Abstract
Black rot, caused by Xanthomonas campestris pv. campestris (Xcc), produces V-shaped chlorotic lesions on the leaves of cabbage (Brassica oleracea var. capitata L.), causing darkened veins and drastically reducing yield and quality. Of the 11 Xcc races identified, races 1, 4, and 6 are predominant globally. In the present study, we aimed to develop a molecular marker linked to black rot resistance against Xcc races 6 and 7. Crossed between black rot-resistant (‘SCNU-C-3470’) and -susceptible (‘SCNU-C-3328’) lines obtained 186 F2 plants. Resistance to Xcc race 6 segregated in a 3:1 (susceptible:resistant) ratio in the F2 population, which is consistent with a monogenic recessive trait. Nucleotide-binding site (NBS) leucine rich repeat (LRR)-encoding resistance (R) genes play a crucial role in plant defenses to various pathogens. The candidate R gene (Bol031422) located on chromosome C08, previously reported by our research group, was cloned and sequenced in resistant and susceptible cabbage lines. The R gene Bol031422 consisted of a single exon with a 3 bp insertion/deletions (InDels), a 292 bp polymorphism (an insertion in the exon of the resistant line relative to the susceptible line) and several single nucleotide polymorphisms (SNPs). Here, we developed the InDel marker BR6-InDel to assess linkage between variation at Bol031422 and resistance to Xcc races 6 and 7. This marker will help cabbage breeders develop cabbage cultivars resistant to Xcc races 6 and 7.
Keywords: Xcc, race 6/7, black rot, molecular marker, cabbage
1. Introduction
Cabbage (Brassica oleracea var. capitata L.) is an important vegetable in Brassica crops worldwide due to its high nutritional quality, good storage properties, and potential health benefits [1,2]. Black rot is the most devastating disease of Brassica vegetables, caused by the aerobic, Gram-negative, and nonsporulating bacterium Xanthomonas campestris pv. campestris (Xcc) [3]. Black rot is mostly a seed-borne and vascular disease [4,5] that can be spread via wind, rainfall, and insects [6]. Typical black rot symptoms include necrotic, darkened leaf veins and a V-shaped chlorotic lesion starting from the leaf margin [3,5]. Black rot substantially reduces the yield and quality of cabbage harvests. To date, 11 pathogenic Xcc races have been identified that can infect Brassica crops [7,8,9]. Among them, races 1, 4, and 6 are the most widespread worldwide and the most aggressive against B. oleracea crops [10].
The occurrence of black rot has also increased in frequency due to recent climate change trends, making it more difficult to mitigate the pathogen effectively via chemical control methods alone. Another, more ecologically benign means to control plant pathogens, especially fungal, bacterial, and viral diseases, is to identify and cultivate resistant cultivars, which can provide very effective control without the need for chemical treatments [11,12]. The cabbage cultivars ‘Early Fuji’ and ‘Hugenot’ were the first reported to be resistant to Xcc [13,14]. Breeders and researchers have also selected resistant genotypes and transferred the underlying resistance loci into other Brassica crops, including Ethiopian mustard (Brassica carinata), brown mustard (Brassica juncea), rapeseed (Brassica napus), black mustard (Brassica nigra), and field mustard (Brassica rapa), as well as cabbage. However, there are no known cabbage cultivars presenting resistance to Xcc in Korea, largely because cabbage cultivars that are resistant to black rot are not adapted to the local environment [9,15,16], creating a pressing need for studies on black rot resistance in cabbage. The development of resistant cabbage cultivars will require the identification and characterization of resistant cabbage germplasm as well as the gene(s) responsible for resistance to black rot.
We previously screened a collection of cabbage germplasm for resistance to black rot, resulting in the identification of resistant cabbage lines as well as R genes encoding nucleotide binding site (NBS) receptors linked to Xcc resistance [17,18]. In plants, most R genes encode NBS-site-leucine rich repeat (NBS-LRR) proteins [19,20]. Previous studies showed that the R gene Bol037156 (Resistance gene to Race 1 of Fusarium oxysporum f. sp. conglutinans, FOC1) confers resistance to the fungal pathogen Fusarium in B. oleracea [21], whereas RESISTANT TO P. SYRINGAE 6 (RPS6) controls resistance to Pseudomonas syringae in Arabidopsis (Arabidopsis thaliana) [22]. NBS-LRR-type R genes are critical for plant responses to various pathogens, including nematodes, bacteria, fungi, and viruses [23]. Recent studies have shown that R genes have also been identified in other plant species, such as Arabidopsis thaliana, melon (Cucumis melo), and rice (Oryza sativa) [24,25,26].
Molecular markers facilitate plant selection based on their genotype without the need for time-consuming phenotypic tests. These genotyping efforts typically rely on sequence-based molecular markers such as simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs), as well as markers based on insertion/deletions (InDels). Our previous study showed susceptibility or resistance to Xcc of 27 cabbage inbred lines [18], highlighting lines ‘SCNU-C-3470’ and ‘SCNU-C-3328’ as being resistant and susceptible to Xcc races 6 and 7, respectively. Our recent study identified 32 differentially expressed R genes distributed across the B. oleracea genome [17]. Among these, one candidate gene (Bol031422) with higher expression in the resistant line than in susceptible line and higher expression in leaves than in other tissues was selected for further characterization. Accordingly, we sequenced the genomic region of Bol031422 and identified several InDels between resistant and susceptible lines. Although efforts have been made to identify R genes conferring resistance to the pathogen [1,17] and develop methods to screen for resistance, very little is known about Xcc-race-specific R genes. In this study, we therefore aimed to develop a specific molecular marker to genotype plants for Bol031422 and then compare these results to tests for resistance or susceptibility to Xcc races 6 and 7 by routine PCR to assess the contribution of Bol031422 to the observed resistance to Xcc.
2. Results
2.1. Inheritance of Xcc Races 6 and 7 Resistance in Cabbage
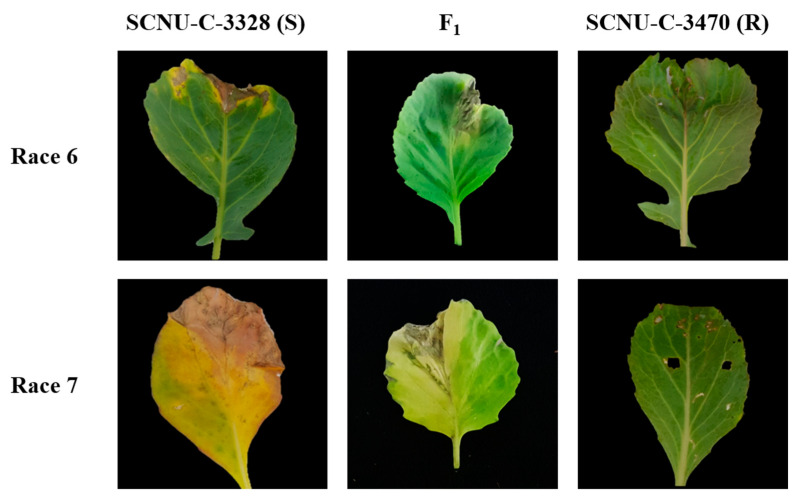
We assessed the inheritance patterns of resistance to Xcc races 6 and 7 using an F2 population derived from a cross between the two cabbage lines ‘SCNU-C-3328’ and ‘SCNU-C-3470’, which differ in their resistance to black rot. F1 plants were all susceptible, suggesting that resistance to Xcc races 6 and 7 is inherited as a recessive trait (Figure 1). A total of 95 F2 plants were inoculated with Xcc race 6, resulting in 21 resistant plants and 72 plants developing symptoms. Excluding two dead plants, only 93 plants were used in this study (Figure S1A). Similarly, we inoculated total of 91 F2 plants with Xcc race 7, with 34 plants showing no symptoms and 55 plants developing a typical black rot disease. Only 89 plants were used in this study, excluding two dead plants (Figure S1B). A χ2 test revealed that resistance to Xcc race 6 follows a 3:1 (susceptible:resistant) segregation ratio, which is consistent with a single recessive gene conferring resistance (Table 1 and Figure S1). By contrast, F2 plants inoculated with race 7 showed a 1.6:1 (susceptible:resistant) segregation ratio, thus ruling out a status as a single monogenic trait.
Figure 1.
Phenotypes of the resistant and susceptible, and their F1 generation two weeks after inoculation with Xanthomonas campestris pv. campestris (Xcc) races 6 and 7.
Table 1.
Inheritance of Xcc races 6 and 7 resistance in cabbage. S; susceptible, R; resistant.
| Crosses | Generation | Susceptible | Resistant | Expected Ratio (S:R) | Chi-Square (χ2) | p |
|---|---|---|---|---|---|---|
| Race 6 | ||||||
| SCNU-C-3328 (S) | P1 | 12 | 0 | |||
| SCNU-C-3470 (R) | P2 | 0 | 12 | |||
| 3328 × 3470 | F1 | 12 | 0 | |||
| 3328 × 3470 | F2 | 72 | 21 | 3:1 | 0.29 | 0.59 |
| Race 7 | ||||||
| SCNU-C-3328 (S) | P1 | 12 | 0 | |||
| SCNU-C-3470 (R) | P2 | 0 | 12 | |||
| 3328 × 3470 | F1 | 12 | 0 | |||
| 3328 × 3470 | F2 | 55 | 34 | 3:1 | 8.27 | 0.004 |
2.2. Selection of Xcc Races 6 and 7 Resistance Gene
The identification of black-rot-resistant genotypes is a prerequisite for the development of cabbage cultivars resistant to black rot. In our previous work, we analyzed 157 NBS-LRR-encoding R genes, some of which were differentially expressed between black rot-resistant and -susceptible cabbage lines [17,20]. Expression levels of these R genes were determined by qRT-PCR (Figure S2). Only two genes, Bol031422 and Bol037412, were more highly expressed in the resistant cabbage line SCNU-C-3470 than in the susceptible line SCNU-C-3328. Both Bol031422 and Bol037412 are intronless genes. Bol031422 was highly expressed specifically in leaf, while Bol037412 was highly expressed only in root, which is not a tissue affected by Xcc [17]. Taken together, these results suggested that Bol031422 might play an important role in resistance to Xcc races 6 and 7 in cabbage.
2.3. Cloning and Sequencing of Candidate Gene
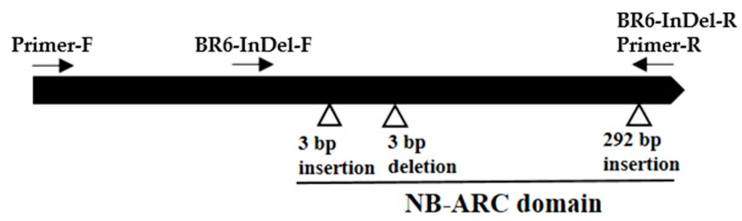
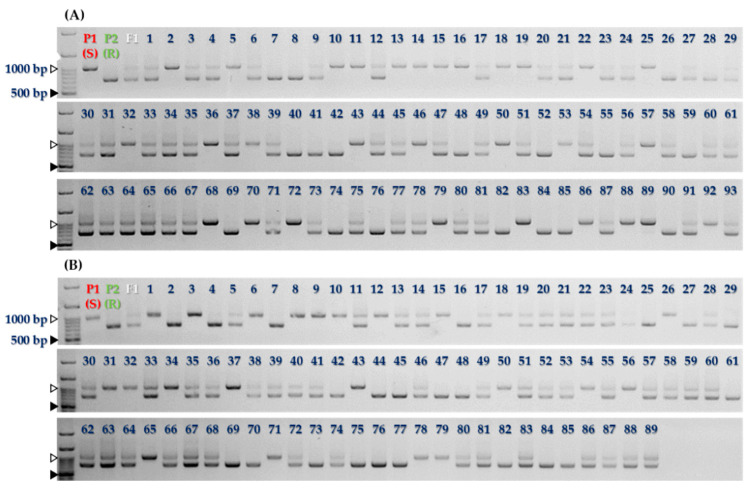
To detect sequence variation in the candidate R gene (Bol031422) for Xcc resistance, gene-specific primers were designed to cover the entire coding sequence (Figure 2 and Table S1). Sequencing of resistant and susceptible lines revealed multiple polymorphisms, consisting of several InDels and SNPs (Figure 3 and Figure S3A). Therefore, missense and non-synonymous mutations were found in the protein sequence (Figure S3B). The susceptible line SCNU-C-3328 showed a 3 bp insertion at nucleotide position 437, as well as a 3 bp deletion at nucleotide position 475. Accordingly, we designed primers to cover each three InDel regions and assess their correlation with Xcc resistance. Only one InDel segregated according to resistance across the resistant and susceptible lines (data not shown). This 292 bp insertion in the susceptible line SCNU-C-3328 at nucleotide position 1087 (Figure 2, Figure 3 and Figure S3A) resulted in a frameshift and premature translation termination (Figure S3B). We designed a new set of primers covering this InDel (Table 2), generating a PCR amplicon of 724 bp in resistant lines and 1013 bp in susceptible lines (Figure 4). The primers also amplified two bands from F1 plants derived from the cross between lines SCNU-C-3328 and SCNU-C-3470, as expected.
Figure 2.
Primer location in R gene used in this study. ORF: Primer-F and Primer-F; Indel marker ‘BR6-InDel’: InDel-F and InDel-R.
Figure 3.
Sequence alignment between resistant (SCNU-C-3470, upper) and susceptible (SCNU-C-3328, lower) cabbage lines and position of InDel marker. The yellow and blue highlights in the alignment of DNA sequences indicate SNP and InDel, respectively.
Table 2.
Primer specifications of InDel marker associated with Xcc resistance in cabbage.
| Gene ID | InDel Marker | Primer (5′-3′) | Tm (°C) | Product Size | |
|---|---|---|---|---|---|
| Bol031422 | BR6-InDel | F | TGGGGTGACTGATGAAACTCCTAT | 60 | 724 bp |
| R | TCACTTCTGATTCATCCTCGTCATCT |
Figure 4.
Banding pattern of InDel marker in resistant, susceptible lines (P1, SCNU-C-3328; P2, SCNU-C-3328), F1 and F2 generation. Inoculated with Xcc race 6 (A), Xcc race 7 (B). R, resistant; S, susceptible. Filled triangle; 1000 bp ladder, unfilled triangle; 500 bp ladder.
2.4. Validation of the InDel Marker
To test the association between the InDel marker BR6-InDel and resistance to black rot, we used two populations inoculated with Xcc races 6 or 7, respectively (Tables S2 and S3). The first population consisted of the F2 population described earlier, and the second population comprised 27 inbred cabbage lines. Genotyping analysis of the F2 population confirmed the close linkage of BR6-InDel with Xcc races 6 and 7 resistance, based on inoculation results (Figure 4 and Figure S4). Of the 27 inbred lines, two cabbage lines (‘SCNU-C-3470’ and ‘SCNU-C-4118’) were resistant to Xcc race 6, while four (‘SCNU-C-107’, ‘SCNU-C-3270’, ‘SCNU-C-3470’, and ‘SCNU-C-4059’) were resistant to race 7 (Table S3) [18]. A comparison of phenotypic results obtained from inoculation with Xcc races 6 or 7 and the genotype results with the InDel marker showed a predictive power of the InDel marker adaptability of 83.9% (83.5% in the F2 population; 85.2% in inbred lines) and 69.0% (67.0% in the F2 population; 76.0% in inbred lines), respectively. These results indicated that Bol031422 might play an important role in resistance to Xcc race 6. In addition, the genotype at BR6-InDel appeared to better predict resistance against Xcc race 6 than to race 7.
3. Discussion
The aim of cabbage breeding is to increase yield and nutritional quality. However, diseases such as black rot, blackleg and clubroot cause severe yield loss in cabbage. Although chemical control is effective for combatting the disease, frequent use of bactericides is detrimental to the environment. In addition, crop pathogens are subjected to selective pressures, such as bactericides. Therefore, it is critical to develop disease-resistant cultivars via molecular breeding. The advancement of high-throughput sequencing technologies has allowed rapid implementation of molecular-marker-assisted selection to detect and utilize polymorphisms. Indeed, molecular markers have been instrumental in the selection of disease-resistant cultivars. R genes are high-confidence candidate genes for conferring resistance to a disease and can easily be turned into DNA markers. In this study, we developed the InDel marker BR6-InDel linked to black rot resistance against Xcc race 6.
Our research group recently reported 157 NBS-LRR-encoding R genes in B. oleracea [20], of which 32 were differentially expressed between leaves, roots, siliques, and stems [17]. Among these, only four genes (Bol009890, Bol022619, Bol042095, and Bol031422) were highly expressed in leaves. We also previously reported that cabbage line SCNU-C-3470 is resistant to Xcc races 6 and 7 [18]. Only two genes (Bol031422 and Bol037412) of the 32 genes above were differentially expressed in SCNU-C-3470 compared to SCNU-C-3328, as determined by qRT-PCR. However, only Bol031422 was highly expressed in leaves, while Bol037412 was highly expressed in roots [17]. In addition, Basic Local Alignment Search Tool (BLAST) searches using the Bol031422 sequence as query against the IRGSP-1.0, TAIR10, and Brapa_1.0 databases revealed sequence similarity to disease resistance genes, such as Os01g0788500 (disease resistance protein domain containing protein), At1g61180 (the LRR and NB-ARC domains containing disease resistance protein UNI), and Bra026923 (NBS-encoding gene in B. rapa).
In this study, the cabbage R gene Bol031422, located on chromosome C08, showed that it can be amplified from both resistant and susceptible lines and displays genotype-specific polymorphisms. Phenotyping of F1 and F2 cabbage plants derived from a cross between resistant and susceptible lines and their parents confirmed that the resistance trait segregates as a single recessive gene (Table 1). Indeed, all F1 plants were susceptible to infection by Xcc races 6 and 7. Furthermore, sequence analysis of Bol031422 identified several InDels and SNPs in the susceptible line SCNU-C-3328 compared to the resistant line (Figure S3). In particular, a 292 bp insertion caused a frameshift and premature termination of translation was detected in SCNU-C-3328 (Figure 3), against which we designed the InDel marker BR6-InDel (Figure 2 and Table 2). The correlation between race 6 resistance phenotypes and the putative InDel marker genotyping results is only 83.9% (Table S2), suggesting Bol031422 might be involved in black rot resistance against Xcc race 6. However, the genetic architecture of resistance to Xcc race 6 appears to be distinct from that against Xcc race 7, as F2 plants inoculated with race 7 showed a ratio of 1.6:1 (susceptible:resistant), and genotype results agreed with the phenotype for only 70% of plants. These results support the hypothesis that resistance to Xcc race 7 in cabbage relies on several genes, although Bol031422 is likely to be a major contributor. More detailed studies of the gene responsible for races 6 and 7 resistance should be performed.
4. Materials and Methods
4.1. Plant Materials
The parental lines that were used in this study were ‘SCNU-C-3470’ and ‘SCNU-C-3328’, which are resistant and susceptible to Xcc races 6 and 7, respectively. ‘SCNU-C-3328’ was crossed with ‘SCNU-C-3470’ to develop the F1 generation. The F2 generation of 186 plants was developed via self-pollination of the F1 population. Of these F2 plants, 95 plants were used in Xcc race 6 and 91 plants were used in race 7. In addition, 27 cabbage inbred lines were also used in this study [18]. Cabbage seeds were sown in trays containing a nursery soil at plant culture room. Twenty-five days after sowing, the seedlings were transferred to pots. All plants were grown in a plant culture room at 24–27 °C under long-day conditions (14 h light/10 h dark cycles) with 60% relative humidity.
4.2. Bacterial Strains and Culture Media
Xanthomonas campestris pv. campestris (Xcc) races 6 and 7, which are causal agent of black rot of cabbage, were provided by the School of Life Sciences, University of Warwick, Coventry, UK (Table 3). Both Xcc strains were grown on petri dish containing 15 mL King’s B (KB) medium for 48 h at 30 °C [27].
Table 3.
Xanthomonas campestris pv. campestris (Xcc) races used in this study.
| Sl. No. | Bacterial Race/Strains | Source | Reference |
|---|---|---|---|
| 1 | Xanthomonas campestris pv. campestris Race 6 (6181) | Portugal | [9] |
| 2 | Xanthomonas campestris pv. campestris Race 7 (8450A) | UK |
4.3. Inoculation Test
Thirty-five days after sowing, the F2 plants and cabbage inbred lines were inoculated. The Xcc races were scraped from the culture plates and subcultured by suspending in liquid KB medium for 48 h at 30 °C. Thereafter, the bacterial suspension was adjusted to a concentration of 1.0 × 108–109 CFU/mL by adding sterile water. Finally, three youngest leaves of each plants were inoculated by mouse-tooth forceps methods at secondary veins (at least 10 inoculation sites per leaf). Then, inoculated plants were covered to maintain high relative humidity [9,28].
4.4. Disease Scoring
To evaluate the disease symptoms of the inoculated leaves, the leaves at 2 weeks after inoculation (WAI) were used. The disease ratings were scored for each leaf at 2 WAI based on a 0–9 scale (Figure 5). Leaf symptoms were scored on the following scale: 0 = no visible symptoms, 1 = chlorosis or small necrosis near the inoculation site, 3 = typical small V-shaped lesion with black veins, 5 = typical lesion half way to the middle vein, 7 = typical lesion succeeding to the middle vein, and 9 = lesion reaching the middle vein as previously described [29]. Scales 0, 1–3, 5–7 and 9 were characterized as highly resistant (HR), resistant (R), susceptible (S) and highly susceptible (HS), respectively (Figure 5).
Figure 5.
The disease rating criteria used in this study for black rot of cabbage. Scales are 0–9. 0, high resistant (HR); 1–3, resistant (R); 5–7, susceptible (S); 9, highly susceptible (HS).
4.5. Isolation of Genomic DNA
Young leaf samples of each cabbage plants were collected and immediately frozen in liquid nitrogen. Genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The purity and integrity of the DNA were assessed by ND-1000 Micro-spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) and gel electrophoresis (0.8% agarose gel), respectively.
4.6. PCR Amplification
PCR amplification was done in 20 μL reaction mixture, comprising 10 μL of 2X Prime Taq premix (Genet Bio, Daejeon, Korea), 1.0 μL of each forward and reverse primers (10 pmoles), 2 μL of 100 ng genomic DNA as template and 6 μL of deionized distilled water. Primers are listed in Table 2 and Table S1. The PCR conditions were as follows: 95 °C for 5 min, followed by amplifications of 30–35 cycles at 95 °C for 30 s, 58 and 60 °C for 30 s (specific Tm to respective primer sets in Table 2 and Table S1), 72 °C for 30 s and 72 °C for 5 min. Then, amplified PCR products were assessed by gel electrophoresis (1.5% agarose gel).
4.7. Cloning and Sequencing
The candidate R gene (Bol031422) was amplified by Phusion High-Fidelity DNA Polymerase (New England Biolabs, EVRY Cedex, France) from the genomic DNA of Xcc resistant and susceptible cabbage lines. Amplified DNA fragments were extracted from the gel and purified using the Wizard SV gel and PCR cleanup system (Promega, Madison, WI, USA). Then, gene cloning was performed using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). The cloned amplicons were sequenced with the universal primers, M13F and M13RpUC, using ABI 3730XL DNA sequencer (Macrogen Co., Seoul, Korea). To remove all ambiguities, each forward and reverse sequence of resistant and susceptible cabbage lines was repeated five times. Gene sequences between the resistant and susceptible lines were compared using the multiple sequence alignment by CLUSTALW (https://www.genome.jp/tools-bin/clustalw, accessed on 4 March 2021).
4.8. RNA Extraction and qRT-PCR Analysis
Total RNAs were extracted from 100 mg of finely ground leaf tissue at 35 days after sowing using RNeasy mini kit (QIAGEN, Hilden, Germany). Total RNA concentration and quality were measured using a ND-1000 Micro-spectrophotometer. SuperScript® III First-Strand Synthesis System kit (Invitrogen, Gaithersburg, MD, USA) was used for cDNA synthesis from total RNA. The cDNA was then used for real-time quantitative PCR with the LightCycler system (Roche, Mannheim, Germany) instrument using qPCRBIO SyGreen Mix Lo-ROX (PCR Biosystems, London, UK) for NBS-encoding genes. Primers were listed in Afrin et al. [17]. Threshold cycle (Ct) values were used to calculate 2−ΔΔCT method, with actin used as an internal control [30].
4.9. Statistical Analysis
A Chi-square (χ2) test for goodness-of-fit was performed to determine deviations of observed data from the expected segregation ratios using XLSTAT software. Data are presented as mean ± standard error of the mean. Statistical differences were assessed by Student’s t-test. The significance of differences between the means was assessed using p-values of <0.01 and <0.05. All the statistical analyses were performed using PRISM 6 software (ver. 6.01, GraphPad Software Inc., San Diego, CA, USA) [31].
5. Conclusions
This study showed that Xcc race 6 resistance in cabbage line ‘SCNU-C-3470’ is regulated by a single recessive gene. The newly developed InDel marker ‘BR6-InDel’ is linked to Xcc races 6 and 7 resistance and located on cabbage chromosome C08, showed consistency with phenotypic results of both the inbred cabbage lines and F2 population. This InDel marker could be valuable for future cabbage breeding programs.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/plants10091940/s1, Figure S1. Disease symptoms of cabbage F2populations 14 days after being inoculation with Xanthomonascampestris pv. campestrisraces 6 (A) and 7 (B).; Figure S2. Mean normalized expression of candidate genes in susceptible (SCNU-C-3328) and resistant (SCNU-C-3470) cabbage lines. Error bar represents ± S.E.M of the means of triplicates. Asterisks indicate significant difference in expression level (* p < 0.05, ** p < 0.01, Student’s t-test).; Figure S3. Sequence information obtained by cloning and sequencing of cabbage resistant (SCNU-C-3470) and susceptible (SCNU-C-3328) lines for a NBS-LRR gene (Bol031422) and their alignments. Genomic DNA (gDNA) sequence and their alignment (A), protein sequence and their alignment (B). Sequence alignments along with the references sequences retrieved from Bolbase (http://www.ocri-genomics.org/bolbase/) considering Brassica oleracea as reference genome database. In sequence alignment asterisks (*) indicate sequence similarity. Absence of asterisks indicates sequence dissimilarity. En dash (–) indicates insertion/deletion of nucleotides. The grey and red highlights in the alignment of genomic DNA sequences indicate the BR6-InDel-F and BR6-InDel-R, respectively. The red text colors in the alignment of genomic sequences indicate InDel regions.; Figure S4. Banding profile of R gene (Bol031422) in 27 inbred cabbage lines using Pimer-F/R (A), BR6-InDel-F/R (B).; Table S1. Primer specifications of candidate Black rot resistance NBS gene Bol031422.; Table S2. Phenotypic and Genotypic screening results for against Xcc race 6 and 7 in cabbage. S; susceptible, H; Hetero, R; Resistant.; Table S3. Comparison of bioassay and molecular screening results for black rot resistance in cabbage inbred lines. S; susceptible, H; hetero, R; resistant.
Author Contributions
I.-S.N., H.-J.J. and M.A.R. conceived the study. J.-E.H. conducted the experiments and wrote the manuscript. M.A.R. and K.S.A. developed F1 and F2 populations, cloned the gene and performed the sequence analysis. M.A.R. edited and critically revised the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by the Golden Seed Project (Center for Horticultural Seed Development) of the Ministry of Agriculture, Food and Rural Affairs in the Republic of Korea (MAFRA) under the grant no. 213007-05-5-CG100.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee J., Izzah N.K., Jayakodi M., Perumal S., Joh H.J., Lee H.-Y., Lee S.-C., Park J.-Y., Yang W.-K., Nou I.-S., et al. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biol. 2015;15:32. doi: 10.1186/s12870-015-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh B., Sharma S. Heterosis for mineral elements in single cross-hybrids of cabbage (Brassica oleracea var. capitata L.) Sci. Hortic. 2009;122:32–36. doi: 10.1016/j.scienta.2009.04.007. [DOI] [Google Scholar]
- 3.Williams P.H. Black rot: A continuing threat to world crucifers. Plant Dis. 1980;64:736–742. doi: 10.1094/PD-64-736. [DOI] [Google Scholar]
- 4.Cook A., Walker J., Larson R. Studies on the disease cycle of black rot of crucifers. Phytopathology. 1952;42:162. [Google Scholar]
- 5.Vicente G.J., Holub E.B. X anthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 2013;14:2–18. doi: 10.1111/j.1364-3703.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma B.B., Kalia P., Singh D., Sharma T.R. Introgression of Black Rot Resistance from Brassica carinata to Cauliflower (Brassica oleracea botrytis Group) through Embryo Rescue. Front. Plant Sci. 2017;8:1255. doi: 10.3389/fpls.2017.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz J., Tenreiro R., Cruz L. Assessment of diversity of Xanthomonas campestris pathovars affecting cruciferous plants in Portugal and disclosure of two novel X. campestris pv. campestris races. J. Plant Pathol. 2017;99:403–414. [Google Scholar]
- 8.Fargier E., Manceau C. Pathogenicity assays restrict the species Xanthomonas campestris into three pathovars and reveal nine races within X. campestris pv. campestris. Plant Pathol. 2007;56:805–818. doi: 10.1111/j.1365-3059.2007.01648.x. [DOI] [Google Scholar]
- 9.Vicente J., Conway J., Roberts S., Taylor J.D. Identification and Origin of Xanthomonas campestris pv. campestris Races and Related Pathovars. Phytopathology. 2001;91:492–499. doi: 10.1094/PHYTO.2001.91.5.492. [DOI] [PubMed] [Google Scholar]
- 10.Singh D., Rathaur P., Vicente J. Characterization, genetic diversity and distribution of Xanthomonas campestris pv. campestris races causing black rot disease in cruciferous crops of India. Plant Pathol. 2016;65:1411–1418. doi: 10.1111/ppa.12508. [DOI] [Google Scholar]
- 11.Lee H.J., Lee J., Oh D.-G. Resistance of pepper cultivars to Ralstonia solanacearum isolates from major cultivated areas of chili peppers in Korea. Hortic. Sci. Technol. 2018:569–576. [Google Scholar]
- 12.Yerasu S.R., Murugan L., Halder J., Prasanna H.C., Singh A., Singh B. Screening tomato genotypes for resistance to early blight and American serpentine leafminer. Hortic. Environ. Biotechnol. 2019;60:427–433. doi: 10.1007/s13580-019-00130-y. [DOI] [Google Scholar]
- 13.Bain D. Reaction of brassica seedlings to blackrot. Phytopathology. 1952;42:316–319. [Google Scholar]
- 14.Bain D. Resistance of Cabbage to black rot. Disappearance of black rot symptoms in Cabbage seedlings. Phytopathology. 1955;45:35–37. [Google Scholar]
- 15.Jensen B.D., Massomo S., Swai I.S., Hockenhull J., Andersen S.B. Field evaluation for resistance to the black rot pathogen Xanthomonas campestris pv. campestris in cabbage (Brassica oleracea) Eur. J. Plant Pathol. 2005;113:297–308. doi: 10.1007/s10658-005-2799-y. [DOI] [Google Scholar]
- 16.Taylor J.D., Conway J., Roberts S., Astley D., Vicente J. Sources and Origin of Resistance to Xanthomonas campestris pv. campestris in Brassica Genomes. Phytopathology. 2002;92:105–111. doi: 10.1094/PHYTO.2002.92.1.105. [DOI] [PubMed] [Google Scholar]
- 17.Afrin K.S., Rahim A., Park J.-I., Natarajan S., Kim H.-T., Nou I.-S. Identification of NBS-encoding genes linked to black rot resistance in cabbage (Brassica oleracea var. capitata) Mol. Biol. Rep. 2018;45:773–785. doi: 10.1007/s11033-018-4217-5. [DOI] [PubMed] [Google Scholar]
- 18.Afrin K.S., Rahim A., Park J.-I., Natarajan S., Rubel M.H., Kim H.-T., Nou A.I.-S. Screening of Cabbage (Brassica oleracea L.) Germplasm for Resistance to Black Rot. Plant Breed. Biotechnol. 2018;6:30–43. doi: 10.9787/PBB.2018.6.1.30. [DOI] [Google Scholar]
- 19.Dangl J.L., Jones J. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 20.Yu J., Tehrim S., Zhang F., Tong C., Huang J., Cheng X., Dong C., Zhou Y., Qin R., Hua W., et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014;15:3. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv H., Fang Z., Yang L., Zhang Y., Wang Q., Liu Y., Zhuang M., Yang Y., Xie B., Liu B., et al. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genom. 2014;15:1094. doi: 10.1186/1471-2164-15-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S.H., Kwon S.I., Saha D., Anyanwu N.C., Gassmann W. Resistance to the Pseudomonas syringae Effector HopA1 Is Governed by the TIR-NBS-LRR Protein RPS6 and Is Enhanced by Mutations in SRFR1. Plant Physiol. 2009;150:1723–1732. doi: 10.1104/pp.109.139238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan H., Yuan W., Bo K., Shen J., Pang X., Chen J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013;14:1–15. doi: 10.1186/1471-2164-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers B., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-Wide Analysis of NBS-LRR–Encoding Genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monosi B., Wisser R.J., Pennill L., Hulbert S.H. Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- 26.Hassan M.Z., Rahim A., Natarajan S., Robin A.H.K., Kim H.-T., Park J.-I., Nou I.-S. Gummy stem blight resistance in melon: Inheritance pattern and development of molecular markers. Int. J. Mol. Sci. 2018;19:2914. doi: 10.3390/ijms19102914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King O.E., Ward M.K., Raney E.D. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 28.Lee J.H., Kim J.C., Jang K.S., Choi Y.H., Ahn K.G., Choi G.J. Development of efficient screening method for resistance of cabbage cultivars to black rot disease caused by Xanthomonas campestris pv. campestris. Res. Plant Dis. 2013;19:95–101. doi: 10.5423/RPD.2013.19.2.095. [DOI] [Google Scholar]
- 29.Vicente J., Taylor J.D., Sharpe A.G., Parkin I.A.P., Lydiate D.J., King G. Inheritance of Race-Specific Resistance to Xanthomonas campestris pv. campestris in Brassica Genomes. Phytopathology. 2002;92:1134–1141. doi: 10.1094/PHYTO.2002.92.10.1134. [DOI] [PubMed] [Google Scholar]
- 30.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Swift M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997;37:411–412. doi: 10.1021/ci960402j. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.