Abstract
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) are part of the same pathophysiological spectrum and have common genetic and cerebrospinal fluid (CSF) biomarkers. Our aim here was to identify causative gene variants in a cohort of Greek patients with FTD, ALS and FTD-ALS, to measure levels of CSF biomarkers and to investigate genotype-phenotype/CSF biomarker associations. In this cohort of 130 patients (56 FTD, 58 ALS and 16 FTD-ALS), we performed C9orf72 hexanucleotide repeat expansion analysis, whole exome sequencing and measurement of “classical” (Aβ42, total tau and phospho-tau) and novel (TDP-43) CSF biomarkers and plasma progranulin. Through these analyses, we identified 14 patients with C9orf72 repeat expansion and 11 patients with causative variants in other genes (three in TARDBP, three in GRN, three in VCP, one in FUS, one in SOD1). In ALS patients, we found that levels of phospho-tau were lower in C9orf72 repeat expansion and MAPT c.855C>T (p.Asp285Asp) carriers compared to non-carriers. Additionally, carriers of rare C9orf72 and APP variants had lower levels of total tau and Aβ42, respectively. Plasma progranulin levels were decreased in patients carrying GRN pathogenic variants. This study expands the genotypic and phenotypic spectrum of FTD/ALS and offers insights in possible genotypic/CSF biomarker associations.
Keywords: frontotemporal dementia, amyotrophic lateral sclerosis, genetics, biomarkers, C9orf72, TARDBP, GRN, VCP
1. Introduction
It has been widely recognized that Frontotemporal Dementia (FTD) and Amyotrophic Lateral Sclerosis (ALS) occupy the two extremes of the same pathophysiological spectrum, sharing several histological features and genetic causes [1]. FTD is a highly inherited disorder, with 30–50% of patients reporting family history of a similar phenotype [2]. For this familial form of FTD, several genes, such as the C9orf72, MAPT, GRN, TARDBP and VCP genes, have been found to harbor pathogenic variants [3,4]. On the other hand, in the 10% of patients with the familial form of ALS and in a proportion of the sporadic cases, causative variants have been identified in an increasing number of genes [5]. Interestingly, many patients display both the FTD and the ALS phenotype, often associated with a specific gene variant [6]. Additionally, a pathogenic FTD/ALS gene variant can cause differing phenotypes (FTD, ALS or both FTD and ALS) in different members of the same family [1].
Recent studies have shown that variants in the FTD/ALS-associated genes or FTD/ALS-specific neuropathological changes may cause neurodegenerative phenotypes beyond the typical FTD/ALS presentation [7,8,9,10]. Inversely, FTD/ALS clinical features may be associated with non-FTD/ALS pathology [11,12]. These cases are clinically indistinguishable; however, biomarkers, such as those derived from the cerebrospinal fluid (CSF), offer clues to their diagnosis. These CSF biomarkers, namely total-tau protein (τT), phosphorylated-tau protein (τP) and β-amyloid peptide with 42 amino acids (Aβ42) have already been successfully incorporated in Alzheimer’s Disease (AD) diagnostic criteria used for research purposes [13,14]. For FTD-ALS, TDP-43 protein in the CSF is an emerging biomarker [15,16]. However, there is still uncertainty about the interplay between genetic variants and CSF biomarkers.
The aim of the present study was to identify causative variants in the FTD and ALS- associated genes in a well-characterized (including CSF biomarker profiling) cohort of Greek patients presenting with FTD, ALS or FTD-ALS phenotypes. Additionally, we aimed to investigate the association of the genotype with the clinical phenotype and the levels of CSF (τT, τP-181, Aβ42, TDP-43) and plasma (progranulin) biomarkers.
2. Materials and Methods
2.1. Participants
A total of 130 patients were included in our study, presenting either to the 1st Department of Neurology of the National and Kapodistrian University of Athens at Eginition Hospital, Athens Greece, the University Hospital of Heraklion, Crete and the Papageorgiou and AHEPA Hospitals, Thessaloniki, Greece. Patients were prospectively enrolled between 2014 and 2019. For inclusion in the study, patients had to receive the diagnosis of FTD, ALS or FTD-ALS according to widely accepted criteria (see below). For the FTD patients, exclusion criterion was the presence of an AD CSF biomarker profile, defined according to the Neurochemistry Unit of the 1st Department of Neurology, University of Athens cutoff values (Aβ42 ≤ 580 pg/mL, τT ≥ 376 pg/mL and τP-181 ≥ 62.5 pg/mL) [16].
The 130 patients were divided into three well-characterized groups:
The FTD group consisted of 56 patients who met the criteria for either the behavioral subtype of FTD (bvFTD) [17] or primary progressive aphasia (PPA), regardless of the subtype [18]. Of the PPA patients who eventually participated in the study, 6 met the semantic variant PPA (svPPA) criteria and 5 met the non-fluent agrammatic PPA (nfaPPA) criteria.
The ALS group included 58 patients who met the Awaji-Shima criteria [19].
The ALS-FTD group included 16 patients who met the criteria of the combined ALS-FTD phenotype [20].
All patients underwent detailed clinical, neuropsychological, biochemical and neuroimaging examination (magnetic resonance imaging [MRI] in all patients and, additionally, single-photon emission computed tomography [SPECT] in most FTD patients) to exclude secondary causes of dementia and to establish the diagnosis of FTD, ALS, or FTD/ALS.
The study was performed according to the ethical guidelines of the 1964 Declaration of Helsinki and had the approval of the Scientific and Ethics Committee of all hospitals involved. Informed consent was obtained from each subject when possible or their authorized caregiver(s).
2.2. Blood Collection and DNA Extraction
For the genetic analyses, whole peripheral blood from participants was collected in ethylenediaminetetraacetic acid (EDTA) tubes, which were stored at −20 °C until DNA extraction and were only thawed once, just before the procedure. For the extraction of the genomic DNA from 400 μL of whole peripheral blood, the QIAamp DNA Blood Mini kit (Qiagen, CA, USA) was used. DNA concentration and purity were assessed spectrophotometrically at 260 and 280 nm. Participant anonymity was ensured through the designation of unique code identifiers for each DNA sample.
2.3. C9orf72 Repeat Expansions
Most of the patients (n = 94) were initially checked for C9orf72 hexanucleotide repeat expansions at the Neurogenetics Laboratory of Eginition Hospital according to a two-step protocol. This protocol included sizing PCR (amplification of the region that contains the hexanucleotide repeat with primers flanking this region), agarose electrophoresis and fragment analysis to identify samples with possible presence of the C9orf72 repeat expansion. This was followed by repeat primed PCR, to verify and separate samples with the C9orf72 repeat expansion, as previously described [21,22]. Samples were then electrophorized on an ABI 310 Capillary Analyzer (Applied Biosystems, Foster City, CA, USA) and analyzed on GeneScan v3.7 (2001, Applied Biosystems, Foster City, CA, USA). In addition, 21 patients were initially analyzed for the presence of C9orf72 repeat expansion by repeat primed PCR amplification and STR (short tandem repeats) PCR analysis at the Diagnostic Service Facility, Laboratory of Neurogenetics, University of Antwerp, Belgium.
2.4. Whole Exome Sequencing
In most patients that did not carry a C9orf72 hexanucleotide repeat expansion, Whole Exome Sequencing (WES) was performed (Figure 1) according to the following procedures:
-
(1)
At Minotech Genomics Facility, Institute of Molecular Biology and Biotechnology (IMBB-FORTH, Crete) with the use of the Illumina NextSeq500 platform (n = 60). In detail, sequencing of 2 × 75 bp DNA fragments with at least 50× coverage, targeting regions of 45.3 Mb size, was performed. Libraries were prepared with the TruSeq® Rapid Exome Library prep kit (Illumina, San Diego, CA, USA). Bioinformatics processing of the data derived from mapping on the hg19 reference genome and quality control of the results (e.g., number of readings and coverage quality) were performed by the BaseSpace® software (Illumina, San Diego, CA, USA). Finally, genetic variation was identified with the VariantStudio® software after comparison with the hg19 reference genome and drawing information from genetic databases, e.g., Human Genome Mutation Database (HGMD), ClinVar® (National Center for Biotechnology Information, Bethesda MD, USA) (Annotation Excel file).
-
(2)
At Macrogen (Seoul, Korea), using the Illumina HiSeq4000 platform (n = 24). In specific, 2 × 100 bp DNA fragments were sequenced with an aim of at least 50x coverage. For the construction of genomic libraries, the Agilent Sure-Select Human All Exon V5 (not including UTRs) Target Enrichment System was used.
-
(3)
At Otogenetics (GA, USA), using the Illumina HiSeq2500 platform (n = 11). In detail, sequencing of 2 × 100 bp DNA fragments was performed aiming at coverage of at least 50× and targeting a region of 45.3 Mb, that represents >98% of the human coding sequence according to the Consensus Coding Sequences (CCDS) and Ensembl. Exon-enriched library preparation was performed with the use of the Agilent V5 (51Mb) Sure-Select Target Enrichment System.
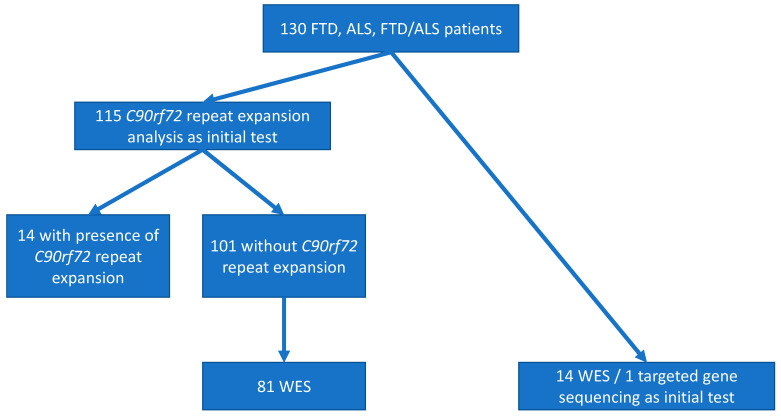
Figure 1.
Flow chart of the genetic characterization of FTD and/or ALS patients in the cohort we analyzed. Most (115 out of the 130) patients were analyzed by C9orf72 repeat expansion analysis as the initial test. Subsequently, 81 of the 101 C9orf72 repeat expansion negative patients were analyzed by WES (whole exome sequencing). For 14 patients, WES was chosen as the initial test, and for 1 (with a pathogenic variant in the VCP gene), targeted gene sequencing was performed initially due to clinical picture and family history highly suggestive of inclusion body myopathy, Paget’s disease and FTD [23].
2.5. Gene Variant Identification and Verification
We specifically searched for variants in the most recognized FTD and ALS related genes, namely C9orf72, GRN, MAPT, TARDBP, FUS, CHMP2B, SQSTM1, UBQLN2, VCP, OPTN, TBK1, SOD1 and CHCHD10, as well as the 3 genes associated with autosomal dominant AD (APP, PSEN1, PSEN2). Single nucleotide polymorphism (SNP) variants reported here were verified by Sanger sequencing to exclude false positive results.
2.6. Measurements of CSF and Plasma Biomarkers
The CSF levels of Aβ42, τT and τP-181 were measured in duplicate by commercially available ELISA kits (Innotest β-amyloid 1–42, hTau antigen and phospho-tau 181; Fujirebio, Gent, Belgium) according to the manufacturer’s instructions. We chose to measure τP-181, as in neurodegenerative disorder research and clinical practice, tau phosphorylated at threonine 181 (τP-181) is the most commonly measured form of phosphorylated tau as a biomarker in the CSF [24] and, recently, in plasma [25]. Additionally, to better evaluate the τP-181 levels, the ratio of τP-181 to τT (τP-181/τT) was calculated [26]. TDP-43 was measured in triplicate by double-sandwich enzyme-linked immunosorbent assay (ELISA) using a commercial kit (Human TAR DNA-binding protein 43 ELISA kit; Cusabio Biotech, Wuhan, China). All determinations were performed using a four-parameter logistic curve and blindly to the clinical diagnosis. Cut-off levels were calculated by ROC (receiver operating characteristic) analysis with the optimal combination of sensitivity and specificity, as previously described [15,16].
Blood samples were collected between 08:00–10:00 a.m. (morning samples), transferred to EDTA-containing tubes and refrigerated until centrifugation (within 3 h) for the plasma isolation. Plasma isolated from these samples was subsequently kept in deep freeze (−80 °C). Plasma progranulin levels were measured using the Human Progranulin ELISA Kit (Adipogen Life Sciences, Liestal, Switzerland) [27].
2.7. Statistical Analysis
All numerical data were tested for normality and homogeneity of variances by the Shapiro–Wilk’s and Brown–Forsyth tests, respectively. When appropriate, differences among groups were tested by one-way analysis of variance (one-way ANOVA) or two-way analysis of covariance (two-way ANCOVA), followed by Bonferroni correction for multiple comparisons. When deviations from normality and/or heterogeneity of variances were noted, Kruskal–Wallis test was performed, followed by Dunn’s post-hoc test. Categorical data were compared among groups by the χ2-test.
3. Results
3.1. Demographic Data
Demographic data are summarized in Table 1. The three patient groups (FTD, ALS, FTD-ALS) had comparable age and sex. Due to violations regarding normality and homogeneity of variances, disease duration was initially compared among groups with Kruskal–Wallis test that showed a significant difference (p < 0.0001). Additionally, Dunn’s post-hoc test revealed significantly longer disease duration for FTD as compared to ALS patients (p < 0.0001).
Table 1.
Demographic and clinical data of the studied groups.
| FTD | ALS | FTD-ALS | p-Value | |
|---|---|---|---|---|
| n (m/f) | 56 (32/24) | 58 (26/32) | 16 (7/9) | NS † |
| Age (y) | 60.2 ± 10.8 | 61.2 ± 11.8 | 60.7 ± 10.7 | NS ‡ |
| Disease Duration (y) | 3.0 (1.3–6.0) a | 1.0 (0.7–2.0) | 3.0 (1.0–4.0) | <0.001 § |
| Family History, 1st degree relative (%) |
16 (28.6) | 9 (15.5) | 4 (25.0) | NS † |
Data are presented as mean values ± standard deviation (SD) for age, or median values (25th–75th percentile) for disease duration. m: males; f: females; y: years; NS: non-significant. † χ2-test, ‡ 1-way ANOVA, § 2-way ANCOVA, a Bonferroni corrected p < 0.001 vs. ALS and 0.066 vs. FTD-ALS.
3.2. Family History
Among the 130 patients within the FTD-ALS spectrum, 28.6% (16/56) of the FTD patients had at least one first degree relative affected with phenotype suggesting either FTD or ALS. For the ALS and FTD-ALS groups, 15.5% (9/58) and 25.0% (4/16), respectively, had positive family history for possible FTD or ALS, with at least one first degree relative affected (Table 1).
3.3. Pathogenic and Likely Pathogenic Variants in FTD-ALS Genes
3.3.1. C9orf72 Repeat Expansion
The percentage of patients positive for C9orf72 repeat expansion (defined as presence of >30 repeats) was 10.4% in patients with FTD (5 out of 48 patients tested), 10.7% in patients with ALS (6/56) and 27.3% in patients with FTD-ALS (3/11). All C9orf72 repeat expansion positive patients in the FTD-ALS group had family history of possible FTD/ALS and presented with a bvFTD phenotype at onset. The clinical and other characteristics of the 14 patients harboring a C9orf72 repeat expansion are shown in Table 2.
Table 2.
Patients with C9orf72 hexanucleotide repeat expansion.
| Patient ID | Sex | Phenotype | Age at Onset | Age at Diagnosis | Family History | MRI | HMPAO-SPECT |
|---|---|---|---|---|---|---|---|
| 1 | F | FTD | 61 | 63 | Sister and 3 cousins ALS |
Mild frontal atrophy and left sylvius and temporal pole. | NA |
| 2 | F | bvFTD | 69 | 70 | Mother with dementia | Mild frontal, temporal (L > R) and parietal atrophy | Frontal hypoperfusion (R > L) and right parietal |
| 3 | F | FTD-psychiatric symptoms | 45 | 54 | Mother FTD-ALS Maternal uncle ALS and aunt dementia |
Mildfrontal atrophy and white matter lesions | Frontal hypoperfusion (L > R) |
| 4 | M | bvFTD | 51 | 58 | Mother ALS |
Bilateral frontal strokes and frontal atrophy | NA |
| 5 | M | bvFTD with psychiatric symptoms | 38 | 41 | No | Frontal, temporal and parietal atrophy | NA |
| 6 | M | ALS | 71 | 72 | No | Mild global atrophy | NA |
| 7 | F | ALS | 59 | 61 | No | Frontal and parietal atrophy | NA |
| 8 | F | ALS | 63 | 64 | Sister ALS |
NA | NA |
| 9 | F | ALS | 42 | 43 | Mother and 2 maternal aunts with ALS | NA | NA |
| 10 | M | FTD-ALS | 48 | 50 | Father with FTD | Diffuse atrophy, temporal > frontal | NA |
| 11 | F | FTD-ALS | 43 | 44 | Grandmother and 7/9 uncles with ALS; mother with dementia |
Frontal, perisylvian atrophy (L > R), mild increase in signal intensity along the corticospinal tract | Diffuse frontal, temporal and parietal hypoperfusion |
| 12 | F | ALS | 56 | 58 | Mother with ALS; maternal cousin with ALS and C9orf72 (+) | Mild ischemic microangiopathy | NA |
| 13 | M | FTD-ALS | 44 | 45 | Mother and 2 maternal uncles with motor disorder | Frontotemporal atrophy | NA (DATSCAN+) |
| 14 | F | ALS | 41 | 43 | Father with dementia; paternal aunt with ALS | Midline cerebellar dysplasia | NA |
ALS: amyotrophic lateral sclerosis; FTD: frontotemporal dementia; bv: behavioral variant; NA: non-available.
3.3.2. Other Causative Variants
From the analysis of the FTD/ALS genes in the WES derived data, causative variants were identified in the TARDBP, GRN, VCP, SOD1 and FUS genes in 11 patients (Table 3). Specifically, in three patients (two with pure ALS and one with FTD-ALS) we found causative variants in the TARDBP gene, namely the p.Met337Val (c.1009A>G), the p.Asn352Ser (c.1055A>G) and the p.Ile383Val (c.1147A>G) variant.
Table 3.
Patients with causative variants other than the C9orf72 repeat expansion.
| Patient ID | Sex | Phenotype | Age at Onset | Age at Diagnosis | Family History | Brain MRI | Gene | Transcript | Variant | gnomAD Frequency (%) | rs | CADD Score | MaxEnt-Scan (Splice Site Loss) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | ALS (bulbar onset) | 67 | 69 | 2 brothers; sister; father with ALS | Unremarkable | TARDBP | NM_007375.4 | p.Met337Val (c.1009A>G) | ≤0.001 | 80356730 | 22.4 | - |
| 2 | F | ALS | 63 | 64 | No | Unremarkable | p.Asn352Ser (c.1055A>G) | 0.000 | 80356734 | 18.6 | - | ||
| 3 | M | FTD-ALS | 57 | 60 | No | Frontal, temporal atrophy | p.Ile383Val (c.1147A>G) | 0.002 | 80356740 | 17.2 | - | ||
| 4 | F | PPA | 60.3 | 61 | No | Frontal, temporal atrophy (L > R) | GRN | NM_002087.4 | c.463-2A>G | 0.000 | - | 33.0 | From 3.77 to −4.19 |
| 5 | M | PPA | 50 | 60 | Yes (grandmother) |
Frontal, temporal, parietal atrophy (L > R) | c.934-1G>A | 0.000 | - | 34.0 | From 9.63 to 0.88 | ||
| 6 | F | PPA | 61 | 62 | No | Perisylvian atrophy (L > R) | p.Cys482Tyr (c.1445G>A) | 0.000 | - | 30.0 | - | ||
| 7 | M | IBM/FTD | 47 | 63 | Yes (brother ALS; brother bvFTD) | Frontal lobe atrophy | VCP | NM_007126.5 | p.Arg159His (c.476G>A) | ≤0.001 | 121909335 | 23.2 | - |
| 8 | M | IBM/FTD | 58 | 68 | Yes (mother ALS) | - | |||||||
| 9 | M | IBM/FTD/PaD | 43 | 53 | No | - | p.Arg155His (c.464G>A) | 0.000 | 121909329 | 24.6 | - | ||
| 10 | M | ALS | 79 | 81 | No | Unremarkable | FUS | NM_001170634.1 | p.Gly506Val (c.1517G>T) | 0.000 | - | 23.7 | - |
| 11 | M | ALS | 36 | 37 | Yes (brother ALS) | Unremarkable | SOD1 | NM_000454.5 | p.Ser106Leu (c.317C>T) | ~0.000 | 1378590183 | 22.4 | - |
ALS: amyotrophic lateral sclerosis; FTD: frontotemporal dementia; IBM: inclusion body myositis; PaD: Paget’s disease.
In three patients with FTD, all with the PPA phenotype, we found three causative variants in the GRN gene (Table 3). The two patients with splice site GRN variants (c.463-2A>G and c.934-1G>A) presented with an svPPA and nfaPPA phenotype, respectively. Both had increased CSF τT and TDP-43 levels and τP-181 and Aβ42 levels within the reference range for healthy controls. Another FTD-PPA patient was detected with the p.Cys482Tyr GRN variant. She had increased CSF levels of τT and τP-181, while the CSF levels of TDP-43 and Aβ42 were within the reference range for healthy controls.
Additionally, in three patients with FTD and Inclusion Body Myopathy (IBM), all from Crete, we found two causative variants in the VCP gene (Table 3). A 63-year-old male patient with the pathogenic p.Arg159His (c.476G>A) heterozygous missense VCP gene variant was identified with the characteristic clinical picture of ΙΒΜ and FTD [23]. The same VCP variant (p.Arg159His) was found in a 68-year-old patient with progressive muscle weakness and atrophies in all four extremities and the trunk and behavioral disturbances. The symptoms of the patient had started at the age of 58 years. Another patient was found to have myopathy, persistently increased Creatine PhosphoKinase (CPK), language deficits and Paget’s disease of bone. He carried the p.Arg155His (c.464G>A) VCP change (Table 3).
Further, we found ALS causative variants in the FUS and the SOD1 genes (Table 3). Specifically, we detected the p.Gly506Val (c.1517G>T) FUS gene variant in a 82-year-old male with a predominately lower motor neuron involvement and no family history. Moreover, we found the p.Ser106Leu (c.317C>T) SOD1 gene variant in a 37-year old patient with personal and family history of ALS.
3.4. Association of Variants in FTD -ALS Genes with CSF Biomarkers
As a next step in our analyses, we proceeded in measuring CSF biomarker levels in this patient cohort and associated these levels with the results of C9orf72 repeat expansion determination and WES based genotyping. Our efforts focused on typical neurodegenerative disease-associated CSF biomarkers (TDP-43, Aβ42, τP-181 and τT) and on genes that either have as their protein product the above biomarkers (TARDBP, APP, MAPT) or are commonly associated with FTD/ALS (C9orf72). Of these four genes, no genotype/CSF biomarker associations were possible for the TARDBP gene since very few patients with variants in this gene were found.
3.4.1. Association of C9orf72 Variants with CSF Biomarkers
Regarding the C9orf72 gene, in the cohort analyzed, besides the pathogenic hexanucleotide repeat expansion (see above), we found five rare intronic variants (c.444+31T>G, c.600+86A>T, c.666-120C>T, c.855+52A>G, c.856-37G>T) and the p.Asn207Ser (p.N207S) exonic variant. These C9orf72 variants differ in their location and nature (repeat expansion, intronic and exonic) and thus possibly have differential effects on gene expression and function. For this reason, these three types of genetic variants were analyzed separately. Thus, for this part of our study, we compared Aβ42, τT, τp-181 and TDP-43 CSF levels and the τP-181/τT ratio in FTD and/or ALS patients carrying the hexanucleotide C9orf72 repeat expansion, rare intronic C9orf72 variants or the p.Asn207Ser (p.N207S) exonic C9orf72 variant to patients without these variants. Analyses were performed separately for FTD patients, ALS patients ± FTD and all patients combined.
These analyses revealed that in patients with ALS (with or without concurrent FTD), the CSF levels of τP-181 were lower in C9orf72 repeat expansion carriers compared to non-carriers, whereas levels of TDP-43, Aβ42 and τT and the τP-181/τT ratio did not differ between the two groups (Figure 2). Additionally, carriers of rare intronic variants in C9orf72 had lower CSF levels of τT (Figure 3). In contrast, this difference in τT levels was not found for the more common C9orf72 p.Asn207Ser (p.N207S) variant (Figure 3). Results on C9orf72-based biomarker level comparisons that did not reveal statistically significant results or results not approaching statistical significance are not shown.
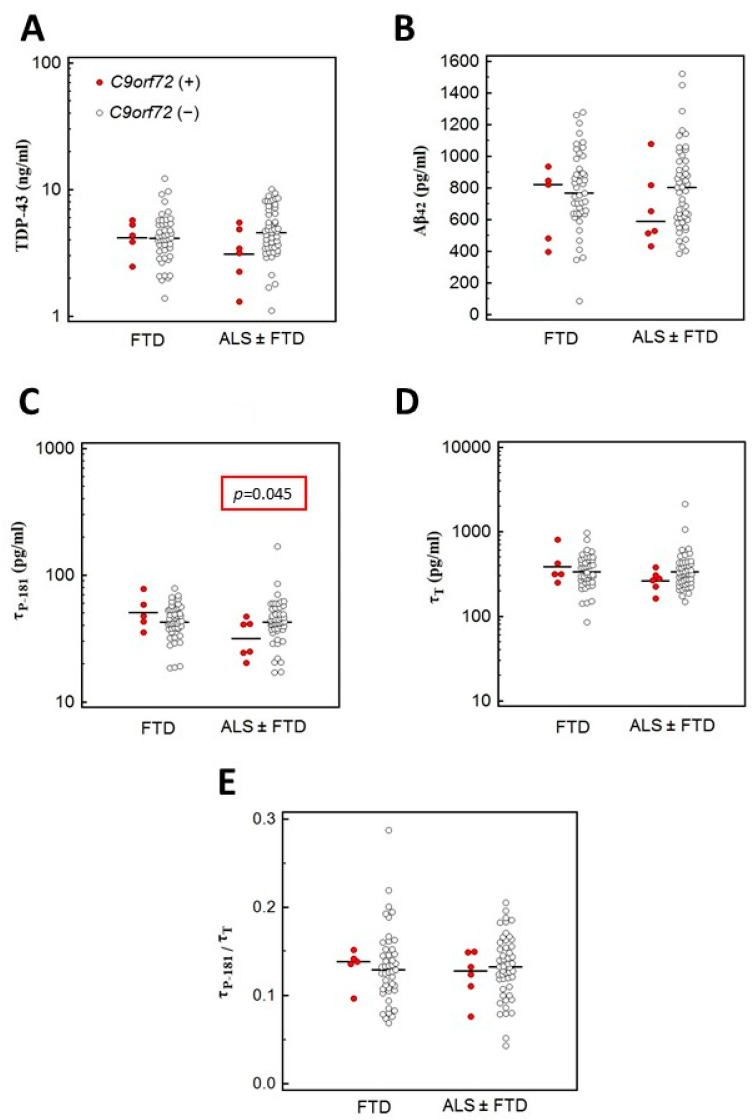
Figure 2.
Comparison of TDP-43 (A); Aβ-42 (B); τP-181 (C) and τT (D) levels and τP-181/τT ratio (E) in C9orf72 hexanucleotide repeat expansion carriers (C9orf72+) and non-carriers (C9orf72−). Analyses were performed separately for FTD patients and ALS (with or without FTD) patients. In patients with ALS (with or without concurrent FTD), levels of τP-181 were lower in C9orf72 carriers compared to non-carriers (C). All other comparisons yielded non-significant results.
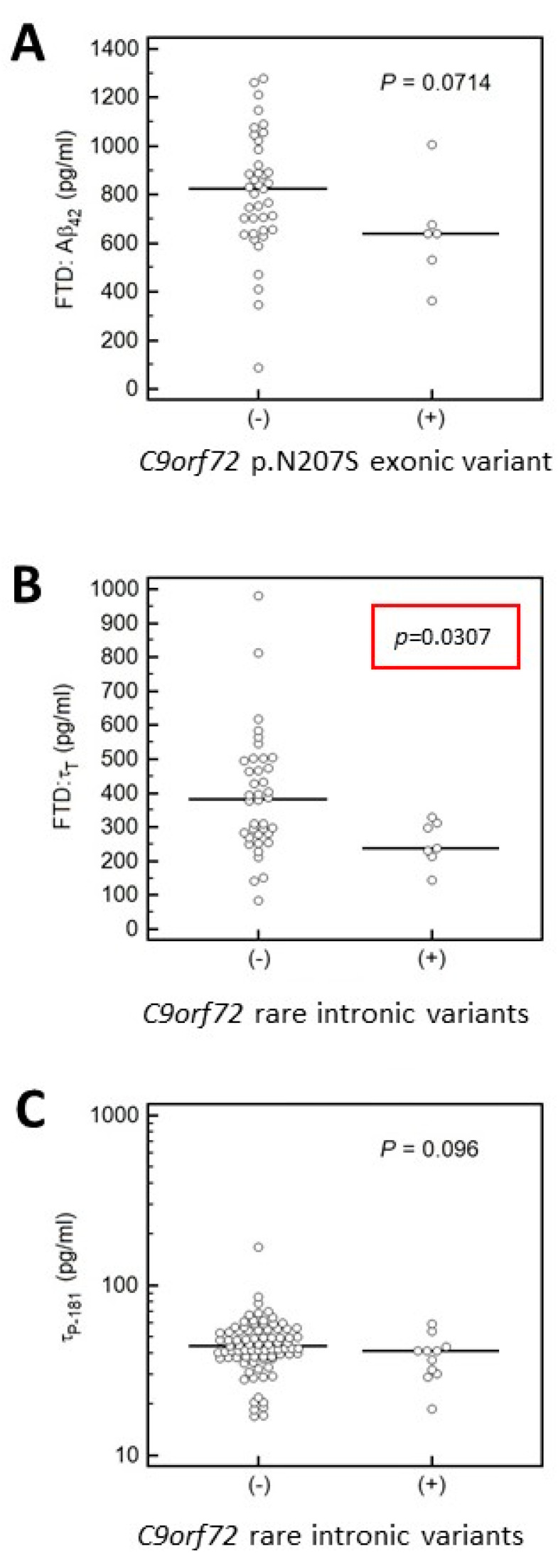
Figure 3.
Aβ42, τT and τp-181 CSF levels in FTD, ALS and FTD/ALS patients carrying either the p.Asn207Ser (p.N207S) exonic C9orf72 variant (A) or the rare intronic C9orf72 variants (B,C). (A) Here we compared Aβ42 CSF levels in FTD patients carrying the p.Asn207Ser (p.N207S) exonic C9orf72 variant to patients without this variant and no statistically significant difference was found. (B) FTD patients that were carriers of rare intronic variants in the C9orf72 gene had lower CSF levels of τT. (C) CSF levels of τp-181 did not differ in FTD and/or ALS patients carrying rare C9orf72 intronic variants.
3.4.2. Association of MAPT and APP Gene Variants with CSF Biomarkers
When we compared levels of CSF biomarkers between groups of patients carrying different MAPT and APP gene variants, we found that the c.855C>T (p.Asp285Asp/p.D285D) synonymous MAPT variant carriers had significantly lower τp-181 (Figure 4). There was no difference found for the other three biomarkers tested (TDP-43, Aβ42 and τT), when comparing carriers of the p.Asp285Asp (p.D285D) change with non-carriers (data not shown). Additionally, all other MAPT variants similarly tested did not show an effect on biomarker levels (data not shown). Additionally, as shown in Figure 4, carriers of rare APP variants (p.Ala11Ala, p.Val118Ile, c.-44C>T/c.-49+321C>T, c.697+50G>A and c.697+11888delT) had lower Aβ42 levels, whereas this was not found for the more common c.1000-31T>C intronic APP variant. Finally, other biomarkers (τT, τp-181, TDP-43) analyzed relative to APP variants did not yield significant results (data not shown).
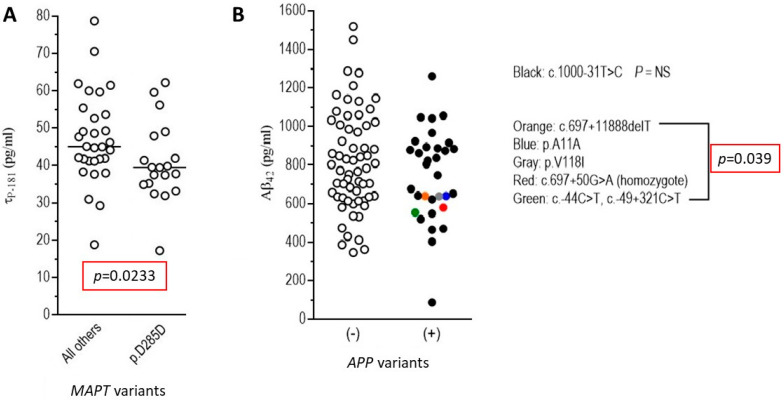
Figure 4.
Association of MAPT and APP gene variants with CSF biomarkers (Aβ42, τT, τp-181, TDP-43) in the entire cohort of FTD and/or ALS patients. (A) As shown here, the c.855C>T (p.Asp285Asp/p.D285D) MAPT synonymous variant carriers had significantly lower τp-181 levels compared to non-carriers; (B) carriers of rare APP variants (p.Ala11Ala/p.A11A, p.Val118Ile/p.V118I, c.-44C>T/c.-49+321C>T, c.697+50G>A and c.697+11888delT) had lower Aβ42 levels. These lower Aβ42 levels were not found for the more common intronic variant c.1000-31T>C.
3.4.3. Correlation of CSF Biomarkers Values
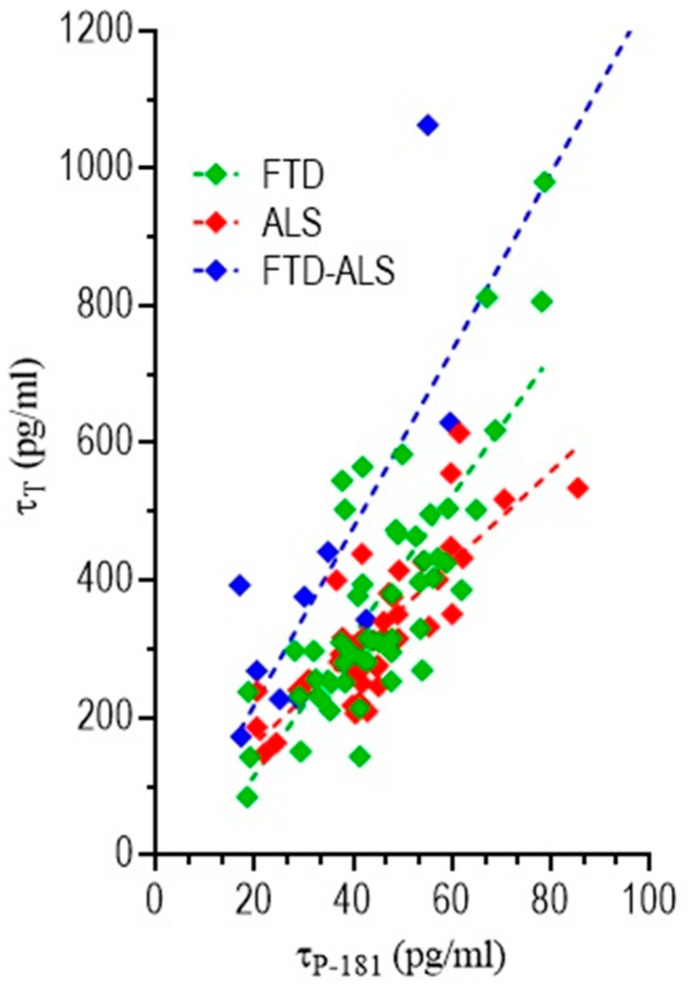
As a next step in our analyses, we performed correlation analyses of CSF biomarker values (Aβ42, τT, τp-181, TDP-43) in pairs across different patient groups. Results of these analyses showed that only τT and τp-181 CSF values were correlated, across all three patient groups (Figure 5). For these analyses, Spearman ρ values were 0.74 (p < 0.0001) for FTD patients, 0.78 (p < 0.0001) for ALS patients and 0.62 (p = 0.031) for FTD/ALS patients. Correlation between other CSF biomarker measurements pairs (τP-181 vs. Aβ42, Aβ42 vs. TDP-43 and TDP-43 vs. τP-181) did not yield statistically significant results (data not shown).
Figure 5.
Correlation of τT and τp-181 CSF values in different patient groups. Results of these analyses showed that τT and τp-181 CSF values were correlated, across all patient groups, with Spearman ρ values being 0.74 (p < 0.0001) for FTD patients, 0.78 (p < 0.0001) for ALS patients and 0.62 (p = 0.031) for FTD/ALS patients.
3.5. Association of Pathogenic Variants in the GRN Gene with Plasma Progranulin Levels
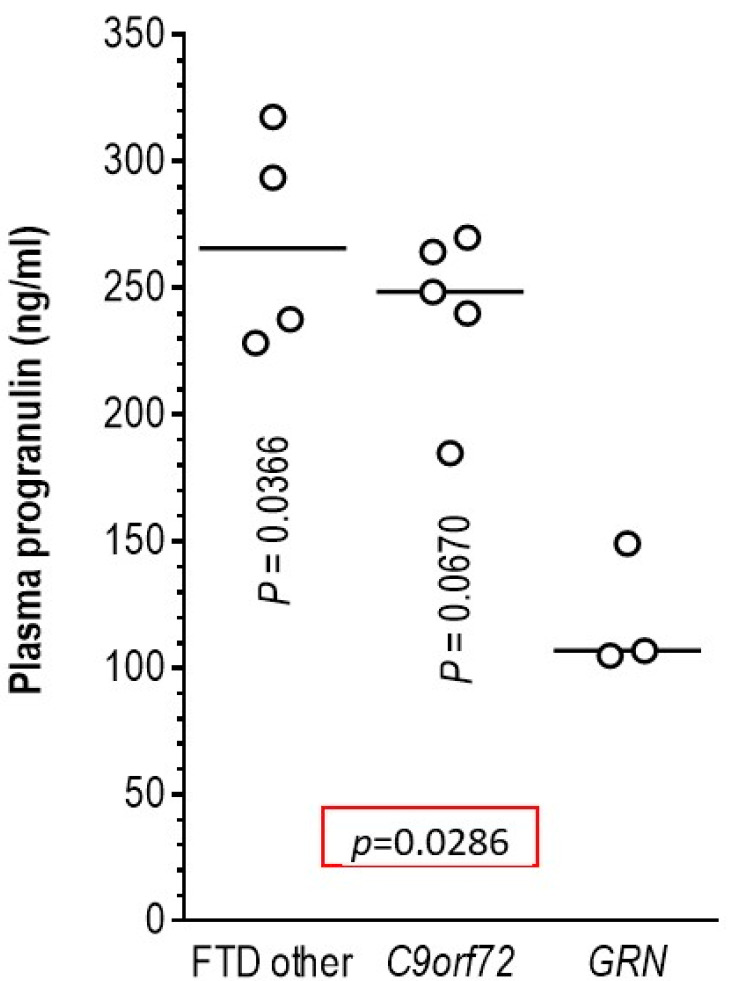
To show that our findings for CSF biomarkers (i.e., that genetic variants affect CSF biomarker levels) are valid also for plasma biomarkers, we performed plasma progranulin quantification in the three patients with the GRN variants, c.463-2A>G, c.934-1G>A and p.Cys482Tyr (c.1445G>A), that were considered causative based on strong in silico predictions. Patients carrying the pathogenic GRN variants had more than two-fold lower plasma progranulin levels compared to FTD patients harboring the C9orf72 repeat expansion and to other FTD patients (Figure 6). These findings verified the pathogenicity of the three novel GRN gene variants.
Figure 6.
Association of GRN pathogenic variants with plasma progranulin levels. Lower plasma progranulin levels were found in the 3 FTD patients with the GRN variants (c.463-2A>G, c.934-1G>A and p.Cys482Tyr) compared to FTD patients harboring the C9orf72 repeat expansion and to other FTD patients.
4. Discussion
Our study presents a clinical series of Greek FTD/ALS spectrum patients by innovatively integrating phenotypic, genotypic and biomarker data. Causative genetic variants (either SNPs or C9orf72 hexanucleotide repeat expansions) were identified in 25 cases (19.2% of the total patient cohort). The frequency of patients carrying causative FTD/ALS gene variants in our series is higher compared to other similar series from Greece, most of them having evaluated either FTD or ALS cohorts separately [28,29,30]. This increased frequency probably reflects a higher proportion of FTD/ALS patients in our study, as well as the exclusion of patients with probable AD pathology through CSF biomarker measurements. Additionally, we have observed several unique associations between genotype and CSF/plasma biomarker levels, with our study being one of the first studies to adopt such an approach.
The most common genetic change detected in the cohort analyzed was, as expected, the C9orf72 repeat expansion (12.2%; 14 out of 115 patients tested). C9orf72 repeat expansions have been reported to 4–40% of patients with ALS [31]. In previously reported Greek ALS cohorts, 5–10% of cases carried the abnormal C9orf72 expansion [28,32], in accordance with our findings. Regarding the FTD group, we found a rate of C9orf72 repeat expansions (10.4%), similar to one of the studies from Greece [29], but almost two-fold higher compared to another Greek study [30]. The latter can be explained by the biomarker supported clinical diagnosis of FTD patients in our study, avoiding the contamination of this group with other pathologies presenting as a frontal-behavioral or language syndrome.
Additionally, using a WES based approach, we identified causative gene variants in 11 more patients: three variants in each of the TARDBP and GRN genes, two variants in the VCP gene (in three patients) and one variant in each of the SOD1 and FUS genes (Table 3). Variants in these genes are commonly described in FTD/ALS cohorts, including the most recent studies [33].
In detail, we detected three patients (two women with ALS and one man with FTD-ALS) with TARDBP pathogenic variants (p.Met337Val, p.Asn352Ser and p.Ile383Val). This is of special importance as TARDBP encodes TDP-43, a protein which is increasingly implicated in neurodegenerative processes and offers an attractive therapeutic target [34]. Pathogenic TARDBP gene variants are considered the cause of 1–7% of familial ALS cases and of several sporadic ALS cases [35,36], but they are less frequently reported in FTD [3,37]. Our 60-year old patient with the p.Ile383Val TARDBP gene variant presented with an FTD-ALS phenotype. This specific variant has been repeatedly described as pathogenic across the FTD/ALS spectrum [33,38,39,40,41]. Additionally, three FTD patients of Greek origin harboring the p.Ile383Val TARDBP gene variant have been recently reported [30]. These data are a call for a systematic study of this variant’s geographic distribution and significance. Furthermore, the c.1009A>G (p.Met337Val) TARDBP variant found in one of our patients with familial ALS has been repeatedly reported to segregate with ALS [42,43,44,45,46], including bulbar onset ALS [47]. The p.Asn352Ser TARDBP variant that we found in another ALS patient has also been often described in the literature [48,49,50] and functionally characterized [51,52].
In three of our patients, we found causative variants in the GRN gene (c.463-2A>G, c.934-1G>A and p.Cys482Tyr), all novel (not previously reported) but with strong evidence in favor of their pathogenicity. Variants in the GRN gene have been emerging as a frequent cause of FTD, with marked phenotypic heterogeneity [53,54,55] but most frequently associated with TDP-43 histopathology and PPA phenotype [56,57,58]. Our data add further support to the association of GRN variants with the PPA phenotype. The two splice site variants (c.463-2A>G, c.934-1G>A) described here have strong in silico evidence that they disrupt splicing, as indicated by their MaxEntScan and CADD scores (Table 3). Additionally, in the GRN gene, this type of variant (splice-site variants) is frequently described as causing FTD. In another FTD-PPA patient, we detected the p.Cys482Tyr GRN variant, which is essentially absent from public variant databases such as gnomAD and has strong computational evidence (CADD score = 32.0) in favor of its pathogenicity (Table 3). All three patients in the cohort we analyzed harboring causative GRN variants showed decreased plasma progranulin levels (Figure 6). This is additional evidence of the pathogenicity of these variants, as it has been shown for other GRN variants [27,59].
Three male patients, all originating from Crete and all presenting with the IBM/FTD phenotype, were found to harbor heterozygous pathogenic VCP variants. These VCP variants were the p.Arg155His (c.464G>A) and, in two apparently unrelated patients, the p.Arg159His (c.476G>A) variant. Pathogenic VCP gene variants are associated with the characteristic clinical picture of IBM, FTD and ALS, often with Paget’s disease of bone. This syndrome can be partially or fully developed depending on the respective causative VCP variant and other factors [60,61]. The p.Arg155His VCP variant has been repeatedly shown to co-segregate with the disease phenotype in multiple affected family members [61,62,63]. This variant is in a mutational hot spot and there are in vitro and in vivo functional studies and strong computational evidence (CADD = 24.6) supportive of its damaging effect. This p.Arg155His VCP missense change occurs at an amino acid residue where different missense changes (p.Arg155Pro, p.Arg155Leu, p.Arg155Cys, p.Arg155Ser) are known to be pathogenic [63]. Regarding the p.Arg159His variant, there are several reports describing patients with this variant in the literature [63], including the clinical description of one of our patients [23]. This variant is not present in population databases such as gnomAD, it has strong in silico evidence of pathogenicity (CADD = 23.2) and affects a conserved arginine residue (Table 3). This amino acid change has been shown to disrupt the function and structure of VCP and can lead to protein aggregates [64,65,66,67].
In addition, we detected the p.Gly506Val (c.1517G>T) FUS gene variant in a patient with ALS. The FUS protein is a widely expressed DNA/RNA-binding protein with functional homology to TDP-43 [68]. It is involved in transcriptional and translational regulation and in mRNA splicing and transport [69]. FUS gene pathogenic variants cause 5% of familial and 1% of sporadic ALS, with predominant lower motor involvement [68,70,71]. The p.Gly506Val FUS gene variant is not present in public databases, such as gnomAD, and there is computational evidence in support of its pathogenicity (CADD = 23.7). Additionally, there are reports on a different amino acid substitution (as p.Gly507Asp) at the same amino acid residue causing ALS [68,70]. Finally, in a 37-year old male with family history of ALS, we identified the p.Ser106Leu SOD1 variant. This variant has already been described in young ALS patients with rather slow disease progression, as was the case in our patient [72,73,74].
There have been several studies on the CSF levels of TDP-43, Aβ42, τT, τP-181 in patients with FTD and ALS [15,75], but very few have focused on the association of these levels with the genotype of these patients. Here we found several interesting genotype/CSF biomarker associations. Specifically, we found that levels of τp-181 were lower in C9orf72 repeat expansion and MAPT c.855C>T (p.Asp285Asp) carriers compared to non-carriers. Additionally, carriers of rare C9orf72 and APP variants had lower levels of τT and Aβ42, respectively.
Regarding C9orf72, it has been reported that 25% of patients with C9orf72 repeat expansion show low Aβ42 levels in the CSF [76], but the effect of this expansion on tau and phospho-tau levels is not clear [77]. Our finding that C9orf72 changes (either the pathogenic repeat expansion or the rare intronic variants) can affect levels of τP-181 and τT, indicates that there is an association between C9orf72 and tau species that needs further assessment.
The effect of the MAPT c.855C>T (p.Asp285Asp) change on levels of τp-181 in the FTD/ALS cohort we studied suggests that this variant could be an indirect modifier of tau phosphorylation (e.g., by affecting mRNA processing) and thus of pathophysiological significance. This is of interest, since the same effect, increased levels of τp-181, has been found for several MAPT variants in patients with AD [78]. Our findings extend the observations about this effect on another neurodegenerative disorder beyond AD.
Finally, the effect of rare APP variants on Aβ42 levels suggests these variants are possibly important for APP processing by secretases, as it has already been shown for known pathogenic and non-pathogenic APP variants in AD and other neurodegenerative disorders [79,80,81].
Strengths and Limitations
The main strength of our work relates to the extent and depth of the genotypic and phenotypic analysis, that included both WES-based genotyping and CSF/plasma biomarker measurements.
One limitation to our work is the rather small cohort size; however, this is counterbalanced by the depth of the genotypic and phenotypic characterization. In addition to the plasma and CSF biomarkers used in our study, there is increasing interest in the possible role of other emerging biomarkers in ALS and FTD, including neurofilaments both in CSF [82,83] and plasma [84,85]. This is of special importance for patients with the FTD/ALS phenotype, given that there is evidence that elevated Neurofilament Heavy chain (NfH) levels could antedate the onset of ALS in FTD patients [83]. Our study was not designed to test the levels of these biomarkers in the plasma and CSF; however, future analyses are panned to include these important biomarkers.
5. Conclusions
In conclusion, we found causative genetic variants (in the C9orf72, TARDBP, GRN, VCP, FUS and SOD1 genes) with a high frequency (19.2%) in a cohort of Greek FTD/ALS spectrum patients. In addition, we observed several potentially important associations between the plasma/CSF biomarker levels and the genotype of several genes. Specifically, we found lower levels of τp-181 in C9orf72 repeat expansion and MAPT c.855C>T (p.Asp285Asp) carriers compared to non-carriers. Additionally, carriers of rare C9orf72 and APP variants had lower levels of τT and Aβ42, respectively. Finally, plasma progranulin levels were decreased in patients with the newly described pathogenic GRN variants.
Our results further expand the genotypic/phenotypic spectrum of FTD/ALS and provide insights in the interaction of the genome with plasma and CSF biomarkers. Taken together, our findings call for an integrated individualized approach in interpreting plasma and CSF biomarker levels, as these can be influenced by the genotype of the patient under study.
Acknowledgments
We would like to thank the patients and their caregivers for their participation in the study.
Author Contributions
Conceptualization, G.P.P., E.K. and I.Z.; methodology, M.B., G.P.P., L.M., G.K. (Georgia Karadima), G.K. (Georgios Koutsis), C.K. ( Christos Kroupis), E.K. and I.Z.; formal analysis, data curation, visualization, M.B., G.P.P., L.M., E.K. and I.Z.; investigation, M.B., G.P.P., M.R., L.M., V.Z., A.B., A.T., V.K.K., V.C., S.Z., M.T., G.P., G.K. (Georgia Karadima), G.K. (Georgios Koutsis), C.K. (Christos Kroupis), C.K. (Chrisoula Kartanou), E.K. and I.Z.; resources, G.P.P., E.K. and I.Z.; writing—original draft preparation, review and editing, M.B., G.P.P., L.M., E.K. and I.Z.; supervision, G.P.P., E.K. and I.Z.; project administration, M.B., L.M.; funding acquisition, E.K. and I.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the “Lilian Voudouri Foundation”.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the University Hospital of Heraklion, Crete (protocol code 5621, 17 June 2015) and the Ethics Committee of the University of Crete (protocol code 22, 22 June 2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weishaupt J.H., Hyman T., Dikic I. Common Molecular Pathways in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Trends Mol. Med. 2016;22:769–783. doi: 10.1016/j.molmed.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Hinz F.I., Geschwind D.H. Molecular Genetics of Neurodegenerative Dementias. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blauwendraat C., Wilke C., Simón-Sánchez J., Jansen I.E., Reifschneider A., Capell A., Haass C., Castillo-Lizardo M., Biskup S., Maetzler W., et al. The wide genetic landscape of clinical frontotemporal dementia: Systematic combined sequencing of 121 consecutive subjects. Genet. Med. 2018;20:240–249. doi: 10.1038/gim.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman J.S., Van Deerlin V.M. Alzheimer’s Disease and Frontotemporal Dementia: The Current State of Genetics and Genetic Testing Since the Advent of Next-Generation Sequencing. Mol. Diagn. Ther. 2018;22:505–513. doi: 10.1007/s40291-018-0347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor J.P., Brown R.H., Cleveland D.W. Decoding ALS: From genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrell J.R., Halliday G.M., Kril J.J., Ittner L.M., Götz J., Kiernan M.C., Hodges J.R. The frontotemporal dementia-motor neuron disease continuum. Lancet. 2016;388:919–931. doi: 10.1016/S0140-6736(16)00737-6. [DOI] [PubMed] [Google Scholar]
- 7.Balendra R., Isaacs A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin S.C., Pastor P., Cooper B., Cervantes S., Benitez B.A., Razquin C., Goate A., Cruchaga C., Ibero-American Alzheimer Disease Genetics Group R. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer’s disease Ibero-American cohort. Alzheimers Res. Ther. 2012;4:34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guven G., Lohmann E., Bras J., Gibbs J.R., Gurvit H., Bilgic B., Hanagasi H., Rizzu P., Heutink P., Emre M., et al. Mutation Frequency of the Major Frontotemporal Dementia Genes, MAPT, GRN and C9ORF72 in a Turkish Cohort of Dementia Patients. PLoS ONE. 2016;11:e0162592. doi: 10.1371/journal.pone.0162592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastor P., Moreno F., Clarimón J., Ruiz A., Combarros O., Calero M., de Munain A.L., Bullido M.J., de Pancorbo M.M., Carro E., et al. MAPT H1 Haplotype is Associated with Late-Onset Alzheimer’s Disease Risk in APOE ε4 Noncarriers: Results from the Dementia Genetics Spanish Consortium. J. Alzheimers Dis. 2016;49:343–352. doi: 10.3233/JAD-150555. [DOI] [PubMed] [Google Scholar]
- 11.Huang W., Zhou Y., Tu L., Ba Z., Huang J., Huang N., Luo Y. TDP-43: From Alzheimer’s Disease to Limbic-Predominant Age-Related TDP-43 Encephalopathy. Front. Mol. Neurosci. 2020;13 doi: 10.3389/fnmol.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson P.T., Dickson D.W., Trojanowski J.Q., Jack C.R., Boyle P.A., Arfanakis K., Rademakers R., Alafuzoff I., Attems J., Brayne C., et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): Consensus working group report. Brain. 2019;142:1503–1527. doi: 10.1093/brain/awz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois B., Feldman H.H., Jacova C., DeKosky S.T., Barberger-Gateau P., Cummings J., Delacourte A., Galasko D., Gauthier S., Jicha G., et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS–ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 14.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourbouli M., Rentzos M., Bougea A., Zouvelou V., Constantinides V.C., Zaganas I., Evdokimidis I., Kapaki E., Paraskevas G.P. Cerebrospinal Fluid TAR DNA-Binding Protein 43 Combined with Tau Proteins as a Candidate Biomarker for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Spectrum Disorders. Dement. Geriatr. Cogn. Disord. 2017;44:144–152. doi: 10.1159/000478979. [DOI] [PubMed] [Google Scholar]
- 16.Paraskevas G.P., Kasselimis D., Kourtidou E., Constantinides V., Bougea A., Potagas C., Evdokimidis I., Kapaki E. Cerebrospinal Fluid Biomarkers as a Diagnostic Tool of the Underlying Pathology of Primary Progressive Aphasia. J. Alzheimers Dis. 2017;55:1453–1461. doi: 10.3233/JAD-160494. [DOI] [PubMed] [Google Scholar]
- 17.Rascovsky K., Hodges J.R., Knopman D., Mendez M.F., Kramer J.H., Neuhaus J., van Swieten J.C., Seelaar H., Dopper E.G.P., Onyike C.U., et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorno-Tempini M.L., Hillis A.E., Weintraub S., Kertesz A., Mendez M., Cappa S.F., Ogar J.M., Rohrer J.D., Black S., Boeve B.F., et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Carvalho M., Dengler R., Eisen A., England J.D., Kaji R., Kimura J., Mills K., Mitsumoto H., Nodera H., Shefner J., et al. Electrodiagnostic criteria for diagnosis of ALS. Clin. Neurophysiol. 2008;119:497–503. doi: 10.1016/j.clinph.2007.09.143. [DOI] [PubMed] [Google Scholar]
- 20.Strong M.J., Abrahams S., Goldstein L.H., Woolley S., McLaughlin P., Snowden J., Mioshi E., Roberts-South A., Benatar M., HortobáGyi T., et al. Amyotrophic lateral sclerosis—Frontotemporal spectrum disorder (ALS-FTSD): Revised diagnostic criteria. Amyotroph. Lateral Scler. Front. Degener. 2017;18:153–174. doi: 10.1080/21678421.2016.1267768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., Van Swieten J.C., Myllykangas L., et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadimas G.K., Paraskevas G.P., Zambelis T., Karagiaouris C., Bourbouli M., Bougea A., Walter M.C., Schumacher N.U., Krause S., Kapaki E. The multifaceted clinical presentation of VCP-proteinopathy in a Greek family. Acta Myol. Myopathies Cardiomyopathies Off. J. Mediterr. Soc. Myol. 2017;36:203–206. [PMC free article] [PubMed] [Google Scholar]
- 24.Kapaki E., Paraskevas G.P., Papageorgiou S.G., Bonakis A., Kalfakis N., Zalonis I., Vassilopoulos D. Diagnostic value of CSF biomarker profile in frontotemporal lobar degeneration. Alzheimers Dis. Assoc. Disord. 2008;22:47–53. doi: 10.1097/WAD.0b013e3181610fea. [DOI] [PubMed] [Google Scholar]
- 25.Thijssen E.H., La Joie R., Strom A., Fonseca C., Iaccarino L., Wolf A., Spina S., Allen I.E., Cobigo Y., Heuer H., et al. Plasma phosphorylated tau 217 and phosphorylated tau 181 as biomarkers in Alzheimer’s disease and frontotemporal lobar degeneration: A retrospective diagnostic performance study. Lancet Neurol. 2021;20:739–752. doi: 10.1016/S1474-4422(21)00214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neddens J., Daurer M., Loeffler T., Alzola Aldamizetxebarria S., Flunkert S., Hutter-Paier B. Constant Levels of Tau Phosphorylation in the Brain of htau Mice. Front. Mol. Neurosci. 2020;13:136. doi: 10.3389/fnmol.2020.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghidoni R., Benussi L., Glionna M., Franzoni M., Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71:1235–1239. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- 28.Sokratous M., Lucia S., Bourinaris T., Marogianni C., Arnaoutoglou M., Patrikiou E., Ralli S., Markou A., Dardiotis E., Houlden H., et al. Prevalence of C9orf72 hexanucleotide repeat expansion in Greek patients with sporadic ALS. Amyotroph. Lateral Scler. Front. Degener. 2020:1–3. doi: 10.1080/21678421.2020.1757115. [DOI] [PubMed] [Google Scholar]
- 29.Kartanou C., Karadima G., Koutsis G., Breza M., Papageorgiou S.G., Paraskevas G.P., Kapaki E., Panas M. Screening for the C9ORF72 repeat expansion in a greek frontotemporal dementia cohort. Amyotroph. Lateral Scler. Front. Degener. 2018;19:152–154. doi: 10.1080/21678421.2017.1400070. [DOI] [PubMed] [Google Scholar]
- 30.Ramos E.M., Koros C., Dokuru D.R., Van Berlo V., Kroupis C., Wojta K., Wang Q., Andronas N., Matsi S., Beratis I.N., et al. Frontotemporal dementia spectrum: First genetic screen in a Greek cohort. Neurobiol. Aging. 2019;75:224.e221–224.e228. doi: 10.1016/j.neurobiolaging.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majounie E., Renton A.E., Mok K., Dopper E.G.P., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J., et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok K.Y., Koutsis G., Schottlaender L.V., Polke J., Panas M., Houlden H. High frequency of the expanded C9ORF72 hexanucleotide repeat in familial and sporadic Greek ALS patients. Neurobiol. Aging. 2012;33:1851.e1851–1851.e1855. doi: 10.1016/j.neurobiolaging.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mol M.O., van Rooij J.G.J., Wong T.H., Melhem S., Verkerk A.J.M.H., Kievit A.J.A., van Minkelen R., Rademakers R., Pottier C., Kaat L.D., et al. Underlying genetic variation in familial frontotemporal dementia: Sequencing of 198 patients. Neurobiol. Aging. 2020 doi: 10.1016/j.neurobiolaging.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Palomo V., Tosat-Bitrian C., Nozal V., Nagaraj S., Martin-Requero A., Martinez A. TDP-43: A Key Therapeutic Target beyond Amyotrophic Lateral Sclerosis. ACS Chem. Neurosci. 2019;10:1183–1196. doi: 10.1021/acschemneuro.9b00026. [DOI] [PubMed] [Google Scholar]
- 35.Zou Z.-Y., Zhou Z.-R., Che C.-H., Liu C.-Y., He R.-L., Huang H.-P. Genetic epidemiology of amyotrophic lateral sclerosis: A systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry. 2017;88:540–549. doi: 10.1136/jnnp-2016-315018. [DOI] [PubMed] [Google Scholar]
- 36.Corcia P., Valdmanis P., Millecamps S., Lionnet C., Blasco H., Mouzat K., Daoud H., Belzil V., Morales R., Pageot N., et al. Phenotype and genotype analysis in amyotrophic lateral sclerosis with TARDBP gene mutations. Neurology. 2012;78:1519–1526. doi: 10.1212/WNL.0b013e3182553c88. [DOI] [PubMed] [Google Scholar]
- 37.Caroppo P., Camuzat A., Guillot-Noel L., Thomas-Antérion C., Couratier P., Wong T.H., Teichmann M., Golfier V., Auriacombe S., Belliard S., et al. Defining the spectrum of frontotemporal dementias associated with TARDBPmutations. Neurol. Genet. 2016;2:e80. doi: 10.1212/NXG.0000000000000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutherford N.J., Zhang Y.-J., Baker M., Gass J.M., Finch N.A., Xu Y.-F., Stewart H., Kelley B.J., Kuntz K., Crook R.J.P., et al. Novel Mutations in TARDBP (TDP-43) in Patients with Familial Amyotrophic Lateral Sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ticozzi N., LeClerc A.L., van Blitterswijk M., Keagle P., McKenna-Yasek D.M., Sapp P.C., Silani V., Wills A.-M., Brown R.H., Landers J.E. Mutational analysis of TARDBP in neurodegenerative diseases. Neurobiol. Aging. 2011;32:2096–2099. doi: 10.1016/j.neurobiolaging.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Y.-W., Lee M.-J., Chen T.-F., Cheng T.-W., Lai Y.-M., Hua M.-S., Chiu M.-J. A single nucleotide TDP-43 mutation within a Taiwanese family: A multifaceted demon. Amyotroph. Lateral Scler. Front. Degener. 2016;17:292–294. doi: 10.3109/21678421.2015.1111905. [DOI] [PubMed] [Google Scholar]
- 41.Ramos E.M., Dokuru D.R., Van Berlo V., Wojta K., Wang Q., Huang A.Y., Deverasetty S., Qin Y., van Blitterswijk M., Jackson J., et al. Genetic screening of a large series of North American sporadic and familial frontotemporal dementia cases. Alzheimer’s Dement. 2020;16:118–130. doi: 10.1002/alz.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu G.-r., Hu W., Zhan L.-L., Wang C., Xu L.-Q., Lin M.-T., Chen W.-J., Wang N., Zhang Q.-J. High frequency of the TARDBP p.M337 V mutation among south-eastern Chinese patients with familial amyotrophic lateral sclerosis. BMC Neurol. 2018;18:35. doi: 10.1186/s12883-018-1028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan S., Shatunov A., Sproviero W., Jones A.R., Shoai M., Hughes D., Al Khleifat A., Malaspina A., Morrison K.E., Shaw P.J., et al. A comprehensive analysis of rare genetic variation in amyotrophic lateral sclerosis in the UK. Brain. 2017;140:1611–1618. doi: 10.1093/brain/awx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang S.Y.-Y., Hsu J.S., Teo K.-C., Li Y., Kung M.H.W., Cheah K.S.E., Chan D., Cheung K.M.C., Li M., Sham P.-C., et al. Burden of rare variants in ALS genes influences survival in familial and sporadic ALS. Neurobiol. Aging. 2017;58:238.e9. doi: 10.1016/j.neurobiolaging.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Narain P., Pandey A., Gupta S., Gomes J., Bhatia R., Vivekanandan P. Targeted next-generation sequencing reveals novel and rare variants in Indian patients with amyotrophic lateral sclerosis. Neurobiol. Aging. 2018;71:265.e9. doi: 10.1016/j.neurobiolaging.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Tamaoka A., Arai M., Itokawa M., Arai T., Hasegawa M., Tsuchiya K., Takuma H., Tsuji H., Ishii A., Watanabe M., et al. TDP-43 M337V Mutation in Familial Amyotrophic Lateral Sclerosis in Japan. Intern. Med. 2010;49:331–334. doi: 10.2169/internalmedicine.49.2915. [DOI] [PubMed] [Google Scholar]
- 48.Kühnlein P., Sperfeld A.-D., Vanmassenhove B., Van Deerlin V., Lee V.M.-Y., Trojanowski J.Q., Kretzschmar H.A., Ludolph A.C., Neumann M. Two German Kindreds With Familial Amyotrophic Lateral Sclerosis Due to TARDBP Mutations. Arch. Neurol. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czell D., Andersen P.M., Morita M., Neuwirth C., Perren F., Weber M. Phenotypes in Swiss Patients with Familial ALS Carrying TARDBP Mutations. Neurodegener. Dis. 2013;12:150–155. doi: 10.1159/000345835. [DOI] [PubMed] [Google Scholar]
- 50.Homma T., Nagaoka U., Kawata A., Mochizuki Y., Kawakami H., Maruyama H., Matsubara S., Komori T. Neuropathological features of Japanese familial amyotrophic lateral sclerosis with p.N352S mutation in TARDBP. Neuropathol. Appl. Neurobiol. 2014;40:231–236. doi: 10.1111/nan.12090. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe S., Kaneko K., Yamanaka K. Accelerated Disease Onset with Stabilized Familial Amyotrophic Lateral Sclerosis (ALS)-linked Mutant TDP-43 Proteins. J. Biol. Chem. 2013;288:3641–3654. doi: 10.1074/jbc.M112.433615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Budini M., Romano V., Avendaño-Vázquez S.E., Bembich S., Buratti E., Baralle F.E. Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 2012;1462:139–150. doi: 10.1016/j.brainres.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 53.Wauters E., Van Mossevelde S., Van der Zee J., Cruts M., Van Broeckhoven C. Modifiers of GRN-Associated Frontotemporal Lobar Degeneration. Trends Mol. Med. 2017;23:962–979. doi: 10.1016/j.molmed.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.-J., et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 55.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 56.Mesulam M.-M. Primary Progressive Aphasia: A 25-year Retrospective. Alzheimer Dis. Assoc. Disord. 2007;21:S8–S11. doi: 10.1097/WAD.0b013e31815bf7e1. [DOI] [PubMed] [Google Scholar]
- 57.Rohrer J.D., Ridgway G.R., Crutch S.J., Hailstone J., Goll J.C., Clarkson M.J., Mead S., Beck J., Mummery C., Ourselin S., et al. Progressive logopenic/phonological aphasia: Erosion of the language network. NeuroImage. 2010;49:984–993. doi: 10.1016/j.neuroimage.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gliebus G., Bigio E.H., Gasho K., Mishra M., Caplan D., Mesulam M.-M., Geula C. Asymmetric TDP-43 distribution in primary progressive aphasia with progranulin mutation. Neurology. 2010;74:1607–1610. doi: 10.1212/WNL.0b013e3181df0a1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sassi C., Capozzo R., Gibbs R., Crews C., Zecca C., Arcuti S., Copetti M., Barulli M.R., Brescia V., Singleton A.B., et al. A Novel Splice-Acceptor Site Mutation in GRN (c.709-2 A>T) Causes Frontotemporal Dementia Spectrum in a Large Family from Southern Italy. J. Alzheimers Dis. 2016;53:475–485. doi: 10.3233/JAD-151170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Zee J., Pirici D., Van Langenhove T., Engelborghs S., Vandenberghe R., Hoffmann M., Pusswald G., Van den Broeck M., Peeters K., Mattheijssens M., et al. Clinical heterogeneity in 3 unrelated families linked to VCP p.Arg159His. Neurology. 2009;73:626–632. doi: 10.1212/WNL.0b013e3181b389d9. [DOI] [PubMed] [Google Scholar]
- 61.Evangelista T., Weihl C.C., Kimonis V., Lochmüller H., Clemen C., Deshaies R., Evangelista T., Eymard B., Greensmith L., Hilton-Jones D., et al. 215th ENMC International Workshop VCP-related multi-system proteinopathy (IBMPFD) 13–15 November 2015, Heemskerk, The Netherlands. Neuromuscul. Disord. 2016;26:535–547. doi: 10.1016/j.nmd.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watts G.D., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 63.Al-Obeidi E., Al-Tahan S., Surampalli A., Goyal N., Wang A.K., Hermann A., Omizo M., Smith C., Mozaffar T., Kimonis V. Genotype-phenotype study in patients with valosin-containing protein mutations associated with multisystem proteinopathy. Clin. Genet. 2018;93:119–125. doi: 10.1111/cge.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gitcho M.A., Strider J., Carter D., Taylor-Reinwald L., Forman M.S., Goate A.M., Cairns N.J. VCP Mutations Causing Frontotemporal Lobar Degeneration Disrupt Localization of TDP-43 and Induce Cell Death. J. Biol. Chem. 2009;284:12384–12398. doi: 10.1074/jbc.M900992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manno A., Noguchi M., Fukushi J., Motohashi Y., Kakizuka A. Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Genes Cells. 2010;15:911–922. doi: 10.1111/j.1365-2443.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- 66.Bartolome F., Wu H.C., Burchell V.S., Preza E., Wray S., Mahoney C.J., Fox N.C., Calvo A., Canosa A., Moglia C., et al. Pathogenic VCP Mutations Induce Mitochondrial Uncoupling and Reduced ATP Levels. Neuron. 2013;78:57–64. doi: 10.1016/j.neuron.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X., Gui L., Zhang X., Bulfer S.L., Sanghez V., Wong D.E., Lee Y., Lehmann L., Lee J.S., Shih P.-Y., et al. Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. USA. 2015;112:E1705–E1714. doi: 10.1073/pnas.1418820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hewitt C., Kirby J., Highley J.R., Hartley J.A., Hibberd R., Hollinger H.C., Williams T.L., Ince P.G., McDermott C.J., Shaw P.J. Novel FUS/TLS Mutations and Pathology in Familial and Sporadic Amyotrophic Lateral Sclerosis. Arch. Neurol. 2010;67:455–461. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- 69.Shang Y., Huang E.J. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016;1647:65–78. doi: 10.1016/j.brainres.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corrado L., Del Bo R., Castellotti B., Ratti A., Cereda C., Penco S., Sorarù G., Carlomagno Y., Ghezzi S., Pensato V., et al. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2010;47:190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 71.Lai S.-L., Abramzon Y., Schymick J.C., Stephan D.A., Dunckley T., Dillman A., Cookson M., Calvo A., Battistini S., Giannini F., et al. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2011;32:550.e551–550.e554. doi: 10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang L., Ma Y., Liu X.L., Chen L., Fan D.S. Better survival in female SOD1-mutant patients with ALS: A study of SOD1-related natural history. Transl. Neurodegener. 2019;8:2. doi: 10.1186/s40035-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen P.M., Sims K.B., Xin W.W., Kiely R., O’Neill G., Ravits J., Pioro E., Harati Y., Brower R.D., Levine J.S., et al. Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: A decade of discoveries, defects and disputes. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. Off. Publ. World Fed. Neurol. Res. Group Mot. Neuron Dis. 2003;4:62–73. doi: 10.1080/14660820310011700. [DOI] [PubMed] [Google Scholar]
- 74.Lysogorskaia E.V., Rossokhin A.V., Abramycheva N., Zakharova M.N., Illarioshkin S.N. SOD1 gene mutations in patients with amyotrophic lateral sclerosis: Potential for the method of molecular. Mol. Biol. 2013;47:861–867. doi: 10.1134/S0026893313050129. [DOI] [PubMed] [Google Scholar]
- 75.Goossens J., Bjerke M., Van Mossevelde S., Van den Bossche T., Goeman J., De Vil B., Sieben A., Martin J.J., Cras P., De Deyn P.P., et al. Diagnostic value of cerebrospinal fluid tau, neurofilament, and progranulin in definite frontotemporal lobar degeneration. Alzheimers Res. Ther. 2018;10:31. doi: 10.1186/s13195-018-0364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kämäläinen A., Herukka S.K., Hartikainen P., Helisalmi S., Moilanen V., Knuuttila A., Jansson L., Tienari P.J., Remes A.M. Cerebrospinal fluid biomarkers for Alzheimer’s disease in patients with frontotemporal lobar degeneration and amyotrophic lateral sclerosis with the C9ORF72 repeat expansion. Dement Geriatr. Cogn. Disord. 2015;39:287–293. doi: 10.1159/000371704. [DOI] [PubMed] [Google Scholar]
- 77.Junttila A., Kuvaja M., Hartikainen P., Siloaho M., Helisalmi S., Moilanen V., Kiviharju A., Jansson L., Tienari P.J., Remes A.M., et al. Cerebrospinal Fluid TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis Patients with and without the C9ORF72 Hexanucleotide Expansion. Dement Geriatr. Cogn Dis. Extra. 2016;6:142–149. doi: 10.1159/000444788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kauwe J.S.K., Cruchaga C., Mayo K., Fenoglio C., Bertelsen S., Nowotny P., Galimberti D., Scarpini E., Morris J.C., Fagan A.M., et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc. Natl. Acad. Sci. USA. 2008;105:8050–8054. doi: 10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bekris L.M., Tsuang D.W., Peskind E.R., Yu C.E., Montine T.J., Zhang J., Zabetian C.P., Leverenz J.B. Cerebrospinal fluid Aβ42 levels and APP processing pathway genes in Parkinson’s disease. Mov Disord. 2015;30:936–944. doi: 10.1002/mds.26172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Etten E.S., Verbeek M.M., van der Grond J., Zielman R., van Rooden S., van Zwet E.W., van Opstal A.M., Haan J., Greenberg S.M., van Buchem M.A., et al. β-Amyloid in CSF: Biomarker for preclinical cerebral amyloid angiopathy. Neurology. 2017;88:169–176. doi: 10.1212/WNL.0000000000003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thordardottir S., Kinhult Ståhlbom A., Almkvist O., Thonberg H., Eriksdotter M., Zetterberg H., Blennow K., Graff C. The effects of different familial Alzheimer’s disease mutations on APP processing in vivo. Alzheimers Res. Ther. 2017;9:9. doi: 10.1186/s13195-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poesen K., De Schaepdryver M., Stubendorff B., Gille B., Muckova P., Wendler S., Prell T., Ringer T.M., Rhode H., Stevens O., et al. Neurofilament markers for ALS correlate with extent of upper and lower motor neuron disease. Neurology. 2017;88:2302–2309. doi: 10.1212/WNL.0000000000004029. [DOI] [PubMed] [Google Scholar]
- 83.Cognat E., De Schaepdryver M., Hugon J., Poesen K., Paquet C. Elevated ALS Biomarker Levels in CSF of a FTD Patient at the Presymptomatic Stage of ALS. Alzheimers Dis. Assoc. Disord. 2018;32:156–157. doi: 10.1097/WAD.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 84.Ashton N.J., Janelidze S., Al Khleifat A., Leuzy A., van der Ende E.L., Karikari T.K., Benedet A.L., Pascoal T.A., Lleó A., Parnetti L., et al. A multicentre validation study of the diagnostic value of plasma neurofilament light. Nat. Commun. 2021;12:3400. doi: 10.1038/s41467-021-23620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rojas J.C., Wang P., Staffaroni A.M., Heller C., Cobigo Y., Wolf A., Goh S.-Y.M., Ljubenkov P.A., Heuer H.W., Fong J.C., et al. Plasma Neurofilament Light for Prediction of Disease Progression in Familial Frontotemporal Lobar Degeneration. Neurology. 2021;96:e2296–e2312. doi: 10.1212/WNL.0000000000011848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.