Abstract
The exquisite sensitivity of the Burkitt’s lymphoma (BL)-derived cell line Daudi to type I interferons has not previously been explained. Here we show that expression of an Epstein-Barr virus (EBV) transcript, designated D-HIT (Y. Gao et al., J. Virol. 71:84–94, 1997), correlates with the sensitivity of different Daudi cell isolates (or that of other EBV-carrying cells, where known) to alpha interferon (IFN-α). D-HIT, transcribed from a GC-rich repetitive region (IR4) of the viral genome, is highly structured, responding to RNase digestion in a manner akin to double-stranded RNA. Comparing EBV-carrying BL cell lines with differing responses to IFN-α, we found the protein levels of the dsRNA-activated kinase, PKR, to be similar, whereas the levels of the autophosphorylated active form of PKR varied in a manner that correlated with endogenous levels of D-HIT expression. In a classical in vitro kinase assay, addition of either poly(I)-poly(C) or an in vitro-transcribed D-HIT homolog stimulated the autophosphorylation activity of PKR from IFN-α-treated cells in both EBV-positive and EBV-negative B lymphocytes. By transfection experiments, these RNAs were shown to reduce cell proliferation and to sensitize otherwise relatively insensitive Raji cells to IFN-α. The data lead to a model wherein the D-HIT viral RNA also serves as a possible transcriptional activator of IFN-α or cellular genes regulated by this cytokine.
Since the discovery that interferons, a family of multifunctional secreted cytokines, might block viral replication, they have been widely studied for their ability to defend eukaryotic cells against infectious agents or to act themselves as antitumor agents. Interferons exert their activities by binding to cell receptors, triggering signals that result in the altered expression of numerous cellular (or viral) genes (reviewed in reference 39). One of the best characterized of the interferon pathways uses a double-stranded RNA (dsRNA)-activated protein kinase, PKR, to phosphorylate and inactivate the peptide chain initiation factor eIF-2, blocking translation. A second pathway has an inducible RNA endonuclease activity (RNase L) capable of cleaving numerous cellular and viral RNAs and regulatory factors, thus modulating the expression of interferons themselves (as reviewed in references 13 and 33). In addition to these well-defined interferon-associated cellular pathways, PKR has been shown to have an effect on cellular pathways other than those directly associated with protein synthesis, increasing the emphasis on this function as a cell growth regulator (as discussed in reference 25 for NF-κB and reference 37 for c-myc).
Some of the earliest studies on interferon focused on Epstein-Barr virus (EBV)-carrying Burkitt’s lymphoma (BL)-derived cell lines. It was shown that several of these, such as the Namalwa line, spontaneously produced alpha interferon (IFN-α) and that different lines varied in their capacities for expressing viral functions. Cell lines that spontaneously produced IFN-α were refractory to superinfection with EBV, suggesting that this protein blocked cellular susceptibility to the virus (1). Other BL-derived lines, among them Daudi and Raji, made undetectable or very low amounts of interferon and could be superinfected with virus or induced to express EBV genes. Daudi cells especially proved to be remarkably sensitive to IFN-α, 5 to 10 U/ml being sufficient to reduce viral gene expression by 50% (as measured by endogenous early antigen expression). More than 500 U/ml was required to produce the same effect with Raji cells (2). Daudi has since become almost axiomatically the cell line of choice for studying mechanism(s) of action of type I interferons, and mutants with various responses to exogenous interferon have been generated (9, 41). In studies using Daudi cell lines of different interferon sensitivities, the levels of expression of the c-myc oncogene, which is translocated and deregulated in BL cells, were found to be decreased in (sensitive) wild-type Daudi cells upon treatment with IFN-α but to be little altered in an (insensitive) mutant line (31). In the former, transcription of c-myc could also be down-regulated by sodium n-butyrate (SB), in a manner independent of interferon-inducible RNase L (36).
Three EBV functions have been studied with regard to the sensitivity of infected cells to interferons.
(i) The EBV nuclear antigen EBNA2, a transactivator function essential for B-cell immortalization, has been described as being at least partly responsible for conferring resistance to the antiproliferative effect of interferon (3). In Daudi cells, where the genome has a deletion that removes the EBNA2 gene (20), superinfection with a virus strain expressing this protein restores resistance to IFN-α (21). The mechanism of action of EBNA2 in this context has not been defined. Notably, Daudi cells can also be converted to interferon-insensitive lines in the absence of EBNA2 (9, 41).
(ii) The EBV small polymerase III-transcribed viral RNAs (the EBERs), as well as adenovirus VA RNAs, have been shown to interact with PKR to limit the activity of the kinase as an inhibitor of protein synthesis (40). Where Daudi cells are concerned, it seems unlikely that the EBER-PKR interaction can explain the differential sensitivities to IFN-α among the various wild-type and mutant lines, however, since in all of them the EBERs are expressed at very high, probably supersaturating, levels for PKR.
(iii) Transcripts from the major EBV genome internal repeat (IR1, or BamHI W fragment) have been suggested to act as regulators of one or other of the interferon pathways, since in vitro they can compete with EBERs for binding to PKR (10); they also regulate kinase activity, although, unlike the EBERs, they do not block its activation by dsRNA. Again, without further evidence, it seems difficult to invoke this RNA as accounting for the observed differential responses to interferon, since all EBV-positive cells, regardless of their interferon sensitivities, carry multiple (10 to 14) copies of the viral IR1 gene.
If EBV itself is involved in regulating the cellular response to interferon, it seemed to us that a viral function(s) other than those described above was playing a role in the differing responses observed among the various natural or mutant BL-derived lines. Here, we describe a viral transcript in Daudi cells, designated D-HIT (Daudi highly inducible transcript), that is a potential candidate for this activity. D-HIT, present in low but detectable levels in wild-type Daudi cells, is transcribed from a region of the EBV genome carrying the GC-rich repetitive sequence, IR4. It is polyadenylated and predicted to have considerable secondary structure (12). D-HIT, or its viral homolog in other EBV-carrying cells, is differentially expressed in the various cell lines, the highest levels of transcription being found in IFN-α-sensitive Daudi cells (as summarized in Table 1). Since this transcript might represent an endogenous double-stranded modulator of interferon pathways, we studied its potential for explaining the differential sensitivity to IFN-α among Daudi isolates and other EBV-carrying cells.
TABLE 1.
Expression levels of EBV D-HIT and homologs in B-cell lines
| Cells | Transcriptiona
|
|
|---|---|---|
| Uninduced | Inducedb | |
| Daudi/ATCC | + | +++ |
| Daudi/ICRF | ++ | +++++ |
| M-ABA | − | + |
| Namalwa | − | − |
| NAD-C15 | − | − |
| P3HR1 | ++ | ++ |
| Raji | − | + |
| Ramos | − | − |
+ to +++++ indicate levels of transcription relative to those observed by Northern blot analyses under comparable conditions. −, no transcript detected. Data are taken in part from previous work (12).
Cells were treated with both TPA and SB.
MATERIALS AND METHODS
Materials.
IFN-α (interferon alfa-2a; Roferon-A) was purchased from Roche Products Ltd., and poly(I)-poly(C) was purchased from Pharmacia Biotech, UK. No length information is provided with the latter product since the two strands are synthesized independently and then annealed together. It is believed to be primarily double stranded (information from the manufacturer).
Cell lines and cultivation.
Daudi, Namalwa, P3HR1, and Raji are EBV-positive BL-derived cell lines, and Ramos is an EBV-negative BL line. NAD-C15, a human lymphoblastoid line, and M-ABA, a marmoset lymphoblastoid line, were established with EBV from nasopharyngeal carcinomas. Daudi/ATCC (CCL 213) cells were obtained from the American Type Culture Collection, and Daudi/ICRF and an interferon-resistant mutant (100K) were from I. M. Kerr (Imperial Cancer Research Fund, London, United Kingdom). All B-cell lines were grown in RPMI 1640 medium (Gibco) supplemented with 10% (vol/vol) fetal calf serum under standard conditions and were passaged twice weekly. Actively dividing cells were treated, at densities of 5 × 105 cells/ml, in growth medium with 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma) at 20 ng/ml for 72 h, and/or SB (BDH) at 3 mM for 72 h or with IFN-α at 500 U/ml for 20 h, unless otherwise stated.
Cell growth assays for IFN-α sensitivity.
BL-derived Daudi or Daudi-mutated (100K) and Raji cell lines were maintained by adjusting the cell concentration to 2 × 105 living cells/ml in fresh growth medium in the continuous presence of different concentrations of interferon. The number of living cells at 7-day intervals was determined by the trypan blue exclusion assay. Each culture was monitored for 4 weeks.
RNA isolation and Northern blotting.
RNA was isolated from the B-cell lines by the guanidinium-cesium chloride method and probed essentially as described elsewhere (12, 22). Polyadenylated RNA was selected by using two sequential oligo(dT) mRNA purification columns (Pharmacia). An actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeper gene probe was routinely used to confirm that comparable quantities of RNAs had been loaded onto gels.
Digestion of RNA with RNases.
Total RNAs from TPA- and SB-induced Daudi/ATCC cells were treated with RNase A (0.1 mg/ml; Sigma) in a digestion buffer composed of 25 mM Tris HCl (pH 7.2), 10 mM MgCl2, and 200 mM NaCl at 20 or 37°C and with RNase V1 (20 U/ml; Pharmacia) in the digestion buffer at 20°C, respectively, over time and then assayed by Northern blot analysis. Blots were hybridized with the BamHI Ia region of the viral genome (4, 17) or with probes for rRNAs.
In vitro synthesis of the Raji D-HIT homolog.
In the absence of cloned fragments from Daudi cells, the D-HIT homolog was synthesized in vitro with a DNA template from Raji cells. A 3.4-kb DNA fragment encompassing the IR4 region, between nucleotide positions 905 and 4340 (22, 23a) was cleaved from the EBV BamHI-Ia clone (4, 17) with restriction enzymes XbaI and SmaI, subcloned into Bluescript (Stratagene), and then transcribed in vitro from a T3 promoter in the vector, using a standard protocol (38).
Transfection of cells with the D-HIT homolog or poly(I)-poly(C) RNAs.
Confluent Raji, Ramos, and Daudi/ICRF cells were harvested, washed with ice-cold phosphate-buffered saline, pelleted, and resuspended in serum-free RPMI 1640 medium plus antibiotics, to give a final concentration of 2 × 105 to 3 × 105 cells/ml. Cell suspensions (300 μl) were placed into 24-well plates and incubated for 2 to 3 h to allow the cells to adhere to the plastic. In a series of control experiments, we assessed the usefulness of two reagents, Effectene and SuperFect, as transfection agents that could be used in small volumes for introducing RNA into lymphocytes without undue toxicity, using conditions recommended by the manufacturer (Qiagen). In our hands, Effectene was the better of the two reagents. Two sets of conditions, optimized for Raji cells, were eventually established. After removal of medium, cells were transfected with either 5 μl of Effectene plus 6.4 μl of Enhancer in 400 μl of EC buffer (conditions 1) or 10 μl of Effectene plus 6.4 μl of Enhancer in 400 μl of EC buffer (conditions 2), with added dsRNA [either 16 or 4 μl of poly(I)-poly(C) or 4 μl of purified IR4 RNA, both at 1 μg/μl], according to the manufacturer’s instructions. After 2 h of incubation, 800 μl of complete tissue culture medium was added to each well to allow the cells to grow and detach from the dish. Following overnight growth, 200-μl aliquots of cells from each of the 24 wells were transferred to 96-well dishes, to provide equal numbers of cells for 4 new wells. In total, 48 wells of each of the three cell lines were used in subsequent experiments aimed at measuring DNA replication and cell survival, as previously described (5). To 32 of these wells, IFN-α (4 μl) was added to produce a concentration of 104 U/ml. Cells were left overnight, and to 20 of the wells, [3H]thymidine (8.0 μCi/well) was added. Control cells were either untransfected, transfected in the absence of dsRNA, or transfected in its presence with tritiated thymidine in the absence of IFN-α. After 5 days, tritium uptake was measured in aliquots of cells as described previously (5). Fresh medium (100 μl) was added to the other wells, and cells from these wells were stained with trypan blue 7 days later. Live (bright colorless) cells were counted on a hemacytometer and compared with dead or dying (blue) cells in control cells receiving IFN-α, with or without Effectene, and in cells transfected with RNA and treated with IFN-α. Three separate counts were made for each experiment in duplicate wells (six for a single set of conditions); when on occasion difficulties in counting were experienced due to cell clustering, further counts (up to eight) were made.
Protein extraction, Western blotting, and immunoprobing.
Proteins from B-cell lines were extracted, their concentrations were determined by the bicinchoninic acid assay (Sigma), and they were separated on a sodium dodecyl sulfate–10% polyacrylamide gel. Products were electrotransferred onto nitrocellulose membranes and then probed with an anti-PKR kinase monoclonal antibody (71/10; Ribogene) as described elsewhere (18, 26). Immunocomplexes were detected by incubation with a horseradish peroxidase-conjugated secondary antibody (Dako) and visualized by enhanced chemiluminescence (ECL reagent; Amersham).
Protein kinase assay.
PKR was immunoprecipitated by incubating protein extracts from IFN-α treated or untreated B-cell lines with monoclonal antibody 71/10 followed by complex binding to protein G-Sepharose (Pharmacia). Washed immunoprecipitates were resuspended in a buffer containing 10 mM Tris-HCl (pH 7.6), 100 mM KCl, 2.5 mM MgCl2, 2.5 mM MnCl2, and 10 mM 2-mercaptoethanol and incubated with 2 mM [γ-32P]ATP (50 Ci/mmol) at 30°C for 20 min, with or without added synthetic poly(I)-poly(C) (0.8 μg/ml; Pharmacia) dsRNA, or the EBV D-HIT homolog (0.8 μg or 8.0 μg/ml) from Raji cells. Phosphorylated PKR was separated by polyacrylamide gel electrophoresis, and products were identified by autoradiography.
RESULTS
Structure of D-HIT RNA.
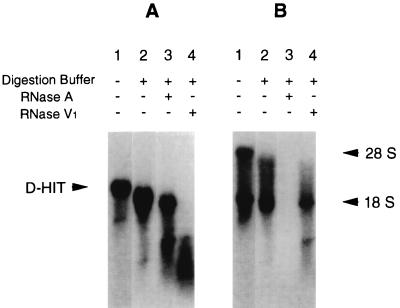
Our previous studies showed that Daudi/ATCC cells endogenously expressed low levels of a transcript (called D-HIT) from a repetitive region of the EBV genome. Induction of these cells (with SB and TPA) led to levels of the RNA that, alongside rRNAs, could even be seen by ethidium bromide staining of gels during separations of total cellular RNA (12). Experiments aimed at assessing translation of D-HIT were carried out with poly(A)+-selected RNA, purified on sucrose gradients, in an in vitro rabbit reticulocyte translation system. Here, not only was there no protein product observed, but control mRNA (supplied by the manufacturer), when mixed with D-HIT RNA from Daudi cells, was also not translated (data not shown). These findings, reminiscent of results obtained when double-stranded or otherwise highly structured RNA is used in in vitro translation systems (19), coupled with sequence analysis (12), prompted us to examine the sensitivity of D-HIT to enzymes specific for single-stranded (RNase A) or double-stranded (RNase V1) RNase. The results (Fig. 1A) show that RNase A activity at 20°C (or even at 37°C [data not shown]) has only a slight effect on D-HIT over time, whereas the dsRNA-specific RNase V1 cleaves it almost instantaneously; the opposite result was obtained on digestion of ribosomal (18S) RNA (Fig. 1B).
FIG. 1.
Action of RNases A and V1 on the 2.8-kb D-HITs. Total RNA from induced Daudi/ATCC cells was treated with excess single (RNase A)- or double (RNase V1)-stranded RNA-specific RNases at 20°C for 30 min. Products were separated by electrophoresis and visualized after transfer to nitrocellulose and hybridization with a 32P-labelled EBV BamHI-Ia probe (A) or probes for rRNAs (B).
Computer-assisted analyses for secondary structure of nucleic acid sequences (43) were also performed. These identified structures which are predicted to be highly stable, both in the 102-bp IR4 repeat region and in its immediate upstream and promoter sequences, where energy calculations give −86.5 kcal/mol (12).
Comparison of interferon sensitivities and levels of D-HIT expression in Daudi cell isolates and other B lymphocytes.
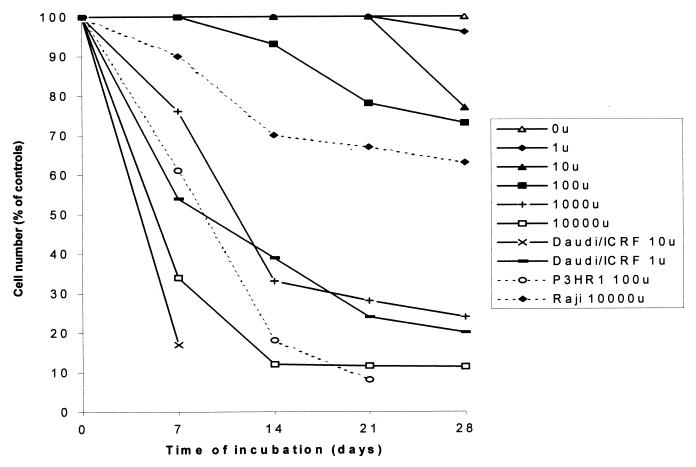
The structural evidence raised the question whether D-HIT, acting as a dsRNA-like RNA, might account for the observed enhanced sensitivity of Daudi cells, compared with some other EBV-carrying cells, to IFN-α (1, 2). To explore this question, two separate Daudi isolates (Daudi/ATCC and Daudi/ICRF), an interferon-insensitive mutant of Daudi (100K), and Raji cells were assessed for their differential responses to various doses of IFN-α and then compared with their corresponding levels of D-HIT. In the interferon sensitivity studies, considerable variation to IFN-α was observed among the parental and mutant Daudi cell lines; Daudi/ATCC cells, for example, were found to be intermediate in response between Daudi/ICRF and its resistant mutant, 100K, the latter being more like Raji cells in its behavior. Plotting cell survival versus time as a function of IFN-α concentration (in units/milliliter), with Daudi/ICRF, 50% cell survival (or death) was achieved after 4 days with 10 U and after about 10 days with 1 U. By comparison with Daudi/ATCC cells, neither of these interferon concentrations results in 50% cell death, even after 28 days. With Daudi/ATCC cells, the 50% figure was reached after 6 days with 104 U and at about 12 days with 103 U. Direct comparisons are complicated by the very nature of the considerable differences in sensitivities and also the fact that the response was not a linear one. With Raji and 100K cells, 50% cell death was not achieved at the maximum IFN-α concentrations (104 U) even after 28 days. In the case of Raji, at 7 days, about 90% of the cells were still alive; at 14 days, this figure had dropped to about 70%, where it remained more or less steady up to 28 days. With 100K, 10% cell death (with 104 U) was observed only after 21 days and may not be wholly attributable to the action of interferon. Figure 2 gives the overall pattern of cell responsiveness of Daudi/ATCC over the range of IFN-α concentrations employed in these studies (1 to 104 U). Superimposed on this pattern, for comparison purposes, are curves obtained for Daudi/ICRF (at 1 and 10 U), Raji (at 104 U), and P3HR1 (at 102 U), where the data are taken from the literature (2).
FIG. 2.
Interferon sensitivities of Daudi/ATCC cells. Cells were grown in the presence of various concentrations of IFN-α over a 28-day period, and the number of live cells (as a percentage of untreated cells) was plotted. The profile of Daudi/ATCC, showing viable cell counts as a percentage of cells grown in the absence of interferon (100%), is taken as a standard for medium IFN-α sensitivity. Superimposed on it are two curves from Daudi/ICRF cells (grown in the presence of 1 and 10 U of IFN-α/ml; high sensitivity), one from Raji cells (grown with 104 U; low sensitivity), and one from P3HR1 cells (102 U; high/medium sensitivity; data taken from the literature [2]), all EBV-positive lines.
D-HIT, IFN-α, and interferon-inducible cellular transcriptional expression in Daudi/ATCC and other EBV-positive B lymphocytes.
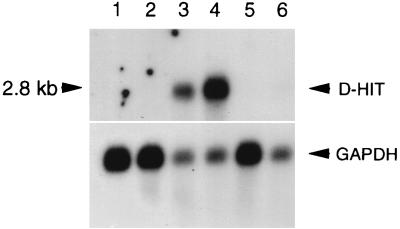
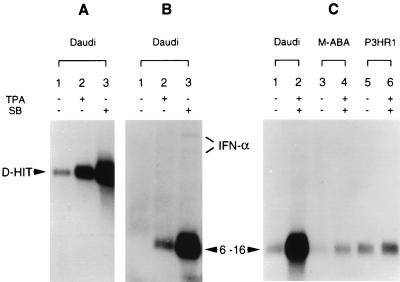
Northern blot analyses of D-HIT expression are shown in Fig. 3 and 4. Endogenous levels of expression of D-HIT, compared with levels induced in Daudi/ATCC cells with either TPA or SB, are shown in Fig. 4A. Both chemical agents induce RNA expression, with the highest levels (track 3) produced by the action of SB. Elevated D-HIT expression was also observed when cells were grown in IFN-α (102 U/ml, for 3 days); levels of the transcript were slightly lower, however, than that observed in the TPA induction experiments (data not shown). To compare with these data, we investigated the RNA levels of dsRNA-inducible IFN-α and interferon-stimulated cellular genes, 6-16 and 9-27, whose transcription is mediated by interferon-stimulated response elements (14). 6-16 transcription levels can be increased more than 100-fold by IFN-α but not significantly by IFN-γ (23). Northern blot data for IFN-α and 6-16 gene expression are shown (Fig. 4B). Corresponding data for 9-27 (not shown) were qualitatively similar, although the overall transcription levels were lower. Endogenous levels of interferon in Daudi cells, reportedly low or possibly absent (1), are consistent with our findings (Fig. 4B, track 1). Upon induction, however, as observed with D-HIT (Fig. 4A), both IFN-α and 6-16 RNA levels were enhanced, SB again being the superior inducing agent (Fig. 4B; compare tracks 2 and 3). These data alone, while revealing parallel transcriptional responses between D-HIT, IFN-α, and 6-16, would not distinguish between induction as a direct consequence of the chemical treatment, or in association with expression of D-HIT or IFN-α, in these cells. Work by others shows that TPA alone does not induce 6-16 transcription (7).
FIG. 3.
Northern blots showing levels of endogenous D-HIT expression in parental Daudi cell lines ATCC and ICRF and two mutants, clone 1 and 100K, with different sensitivities to IFN-α (Fig. 2), compared with expression of homologs in two other BL-derived lines, Namalwa and Raji. Tracks 1 to 6 contain poly(A)+ RNA (7 μg) from Namalwa, Raji, Daudi/ATCC, Daudi/ICRF, Daudi/clone 1, and Daudi/100K cells, respectively. The upper blot was deliberately overexposed in an attempt to look for transcripts in apparently negative lines. A GAPDH probe was used as a control for RNA loading.
FIG. 4.
Northern blot analysis of D-HIT, IFN-α, or interferon-inducible (6-16) gene transcripts in B cells. Poly(A)+ RNA (5 μg) from uninduced or TPA- and SB-induced cells (as indicated) was loaded onto gels and detected by hybridization with 32P-radiolabelled probes from EBV IR4 DNA (A) or cDNAs for IFN-α (B) or 6-16 (B and C). The sources of the RNA and the nature of the inducing agents (TPA and SB), if used, are indicated, and predicted locations of the individual RNAs are marked. Although the levels of IFN-α in panel B are low, they mimic those observed for D-HIT (A) and 6-16 (B), where increased levels are observed with TPA-treated cells and even higher levels are detected with SB-treated cells.
Attempting to answer this query, and also to examine whether the data might be related to the absence of EBNA2 in Daudi cells, we examined 6-16 expression in two other EBV-carrying B-cell lines. One of them, M-ABA, contains both the IR4 sequence and the gene for EBNA2, and the other, P3HR1, is similar to Daudi in retaining IR4 but having a deletion that removes the EBNA2 gene (20). Both show endogenous expression of the RNA at levels which are not greatly affected by induction compared with Daudi cells. The data (Fig. 4C) show that whereas low levels of endogenously expressed 6-16 transcripts are seen in all three cell lines (tracks 1, 3, and 5), high-level transcription after induction is most notable in Daudi cells (track 2). Overall, they suggest that 6-16 expression levels not only correlate with that of D-HIT, rather than depending on treatment of cells with TPA and SB, but also are not strictly tied to the presence or absence of EBNA2.
Expression of PKR in B cells in response to IFN-α or chemical inducing agents.
Since Northern blot data showed comparable levels of p68 transcripts in Daudi cell lines of various sensitivities (data not shown), we examined whether varying control over translation might have been influencing interferon sensitivities. Expression of the IFN-inducible, dsRNA-activated protein kinase, PKR, was thus examined in the Daudi lines, using a monoclonal antibody to PKR (26). In the first set of experiments (Fig. 5A), the levels of the endogenously expressed protein were observed to be roughly the same in all of the EBV-positive cell lines examined (Daudi/ATCC, Daudi/ICRF, and Daudi/100K [tracks 3, 5, and 7, respectively]) and Raji cells (track 9), although below the level of detection in the EBV-negative Ramos B-cell line (track 1). In interferon-treated cells, this level, as anticipated, was increased (even in Ramos), except in the case of the insensitive Daudi cell line, 100K (track 8). A similar pattern was observed in Fig. 5B, the levels of protein in uninduced cells (tracks 3, 5, 7, and 9) being enhanced upon chemical induction with TPA and SB (tracks 4, 6, and 10), except again in the interferon-insensitive 100K line, where the levels remained essentially constant (track 8). Unlike the findings with IFN-α, chemical induction had little or no effect on PKR levels in Ramos cells (compare tracks 1 and 2 in Fig. 5A and B). In EBV-carrying cells, TPA and SB seem to mimic the PKR stimulatory effects of IFN-α, but whether or not this is the case in EBV-negative cells needs to be further explored. Notably, the protein levels themselves do not fully account for the different sensitivities to IFN-α observed among the various cell lines. For example, the virtually insensitive 100K line contains reasonable levels of protein.
FIG. 5.
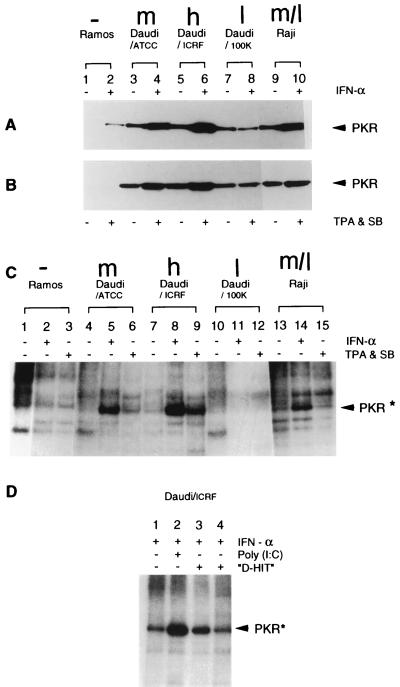
Expression and activity of PKR. (A and B) Western blot analyses of PRK expression in untreated or IFN-α treated cells (A) and untreated or TPA- and SB-induced cells (B). Comparable levels of total protein were added to all tracks, and the presence of PKR was detected with monoclonal antibody 71/10. (C) Kinase activity of PKR in various B-cell lines. PKR was immunoprecipitated with 71/10, and kinase activity assessed in the presence of [γ-32P]ATP. The cells used in the assays were untreated or treated with IFN-α or with TPA and SB, respectively, as noted, prior to protein isolation. The data shown are from the same autoradiogram but reorganized so that the controls (PKR from Ramos and Raji cells) flank the Daudi data. (D) The same assay as in panel C, with either poly(I)-poly(C) or an in vitro-transcribed D-HIT homolog (both at 0.8 μg/ml) added to the phosphorylation reaction of PKR isolated from IFN-α-treated Daudi cells, as noted. Track 4 contains a 10-fold-higher concentration of viral RNA (8.0 μg/ml) than used in track 3. Arrowheads indicate the positions of PKR and phosphorylated PKR (PRK*). The abbreviations h (high), m (medium), and l (low or undetectable), and m/l (medium/low) represent the relative levels of D-HIT present in the EBV-positive cells (Table 1).
PKR kinase activity.
As the biological activity of PKR depends on its autophosphorylation and its ability to phosphorylate a subunit of the protein initiation factor eIF-2α, phosphorylation of PKR in Daudi, Raji, and Ramos cell lines was investigated. The protein kinase from various cell extracts was immunoprecipitated with monoclonal antibody 71/10 (26), and then autophosphorylation was monitored in the presence of [γ-32P]ATP (18). Figure 5C shows data for radiolabelled proteins derived from untreated Ramos, Daudi/ATCC, Daudi/ICRF, Daudi/100K, and Raji cells (tracks 1, 4, 7, 10, and 13, respectively). Other tracks contain materials from the same cells after induction with IFN-α (tracks 2, 5, 8, 11, and 14) or treatment with TPA and SB (tracks 3, 6, 9, 12, and 15). The results show that the PKR protein is phosphorylated in every case except in interferon-insensitive Daudi/100K or EBV-negative Ramos cells. Further, whereas IFN-α treatment produces enhanced levels of phosphorylated PKR in all EBV-positive cells (tracks 5, 8, and 14), the effect was greatest for the most sensitive Daudi/ICRF line (track 8). Comparable results (compare with data in Fig. 5B) were observed with chemically induced Daudi/ATCC and Daudi/ICRF cells. But with Ramos and Raji cells, chemical induction was only marginally effective as a means of stimulating PKR activity. Notably, from previous studies (12), the levels of D-HIT in Raji cells were little altered by chemical induction. With regard to Daudi/100K, there is no evidence, from this experiment, of a phosphorylated PKR in cells treated with either IFN-α or the chemical reagents (Fig. 5C, tracks 11 and 12), although the protein is expressed (Fig. 5A and B).
Finally, in similar studies, we examined whether D-HIT (using its homolog from Raji cells), like the synthetic dsRNA poly(I)-poly(C), could directly stimulate autophosphorylation of PKR. The data, reproduced on several occasions and illustrated in Fig. 5D, show levels of phosphorylated PKR in Daudi cells to be 4.6 (compare tracks 1 and 2) and 2.5 (compare tracks 1 and 3), respectively, times higher in the presence of comparable amounts of the synthetic polymer and the viral transcript than in the control with no added RNA (track 1). In this in vitro assay, higher levels of the viral transcript (track 4) appeared to be inhibitory, as noted elsewhere for poly(I)-poly(C) (6, 11). Similar data (not shown) were obtained with Ramos cells, where levels of activation achieved with the viral and nonviral RNAs were similar, although here the bands of phosphorylated PKR were not so sharply defined.
The same cells were treated with the inhibitor 2-aminopurine in an attempt to block phosphorylation, but we, like others (23a), observed no effect even with high concentrations of this reagent (data not shown).
Comparison of IFN-α sensitivities in Raji cells transfected with poly(I)-poly(C) or the D-HIT homolog. (i) Cell survival.
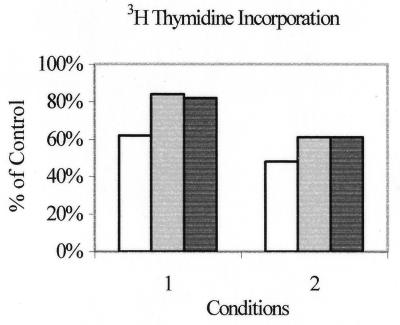
Cells were transfected with the different RNAs by using Effectene, as described in Materials and Methods, in the presence or absence of IFN-α. Ramos cells proved highly sensitive to the transfection protocol in that only about 10% of cells survived conditions 1 (in the absence of dsRNA) and fewer than this survived conditions 2. Thus, the bulk of the experiment was carried out on Raji cells, where on average on day 1, 46% cells had survived conditions 1 and 56% survived conditions 2, compared with untreated controls. In the first sets of experiments, proliferation of cells grown 5 days in the presence of tritiated thymidine was assessed essentially as described previously (5). The data under the two sets of conditions are shown in Fig. 6. The control, taken as 100%, was an average of the tritium count in Raji cells treated with IFN-α with or without added Effectene. This control was adopted since it was observed that Effectene reagent was actually less toxic to cells when mixed with dsRNA. Under both sets of conditions, decrease in the proliferating cell population was observed to correlate with the quantity of added dsRNA, rather than to its composition, the most significant increase (50%) being observed under conditions 2 with 16 μg of poly(I)-poly(C). With 4 μg, the synthetic dsRNA and the D-HIT homolog gave comparable (40%) reductions.
FIG. 6.
Proliferation assay. Raji cells were treated with either 16 μg (white bars) or 4 μg (grey bars) of poly(I)-poly(C) or with 4 μg of the EBV D-HIT homolog (black bars), and thymidine incorporation was measured 7 days posttransfection, using conditions 1 or 2 (see text for definition), as noted, in the presence of 104 U of IFN-α/ml. Cell survival is presented as percentage of control cells without added exogenous RNA.
(ii) Cell viability.
Cell viability was determined microscopically after a total of 14 days (from the time of transfection) by the trypan blue exclusion assay. In each case, the numbers of cells from both experimental conditions were further evaluated to determine the effects of 104 U of IFN-α on survival, taking the average viable cell count of treated compared with untreated cells (without added dsRNA) as 100%. Note that with the control Daudi/ICRF cells, in the presence of 104 U of IFN-α, virtually all the cells were killed within a few days. With conditions 1, where comparable amounts of poly(I)-poly(C) and the D-HIT homolog were used in the transfection mix, the survival rates at this time were evaluated as 29 and 26%, respectively; with a fourfold higher concentration of poly(I)-poly(C), this figure was 34%. Under conditions 2, the corresponding figures by this technique were 27, 48, and 39%, respectively. No significance is placed on the precise figures here, as mixtures of live and dead cells showed a tendency to clump, reducing the count accuracy, especially when cell numbers were decreasing. Comparison of these data with those for Raji cells (Fig. 2)—where at 14 days in the presence of 104 U of IFN-α, 60 to 70% of the cells were still alive—shows that cells had been sensitized to the interferon by the addition of the dsRNAs, whether the latter was of synthetic or EBV origin.
DISCUSSION
The high degree of secondary structure predicted and apparent in an EBV transcript, designated D-HIT, and its variation in expression levels among different EBV-carrying cell lines (12) stimulated us to investigate a possible association between this RNA and the known differential sensitivities of certain EBV-positive B lymphocytes to type I interferons (1, 2). D-HIT was found to behave in the presence of RNases essentially as a dsRNA (Fig. 1), and its constitutive expression was observed by Northern blots at low but detectable levels, particularly in interferon-sensitive Daudi cells (Fig. 3 and 4A; Table 1), suggesting that it might be an effector of interferon pathways responsive to dsRNAs. Notably, interferon sensitivities in non-chemically induced cells (Fig. 2) appeared to parallel the endogenous levels of expression of D-HIT. This viral transcript can be expressed at remarkably high levels when certain Daudi lines, but not all other EBV-containing B-cell lines investigated, are chemically induced. In Daudi cells, the transcriptional expression of two interferon-sensitive human cellular genes, 6-16 and 9-27, and to a much lesser extent that of the IFN-α gene itself, were also shown to be up-regulated following treatment with TPA or SB (Fig. 4B).
Parental and mutant Daudi cells, with different sensitivities to IFN-α, show no apparent difference in RNase L and 2,5-A synthetase activities (41). We thus explored D-HIT expression as it might affect the other major interferon pathway, that involving the cellular protein kinase PKR, which regulates the transcriptional and translational machinery of the cell. PKR is activated by a mechanism that involves dsRNA binding in a sequence-independent manner (32). In a series of experiments, typified by data shown in Fig. 5, using two different primary Daudi cell lines, a mutant (100K) from one of these, and as controls the relatively IFN-α insensitive line Raji and the EBV-negative line Ramos, we compared protein levels and autophosphorylation of PKR in cells treated either with IFN-α or with TPA and SB. PKR protein levels were found to be essentially indistinguishable among the IFN-sensitive lines (compare Fig. 5A and B), with the chemical inducing agents mimicking the effects of interferon in their ability to up-regulate expression. (Neither reagent had much effect, however, on PKR levels in the Daudi interferon-insensitive mutant line, 100K.) We observed little difference in endogenous PKR levels among Raji, all Daudi, and Ramos cells. In assays investigating the kinase activity of PKR (Fig. 5C), the overall levels of phosphorylated PKR were considerably increased when inducing agents (IFN-α; TPA and SB) were added to the cells, with the chemical inducing agents again mimicking the action of interferon in Daudi, but not in Raji, cells. (In the case of 100K, no kinase activity was observed, although a 68-kDa-migrating protein is clearly present in these cells [Fig. 5A and B], suggesting that an inhibitor of phosphorylation may have been coimmunoprecipitated together with PKR in this case, or alternatively, the protein was already phosphorylated.) Our data suggest that D-HIT, acting as a dsRNA, directly stimulates the activity of PKR. They support the supposition that D-HIT per se, and not the chemical inducing agents, is responsible for the observed effects; that is, whereas IFN-α stimulates the expression of PKR in the EBV-negative cell line Ramos, this is not observed to be the case when TPA and SB are used (Fig. 5). To examine this argument further, poly(I)-poly(C) dsRNA or an in vitro-transcribed homolog of D-HIT was added to the in vitro PKR kinase assay. Here, using comparable amounts of material, we found both poly(I)-poly(C) and the viral transcript to stimulate autophosphorylation of PKR. Increasing the concentration of the latter 10-fold nullified this effect, as observed earlier for poly(I)-poly(C) (11) and other potentially double-stranded RNAs (6, 35).
Although B cells often prove problematic in transfection experiments, they were chosen for the next set of experiments as the most appropriate model for examining the effect of ds-RNA, including D-HIT, on cell proliferation. The poly(I)-poly(C) and D-HIT RNAs were transfected directly into Raji cells and proliferation was examined by two assays, as described elsewhere (5). One of these used tritiated thymidine, measuring radioactive uptake over a 5-day period, 7 days after introduction of the RNAs into in vitro cultures of Raji cells in the presence of 104 U of IFN-α. In the absence of added dsRNA, this technique had identified no differences over time (4 and 6 days) in Raji cells treated with various concentrations of IFN-γ compared with untreated Raji or other B-cells (5). As shown in Fig. 6, however, when Raji cells were transfected with either the synthetic polymer or the D-HIT homolog, the number of proliferating cells was reduced in the presence of IFN-α, in a manner that corresponded to the amount of exogenous RNA added rather than to the nature of the RNA, consistent with other studies (32). Our data showed a 40 to 50% reduction in tritium uptake in the presence of 16 μg of poly(I)-poly(C) and 22 to 40% (depending on conditions) reduction in assays using 4 μg of either this RNA or the D-HIT homolog. Further work to optimize these conditions might increase these levels of sensitivity. Notably, as suggested elsewhere (29), it may prove advantageous to increase the RNA concentrations. In the second assay, cell survival in the presence of 104 U of IFN-α was measured. Where previous data (Fig. 2) showed that after 14 days, about 70% of Raji cells had survived in the absence of added exogenous RNA, this figure was reduced to values ranging between 26 to 48% when the dsRNAs were added (Fig. 6). Again, these experiments clearly indicate that D-HIT can mimic poly(I)-poly(C) in sensitizing cells to the effects of interferon. In future, it may prove advantageous to generate a library of EBV clones from Daudi cells that will allow us to isolate D-HIT per se, since the viral gene sequence in these cells (12) has a small change relative to that used in the current experiments, which conceivably can influence the stability of the dsRNA-like RNA and thus its overall effect on cells.
Throughout our investigations, we observed variation between the two parental Daudi cell lines (ATCC and ICRF), both with regard to their sensitivity to IFN-α and with regard to their endogenous and inducible levels of D-HIT, growth habits, and even responses to antibodies (13). These differences, which probably reflect alterations that occurred at different times in the development and treatment of the BL (24), have proved useful here in further allowing correlations between interferon response and D-HIT expression levels to be made. For Daudi/ICRF and to a lesser extent Daudi/ATCC cells, the enhanced sensitivity to IFN-α may be a consequence of the fact that they contain detectable (although different) levels of endogenous D-HIT. This suggestion is substantiated by the data shown in Fig. 2, 3, and 4A and is supported by those for another interferon-sensitive BL line, P3HR1 (2), which also expresses detectable endogenous levels of this transcript. These findings (summarized in Table 1) further indicate a correlation between endogenous D-HIT expression and the relative interferon sensitivities observed in EBV-carrying B-cell lines (Fig. 2).
The viral function EBNA2, shown to activate transcription of at least one other viral protein (16), has been invoked as conferring interferon resistance to cells (3). In the two most interferon sensitive EBV lines assessed, Daudi and P3HR1, deletions remove the gene for EBNA2 (20). The EBNA2 function is silent in most EBV-associated tumors, and yet interferon has proved disappointing as a therapeutic agent. We postulated that another viral (or host) function, expressed in cell lines but presumably largely silent in tumors, may be relevant in understanding interferon responsiveness. Our data suggest a direct role for the EBV transcript D-HIT (or its homolog in other cells) in IFN-α induction or activities associated with cellular responses to interferon, or both, that is, in sensitizing cells to interferon. An alternative role can be suggested for EBNA2. That is, were it acting as a down-regulator of transcriptional expression of the D-HIT promoter, then sensitivity would be lost. This has not been explored. Another, obvious mechanism for down-regulation of transcriptional expression would involve epigenetic events, notably down-regulation by methylation. Table 2 presents data which show that over the promoter/transcriptional initiation region of D-HIT, the numbers of CpG and GpC dinucleotides are both high and comparable. This strongly suggests that the promoter behaves as a CpG island, many of which act as methylation hot spots (8) and control transcriptional expression.
TABLE 2.
CpG and GpC dinucleotides and HpaII sites in the promoter/initiation region of D-HIT
| Position (5′-3′)a | No.
|
||
|---|---|---|---|
| CpG | GpC | HpaII (CCGG) sites | |
| 4609–3959b | 17 | 29 | 2 |
| 4609–3700c | 25 | 39 | 3 |
| 4609–3562d | 39 | 46 | 6 |
| 4609–3475e | 51 | 51 | 12 |
From the EBV sequence in Raji cells (34).
From the start site of the direct repeat, DR, to the initiation site of D-HIT.
From the corresponding sequence in the M-ABA strain (27).
From the start of DR to the first IR4 repeat.
From the start of DR to the first PstI restriction enzyme site in IR4.
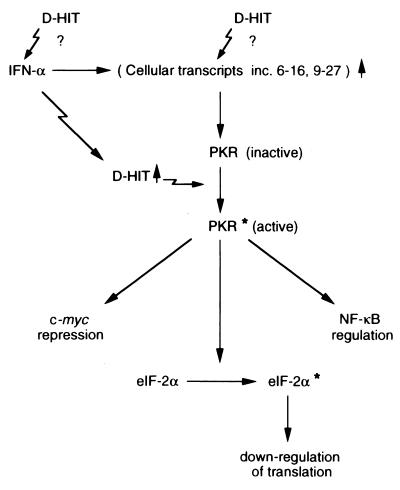
The normal function of D-HIT and its homologs in EBV-infected cells and associated tumors has not been defined, although the fact that its promoter may incorporate a lytic origin of replication (12, 15) suggests a regulatory role in the viral life cycle. In most infected cells and EBV-associated tumors, however, little or no lytic replication is observed, although in some cell lines it can be stimulated by TPA and SB. Levels of endogenous expression of D-HIT, or homologs, in tumors are very low, detectable only by reverse transcription-PCR methods (our unpublished observations). The significance of the present findings, where D-HIT may act to sensitize cells to interferon, suggests that understanding the regulation of this transcript could have practical consequences for growth control of EBV-associated malignancies by this cytokine. It is predicted that the viral transcript should be relatively insensitive to the 2,5-A dependent RNase, which chiefly acts on single-stranded RNA (30), and in stimulating the autophosphorylation of PKR (Fig. 5D), D-HIT may offer a natural, valuable molecule for studying interferon action, as proposed (Fig. 7).
FIG. 7.
The PKR interferon pathway. Sites in the pathway identified as, or postulated to be, stimulated by D-HIT expression are indicated by jagged arrows. Asterisks indicate phosphorylation. Parentheses are used for the cellular RNAs (6-16, 9-27 etc.) since work on another mRNA, 561, implies that these may not be dependent solely on interferon for activation but directly induced efficiently by dsRNA (42).
ACKNOWLEDGMENTS
We acknowledge support provided by the Cancer Research Campaign, United Kingdom (to Y.G.), the European Community (contract 1C18-CT96-0132, to S.A.X.) and The Leverhulme Trust (to B.E.G.) for this work.
We thank I. M. Kerr for critical comments on the manuscript and Daniel Holleyman for help with the figures.
REFERENCES
- 1.Adams A, Lidin B, Strander H, Cantell K. Spontaneous interferon production and Epstein-Barr virus antigen expression in human lymphoid cell lines. J Gen Virol. 1975;28:219–223. doi: 10.1099/0022-1317-28-2-219. [DOI] [PubMed] [Google Scholar]
- 2.Adams A, Strander H, Cantell K. Sensitivity of the Epstein-Barr virus transformed human lymphoid cell lines to interferon. J Gen Virol. 1975;28:207–217. doi: 10.1099/0022-1317-28-2-207. [DOI] [PubMed] [Google Scholar]
- 3.Åman P, von Gabain A. An Epstein-Barr virus immortalization associated gene segment interferes specifically with the IFN-induced antiproliferative response in human B-lymphoid cell lines. EMBO J. 1990;9:147–152. doi: 10.1002/j.1460-2075.1990.tb08090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrand J R, Rymo L, Walsh J E, Björck E, Lindahl T, Griffin B E. Molecular cloning of the complete Epstein-Barr virus genome as a set of overlapping restriction endonuclease fragments. Nucleic Acids Res. 1981;9:2999–3014. doi: 10.1093/nar/9.13.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomhoff H K, Davies C, Ruud E, Funderud S, Godal T. Distinct effects of γ interferon on human B-lymphocyte precursor cell lines. Scand J Immunol. 1985;22:611–617. doi: 10.1111/j.1365-3083.1985.tb01923.x. [DOI] [PubMed] [Google Scholar]
- 6.Chu W-M, Ballard R, Carpick B W, Williams B R G, Schmid C W. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol Cell Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens M J, Tilleray V J, James R, Gewert D R. Relationship of cellular oncogene expression in interferon-treated cells. Implications for the regulation of protein synthesis and the antiviral state. J Cell Biochem. 1988;38:251–259. doi: 10.1002/jcb.240380404. [DOI] [PubMed] [Google Scholar]
- 8.Cross S H, Bird A P. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 9.Dron M, Tovey M G. Isolation of Daudi cells with reduced sensitivity to interferon. I. Characterization. J Gen Virol. 1983;64:2641–2647. doi: 10.1099/0022-1317-64-12-2641. [DOI] [PubMed] [Google Scholar]
- 10.Elia A, Laing K G, Schofield A, Tilleray V J, Clemens M J. Regulation of the double-stranded RNA-dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein-Barr virus genome. Nucleic Acids Res. 1996;24:4471–4478. doi: 10.1093/nar/24.22.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galabru J, Katze M G, Robert N, Hovanessian A G. Autophosphorylation of the protein kinase dependent on double-stranded RNA. Euro J Biochem. 1989;178:581–589. doi: 10.1111/j.1432-1033.1989.tb14485.x. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Smith P R, Karran L, Lu Q L, Griffin B E. Induction of an exceptionally high-level, nontranslated, Epstein-Barr virus-encoded polyadenylated transcript in the Burkitt’s lymphoma line Daudi. J Virol. 1997;71:84–94. doi: 10.1128/jvi.71.1.84-94.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y. Properties of the highly inducible EBV IR4 transcript in Daudi cells and possible association with interferon sensitivity. Ph.D. thesis. London, England: University of London; 1998. [Google Scholar]
- 14.Guille M J, Laxton C D, Rutherford M N, Williams B R G, Kerr I M. Functional differences in the promoters of the interferon-inducible (2′-5′)A oligoadenylate synthetase and 6-16 genes in interferon-resistant Daudi cells. Eur J Biochem. 1994;219:547–553. doi: 10.1111/j.1432-1033.1994.tb19970.x. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 16.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitt M M, Allday M, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovanessian A G, Galabru J, Meurs E, Buffet-Janvresse C, Svab J, Robert N. Rapid decrease in the double-stranded RNA-dependent protein kinase during virus infections. Virology. 1987;159:126–136. doi: 10.1016/0042-6822(87)90355-2. [DOI] [PubMed] [Google Scholar]
- 19.Hunter T, Hunt T, Jackson R J. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975;25:409–417. [PubMed] [Google Scholar]
- 20.Jones M D, Foster L, Sheddy T, Griffin B E. The EB virus genome in Daudi Burkitt’s lymphoma cells has a deletion similar to that observed in a non-transforming strain (P3HR1) of the virus. EMBO J. 1984;3:813–821. doi: 10.1002/j.1460-2075.1984.tb01890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanda K, Decker T, Åman P, Wahlstrõm M, von Gagain A, Kallin B. The EBNA2-related resistance towards alpha interferon (IFN-α) in Burkitt’s lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol Cell Biol. 1992;12:4930–4936. doi: 10.1128/mcb.12.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karran L, Gao Y, Smith P R, Griffin B E. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly J M, Gilbert C S, Stark G R, Kerr I M. Differential regulation of interferon-induced mRNAs and c-myc mRNA by alpha- and gamma-interferons. Eur J Biochem. 1985;153:367–371. doi: 10.1111/j.1432-1033.1985.tb09312.x. [DOI] [PubMed] [Google Scholar]
- 23a.Kerr, I. M. Personal communication.
- 24.Klein E, Klein G, Nadkarni J S, Nadkarni J J, Wigzell H, Clifford P. Surface IgM specificity on cells derived from a Burkitt lymphoma. Cancer Res. 1968;28:1300–1310. [PubMed] [Google Scholar]
- 25.Kumar A, Haque J, Lacoste L, Hisott J, Williams B R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent A G, Krust B, Balabru J, Svab J, Hovanessian A G. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laux G, Freese U K, Bornkamm G W. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J Virol. 1985;56:987–995. doi: 10.1128/jvi.56.3.987-995.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luvrukhina L A, Ershov F I. Antiproliferative effect of interferon inducers. Vopr Virusol. 1985;30:446–449. [PubMed] [Google Scholar]
- 30.Maran A, Maitra R K, Kumar A, Dong B, Wei X, Li G, Williams B R G, Torrence R F, Silverman R H. Blockage of NF-κB signalling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 31.McMahon M, Stark G R, Kerr I M. Interferon-induced gene expression in wild-type and interferon-resistant human lymphoblastoid (Daudi) cells. J Virol. 1986;57:362–366. doi: 10.1128/jvi.57.1.362-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanduri S, Carpick B W, Yang Y, Williams B R G, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-medicated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offermann M K, Zimring J, Mellits K H, Hagan M K, Shaw R, Medford R M, Mathews M B, Goodbourn S, Jagus R. Activation of the double-stranded RNA-activated protein kinase and induction of vascular cell adhesion molecule-1 by poly(I):poly(C) in endothelial cells. Eur J Biochem. 1995;232:28–36. doi: 10.1111/j.1432-1033.1995.tb20777.x. [DOI] [PubMed] [Google Scholar]
- 34.Parker B D, Bankier A, Satchwell S, Barrell B, Farrell P J. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179:339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- 35.Pe’ery T, Mathews M B. Synthesis and purification of single-stranded RNA for use in experiments with PKR and in cell-free translation systems. Methods. 1997;11:371–381. doi: 10.1006/meth.1996.0435. [DOI] [PubMed] [Google Scholar]
- 36.Polack A, Eick D, Koch E, Bornkamm G W. Truncation does not abrogate transcriptional downregulation of the c-myc gene by sodium butyrate in Burkitt’s lymphoma cells. EMBO J. 1987;6:2959–2964. doi: 10.1002/j.1460-2075.1987.tb02601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raveh T, Hovanessian A G, Meuro E F, Sonenberg N, Clemens M J. Double-stranded RNA-dependent protein kinase mediates c-myc suppression induced by type I interferons. J Biol Chem. 1996;271:25479–25484. doi: 10.1074/jbc.271.41.25479. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 10.27–10.37. [Google Scholar]
- 39.Sen G C. Transcriptional regulation of interferon-inducible genes. In: Cohen P, Foulkes J G, editors. The hormonal control of gene transcription. Amsterdam, The Netherlands: Elsevier; 1991. pp. 349–374. [Google Scholar]
- 40.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4491. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman R H, Watling D, Balkwill F R, Trowsdale J, Kerr I M. The ppp(A2′p)nA and protein kinase systems in wild type and interferon-resistant Daudi cells. Eur J Biochem. 1982;126:333–341. doi: 10.1111/j.1432-1033.1982.tb06783.x. [DOI] [PubMed] [Google Scholar]
- 42.Tiwari R K, Kusari J, Sen G C. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. On finding all suboptimal foldings of an mRNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]