Abstract
Elastin, an extracellular component of arteries, lung, and skin, is produced during fetal and neonatal growth. We reported previously that the cessation of elastin production is controlled by a posttranscriptional mechanism. Although tropoelastin pre-mRNA is transcribed at the same rate in neonates and adults, marked instability of the fully processed transcript bars protein production in mature tissue. Using RNase protection, we identified a 10-nucleotide sequence in tropoelastin mRNA near the 5′ end of the sequences coded by exon 30 that interacts specifically with a developmentally regulated cytosolic 50-kDa protein. Binding activity increased as tropoelastin expression dropped, being low in neonatal fibroblasts and high in adult cells, and treatment with transforming growth factor β1 (TGF-β1), which stimulates tropoelastin expression by stabilizing its mRNA, reduced mRNA-binding activity. No other region of tropoelastin mRNA interacted with cellular proteins, and no binding activity was detected in nuclear extracts. The ability of the exon-30 element to control mRNA decay and responsiveness to TGF-β1 was assessed by three distinct functional assays: (i) insertion of exon 30 into a heterologous gene conferred increased reporter activity after exposure to TGF-β1; (ii) addition of excess exon 30 RNA slowed tropoelastin mRNA decay in an in vitro polysome degradation assay; and (iii) a mutant tropoelastin cDNA lacking exon 30, compared to wild-type cDNA, produced a stable transcript whose levels were not affected by TGF-β1. These findings demonstrate that posttranscriptional regulation of elastin production in mature tissue is conferred by a specific element within the open reading frame of tropoelastin mRNA.
Elastin is a resilient connective tissue protein present in the extracellular matrix of most terrestrial vertebrate tissues but, because of its unique elastomeric properties, it is especially abundant in the interstitium of tissues that undergo repeated physical deformations, such as lungs, blood vessels, and skin (43). Elastic fibers are assembled extracellularly and are comprised of elastin and microfibrillar proteins (40). Elastin itself is a polymer of enzymatically cross-linked tropoelastin monomers, the secreted, soluble precursor protein, and constitutes ca. 90% of the mass of elastic fibers.
Like other structural extracellular matrix proteins, the bulk of elastin production is limited to a narrow window of development. In most tissues, elastogenesis begins around the time of midgestation, peaks near birth and during early neonatal periods, drops sharply thereafter, and is nearly completely repressed by maturity (46). Because elastin is an extremely durable polymer and essentially does not turn over in healthy tissues (56, 63), fiber function and tissue integrity are not compromised by this limited pattern of production. While transcriptional regulation controls both the turning on and turning off of many developmentally regulated, tissue-specific genes, we determined previously that tropoelastin production is governed by distinct mechanisms acting at different stages of growth (69). Whereas gene transcription controls the induction of tropoelastin expression in utero, a posttranscriptional mechanism mediating rapid decay of the mRNA regulates the dwindling tropoelastin expression during postnatal growth and maintains protein production at undetectable levels in adult tissue (43).
In addition to our in vivo studies, regulation of mRNA turnover has been shown to control the repression and reinitiation of tropoelastin expression in a variety of cell models. We reported that vitamin D3 and phorbol ester (phorbol myristate acetate) potently repress tropoelastin expression in rodent and bovine cells by mediating an accelerated decay of its mRNA with no effect on gene transcription (45, 51). Similarly, downregulation of tropoelastin mRNA levels mediated by glucocorticoids or aprotinin or that which occurs in freshly isolated tissue is controlled solely by a reduction in the mRNA half-life (23, 25, 37). In addition, transforming growth factor β1 (TGF-β1) stimulates the low levels of tropoelastin production by adult human and rat fibroblasts from various tissues by increasing the stability of tropoelastin mRNA (24, 30, 36, 71). Thus, modulation of mRNA turnover regulates elastin production in vivo, ex vivo, and in cell-based models, but the precise mechanism controlling transcript decay is not known.
The half-life of mRNA transcripts is influenced by poly(A) tail length and by regulatory sequences located in the 5′ or 3′ untranslated regions (UTRs) or within the open reading frame (5, 7, 58), and these elements interact with specific RNA-binding proteins (2, 49). The heterogeneous localization of regulatory elements suggests that mRNA decay is not mediated by a common pathway. Tropoelastin mRNA does not contain any sequences that have been demonstrated or suggested to mediate degradation of other transcripts, such as AU-rich regions and, thus, decay of tropoelastin mRNA may be controlled by unique cis-acting sequences. Typically, the rate at which an mRNA is degraded is determined by the activity of destabilizing sequences and not by stabilization sequences (59), though stabilization sequences have been identified in many transcripts (16, 31, 39, 66). As reported here, we have identified an element in the translated portion of tropoelastin mRNA that specifically binds a cytosolic protein. The level of this binding activity increases as tropoelastin expression declines with age. In addition, binding activity decreases in response to TGF-β1, which, as mentioned above, is known to mediate the stabilization of tropoelastin mRNA. Our findings indicate that the interaction of this cytosolic factor with tropoelastin mRNA element controls elastin production in growing and mature tissues.
MATERIALS AND METHODS
Animals and cell culture.
Sprague-Dawley rats were obtained from Charles River Laboratories (Cambridge, Mass.). Animals were killed with sodium pentobarbital, and lungs were removed from 19-day fetuses, 3- and 11-day-old neonates, 6-month-old mothers, and older adult rats for RNA isolation. Lung interstitial fibroblasts were isolated by explant culture from 3-day-old neonates and 6-month-old adults as described earlier (33). Neonatal and adult lung fibroblasts (NLFs and ALFs) were grown to visual confluency, split 1:4, and used at passage 2. Other cells used in these studies were RFL-6, an elastogenic fibroblast cell line derived from lung tissue of normal 18-day-gestation Sprague-Dawley rat fetus (CCL-192; American Type Culture Collection, Manassa, Va.). Human fibroblasts derived from neonatal foreskin and adult dermis were isolated and cultured as described previously (12) and were used at passage 4. A human pigmented epithelial (PE) cell line was kindly provided by Martin Wax (Washington University School of Medicine). PE cells were grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum (14). In some experiments, cells were treated with 50 pM TGF-β1 (R&D Systems, Minneapolis, Minn.) for 48 h or with 50 μM 5,6-dichloro-1-β-ribofuranosylbenzimidazole (DRB; Sigma, St. Louis, Mo.) or 10 μg of actinomycin D (Sigma) per ml for the times indicated.
mRNA quantification.
Total RNA was isolated from lungs of individual animals and from cultured cells by using RNAzol B (Tel-Test, Inc., Friendswood, Tex.). If destined for reverse transcription-PCR (RT-PCR), RNA samples were treated with RQ1 RNase-free DNase (Promega, Madison, Wis.) in the presence of 1 U of RNasin (Promega) as described earlier (69). As determined by UV spectrophotometry, ethidium bromide staining, and Northern hybridization, RQ1-DNase treatment does not affect total RNA recovery or integrity or the relative amount of tropoelastin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs (69). Northern hybridization and washes were done under stringent conditions as described previously (47). Gel-purified cDNA fragments for rat tropoelastin and GAPDH were labeled by random priming with [α-32P]dCTP. Autoradiographic signal was quantified by densitometry and normalized to the relative amounts of GAPDH mRNA.
Pre-mRNA assay.
Tropoelastin pre-mRNA, used as an indicator of ongoing transcription in intact tissues, was detected by RT-PCR amplification of intron sequences present in the primary transcript as described in detail earlier (69). DNase-treated total RNA (0.1 μg) was reverse transcribed with random hexamers, and the resultant cDNA was amplified for 25 cycles with intron-specific primers. After amplification, RT-PCR products were extracted with chloroform-isoamyl alcohol, and 40 μl of each sample was directly resolved by electrophoresis through 2% SeaKem ME agarose (FMC, Rockland, Maine). After transfer onto nylon membranes, blots were hybridized with 32P-labeled intron-specific oligomer probes as described earlier (69). As we determined previously, production of pre-mRNA cDNA was proportional to the amount of input RNA. With 25 cycles of amplification, the signal for intron-specific cDNA increased linearly at between 0.0125 and 0.4 μg of total RNA. With 0.1 μg of RNA, signal for the intron product increased exponentially between 21 and 29 cycles of amplification. For each sample, a parallel reaction was run with no reverse transcriptase. The sequence and preparation of PCR primers and oligomer probes for tropoelastin pre-mRNA were done as described previously (69).
RNA protein binding assay.
We modified an RNA protection assay (1) to identify potential cis elements in tropoelastin mRNA. Restriction or PCR-generated fragments of full-length human and rat tropoelastin cDNAs were subcloned into a transcription vector (pCRII, Invitrogen, Carlsbad, Calif.). Constructs were linearized to transcribe 32P-labeled RNA or unlabeled transcripts over a desired region in either the sense or the antisense orientation. Synthesized RNA was purified by phenol extraction and ethanol precipitation. Nuclear and cytoplasmic extracts were isolated as described previously (53). A fixed amount of nuclear or cytoplasmic extract (10 to 30 μg) was incubated at 30°C for 30 min with 1 × 105 to 2 × 105 cpm of 32P-labeled RNA probe in 12 mM HEPES (pH 7.9) containing 15 mM KCl, 0.25 mM EDTA, 0.25 mM dithiothreitol, 5 mM MgCl2, 5% glycerol, and 200 ng of yeast tRNA per ml. T1 RNase (100 U) was added to digest unprotected RNA, and the mixture was incubated for 30 min at 37°C. Then, 5 mg of heparin per ml was added to disrupt nonspecific binding and to inhibit endogenous RNases. The reaction products, which consisted of the radiolabeled RNA element and bound extract factor, were resolved under nondenaturing conditions through an 8% polyacrylamide gel, and the protected products were detected by autoradiography. To characterize the nature of the RNA-binding factor and to determine the specific interaction with tropoelastin mRNA, cytosolic extracts were treated for 30 min at 30°C with 2.5 mg of proteinase K per ml before the addition of 32P-labeled RNA probe or else incubations included various amounts (20- to 100-fold molar excess) of unlabeled tropoelastin sense or pGEM4z RNA.
Analysis of protected RNA fragment.
Radiolabeled-RNA–cytosolic-protein complexes were resolved by nondenaturing electrophoresis and detected by autoradiography. The corresponding area of the gel was excised, and products were eluted in buffer containing 0.5 M NH4 acetate and 1 mM EDTA. After it was denatured for 5 min at 100°C, the protein was extracted with phenol-chloroform, and the remaining 32P-labeled RNA was precipitated in ethanol with 5 μg of yeast tRNA per ml. The RNA fragment was resolved by electrophoresis through a 20% denaturing polyacrylamide gel containing 8 M urea. The size of RNA fragment was estimated by comparison with single-stranded DNA markers.
UV cross-linking.
RNA-protein binding reactions and RNase treatment were done as described above. After the addition of heparin, reaction mixtures were transferred to a 96-well microtiter plate and irradiated at 4°C with 2,500 μJ for 10 min by using a UV cross-linking chamber apparatus (254 nm; Fisher Scientific, Pittsburgh, Pa.). The samples were then boiled for 5 min, and products were separated by electrophoresis in a 10% polyacrylamide-sodium dodecyl sulfate (SDS) gel under reducing conditions. The gel was dried, and components complexed to the 32P-labeled RNA element were visualized by autoradiography.
3′-End labeling of synthetic RNA oligonucleotides.
RNA oligonucleotides were synthesized by Oligo, Etc. (Wilsonville, Oreg.). Ribonucleotides were labeled at their 3′ ends with T4 RNA ligase by using [32P]biphosphate ribonucleotide (32pCp; Dupont-NEN, Boston, Mass.) (9). The 3′-end-labeled RNA oligomers were extracted in phenol, reconstituted in RNase-free water, and stored at −70°C. Binding reactions with RNA oligomers were done as described above.
RNA structure.
The potential secondary structure of the 5′ end of tropoelastin exon 30 was derived using MFOLD, version 3.0, developed by Michael Zuker, Institute for Biomedical Computing, Washington University (73).
Reporter constructs and transfection.
We made a luciferase reporter construct under control of a herpes simplex virus thymidine kinase (hsvTK) promoter with an engineered SmaI site between the translation stop sequence and the 3′ UTR of the luciferase cDNA (see Fig. 5). This design allowed us to blunt ligate any fragment in a sense or antisense orientation into the transcribed region of the luciferase cDNA without disrupting the open reading frame. The luciferase cassette, including a short run of plasmid sequences located 3′ of the polyadenylation site, was removed from pT3/T7-luciferase (Clontech Laboratories, Palo Alto, Calif.) by digestion with BamHI. This fragment was ligated into a BglII site between the hsvTK promoter and a chloramphenicol acetyltransferase (CAT) gene. By using EcoRV-KpnI digestion, a cassette containing a 3′ fragment of the luciferase coding sequence, its 3′ UTR, the plasmid sequence junction, and the CAT gene with its simian virus 40 (SV40) polyadenylation site were isolated and discarded. We replaced the missing 3′-end luciferase sequences with a modified fragment generated by splicing of overlapping ends (SOE) (19). For this insert, we generated two overlapping PCR fragments of the luciferase gene. The 5′ fragment began upstream of the EcoRV site and ended with an engineered SmaI site located just 3′ of the translation stop codon (TAA). The 3′ PCR fragment began at the engineered SmaI site and ended with a complete KpnI site ca. 40 bp 3′ of the luciferase polyadenylation signal. The two PCR fragments were subjected to SOE by using the 5′ primer of the upstream PCR fragment and the 3′ KpnI primer of the downstream fragment. The resulting product was gel purified, cut with EcoRV-KpnI, and ligated into the linearized luciferase plasmid.
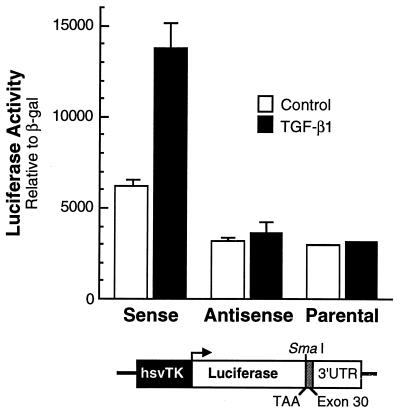
FIG. 5.
Insertion of exon 30 increases expression of a heterologous gene in response to TGF-β1. Rat tropoelastin exon-30 cDNA was inserted into a SmaI site in either a sense or an antisense orientation into a luciferase gene just 3′ of the translation stop codon (TAA). Adult lung fibroblasts were transiently transfected with these constructs or the parental plasmid and a CMV–β-Gal (β-gal) construct and, 24 h later, half of the dishes were treated with 50 pM TGF-β1. After an additional 24 h, cells were harvested, and lysates were assayed for reporter gene activity. The data shown are the mean ± the standard deviation of triplicate dishes for each condition from three separate experiments.
The rat tropoelastin fragment from base 2192 to base 2334, covering all 144 nucleotides (nt) of exon 30, was generated by PCR. The forward and reverse primers used for amplification were AGTACGGTCTTGGTGGAGCT and CTCCAAGGCCTACAGCACCA, respectively. The PCR fragment was blunted by RsaI and StuI at both ends and then ligated to the SmaI site in the reporter plasmid. The orientation of the chimeric clones was verified by restriction mapping and sequencing. Expression constructs (2 μg) were transiently transfected into NLFs and ALFs with PerFect Lipid (Invitrogen) according to the manufacturer’s protocol and, 24 h later, cells were treated with 50 pM TGF-β1 and then harvested 24 h after that. Control cells were transfected with the parental ptk-LUC construct and with expression constructs containing the potential cis element in the antisense orientation. At the indicated times, cells were harvested, washed, and lysed in 200 μl of 250 mM Tris (pH 7.8) by freeze-thawing. Lysates were incubated at 65°C for 5 min and then cleared of debris by centrifugation. Equivalent amounts (25 to 100 μg) of total protein (Bradford Protein Assay; Bio-Rad Laboratories, Palo Alto, Calif.) were assayed for luciferase activity as described earlier (68). Transfection efficiency was determined by cotransfection with a cytomegalovirus (CMV)–β-galactosidase (β-Gal) expression plasmid. β-Gal activity was assayed as described earlier (45).
Polysome-mRNA degradation assay.
Confluent NLFs and ALFs were collected and chilled rapidly on ice to inhibit runoff of nascent transcripts from the ribosomes. The cells were washed in ice-cold phosphate-buffered saline, suspended in 3 volumes of disruption buffer (100 mM potassium acetate; 20 mM HEPES, pH 7.4; 1 mM MgCl2; 2 mM dithiothreitol), and disrupted by 20 vigorous strokes with a tight-fitting Teflon pestle homogenizer (0.1-mm clearance; Thomas Scientific, Swedesboro, N.J.). The cell lysate was centrifuged at 10,000 × g for 10 min to remove nuclei and cell debris. Polysomes were isolated from cytosolic supernatant by centrifuging twice in disruption buffer at 100,000 × g for 1 h in a TLS Rotor (Beckman Instruments, Fullerton, Calif.). Supernatants from the first high-speed spin were saved and used as S100 extracts. The pellets were gently suspended in disruption buffer and centrifuged again. The supernatants from the second high-speed spin were discarded, and the polysome pellets were suspended gently in a small volume of buffer. Polysomes were quantified by an absorbance at 260 nm, and the protein concentration of the S100 extract was determined by using the Bio-Rad protein assay.
In vitro RNA decay reactions were modified from previously described methods (8, 32). Reactions were done in a total volume of 30 μl at 20°C containing 8 μg of S100 extract from NLF or ALF cells and 0.07 A260 U of isolated polysomes in 10 mM Tris-HCl (pH 7.5), 100 mM potassium phosphokinase, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine, and 1 U of RNasin. Specific competitor RNA made by in vitro transcription was added to some reactions. After incubation for 0 to 10 h, the reactions were stopped by adding 4 volumes of ice-cold 0.1 M Tris-HCl (pH 7.0) containing 0.5% SDS, and the samples were deproteinized with 5 volumes of chilled aqueous phenol. The RNA was recovered by ethanol precipitation, and tropoelastin mRNA was quantified by RT-PCR.
Bovine tropoelastin cDNA.
Full-length (BTEWT) and truncated (BTENotI) bovine tropoelastin cDNAs were inserted into pCLneo (Promega) at the MluI and XbaI sites. The pCLneo plasmid contains a CMV promoter, a chimeric intron from the human β-globulin gene and the human immunoglobulin heavy-chain gene, and an SV40 polyadenylation site. The full-length construct (pCLneoBTEWT) extended to 156 bp 3′ of the translation stop codon, and the truncated cDNA (pCLneoBTENotI) stopped at the 3′ end of exon 29. To selectively delete exon 30, a 113-bp fragment was generated by PCR amplification of BTEWT cDNA by using a forward primer to exon 28 (GGAATTCAGATCTTGGTGGAGCCG) and reverse primer that included 4 bases of exon 29 and the beginning of exon 31 (AACT-GCAGCTGGAGACACACCAAATTGGGCGGCTTTGGCGGC). The PCR product was digested with EcoRI and PstI, and the resultant fragment was ligated into EcoRI/PstI-cut pUC28-36 (containing exons 28 to 36 of bovine tropoelastin cDNA). The new plasmid pUCΔ30 was cut with NotI and XbaI and ligated in the NotI/XbaI-restricted site of pCLneoBTEWT. The resultant plasmid, pCLneoΔ30, contains an in-frame deletion of exon 30. The composition of all plasmids was verified by sequence analysis.
Human PE cells were transfected with either construct, and stable lines were amplified and pooled under G418 selection. Relative transgene levels were compared among pools by Southern hybridization, and pools with essentially identical levels were selected for study. For our experiments, cells were treated with 50 pM TGF-β1 for 48 h, followed by the addition of 10 μg of actinomycin D per ml. Total RNA was isolated at various times thereafter. Expression of the bovine transgenes and endogenous GAPDH was assessed by Northern hybridization or by RT-PCR and Southern hybridization. The forward and reverse primers for bovine tropoelastin were TCCTGCTGTGCATCCTCCAG and GCTTTATAGGCTGCAGCAGC, respectively.
RESULTS
Developmental pattern of tropoelastin expression.
We demonstrated previously that the cessation of tropoelastin expression in normal tissue is controlled primarily, if not solely, by a posttranscriptional mechanism (69). For these in vivo studies, we developed an RT-PCR assay to quantify tropoelastin pre-mRNA levels as an indicator of ongoing transcription. Our assay is based on the detection of intron sequences in newly transcribed pre-mRNA. Because intron sequences are rapidly degraded once they are spliced from the primary transcript (4, 21) and because pre-mRNAs are retained in the nucleus until splicing is completed (29), assessment of the relative steady-state levels of preprocessed mRNA provides a reliable estimate of the rate of active transcription. The data provided in Fig. 1 are representative of the more-extensive study we reported earlier (69). Several controls were done in the earlier study to confirm the reliability of the RT-PCR assay and the veracity of the results.
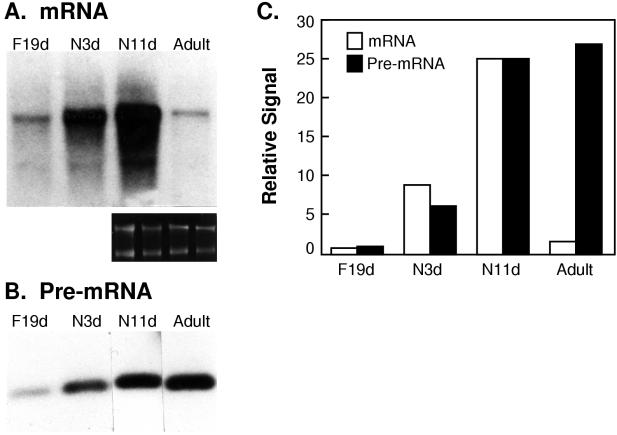
FIG. 1.
Expression of tropoelastin pre-mRNA persists in adult tissue. (A) Total RNA was isolated from lungs of 19-day fetuses, 3- and 11-day-old neonates, and 6-month-old adult rats and analyzed by Northern hybridization for tropoelastin and GAPDH mRNAs. Ethidium bromide staining of the gel before transfer (smaller panel) confirmed equivalent loading among lanes. (B) The same RNA samples were amplified by RT-PCR with intron-35 primers. Products were detected by Southern hybridization with 32P-labeled intron-specific oligomeric probe. No signal was detected in samples processed without reverse transcriptase (data not shown). (C) Autoradiographic signal for tropoelastin mRNA and pre-mRNA was quantified and normalized to the signal for the 11-day-old neonate sample, which was arbitrarily set at 25. The autoradiographic signal density of tropoelastin mRNA is expressed relative to that of GAPDH mRNA (data not shown).
We isolated total lung RNA from 19-day fetal, 3- and 11-day-old neonatal, and 6-month-old adult rats. These ages represent distinct stages of tropoelastin expression, namely, the onset, peak, and cessation of elastin production. In agreement with earlier observations from us and others (10, 42, 69), steady-state levels of tropoelastin mRNA, assayed by Northern hybridization, were low in the 19-day fetal lung, shortly after tropoelastin expression begins in the rat lung, then increased markedly during the neonatal period, and were markedly repressed in the adult (Fig. 1A and C), when active protein deposition is at undetectable levels.
Tropoelastin transcription persists in adult tissues.
Low levels of tropoelastin pre-mRNA were detected in 19-day fetal samples and much higher levels were seen in neonatal samples (Fig. 1B). The tight correlation between mRNA and pre-mRNA levels in the fetal and neonatal samples (Fig. 1C) indicates that modulation of gene transcription controls elastin production during these periods of rapid lung development. In contrast, the levels of tropoelastin pre-mRNA remained elevated in adult lung samples (Fig. 1B and C), even though steady-state mRNA levels were reduced by at least 20-fold in the mature tissue (Fig. 1A and C). In our previous report, we demonstrated that transcription of the tropoelastin gene persists in much older rats (up to 18 months) when mRNA levels have dropped about 80- to 100-fold relative to the levels in neonates (69). Together, these findings indicate that tropoelastin transcription does not turn off at the end of elastin production and that a posttranscriptional mechanism regulates the low levels of tropoelastin mRNA in the mature tissue throughout postnatal life.
Posttranscriptional regulation of elastin production occurs in the cytosol.
To study the posttranscriptional control of tropoelastin expression, we used interstitial fibroblasts isolated by explant culture of lung tissue from 3-day-old neonates (NLFs) and from 6-month-old adult mothers (ALFs). As we established earlier (69), the mechanisms controlling tropoelastin expression in vivo are retained in early-passage fibroblasts derived from tissues at different stages of development. Because tropoelastin pre-mRNA expression is maintained at high levels in adult lung tissue and in fibroblasts isolated from adult tissue, accelerated decay of the transcript is likely responsible for maintaining the steady-state mRNA at a low level. However, these data do not tell us whether the nuclear pre-mRNA or the fully processed cytosolic mRNA is the target of posttranscriptional regulation. To assess these possibilities, we treated NLFs and ALFs with DRB, a specific inhibitor of RNA polymerase II, or actinomycin D, an inhibitor of all RNA polymerases, and isolated total RNA at various times thereafter. RT-PCR with intron primers demonstrated that tropoelastin pre-mRNA in both neonatal and adult cells declined rapidly, with a half-life of ca. 15 to 30 min (Fig. 2A), a result consistent with rapid processing and transport of pre-mRNA.
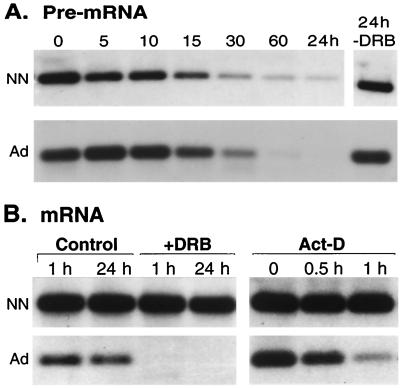
FIG. 2.
Turnover of tropoelastin pre-mRNA and mRNA. (A) Second-passage neonatal (NN) and adult (Ad) rat lung fibroblasts were grown to confluence and were treated with 50 μM DRB for 0 to 60 min or 24 h. At the indicated times, total RNA was isolated, and levels of tropoelastin pre-mRNA were determined by RT-PCR. As seen in intact tissue (Fig. 1), equivalent levels of pre-mRNA were detected in control (0- and 24-h-DRB) neonatal and adult fibroblasts. In the presence of DRB, tropoelastin pre-mRNA levels decayed at about the same rate (t1/2 = ∼15 min) in both neonatal and adult fibroblasts. (B) Neonatal and adult rat lung fibroblasts were treated with DRB for 1 or 24 h, and total RNA was isolated and analyzed by Northern hybridization. Tropoelastin mRNA in neonatal fibroblasts did not decay over the 24-h treatment. In contrast, tropoelastin mRNA in adult fibroblasts decayed rapidly to undetectable levels by 1 h post-DRB. In another experiment, fibroblasts were treated with 10 μg of actinomycin D (Act-D) per ml for 0.5 or 1 h. As with DRB, tropoelastin mRNA was stable in neonatal fibroblasts but decayed rapidly in adult cells.
Because the kinetics of pre-mRNA clearance was the same in neonatal and adult fibroblasts, posttranscriptional regulation of tropoelastin is likely directed towards the fully processed mRNA in the cytosol. Indeed, tropoelastin mRNA from neonatal fibroblasts was quite stable and did not decay appreciably during the 24-h DRB treatment (Fig. 2B). In contrast, tropoelastin mRNA from adult cells decayed rapidly and was not detected 1 h after exposure to DRB (Fig. 2B). Similar data were obtained with other strains of NLFs and ALFs treated with actinomycin D (Fig. 2B). The age-dependent differences in tropoelastin mRNA turnover rates were consistently seen in all cell strains tested (six of each age), regardless of the assay (e.g., see Fig. 6). These data indicate that the half-life of tropoelastin mRNA is greater than 24 h in NLFs and is less than 0.5 h in ALFs. In other words, the rate of tropoelastin transcript turnover increases at least 50-fold in adult fibroblasts compared to the slow decay in neonatal cells.
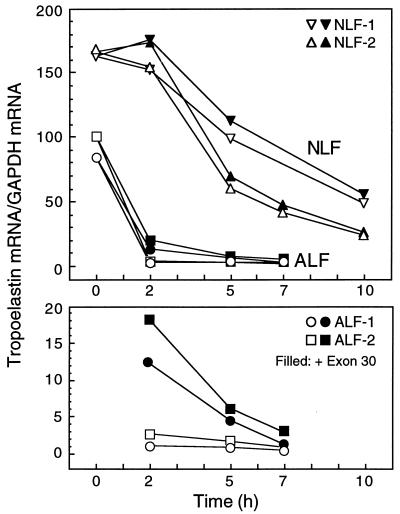
FIG. 6.
Competition of tropoelastin mRNA degradation by exon 30. Polysomes and S100 extracts were isolated from two lines of neonatal lung fibroblasts (NLF-1 and -2) and two lines of adult lung fibroblasts (ALF-1 and -2) and were combined and incubated with (solid symbols) or without (open symbols) an excess of in vitro-transcribed exon-30 RNA. Samples were harvested, and mRNA levels for tropoelastin and GAPDH were assessed by RT-PCR. For tropoelastin mRNA, sequences coded by exons 35 and 36 were amplified. Tropoelastin mRNA in NLFs degraded with a half-life of ca. 6 h, and this rate was only minimally increased in the presence of exon-30 RNA. In contrast, tropoelastin mRNA degraded rapidly in ALFs with a half-life of less than 1 h and was essentially undetectable by 2 h. In the presence of excess exon-30 RNA, degradation of tropoelastin mRNA in ALF slowed but was nearly completely degraded by 7 h. The bottom panel shows the 2-, 5-, and 7-h ALF datum points graphed on an expanded y axis.
Identification of a cis element in tropoelastin mRNA.
Regulated degradation of a mRNA implies that a trans factor or complex interacts with a specific site in the target transcript. Because tropoelastin pre-mRNA is ca. 45 kb, we were pleased that the decay data indicated that the considerably smaller and, hence, more easily mapped 3.5-kb mRNA was the target of posttranscriptional regulation. Although poly(A) tail length can affect transcript stability (58), we found, by using a variety of RNase protection, RNase H digestion, and RT-PCR techniques, no age-related difference in the average length of the poly(A) tail in tropoelastin mRNA or in frequency of usage of the two different polyadenylation signals (data not shown).
We modified an RNA protection assay to identify potential cis elements in rat tropoelastin mRNA. Radiolabeled RNA probes transcribed from various regions of tropoelastin cDNA (Fig. 3A) in either the sense or antisense orientation were incubated with nuclear and cytoplasmic extracts and were then treated with T1 RNase to digest unprotected RNA. Heparin was added to disrupt nonspecific binding and to inhibit endogenous RNases. The reaction products, which consisted of the radiolabeled RNA element and bound extract factor, were resolved under nondenaturing conditions, and protected products were detected by autoradiography. For these initial mapping studies, we used ALF extracts, since we believed that tropoelastin mRNA-binding factor(s) or activity would be more abundant during periods of accelerated transcript decay.
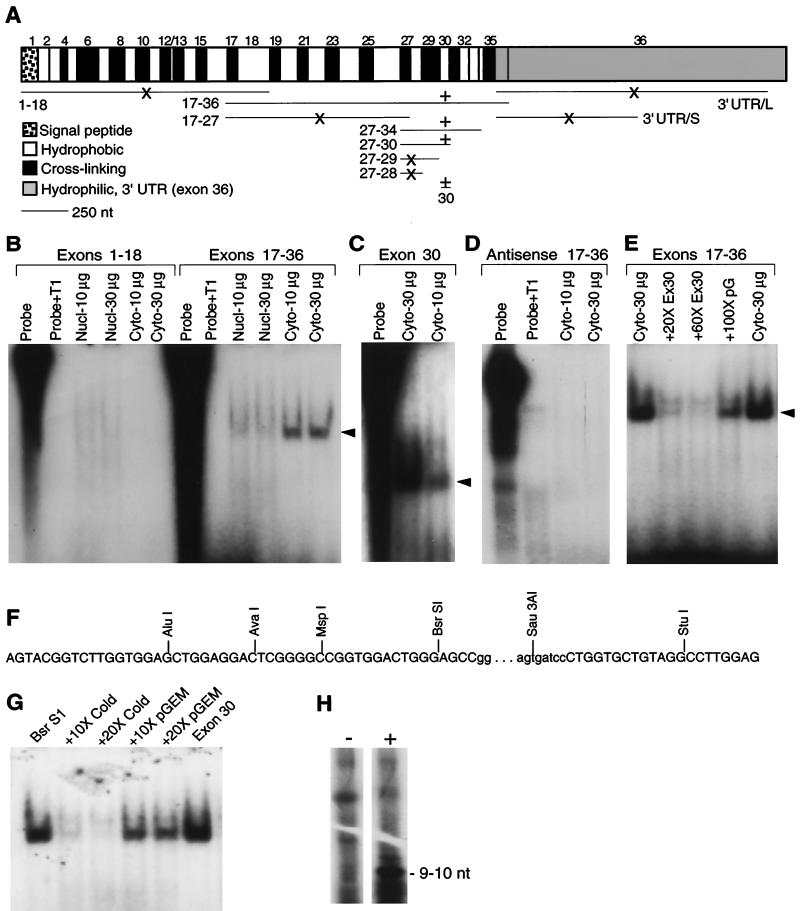
FIG. 3.
Sequences in exon 30 bind a cytosolic factor present in adult lung fibroblasts. (A) Map of tropoelastin mRNA and summary of the binding data. Tropoelastin mRNA is transcribed from 36 exons, which alternate between regions coding for hydrophobic or cross-linking domains. Exon 36 codes for a conserved hydrophilic domain and the fairly large 3′ UTR. The horizontal lines under the mRNA map indicate the RNA probes used protection assays; the numbers designate which exons the probes represent. For exon 36, two probes were made: 3′ UTR/L include all of the exon-36 sequences up to the first polyadenylation signal; 3′ UTR/S is a truncated version of 3′ UTR/L. +, Protected band was detected; ×, no detectable binding. (B) 32P-labeled RNA probes were transcribed in vitro and incubated with fixed amounts (10 or 30 μg of total protein) of nuclear or cytosolic extracts from adult lung fibroblasts. After a 30-min incubation, unbound RNA was digested by T1 RNase, and protected products were resolved by electrophoresis and visualized by autoradiography. No protected fragment or residual probe was seen in reactions containing T1 RNase without cytosolic extract (Probe+T1). A protected band (arrowhead) was produced with an RNA probe covering exons 17 to 36 incubated with cytosolic extract (Cyto) but not when incubated with nuclear extracts (Nucl). In contrast, no protected band was detected with an RNA probe covering exons 1 to 18. (C) A protected band was detected with exon-30 RNA and cytosolic extract. Because gels were not all the same dimension or run for the same time, the migration of the protected band differs among experiments. (D) No protected bands were detected with antisense RNA probes. (E) Yield of the protected band produced with RNA probe 17-36 was inhibited with a 20- or 60-fold excess of unlabeled exon 30 RNA (Ex30). Unlabeled RNA transcribed from pGEM plasmid sequences (pG) did not inhibited production of the protected band. (F) Sequence of rat tropoelastin exon 30. The bases in lowercase letters represent a 72-bp insert found only in the rodent gene. Progressively smaller 32P-labeled RNA probes were transcribed from insert linearized with the indicated restriction enzymes and were incubated with adult fibroblast cytosolic extract. All probes produced the same protected fragment, and an example is shown in the next panel. (G) Incubating 32P-labeled RNA probe transcribed from BsrSI-linearized exon-30 cDNA with 30 μg of cytosolic extract from adult lung fibroblasts produced a protected fragment. The size of the protected fragment was identical to that produced with a full-length exon-30 RNA probe. Binding was specifically competed with excess cold exon-30 RNA but not with RNA transcribed from pGEM plasmid sequences. (H) RNA probe 27-34 was incubated with (+) or without (−) adult fibroblast cytosolic extract before addition of T1 RNase. The protected products were resolved by electrophoresis, excised from the gel, and extracted in phenol-chloroform. The resulting 32P-labeled RNA fragment was resolved on a sequencing gel, and its size was determined by comparison to the migration of single-stranded DNA markers. One prominent band migrating at ca. 9 to 11 nt was detected. Other bands common to both lanes likely represent undigested RNA.
A protected band was detected only with RNA fragments containing sequences coded by exon 30 incubated with cytoplasmic extract from ALFs (Fig. 3A). No binding activity was detected with RNA probes covering exons 1 to 18 (Fig. 3B) or the 3′ UTR (data not shown). In contrast, a prominent band was seen with an RNA probe transcribed from exons 17 to 36 (Fig. 3B). In agreement with the selective, accelerated degradation of fully processed tropoelastin mRNA (Fig. 2B), binding activity was only seen with RNA probes incubated with cytosolic extract (Fig. 3B). A weak protected band with the same mobility as that produced with cytoplasmic extract was detected with RNA from exons 17 to 36 incubated with nuclear extracts (Fig. 3B), but this binding activity was likely due to some carryover of cytoplasmic components during nuclear isolation. Incubation of progressively smaller RNA probes indicated that binding activity was conferred by sequences coded by exon 30 (Fig. 3A and C). No binding activity was detected with radiolabeled antisense RNA transcribed from exons 17 to 36 (Fig. 3D).
The specificity of binding to exon 30 was demonstrated by competition with unlabeled RNA. Binding activity to radiolabeled RNA from exons 17 to 36 was effectively inhibited by a 20-fold or 60-fold molar excess of cold exon-30 RNA but was only minimally reduced by a 100-fold excess of cold plasmid RNA (Fig. 3E). In addition, no protected bands were seen with RNA probes transcribed in either direction from linearized parental plasmid (data not shown). Occasionally, the protected band appeared as a doublet (e.g., Fig. 3E), which may represent incomplete digestion of the RNA target. These observations were fully reproducible among numerous experiments with extracts from at least seven different ALF cell strains. Together, these data demonstrate the specificity of the binding interaction with sequences in exon 30. We then used similar techniques to map the binding region in the mRNA sequences coded by exon 30.
In the rat tropoelastin gene, exon 30 contains a 72-bp insertion not found in higher mammals (Fig. 3F, lowercase letters) (50). The bases flanking this insert, however, are conserved among species (20). Using different restriction enzymes, we were able to transcribe progressively smaller RNAs of exon 30. Binding activity was retained in exon-30 RNA probes lacking the 3′ 22-nt conserved region or the 72-nt rat-specific insert (Fig. 3F), and the protected band produced with the smaller RNAs was identical in size to that produced by intact exon-30 RNA (Fig. 3G). Equivalent binding activity was detected in all exon-30 RNA probes, including RNA that extended to the AluI restriction enzyme site (Fig. 3F).
To assess the size of the binding element, we performed binding reactions with exon-30 RNA probe. After digestion with T1 RNase, the samples were extracted with phenol-chloroform to remove bound and soluble proteins, and the radiolabeled protected RNA fragment was resolved in a 20% polyacrylamide–7.5 M urea gel. After autoradiography, we detected one prominent band that, based on the migration of standards, was 9 to 10 nt (Fig. 3H). Thus, we conclude that the cis-regulatory region in tropoelastin mRNA is a 9- to 10-nt element that resides within 18 nt at the 5′ end of exon 30.
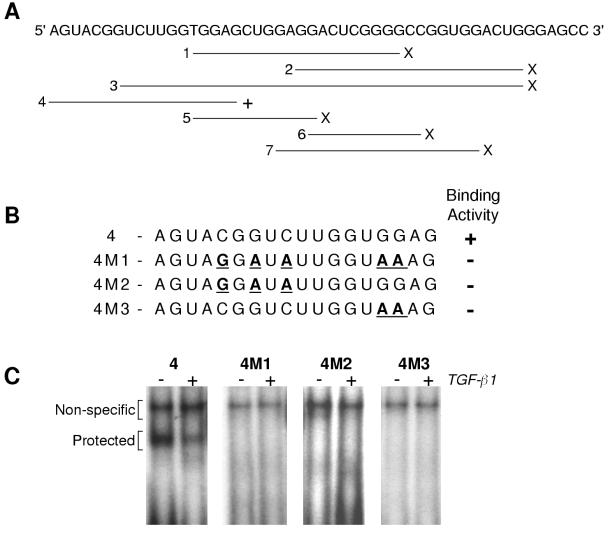
Using synthetic, 32P-labeled RNA probes, we confirmed binding activity to this 18-nt region (Fig. 4A and C). Only the oligomer that contained all 18 nt (oligomer 4), which was equivalent to the AluI probe used in Fig. 3F, showed specific binding and yielded a protected product identical to that produced with larger RNA probes. No specific protected band was detected with oligomers to regions 3′ of this element (oligomers 2, 6, and 7; Fig. 4A) or that overlapped with portions of the 5′ end of oligomer 4 (oligomers 1, 2, and 5S; Fig. 4A).
FIG. 4.
Binding of the tropoelastin mRNA-binding protein relies more on RNA sequence than structure. (A) RNA oligomers 1 to 7 of various lengths were synthesized to cover overlapping regions of the 5′ 50-nt of rat tropoelastin exon 30. The oligomer lengths are as follows: 1, 21 nt; 2, 23 nt; 3, 40 nt; 4, 18 nt; 5, 13 nt; 6, 12 nt; and 7, 20 nt. A protected product was produced only with oligomer 4 (+), which is identical to the AluI RNA probe used in the experiments summarized in Fig. 3F. (B) Sequences of oligomer 4 and three mutant RNA oligomers (M1, M2, and M3), with the mutated bases underlined. (C) 32P-labeled RNA oligomers were incubated with cytosolic extract from control adult lung fibroblasts (−) or cells treated with 50 pM TGF-β1 (+) for 48 h. A specific protected band was detected only with wild-type oligomer 4, and binding activity was decreased by treatment with TGF-β1.
Many RNA regulatory elements have a secondary structure of stems, bulges, and loops. Using an RNA folding program, we found that the first 50 nt of exon 30, extending up to the beginning of the rat-specific insert (Fig. 3F), can potentially form a stem with two intermediate bulges and a looped end and with a free energy of −13.6 kcal (not shown). However, because a cytosolic factor interacts with the first 18 nt of this region (Fig. 3F, oligomer 4) and because these 18 nt cannot form a similar or any potentially stable structure, we do not believe that secondary mRNA structure is necessary for factor interaction. We have begun mutational analysis of oligomer 4 (Fig. 4B and C). Our initial findings indicate that adenosines at the 3′ end of this element (oligomer 4M3) and adenosines and guanines near the middle (oligomer 4M2) are required for binding. In addition, treatment of ALFs with TGF-β1, which stimulates tropoelastin expression by stabilization of the mRNA, decreased the specific cytosolic binding activity detected with oligomer 4 (Fig. 4C).
Functional studies of exon-30 sequences.
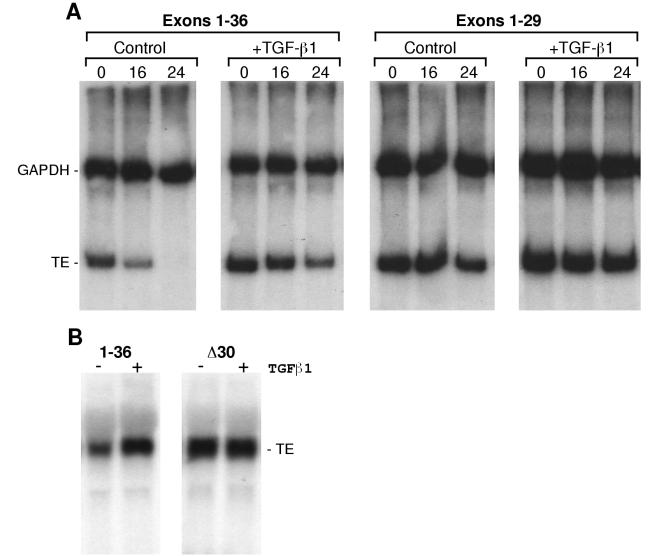
We used three assays to assess the functional role of exon 30 in regulating transcript stability. First, rat tropoelastin exon-30 sequences were inserted in both the sense and antisense orientations 3′ of the translation stop codon of a luciferase expression construct. The tropoelastin sequences were placed outside of the luciferase coding region to prevent any interference of reporter translation. Because we thought that the trans factors controlling turnover of tropoelastin mRNA may be limiting, we used the relatively weak hsvTK promoter to drive transcription of the luciferase gene. Constructs were transfected into ALFs and, 24 h later, cultures were treated with 50 pM TGF-β1 for 48 h. Luciferase activity was not affected by TGF-β1 in ALF cultures transfected with parental plasmid or with expression constructs containing exon-30 sequences in the antisense orientation, but reporter activity was stimulated by about threefold in ALFs transfected with constructs containing this element in the sense orientation (Fig. 5). Similar results were obtained with transfected NLFs (data not shown). Consistent with the idea that exon-30 sequences conferred stability to the reporter gene transcript in response to this cytokine, the increase we detected in reporter activity is approximately the same as the stimulation of tropoelastin expression mediated by TGF-β1 in these and other adult fibroblasts (24, 30, 36, 71).
Because mRNAs and mRNA-degrading enzymes associate with polysomes (39), we developed an in vitro degradation assay to assess the turnover of tropoelastin mRNA in these organelles. Polysomes were isolated from NLFs and ALFs and then incubated in matched cytosolic extracts, which contained little tropoelastin mRNA (data not shown), with or without excess in vitro-transcribed exon-30 RNA. At various times, total RNA was isolated from the samples, and the kinetics of tropoelastin mRNA turnover were assessed by RT-PCR and Southern hybridization. During the first 2 h, tropoelastin mRNA remained stable in polysomes from NLFs but degraded progressively thereafter (Fig. 6, open symbols). At 10 h, tropoelastin mRNA levels in NLF polysomes had dropped ca. threefold compared to 0-h levels. In contrast, tropoelastin mRNA in polysomes from ALFs degraded rapidly and nearly completely by 2 h (an ∼100-fold decrease). Addition of excess exon 30 slowed slightly the decay of tropoelastin mRNA in NLF polysomes in both experiments (Fig. 6). However, in polysomes from ALFs, the addition of excess exon 30 led to a nearly 10-fold increase in tropoelastin mRNA levels at 2 h and to an approximately 3-fold increase at 5 h (Fig. 6, bottom panel). These observations support the idea that binding of a cytosolic factor in ALF cells to exon 30 leads to rapid degradation of tropoelastin mRNA.
For the third functional assay, we assessed the expression and turnover of tropoelastin mRNA in human PE cells stably transfected with a full-length bovine tropoelastin cDNA or with a mutant bovine cDNAs lacking exons 30 to 36 or only exon 30 (Δ30; Fig. 7). Expression of these cDNAs was controlled by a CMV promoter. PE cells do not endogenously express tropoelastin (14). Three observations on the expression and turnover of tropoelastin mRNA in PE cells support the idea that sequences in exon 30 regulate transcript stability and responsiveness to TGF-β1. First, basal expression of wild-type tropoelastin mRNA was less than that of either mutant transcript lacking exon 30 (Fig. 7), and this effect was presumably due to exon-30-mediated degradation of the wild-type transcript (see below). Second, whereas the wild-type mRNA turned over with a half-life of about 16 h, the 30-36 mutant mRNA was quite stable, with no appreciable degradation during actinomycin D exposure (Fig. 7B). Third, TGF-β1 stimulated the steady-state levels of the wild-type transcript by stabilization of the mRNA (Fig. 7B), but this cytokine had no effect on the steady-state levels or stability of the mutant transcripts lacking exon 30 (Fig. 7). Southern hybridization confirmed that the pooled stable cell lines contained the same copy number of integrated cDNAs; thus, the difference in steady-state mRNA levels cannot be attributed to a difference in the number of transgenes. Furthermore, the levels of secreted tropoelastin protein paralleled the levels of the mRNAs (data not shown), indicating that translational efficiency was not affected by the exclusion of exon 30. Thus, considered together, data from the three distinct function assays—expression of a heterologous transcript, competition of mRNA degradation, and mutant tropoelastin cDNAs lacking exon 30—demonstrate that sequences coded by exon 30 regulate tropoelastin mRNA turnover.
FIG. 7.
Exon 30 confers regulated mRNA turnover and responsiveness to TGF-β1. (A) Human PE cells were transfected a full-length (exons 1 to 36) or a truncated (exons 1 to 29) bovine tropoelastin cDNA under the control of a CMV promoter. Stable cell lines were selected and pooled. For the experiment shown, some cells of each group were treated with 50 pM TGF-β1 and, 24 h later, some control and treated cells were harvested (0 h). The remaining cells were treated with 10 μg of actinomycin D per ml with or without TGF-β1 and were harvested 16 and 24 h later. Tropoelastin and GAPDH mRNAs were assayed by RT-PCR and Southern hybridization. (B) PE cells were transfected with full-length (1-36) bovine tropoelastin cDNA or with a deletion mutant lacking exon 30 (Δ30), and stable cell lines were selected and pooled. Cells of each group were treated with 50 pM TGF-β1 and, 24 h later, RNA was isolated. Tropoelastin mRNA was assessed by Northern hybridization. Loading equivalence among lanes was demonstrated by ethidium bromide staining (not shown).
Initial characterization of the tropoelastin mRNA-binding protein.
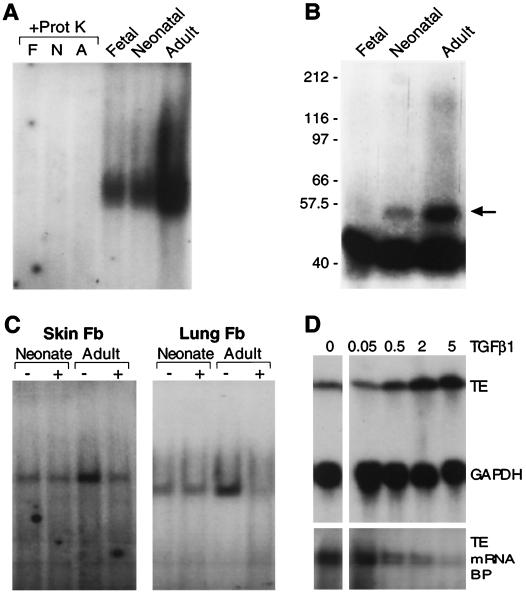
Our findings indicate that posttranscriptional regulation of tropoelastin expression is mediated by a cytosolic factor or factors. If this factor is involved in the developmental regulation of tropoelastin expression and if it mediates degradation of the mRNA, then we would expect its binding activity would increase as elastin production declines with age. Indeed, we found that the exon-30 binding activity was low in extracts of fetal and neonatal lung fibroblasts, in which tropoelastin is actively expressed, but high in cytosolic extracts from ALFs (Fig. 8A and B), which produce little to no tropoelastin. Furthermore, the exon-30 binding activity was decreased in adult rat skin or lung fibroblasts exposed to TGF-β1 (Fig. 8C). TGF-β1 had little effect on the low level of binding activity detected in extracts from neonatal fibroblasts. These same age-specific patterns in tropoelastin expression, exon-30 binding activity, and responsiveness to TGF-β1 were seen in fibroblasts isolated from neonatal and adult human skin (data not shown). In addition, binding activity to exon dropped as tropoelastin mRNA levels in ALFs rose in response to increasing concentration of TGF-β1 (Fig. 8D).
FIG. 8.
Characterization of the tropoelastin mRNA-binding protein. (A) Cytoplasmic extracts from fetal (F), neonatal (N), and adult (A) rat lung fibroblasts were incubated with RNA probe 17-36. Before T1 RNase digestion, half of the samples were incubated with proteinase K (+Prot K), which eliminated all binding activity. In addition, binding activity was developmentally regulated, being greatest in extracts from adult fibroblasts. (B) Cytoplasmic extracts from fetal, neonatal, and adult rat lung fibroblasts were incubated with RNA probe 17-36 and treated with T1 RNase. The samples were then exposed to UV light, and cross-linked products were resolved by denaturing SDS-PAGE. A band migrating at ca. 53 kDa (arrow) was seen in all lanes, and its abundance increased with age, being most prominent in adult fibroblasts extracts. The large, faster-migrating band seen in all lanes likely represents non-cross-linked probe and nonspecific cross-linked products. (C) Fibroblasts were isolated from neonatal and adult rat skin and lung and were treated with 50 pM TGF-β1 for 48 h before the isolation of cytosolic extracts. Extracts were incubated with RNA probe 17-36. Binding activity was greater in adult fibroblast extracts and was reduced after exposure to TGF-β1, but only in adult extracts. (D) Adult rat lung fibroblasts were treated with 0, 0.05, 0.5, 2, or 5 pM TGF-β1. After a 48-h incubation, cells were harvested and used for isolation of total RNA and cytosolic extract. Levels of tropoelastin (TE) and GAPDH mRNAs were assessed by Northern hybridization (upper panels), and tropoelastin mRNA-binding protein (TE mRNA BP) activity was assessed by incubation with labeled exon-30 RNA (lower panels).
To assess the nature of the exon-30 binding factor, we treated cytosolic extracts with proteinase K before the RNA-binding reaction. In all samples, the binding activity was completely abolished by the proteinase K treatment (Fig. 8A), suggesting that the cytosolic factor that interacts with exon 30 is a protein. In addition, cell extract-RNA reactions were exposed to UV light to cross-link interacting components, and the products were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) under denaturing conditions. We detected a single cross-linked product which was more abundant in extracts of ALFs than in extracts from fetal or neonatal lung cells (Fig. 8B). In comparison to the migration of molecular mass standards, and after subtraction of the weight of the 10-nt protected RNA fragment (ca. 3 kDa), we estimate the size of the cytosolic factor to be ca. 50 kDa.
DISCUSSION
Whereas transcription regulates both the induction and repression of most developmentally regulated, tissue-specific genes, our data demonstrate that tropoelastin production is governed by different mechanisms acting at distinct stages of growth. As we and others have reported (22, 52, 69), induction of tropoelastin expression is controlled by transcriptional activation, which is not surprising for a developmentally regulated gene. Furthermore, in these previous studies we found that stimulation of ongoing tropoelastin expression is also transcriptionally regulated, but only during periods of active production, i.e., during fetal and neonatal growth. In contrast, repression of tropoelastin expression postnatally, as well as maintaining no production in adult tissue, is controlled by a posttranscriptional mechanism mediating rapid decay of the mRNA. In essentially all tissues and species, production and release of tropoelastin protein correlate with steady-state mRNA levels, indicating no significant regulation of translation or secretion.
Our data demonstrate a marked difference in the turnover rate of tropoelastin mRNA during periods of active protein production compared to the rate in mature tissues. In NLFs, tropoelastin mRNA was quite stable and did not decay appreciably even after 24 h in the presence of RNA polymerase inhibitors (Fig. 2). Because DRB and actinomycin D are cytotoxins, we prefer to limit such experiments to a 9-h exposure, as we have done in other studies (15, 45, 57), but even at 48 h, we detected little, if any, decay of tropoelastin mRNA in NLFs (data not shown). In contrast, tropoelastin mRNA in ALFs was very unstable and, in most adult cell lines, was completely degraded by 1 h after inhibition of transcription (e.g., Fig. 2B). The relative difference in tropoelastin mRNA steady-state mRNA between NLFs and ALFs was not a great as that seen between neonatal and adult lung (compare Fig. 2B and Fig. 6 to Fig. 1A and C). As we have shown before, once removed from tissue, tropoelastin expression declines in cultured fetal and neonatal elastogenic cells but remains relatively constant in cells from adult tissues (48, 69). We do not know what regulates the drop in tropoelastin mRNA in response to culture, but it may be associated with a decline in mRNA stability. Similarly, in the chick aorta, tropoelastin expression declines as the tissue is removed from the animal and placed in medium, and this downregulation is mediated by enhanced mRNA turnover (23). Although we estimated that the turnover rate of tropoelastin mRNA in neonatal cells was ca. 50-fold slower than that in adult cell, the actual difference is likely greater, approaching 100-fold, and may be even more so in intact tissues. Since tropoelastin transcription remains fully active in mature tissue (69), a potent posttranscriptional mechanism would be needed to prevent excess accumulation of elastin matrix after growth is complete.
Multiple mechanisms can participate in the control of gene expression (28), but production of most structural proteins is primarily regulated at the level of transcription. There are, however, numerous examples of proteins whose production is primarily or significantly regulated by a posttranscriptional mechanism (55, 59, 62). Many of these products, such as cytokines, iron metabolism proteins, oncogenes, and cytoskeletal proteins, are expressed during physiologic transitions or for brief periods during developmental processes, and changes in the stability of the mRNA provides a mechanism to rapidly govern protein synthesis and activity. In contrast, once the growth of elastic tissue is complete, new elastin production, under normal conditions, is not needed since the protein is extremely durable (63). Thus, the posttranscriptional control we describe is a novel mechanism to control the expression of a stable structural protein. Although continual production of large pre-mRNA is seemingly an inefficient mechanism, sustained transcription of the tropoelastin gene does not create a significant energy drain on the cell. As determined by [3H]uridine incorporation and nuclear runoff assay, total transcriptional activity is not noticeably different between neonatal and adult cells (45, 51, 69). In addition, turning off transcription and keeping it turned off requires energy. Numerous and diverse proteins must be produced to maintain genes and chromosomes in quiescent or inactive states.
Our findings demonstrate that the posttranscriptional control of tropoelastin expression is conferred by an element within the 5′ 18 nt of the sequences coded by exon 30. Not only was this fragment the only part of tropoelastin mRNA that interacted with a cytosolic protein, but this interaction increased as elastin production waned and as the half-life of tropoelastin mRNA plummeted. Interestingly, point mutations have been found near the 5′ end of exon 30 of the human tropoelastin gene in several individuals of two families with inherited cutis laxa (72), an elastin-related disease. This frameshift mutation is located within sequences that are homologous to those coding for the mRNA cis element we identified in the rat gene. Related to the findings we report here, this mutation is associated with a marked change in tropoelastin mRNA stability (71, 72). Using synthetic RNA probes, we assessed whether binding of the tropoelastin mRNA-binding protein would be affected by this mutation in the human sequence, but no overt difference was detected (data not shown). However, the protein-RNA interaction that yields a protected RNA fragment may be distinct from the RNase activity. The degradative activity may be mediated by a different protein that recognizes the tropoelastin mRNA-protein complex. The cutis laxa mutation may alter this secondary interaction leading to the observed changes in tropoelastin mRNA turnover (72); however, more work is needed to address this speculation.
Although we did not detect any other protected elements over the length of tropoelastin mRNA, our assay conditions were intentionally stringent to select specific interactions, and our probes lacked a polyadenylated tail, which would predictably interact with poly(A)-binding proteins. It is quite likely that tropoelastin mRNA interacts with other cellular proteins associated with transcript processing and transport. When we first began to examine the mechanism of tropoelastin mRNA turnover, we focused on the 3′ UTR. Of the many genes whose production is controlled by a posttranscriptional mechanism, regulatory sequences have been localized to the 3′ UTR, such as the iron response element in transferrin mRNA and the AU-rich region in many cytokine transcripts (41, 64). The 3′ UTR of tropoelastin mRNA contains two highly conserved regions near its 5′ end which can form secondary structures (18), which led us and others to speculate that these regions confer posttranscriptional regulation (18, 44, 47). However, binding activity was not seen in protection assays with the rat (Fig. 3) or human (data not shown) 3′ UTRs. In addition, we saw no modulation of luciferase activity from transfected expression constructs containing either the entire or 5′ half of tropoelastin 3′ UTR inserted in the sense or antisense orientation (data not shown). The conserved regions in tropoelastin 3′ UTR likely confer some regulatory function, such as directing the cytoplasmic localization of the transcripts, that is similar to the role of the 3′ UTR of other transcripts (26, 34).
We determined that the protected RNA fragment in exon 30 is ca. 9 to 10 nt, which is a common size for a cis element in mRNA (59), although much longer elements have been identified (13, 65). The method we used determined the size of the area directly protected by a binding protein and, hence, the complete functional element may include bases flanking this region. Our binding studies with smaller probes and RNA oligomers support this idea, but more extensive functional assays with mutant elements will be needed to accurately map the cis element in exon 30 and, possibly, into exon 29. Regulatory elements in many mRNAs form stable secondary structures and, using computer modeling, we found a potential, though weakly stable stem-loop within the 5′ end of exon 30. However, if such a structure does form in cells, we do not believe that it is required for binding of the cytosolic factor we have identified. The most compelling data in support of this conclusion are the findings that the cytosolic factor interacts with an 18-nt fragment (Fig. 3F and 4) that cannot form any potentially stable structure. In addition, the observation that antisense exon-30 RNA was fully degraded by T1 RNase in the presence of cytosolic extract indicates further that secondary structure is not a critical determinate for protein binding. Thus, we predict that the tropoelastin mRNA-binding activity relies more on primary transcript sequence than on potential secondary structure. This idea is not without precedent. The bacterial RNA-binding protein TRAP recognizes a linear RNA sequence, not secondary structure (3).
Using different functional assays, we demonstrated that exon-30 sequences conferred transcript stability and responsiveness to TGF-β1. We were somewhat perplexed that luciferase activity from constructs containing antisense exon 30 was consistently less than that produced by the sense constructs (Fig. 5). Although one may have predicted that inclusion of exon-30 sequences would have led to diminished basal luciferase activity due to enhanced mRNA destabilization, the addition of any element into a heterologous cDNA produces a structurally distinct transcript. Thus, a direct comparison of the absolute levels of reporter gene activity among constructs may not be valid. To understand fully the influence of an inserted element in a heterologous gene, numerous controls are needed to assess potential transcriptional enhancer activity, changes in pre-mRNA processing and transport, the transcript stability, and the translational efficiency, among other effects. Thus, we elected to assess the function of exon 30 by more direct means. Still, the exon-30-containing luciferase construct was affected by TGF-β1, a finding consistent with other findings reported here.
The in vitro polysome degradation assay provided further evidence of the marked instability of tropoelastin mRNA in adult cells (Fig. 6). Furthermore, these observations indicate that decay of tropoelastin mRNA occurs after the transcript has been delivered and docked to ribosomes and suggests that tropoelastin transcript degradation occurs during translation, as it does for procyclin, tubulin, and other mRNAs (17, 70). Indeed, we detected very little tropoelastin mRNA in cytosolic extracts cleared of the polysome fraction (data not shown). Tropoelastin mRNA in NLF polysomes degraded with a half-life of about 6 h in an in vitro assay, much faster than it did in intact cells (Fig. 2). However, the disruption of cellular compartments may have allowed nonspecific RNases in the cytosolic extract to act on the transcript.
Our initial characterization of the tropoelastin mRNA-binding protein shows that it is a cytosolic factor of about 50 kDa. As stated, we do not yet know whether modulation of the binding activity of this protein that occurs with age and in response to TGF-β1 is controlled by expression or by posttranslational modification. In addition, we do not know whether this factor has intrinsic RNase activity. Most known mRNA-binding proteins that have been implicated in transcript degradation are not RNases, and we predict the same is true for the tropoelastin mRNA-binding protein. As suggested above, the tropoelastin mRNA-binding protein may be required to target and/or activate an RNase, which initiates degradation of the transcript. However, we cannot determine how this factor functions until we have isolated and characterized it and, clearly, this goal is the current focus of our efforts. We also predict that the tropoelastin mRNA-binding protein is not dedicated to regulating tropoelastin mRNA turnover. The pigmented epithelial cells used for the functional assays (Fig. 7) do not transcribe tropoelastin pre-mRNA (data not shown), yet they express the mRNA-binding protein. Thus, it will be of interest to identify other transcripts regulated by this factor and, possibly, other activities not related to mRNA turnover.
In addition to being developmentally regulated, the activity of the mRNA-binding protein was reduced by TGF-β1, which stimulates tropoelastin production by transcript stabilization. We have not yet determined if the expression or binding activity of the trans factor is affected by age or TGF-β1, and such information will require more knowledge of the protein. TGF-β1 is among the more effective stimulators of tropoelastin expression, but it is particularly potent in fibroblasts from adult tissues. In neonatal fibroblasts, TGF-β1 upregulates tropoelastin expression less than 2-fold (38), but in adult fibroblasts expression increases ca. 10-fold (24, 30, 71). The age-specific response to TGF-β1 agrees with our findings. In neonatal fibroblasts, we found a low level of the tropoelastin mRNA-binding protein activity, which was only minimally reduced by TGF-β1 (Fig. 8). In contrast, the binding activity was much greater in adult fibroblasts and was reduced ca. 10-fold by TGF-β1 (Fig. 8). Thus, TGF-β1 may stimulate elastin production by repressing the activity or expression of the mRNA-binding protein, thereby allowing steady-state mRNA levels to build up and protein production to resume. Analogously, TGF-β1 modulates expression of other mRNA-binding proteins that, in turn, regulate specific genes during development. For example, the expression of CRD-BP, an RNA-binding protein implicated in the stabilization of c-myc mRNA, parallels the expression of c-myc during liver development (31). Similar to its effect on tropoelastin, TGF-β1 increases the stability of C-α mRNAs during immunoglobulin isotype switching in B cells by reducing the binding activity of a 45-kDa mRNA-binding protein (16).
Thinking teleologically, reliance on a posttranscriptional mechanism to bar production of a protein, such as elastin, in fully developed tissues does not provide any apparent advantage. Because reinitiation of elastin production is typically a late event in many injury and diseased conditions, such as burn wounds, arterial restonisis, and lung fibrosis (27, 35, 54, 67), posttranscriptional regulation of tropoelastin does not seem to allow cells to restore rapidly damaged matrix. This argument, however, assumes that evolution of the elastic phenotype is complete. As for any cellular processes, we have uncovered the regulatory mechanisms that are operative now. The tropoelastin gene developed relatively recently, having evolved along with high-pressure circulatory systems and lungs (6). Elastin is not found in cartilaginous fish (60, 61), and that expressed by bony fish is quite different from terrestrial elastin (11). Thus, compared to more-ancient extracellular matrix proteins, such as the collagens and fibronectin, which are found throughout the animal kingdom, unique regulatory mechanisms may have evolved in the elastin gene, or alternatively, more “conventional” mechanisms may not yet have evolved.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant HL48762.
We thank Mei Swee for her early work leading to this project and Martin Wax, Department of Ophthalmology, Washington University School of Medicine, for the human pigmented epithelial cells.
REFERENCES
- 1.Amara F M, Chen F Y, Wright J A. A novel transforming growth factor-β1 responsive cytoplasmic trans-acting factor binds selectively to the 3′-untranslated region of mammalian ribonucleotide reductase R2 mRNA: role in message stability. Nucleic Acids Res. 1993;21:4803–4809. doi: 10.1093/nar/21.20.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandziulis R J, Swanson M S, Dreyfus G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 3.Baumann C, Xirasagar S, Gollnick P. The trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis binds to unstacked trp leader RNA. J Biol Chem. 1997;272:19863–19869. doi: 10.1074/jbc.272.32.19863. [DOI] [PubMed] [Google Scholar]
- 4.Baurén G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 5.Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. [Google Scholar]
- 6.Boyd C D, Christiano A M, Pierce R A, Stolle C A, Deak S B. Mammalian tropoelastin: multiple domains of the protein define an evolutionarily divergent amino acid sequence. Matrix. 1991;11:235–241. doi: 10.1016/s0934-8832(11)80230-1. [DOI] [PubMed] [Google Scholar]
- 7.Brawerman G. Determinants of messenger RNA stability. Cell. 1987;48:5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- 8.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruce A G, Uhlenbeck O C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978;5:3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce M C. Developmental changes in tropoelastin mRNA levels in rat lung: evaluation by in situ hybridization. Am J Respir Cell Mol Biol. 1991;5:344–350. doi: 10.1165/ajrcmb/5.4.344. [DOI] [PubMed] [Google Scholar]
- 11.Chow M, Boyd C D, Iruela-Arispe M-L, Wrenn D S, Mecham R P, Sage E H. Characterization of elastin protein and mRNA from salmonid fish (oncorhynchus kisutch) Comp Biochem Physiol. 1989;93B:835–845. doi: 10.1016/0305-0491(89)90055-2. [DOI] [PubMed] [Google Scholar]
- 12.Clark S D, Kobayashi D K, Welgus H G. Regulation of the expression of tissue inhibitor of metalloproteinases and collagenase by retinoids and glucocorticoids in human fibroblasts. J Clin Investig. 1987;80:1280–1288. doi: 10.1172/JCI113203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clerch L B, Wright A, Slobodyansky E, Wang W, Mouradian M M, Jose P. Kidney extracts from spontaneously hypertensive rats (SHR) have greater dopamine 1A receptor RNA-binding activity than extracts from normotensive Wistar-Kyoto (WKY) rats. Clin Exp Hypertens. 1997;19:1009–1021. doi: 10.3109/10641969709083202. [DOI] [PubMed] [Google Scholar]
- 14.Davis, E. C., H. Wachi, T. Schaub, B. W. Robb, and R. P. Mecham. Characterization of an in vitro model of elastic fiber assembly. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 15.Doyle G A R, Saarialho-Kere U K, Parks W C. Distinct mechanisms regulate TIMP-1 expression at different stages of phorbol ester-mediated differentiation of U937 cells. Biochemistry. 1997;36:2492–2400. doi: 10.1021/bi962161e. [DOI] [PubMed] [Google Scholar]
- 16.Edmiston J S, Lebman D A. A transforming growth factor-beta regulable RNA-binding protein interacts specifically with germline Ig alpha transcripts. Int Immunol. 1997;9:427–433. doi: 10.1093/intimm/9.3.427. [DOI] [PubMed] [Google Scholar]
- 17.Furger A, Schurch N, Kurath U, Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hew Y, Grzelczak Z, Lau C, Keeley F W. Identification of a large region of secondary structure in the 3′-untranslated region of chicken elastin mRNA with implications for the regulation of mRNA stability. J Biol Chem. 1999;274:14415–14421. doi: 10.1074/jbc.274.20.14415. [DOI] [PubMed] [Google Scholar]
- 19.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chair reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 20.Indik Z, Yeh H, Ornstein-Goldstein N, Rosenbloom J. Structure of the elastin gene and alternative splicing of elastin mRNA. In: Sandell L, Boyd C D, editors. Extracellular matrix genes. New York, N.Y: Academic Press, Inc.; 1990. pp. 221–250. [Google Scholar]
- 21.Jakubowski M, Roberts J L. Processing of gonadotropin-releasing hormone gene transcripts in the rat brain. J Biol Chem. 1994;269:4078–4083. [PubMed] [Google Scholar]
- 22.James M F, Rich C B, Trinkaus-Randall V, Rosenbloom J, Foster J A. Elastogenesis in the developing chick lung is transcriptionally regulated. Dev Dyn. 1998;213:170–181. doi: 10.1002/(SICI)1097-0177(199810)213:2<170::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D J, Robson P, Hew Y, Keeley F W. Decreased elastin synthesis in normal development and in long-term aortic organ and cell cultures is related to rapid and selective destabilization of mRNA for elastin. Circ Res. 1995;77:1107–1113. doi: 10.1161/01.res.77.6.1107. [DOI] [PubMed] [Google Scholar]
- 24.Kähäri V-M, Olsen D R, Rhudy R W, Carillo P A, Chen Y Q, Uitto J. Transforming growth factor-β upregulates elastin gene expression in human skin fibroblast: evidence for posttranscriptional modulation. Lab Investig. 1992;66:580–588. [PubMed] [Google Scholar]
- 25.Kähäri V M. Dexamethasone suppresses elastin gene expression in human skin fibroblasts in culture. Biochem Biophys Res Commun. 1994;201:1189–1196. doi: 10.1006/bbrc.1994.1831. [DOI] [PubMed] [Google Scholar]
- 26.Kislauskis E H, Li Z, Singer R H, Taneja K L. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh D W, Roby J D, Starcher B, Senior R M, Pierce R A. Postpneumonectomy lung growth: a model of reinitiation of tropoelastin and type I collagen expression in normal adult rat lung. Am J Respir Cell Mol Biol. 1996;15:611–623. doi: 10.1165/ajrcmb.15.5.8918368. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. A profusion of controls. J Cell Biol. 1988;107:1–7. doi: 10.1083/jcb.107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krug R M. The regulation of export of mRNA from nucleus to cytoplasm. Curr Opin Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 30.Kucich U, Rosenbloom J C, Abrams W R, Bashir M M, Rosenbloom J. Stabilization of elastin mRNA by TGF-beta: initial characterization of signaling pathway. Am J Respir Cell Mol Biol. 1997;17:10–16. doi: 10.1165/ajrcmb.17.1.2816. [DOI] [PubMed] [Google Scholar]
- 31.Leeds P, Kren B T, Boylan J M, Betz N A, Steer C J, Gruppuso P A, Ross J. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–1286. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- 32.Liang H M, Jost J P. An estrogen-dependent polysomal protein binds to the 5′ untranslated region of the chicken vitellogenin mRNA. Nucleic Acids Res. 1991;19:2289–2294. doi: 10.1093/nar/19.9.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Harvey C S, McGowan S E. Retinoic acid increases elastin in neonatal rat lung fibroblast cultures. Am J Physiol. 1993;265:L430–L437. doi: 10.1152/ajplung.1993.265.5.L430. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald P M, Kerr K, Smith J L, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 35.Mariani T J, Crouch E, Roby J D, Starcher B, Pierce R A. Increased elastin production in experimental granulomatous lung disease. Am J Pathol. 1995;147:988–1000. [PMC free article] [PubMed] [Google Scholar]
- 36.McGowan S E, Jackson S K, Olson P J, Parekh T, Gold L I. Exogenous and endogenous transforming growth factors-beta influence elastin gene expression in cultured lung fibroblasts. Am J Respir Cell Mol Biol. 1997;17:25–35. doi: 10.1165/ajrcmb.17.1.2686. [DOI] [PubMed] [Google Scholar]
- 37.McGowan S E, Liu R, Harvey C S, Jaeckel E C. Serine proteinase inhibitors influence the stability of tropoelastin mRNA in neonatal rat lung fibroblast cultures. Am J Physiol. 1996;270:L376–L385. doi: 10.1152/ajplung.1996.270.3.L376. [DOI] [PubMed] [Google Scholar]
- 38.McGowan S E, McNamer R. Transforming growth factor-β increases elastin production by neonatal rat lung fibroblasts. Am J Respir Cell Mol Biol. 1990;3:369–376. doi: 10.1165/ajrcmb/3.4.369. [DOI] [PubMed] [Google Scholar]
- 39.McLaren R S, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mecham R P, Davis E C. Elastic fiber structure and assembly. In: Yurchenco P D, Birk D E, Mecham R P, editors. Extracellular matrix assembly and structure. San Diego, Calif: Academic Press, Inc.; 1994. pp. 281–314. [Google Scholar]
- 41.Müllner E W, Kühn L C. A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- 42.Noguchi A, Samaha H, deMello D E. Tropoelastin gene expression in the rat pulmonary vasculature: a developmental study. Pediatr Res. 1992;31:280–285. doi: 10.1203/00006450-199203000-00018. [DOI] [PubMed] [Google Scholar]
- 43.Parks W C. Posttranscriptional regulation of lung elastin production. Am J Respir Cell Mol Biol. 1997;17:1–2. doi: 10.1165/ajrcmb.17.1.f135. [DOI] [PubMed] [Google Scholar]
- 44.Parks W C, Deak S B. Tropoelastin heterogeneity: implications for protein function and disease. Am J Respir Cell Mol Biol. 1990;2:399–406. doi: 10.1165/ajrcmb/2.5.399. [DOI] [PubMed] [Google Scholar]
- 45.Parks W C, Kolodziej M E, Pierce R A. Phorbol ester-mediated downregulation of tropoelastin expression is controlled by a posttranscriptional mechanism. Biochemistry. 1992;31:6639–6645. doi: 10.1021/bi00144a003. [DOI] [PubMed] [Google Scholar]
- 46.Parks W C, Pierce R A, Lee K A, Mecham R P. Elastin. In: Kleinman H K, editor. Advances in molecular and cell biology. Vol. 6. Greenwich, Conn: JAI Press, Inc.; 1993. pp. 133–182. [Google Scholar]
- 47.Parks W C, Secrist H, Wu L C, Mecham R P. Developmental regulation of tropoelastin isoforms. J Biol Chem. 1988;263:4416–4423. [PubMed] [Google Scholar]
- 48.Parks W C, Whitehouse L A, Wu L C, Mecham R P. Terminal differentiation of nuchal ligament fibroblasts: characterization of synthetic properties and responsiveness to external stimuli. Dev Biol. 1988;129:555–564. doi: 10.1016/0012-1606(88)90400-9. [DOI] [PubMed] [Google Scholar]
- 49.Peltz S W, Jacobson A. mRNA stability: in trans-it. Curr Opin Cell Biol. 1992;4:979–983. doi: 10.1016/0955-0674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 50.Pierce R A, Deak S B, Stolle C A, Boyd C D. Heterogeneity of rat tropoelastin mRNA revealed by cDNA cloning. Biochemistry. 1990;29:9677–9683. doi: 10.1021/bi00493a024. [DOI] [PubMed] [Google Scholar]
- 51.Pierce R A, Kolodziej M E, Parks W C. 1,25-Dihydroxyvitamin D3 represses tropoelastin expression by a posttranscriptional mechanism. J Biol Chem. 1992;267:11593–11599. [PubMed] [Google Scholar]
- 52.Pierce R A, Mariencheck W, Sandefur S, Crouch E C, Parks W C. Glucocorticoid-mediated upregulation of tropoelastin expression during late stages of fetal lung development. Am J Physiol (Lung Cell Mol Physiol) 1995;268:L491–L500. doi: 10.1152/ajplung.1995.268.3.L491. [DOI] [PubMed] [Google Scholar]
- 53.Pierce R A, Sandefur S, Doyle G A R, Welgus H G. Monocytic cell type-specific transcriptional induction of collagenase. J Clin Investig. 1996;97:1890–1899. doi: 10.1172/JCI118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raghunath M, Bachi T, Meuli M, Altermatt S, Gobet R, Bruckner-Tuderman L, Steinmann B. Fibrillin and elastin expression in skin regenerating from cultured keratinocyte autografts: morphogenesis of microfibrils begins at the dermo-epidermal junction and precedes elastic fiber formation. J Investig Dermatol. 1996;106:1090–1095. doi: 10.1111/1523-1747.ep12339373. [DOI] [PubMed] [Google Scholar]
- 55.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rucker R B, Dubick M A. Elastin metabolism and chemistry: potential roles in lung development and structure. Environ Health Perspect. 1984;53:179–191. doi: 10.1289/ehp.8455179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saarialho-Kere U K, Welgus H G, Parks W C. Divergent mechanisms regulate interstitial collagenase and 92 kDa gelatinase expression in human monocytic-like cells exposed to bacterial endotoxin. J Biol Chem. 1993;268:17354–17361. [PubMed] [Google Scholar]
- 58.Sachs A. The role of poly(A) in the translation and stability of mRNA. Curr Opin Cell Biol. 1991;2:1092–1098. doi: 10.1016/0955-0674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 59.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 60.Sage H, Gray W R. Studies on the evolution of elastin-III. The ancestral protein. Comp Biochem Physiol. 1981;68B:473–480. [Google Scholar]
- 61.Sage H, Gray W R. Studies on the evolution of elastin. I. Phylogenetic distribution. Comp Biochem Physiol. 1979;64B:313–327. doi: 10.1016/0305-0491(79)90277-3. [DOI] [PubMed] [Google Scholar]
- 62.Shapiro D J, Blume J E, Nielsen D A. Regulation of mRNA stability in eukaryotic cells. Bioessays. 1987;6:221–226. doi: 10.1002/bies.950060507. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro S D, Endicott S K, Province M A, Pierce J A, Campbell E J. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of d-aspartate and nuclear weapons-related radiocarbon. J Clin Investig. 1991;87:1828–1834. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 65.Shetty S, Kumar A, Idell S. Posttranscriptional regulation of urokinase receptor mRNA: identification of a novel urokinase receptor mRNA binding protein in human mesothelioma cells. Mol Cell Biol. 1997;17:1075–1083. doi: 10.1128/mcb.17.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sozhamannan S, Stitt B L. Effects on mRNA degradation by Escherichia coli transcription termination factor Rho and pBR322 copy number control protein Rop. J Mol Biol. 1997;268:689–703. doi: 10.1006/jmbi.1997.1004. [DOI] [PubMed] [Google Scholar]
- 67.Strauss B H, Chisholm R J, Keeley F W, Gotlieb A I, Logan R A, Armstrong P W. Extracellular matrix remodeling after balloon angioplasty injury in a rabbit model of restenosis. Circ Res. 1994;75:650–658. doi: 10.1161/01.res.75.4.650. [DOI] [PubMed] [Google Scholar]
- 68.Sudbeck B D, Pilcher B K, Welgus H G, Parks W C. Induction and repression of collagenase-1 by keratinocytes is controlled by distinct components of different extracellular matrix compartments. J Biol Chem. 1997;272:22103–22110. doi: 10.1074/jbc.272.35.22103. [DOI] [PubMed] [Google Scholar]
- 69.Swee M H, Parks W C, Pierce R A. Developmental regulation of elastin production. Expression of tropoelastin pre-mRNA persists after downregulation of steady-state mRNA levels. J Biol Chem. 1995;270:14899–14906. doi: 10.1074/jbc.270.25.14899. [DOI] [PubMed] [Google Scholar]
- 70.Theodorakis N G, Cleveland D W. Translationally coupled degradation of tubulin mRNA. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 219–238. [Google Scholar]
- 71.Zhang M C, Giro M, Quaglino D, Jr, Davidson J M. Transforming growth factor-beta reverses a posttranscriptional defect in elastin synthesis in a cutis laxa skin fibroblast strain. J Clin Investig. 1995;95:986–994. doi: 10.1172/JCI117808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang M C, He L, Giro M G, Yong S L, Tiller G E, Davidson J M. Cutis laxa arising from frame shift mutations in exon 30 of the elastin gene (ELN) J Biol Chem. 1999;274:981–986. doi: 10.1074/jbc.274.2.981. [DOI] [PubMed] [Google Scholar]
- 73.Zuker, M. 1 May 1999, revision date. [Online.] mfold version 3.0. Institute for Biomedical Computing, Washington University School of Medicine, St. Louis, Mo. http://mfold2.wustl.edu/∼mfold/RNA/form1.cgi. [8 September 1999, last date accessed.]