay. PLoS Pathog. 2017;13:e1006303. doi: 10.1371/journal.ppat.1006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paranaiba L.F., de Assis R.R., Nogueira P.M., Torrecilhas A.C., Campos J.H., Silveira A.C., Martins-Filho O.A., Pessoa N.L., Campos M.A., Parreiras P.M., et al. Leishmania enriettii: Biochemical characterisation of lipophosphoglycans (LPGs) and glycoinositolphospholipids (GIPLs) and infectivity to Cavia porcellus. Parasit. Vectors. 2015;8:31. doi: 10.1186/s13071-015-0633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Assis R.R., Ibraim I.C., Nogueira P.M., Soares R.P., Turco S.J. Glycoconjugates in New World species of Leishmania: Polymorphisms in lipophosphoglycan and glycoinositolphospholipids and interaction with hosts. Biochim. Biophys. Acta. 2012;1820:1354–1365. doi: 10.1016/j.bbagen.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Albanaz A.T.S., Gerasimov E.S., Shaw J.J., Sádlová J., Lukeš J., Volf P., Opperdoes F.R., Kostygov A.Y., Butenko A., Yurchenko V. Genome analysis of Endotrypanum and Porcisia spp.; closest phylogenetic relatives of Leishmania, highlights the role of amastins in shaping pathogenicity. Genes. 2021;12:444. doi: 10.3390/genes12030444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Imamura H., Monsieurs P., Jara M., Sanders M., Maes I., Vanaerschot M., Berriman M., Cotton J.A., Dujardin J.C., Domagalska M.A. Evaluation of whole genome amplification and bioinformatic methods for the characterization of Leishmania genomes at a single cell level. Sci. Rep. 2020;10:15043. doi: 10.1038/s41598-020-71882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dia A., Cheeseman I.H. Single-cell genome sequencing of protozoan parasites. Trends Parasitol. 2021 doi: 10.1016/j.pt.2021.05.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akhoundi M., Kuhls K., Cannet A., Votýpka J., Marty P., Delaunay P., Sereno D. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl. Trop. Dis. 2016;10:e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klatt S., Simpson L., Maslov D.A., Konthur Z. Leishmania tarentolae: Taxonomic classification and its application as a promising biotechnological expression host. PLoS Negl. Trop. Dis. 2019;13:e0007424. doi: 10.1371/journal.pntd.0007424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Telford S.R. Hemoparasites of the Reptilia: Color Atlas and Text. Volume xv. CRC Press; Boca Raton, FL, USA: 2009. 376p [Google Scholar]
- 84.Espinosa O.A., Serrano M.G., Camargo E.P., Teixeira M.M., Shaw J.J. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology. 2018;145:430–442. doi: 10.1017/S0031182016002092. [DOI] [PubMed] [Google Scholar]
- 85.Warren W.C., Akopyants N.S., Dobson D.E., Hertz-Fowler C., Lye L.F., Myler P.J., Ramasamy G., Shanmugasundram A., Silva-Franco F., Steinbiss S., et al. Genome assemblies across the diverse evolutionary spectrum of Leishmania protozoan parasites. bioRxiv. 2021 doi: 10.1101/2021.05.29.446295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Batra D., Lin W., Narayanan V., Rowe L.A., Sheth M., Zheng Y., Loparev V., de Almeida M. Draft genome sequences of Leishmania (Leishmania) amazonensis, Leishmania (Leishmania) mexicana, and Leishmania (Leishmania) aethiopica, potential etiological agents of diffuse cutaneous leishmaniasis. Microbiol. Resour. Announc. 2019;8:e00269-19. doi: 10.1128/MRA.00269-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Almutairi H., Urbaniak M.D., Bates M.D., Jariyapan N., Al-Salem W.S., Dillon R.J., Bates P.A., Gatherer D. Chromosome-scale assembly of the complete genome sequence of Leishmania (Mundinia) martiniquensis, isolate LSCM1, strain LV760. Microbiol. Resour. Announc. 2021;10:e0005821. doi: 10.1128/MRA.00058-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin W., Batra D., Narayanan V., Rowe L.A., Sheth M., Zheng Y., Juieng P., Loparev V., de Almeida M. First draft genome sequence of Leishmania (Viannia) lainsoni strain 216–34, isolated from a Peruvian clinical case. Microbiol. Resour. Announc. 2019;8:e01524-18. doi: 10.1128/MRA.01524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Runckel C., DeRisi J., Flenniken M.L. A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae. PLoS ONE. 2014;9:e95057. doi: 10.1371/journal.pone.0095057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmid-Hempel P., Aebi M., Barribeau S., Kitajima T., du Plessis L., Schmid-Hempel R., Zoller S. The genomes of Crithidia bombi and C. expoeki, common parasites of bumblebees. PLoS ONE. 2018;13:e0189738. doi: 10.1371/journal.pone.0189738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gerasimov E., Zemp N., Schmid-Hempel R., Schmid-Hempel P., Yurchenko V. Genomic variation among strains of Crithidia bombi and C. expoeki. mSphere. 2019;4:e00482-19. doi: 10.1128/mSphere.00482-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghosh S., Banerjee P., Sarkar A., Datta S., Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J. Clin. Microbiol. 2012;50:2774–2778. doi: 10.1128/JCM.00966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flegontov P., Butenko A., Firsov S., Kraeva N., Eliáš M., Field M.C., Filatov D., Flegontova O., Gerasimov E.S., Hlaváčová J., et al. Genome of Leptomonas pyrrhocoris: A high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci. Rep. 2016;6:23704. doi: 10.1038/srep23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Filosa J.N., Berry C.T., Ruthel G., Beverley S.M., Warren W.C., Tomlinson C., Myler P.J., Dudkin E.A., Povelones M.L., Povelones M. Dramatic changes in gene expression in different forms of Crithidia fasciculata reveal potential mechanisms for insect-specific adhesion in kinetoplastid parasites. PLoS Negl. Trop. Dis. 2019;13:e0007570. doi: 10.1371/journal.pntd.0007570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghobakhloo N., Motazedian M.H., Naderi S., Sepideh E. Isolation of Crithidia spp. from lesions of immunocompetent patients with suspected cutaneous leishmaniasis in Iran. Trop. Med. Int. Health. 2018;24:116–126. doi: 10.1111/tmi.13042. [DOI] [PubMed] [Google Scholar]
- 96.Zakharova A., Saura A., Butenko A., Podešvová L., Warmusová S., Kostygov A.Y., Nenarokova A., Lukeš J., Opperdoes F.R., Yurchenko V. A new model trypanosomatid Novymonas esmeraldas: Genomic perception of its "Candidatus Pandoraea novymonadis" endosymbiont. mBio. 2021;12:e01606-21. doi: 10.1128/mBio.01606-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Votýpka J., Klepetková H., Yurchenko V.Y., Horák A., Lukeš J., Maslov D.A. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist. 2012;163:616–631. doi: 10.1016/j.protis.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 98.Kostygov A.Y., Grybchuk-Ieremenko A., Malysheva M.N., Frolov A.O., Yurchenko V. Molecular revision of the genus Wallaceina. Protist. 2014;165:594–604. doi: 10.1016/j.protis.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 99.Ganyukova A.I., Zolotarev A.V., Frolov A.O. Geographical distribution and host range of monoxenous trypanosomatid Crithidia brevicula (Frolov et Malysheva, 1989) in the northern regions of Eurasia. Protistology. 2020;14:70–78. doi: 10.21685/1680-0826-2020-14-2-3. [DOI] [Google Scholar]
- 100.Kostygov A.Y., Yurchenko V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae) Folia Parasitol. 2017;64:020. doi: 10.14411/fp.2017.020. [DOI] [PubMed] [Google Scholar]
- 101.Marin C., Fabre S., Sanchez-Moreno M., Dollet M. Herpetomonas spp. isolated from tomato fruits (Lycopersicon esculentum) in southern Spain. Exp. Parasitol. 2007;116:88–90. doi: 10.1016/j.exppara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 102.Fiorini J.E., Takata C.S., Teofilo V.M., Nascimento L.C., Faria-e-Silva P.M., Soares M.J., Teixeira M.M., De Souza W. Morphological, biochemical and molecular characterization of Herpetomonas samuelpessoai camargoi n. subsp.; a trypanosomatid isolated from the flower of the squash Cucurbita moschata. J. Eukaryot. Microbiol. 2001;48:62–69. doi: 10.1111/j.1550-7408.2001.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 103.Morio F., Reynes J., Dollet M., Pratlong F., Dedet J.P., Ravel C. Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. J. Clin. Microbiol. 2008;46:3845–3847. doi: 10.1128/JCM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Porcel B.M., Denoeud F., Opperdoes F.R., Noel B., Madoui M.-A., Hammarton T.C., Field M.C., Da Silva C., Couloux A., Poulain J., et al. The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLoS Genet. 2014;10:e1004007. doi: 10.1371/journal.pgen.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kořený L., Sobotka R., Kovářová J., Gnipová A., Flegontov P., Horváth A., Oborník M., Ayala F.J., Lukeš J. Aerobic kinetoplastid flagellate Phytomonas does not require heme for viability. Proc. Natl. Acad. Sci. USA. 2012;109:3808–3813. doi: 10.1073/pnas.1201089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Butler C.E., Jaskowska E., Kelly S. Genome sequence of Phytomonas françai, a cassava (Manihot esculenta) latex parasite. Genome Announc. 2017;5:e01266-16. doi: 10.1128/genomeA.01266-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sloan M.A., Brooks K., Otto T.D., Sanders M.J., Cotton J.A., Ligoxygakis P. Transcriptional and genomic parallels between the monoxenous parasite Herpetomonas muscarum and Leishmania. PLoS Genet. 2019;15:e1008452. doi: 10.1371/journal.pgen.1008452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frolov A.O., Malysheva M.N., Ganyukova A.I., Spodareva V.V., Yurchenko V., Kostygov A.Y. Development of Phytomonas lipae sp. n. (Kinetoplastea: Trypanosomatidae) in the true bug Coreus marginatus (Heteroptera: Coreidae) and insights into the evolution of life cycles in the genus Phytomonas. PLoS ONE. 2019;14:e0214484. doi: 10.1371/journal.pone.0214484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seward E.A., Votýpka J., Kment P., Lukeš J., Kelly S. Description of Phytomonas oxycareni n. sp. from the salivary glands of Oxycarenus lavaterae. Protist. 2017;168:71–79. doi: 10.1016/j.protis.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 110.Yurchenko V., Kostygov A., Havlová J., Grybchuk-Ieremenko A., Ševčíková T., Lukeš J., Ševčík J., Votýpka J. Diversity of trypanosomatids in cockroaches and the description of Herpetomonas tarakana sp. n. J. Eukaryot. Microbiol. 2016;63:198–209. doi: 10.1111/jeu.12268. [DOI] [PubMed] [Google Scholar]
- 111.Frolov A.O., Malysheva M.N., Yurchenko V., Kostygov A.Y. Back to monoxeny: Phytomonas nordicus descended from dixenous plant parasites. Eur. J. Protistol. 2016;52:1–10. doi: 10.1016/j.ejop.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 112.Ganyukova A.I., Frolov A.O., Malysheva M.N., Spodareva V.V., Yurchenko V., Kostygov A.Y. A novel endosymbiont-containing trypanosomatid Phytomonas borealis sp. n. from the predatory bug Picromerus bidens (Heteroptera: Pentatomidae) Folia Parasitol. 2020;67:4. doi: 10.14411/fp.2020.004. [DOI] [PubMed] [Google Scholar]
- 113.Motta M.C., Martins A.C., de Souza S.S., Catta-Preta C.M., Silva R., Klein C.C., de Almeida L.G., de Lima Cunha O., Ciapina L.P., Brocchi M., et al. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS ONE. 2013;8:e60209. doi: 10.1371/journal.pone.0060209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morales J., Kokkori S., Weidauer D., Chapman J., Goltsman E., Rokhsar D., Grossman A.R., Nowack E.C. Development of a toolbox to dissect host-endosymbiont interactions and protein trafficking in the trypanosomatid Angomonas deanei. BMC Evol. Biol. 2016;16:247. doi: 10.1186/s12862-016-0820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Skalický T., Alves J.M.P., Morais A.C., Režnarová J., Butenko A., Lukeš J., Serrano M.G., Buck G.A., Teixeira M.M.G., Camargo E.P., et al. Endosymbiont capture, a repeated process of endosymbiont transfer with replacement in trypanosomatids Angomonas spp. Pathogens. 2021;10:702. doi: 10.3390/pathogens10060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Skalický T., Dobáková E., Wheeler R.J., Tesařová M., Flegontov P., Jirsová D., Votýpka J., Yurchenko V., Ayala F.J., Lukeš J. Extensive flagellar remodeling during the complex life cycle of Paratrypanosoma, an early-branching trypanosomatid. Proc. Natl. Acad. Sci. USA. 2017;114:11757–11762. doi: 10.1073/pnas.1712311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Opperdoes F.R., Butenko A., Flegontov P., Yurchenko V., Lukeš J. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J. Eukaryot. Microbiol. 2016;63:657–678. doi: 10.1111/jeu.12315. [DOI] [PubMed] [Google Scholar]
- 118.Opperdoes F.R., Butenko A., Zakharova A., Gerasimov E.S., Zimmer S.L., Lukeš J., Yurchenko V. The remarkable metabolism of Vickermania ingenoplastis: Genomic predictions. Pathogens. 2021;10:68. doi: 10.3390/pathogens10010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frolov A.O., Kostygov A.Y., Yurchenko V. Development of monoxenous trypanosomatids and phytomonads in insects. Trends Parasitol. 2021;37:538–551. doi: 10.1016/j.pt.2021.02.004. [DOI] [PubMed] [Google Scholar]
- 120.Frolov A.O., Malysheva M.N., Ganyukova A.I., Spodareva V.V., Kralova J., Yurchenko V., Kostygov A.Y. If host is refractory, insistent parasite goes berserk: Trypanosomatid Blastocrithidia raabei in the dock bug Coreus marginatus. PLoS ONE. 2020;15:e0227832. doi: 10.1371/journal.pone.0227832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frolov A.O., Malysheva M.N., Ganyukova A.I., Yurchenko V., Kostygov A.Y. Obligate development of Blastocrithidia papi (Trypanosomatidae) in the Malpighian tubules of Pyrrhocoris apterus (Hemiptera) and coordination of host-parasite life cycles. PLoS ONE. 2018;13:e0204467. doi: 10.1371/journal.pone.0204467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hamilton P.T., Votýpka J., Dostalova A., Yurchenko V., Bird N.H., Lukeš J., Lemaitre B., Perlman S.J. Infection dynamics and immune response in a newly described Drosophila-trypanosomatid association. mBio. 2015;6:e01356-15. doi: 10.1128/mBio.01356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Svobodová M., Zídková L., Čepička I., Oborník M., Lukeš J., Votýpka J. Sergeia podlipaevi gen. nov.; sp. nov. (Trypanosomatidae, Kinetoplastida), a parasite of biting midges (Ceratopogonidae, Diptera) Int. J. Syst. Evol. Microbiol. 2007;57(Pt 2):423–432. doi: 10.1099/ijs.0.64557-0. [DOI] [PubMed] [Google Scholar]
- 124.Dvorák V., Shaw J.J., Volf P. Parasite Biology: The Vectors. In: Bruschi F., Gradoni L., editors. The leishmaniases: Old Neglected Tropical Diseases. Springer; Cham, Switzerland: 2018. pp. 31–77. [Google Scholar]
- 125.Jackson A.P. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol. Biol. Evol. 2010;27:33–45. doi: 10.1093/molbev/msp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Durante I.M., Butenko A., Rašková V., Charyyeva A., Svobodová M., Yurchenko V., Hashimi H., Lukeš J. Large-scale phylogenetic analysis of trypanosomatid adenylate cyclases reveals associations with extracellular lifestyle and host-pathogen interplay. Genome Biol. Evol. 2020;12:2403–2416. doi: 10.1093/gbe/evaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Souza D.A.S., Pavoni D.P., Krieger M.A., Ludwig A. Evolutionary analyses of myosin genes in trypanosomatids show a history of expansion, secondary losses and neofunctionalization. Sci. Rep. 2018;8:1376. doi: 10.1038/s41598-017-18865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bianchi C., Kostygov A.Y., Kraeva N., Záhonová K., Horáková E., Sobotka R., Lukeš J., Yurchenko V. An enigmatic catalase of Blastocrithidia. Mol. Biochem. Parasitol. 2019;232:111199. doi: 10.1016/j.molbiopara.2019.111199. [DOI] [PubMed] [Google Scholar]
- 129.Kraeva N., Horáková E., Kostygov A., Kořený L., Butenko A., Yurchenko V., Lukeš J. Catalase in Leishmaniinae: With me or against me? Infect. Genet. Evol. 2017;50:121–127. doi: 10.1016/j.meegid.2016.06.054. [DOI] [PubMed] [Google Scholar]
- 130.Jackson A.P., Vaughan S., Gull K. Evolution of tubulin gene arrays in trypanosomatid parasites: Genomic restructuring in Leishmania. BMC Genom. 2006;7:261. doi: 10.1186/1471-2164-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Silva Pereira S., Jackson A.P. UDP-glycosyltransferase genes in trypanosomatid genomes have diversified independently to meet the distinct developmental needs of parasite adaptations. BMC Evol. Biol. 2018;18:31. doi: 10.1186/s12862-018-1149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dobson D.E., Scholtes L.D., Valdez K.E., Sullivan D.R., Mengeling B.J., Cilmi S., Turco S.J., Beverley S.M. Functional identification of galactosyltransferases (SCGs) required for species-specific modifications of the lipophosphoglycan adhesin controlling Leishmania major-sand fly interactions. J. Biol. Chem. 2003;278:15523–15531. doi: 10.1074/jbc.M301568200. [DOI] [PubMed] [Google Scholar]
- 133.Beverley S.M., Turco S.J. Lipophosphoglycan (LPG) and the identification of virulence genes in the protozoan parasite Leishmania. Trends Microbiol. 1998;6:35–40. doi: 10.1016/S0966-842X(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 134.Sacks D.L. Leishmania-sand fly interactions controlling species-specific vector competence. Cell Microbiol. 2001;3:189–196. doi: 10.1046/j.1462-5822.2001.00115.x. [DOI] [PubMed] [Google Scholar]
- 135.Soares R.P., Margonari C., Secundino N.C., Macedo M.E., da Costa S.M., Rangel E.F., Pimenta P.F., Turco S.J. Differential midgut attachment of Leishmania (Viannia) braziliensis in the sand flies Lutzomyia (Nyssomyia) whitmani and Lutzomyia (Nyssomyia) intermedia. J. Biomed. Biotechnol. 2010;2010:439174. doi: 10.1155/2010/439174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Azevedo L.G., de Queiroz A.T.L., Barral A., Santos L.A., Ramos P.I.P. Proteins involved in the biosynthesis of lipophosphoglycan in Leishmania: A comparative genomic and evolutionary analysis. Parasit. Vectors. 2020;13:44. doi: 10.1186/s13071-020-3914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Butenko A., Vieira T.D.S., Frolov A.O., Opperdoes F.R., Soares R.P., Kostygov A.Y., Lukeš J., Yurchenko V. Leptomonas pyrrhocoris: Genomic insight into parasite’s physiology. Curr. Genom. 2018;19:150–156. doi: 10.2174/1389202918666170815143331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Díaz-Viraqué F., Greif G., Berna L., Robello C. Nanopore long read DNA sequencing of protozoan parasites: Hybrid genome assembly of Trypanosoma cruzi. In: de Pablos L.M., Sotillo J., editors. Parasite Genom. Humana; New York, NY, USA: 2021. pp. 3–13. [DOI] [PubMed] [Google Scholar]
- 139.Müller L.S.M., Cosentino R.O., Förstner K.U., Guizetti J., Wedel C., Kaplan N., Janzen C.J., Arampatzi P., Vogel J., Steinbiss S., et al. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature. 2018;563:121–125. doi: 10.1038/s41586-018-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davey J.W., Catta-Preta C.M.C., James S., Forrester S., Motta M.C.M., Ashton P.D., Mottram J.C. Chromosomal assembly of the nuclear genome of the endosymbiont-bearing trypanosomatid Angomonas deanei. G3 (Bethesda) 2021;11:jkaa018. doi: 10.1093/g3journal/jkaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Grybchuk D., Akopyants N.S., Kostygov A.Y., Konovalovas A., Lye L.F., Dobson D.E., Zangger H., Fasel N., Butenko A., Frolov A.O., et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl. Acad. Sci. USA. 2018;115:E506–E515. doi: 10.1073/pnas.1717806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gerasimov E.S., Gasparyan A.A., Kaurov I., Tichý B., Logacheva M.D., Kolesnikov A.A., Lukeš J., Yurchenko V., Zimmer S.L., Flegontov P. Trypanosomatid mitochondrial RNA editing: Dramatically complex transcript repertoires revealed with a dedicated mapping tool. Nucleic Acids Res. 2018;46:765–781. doi: 10.1093/nar/gkx1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cooper S., Wadsworth E.S., Ochsenreiter T., Ivens A., Savill N.J., Schnaufer A. Assembly and annotation of the mitochondrial minicircle genome of a differentiation-competent strain of Trypanosoma brucei. Nucleic Acids Res. 2019;47:11304–11325. doi: 10.1093/nar/gkz928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li S.J., Zhang X., Lukeš J., Li B.Q., Wang J.F., Qu L.H., Hide G., Lai D.H., Lun Z.R. Novel organization of mitochondrial minicircles and guide RNAs in the zoonotic pathogen Trypanosoma lewisi. Nucleic Acids Res. 2020;48:9747–9761. doi: 10.1093/nar/gkaa700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Greif G., Rodriguez M., Reyna-Bello A., Robello C., Alvarez-Valin F. Kinetoplast adaptations in American strains from Trypanosoma vivax. Mutat. Res. 2015;773:69–82. doi: 10.1016/j.mrfmmm.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 146.Callejas-Hernández F., Herreros-Cabello A., Del Moral-Salmoral J., Fresno M., Gironès N. The complete mitochondrial DNA of Trypanosoma cruzi: Maxicircles and minicircles. Front. Cell Infect. Microbiol. 2021;11:672448. doi: 10.3389/fcimb.2021.672448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Camacho E., Rastrojo A., Sanchiz A., Gonzalez-de la Fuente S., Aguado B., Requena J.M. Leishmania mitochondrial genomes: Maxicircle structure and heterogeneity of minicircles. Genes. 2019;10:758. doi: 10.3390/genes10100758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gerasimov E.S., Gasparyan A.A., Afonin D.A., Zimmer S.L., Kraeva N., Lukeš J., Yurchenko V., Kolesnikov A. Complete minicircle genome of Leptomonas pyrrhocoris reveals sources of its non-canonical mitochondrial RNA editing events. Nucleic Acids Res. 2021;49:3354–3370. doi: 10.1093/nar/gkab114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rusman F., Floridia-Yapur N., Tomasini N., Diosque P. Guide RNA repertoires in the main lineages of Trypanosoma cruzi: High diversity and variable redundancy among strains. Front. Cell Infect. Microbiol. 2021;11:663416. doi: 10.3389/fcimb.2021.663416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gerasimov E.S., Kostygov A.Y., Yan S., Kolesnikov A.A. From cryptogene to gene? ND8 editing domain reduction in insect trypanosomatids. Eur. J. Protistol. 2012;48:185–193. doi: 10.1016/j.ejop.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 151.Králová J., Grybchuk-Ieremenko A., Votýpka J., Novotný V., Kment P., Lukeš J., Yurchenko V., Kostygov A.Y. Insect trypanosomatids in Papua New Guinea: High endemism and diversity. Int. J. Parasitol. 2019;49:1075–1086. doi: 10.1016/j.ijpara.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 152.Týč J., Votýpka J., Klepetková H., Šuláková H., Jirků M., Lukeš J. Growing diversity of trypanosomatid parasites of flies (Diptera: Brachycera): Frequent cosmopolitism and moderate host specificity. Mol. Phylogenet. Evol. 2013;69:255–264. doi: 10.1016/j.ympev.2013.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.