Abstract
During our ongoing studies of bambusicolous fungi in southwest China and Thailand, three saprobic pleosporalean taxa were discovered on bamboos in Yunnan Province of China. Occultibambusa hongheensis and Seriascoma bambusae spp. nov. are introduced based on morphological characteristics coupled with multi-locus phylogenetic analyses of combined LSU, SSU, TEF1-α, RPB2 and ITS sequence data. Occultibambusa kunmingensis is also reported from a terrestrial habitat for the first time. Comprehensive descriptions, color photo plates of micromorphology, and a phylogenetic tree showing the placements of these three taxa are provided. In addition, synopsis tables of Occultibambusa and Seriascoma with morphological features are also provided.
Keywords: Occultibambusa, one new record, Seriascoma, taxonomy, two new taxa
1. Introduction
Occultibambusaceae is a well-resolved family with a strong morpho-molecular basis. The family accommodates five genera viz. Brunneofusispora S.K. Huang & K.D. Hyde, Neooccultibambusa Doilom & K.D. Hyde, Occultibambusa D.Q. Dai & K.D. Hyde, Seriascoma Phookamsak, D.Q. Dai & K.D. Hyde, and Versicolorisporium Sat. Hatak., Kaz. Tanaka & Y. Harada [1,2,3]. Occultibambusaceae was introduced by Dai et al. [4] to accommodate Neooccultibambusa, Occultibambusa, Seriascoma, and Versicolorisporium, with Occultibambusa as the type genus. Brunneofusispora became a new member of Occultibambusaceae [5]. As a result of thriving molecular techniques, all genera in Occultibambusaceae have been resolved using multi-gene phylogeny [4,5,6,7].
Occultibambusaceae is characterized by solitary, immersed, subglobose to conical, greyish to dark brown, uni- or multi-loculate ascostromata, scattered or in small groups, with papillate, or protruding ostioles, bitunicate, fissitunicate, (6)–8-spored, cylindrical to clavate asci with short, furcate or bulb-like pedicels, and 1–3-seriate, fusiform, hyaline, or pale brown to dark brown, 1–3-septate ascospores with or without a sheath [1,4,5,6,7,8,9,10,11,12,13]. Brunneofusispora, Occultibambusa, Seriascoma, and Versicolorisporium were reported to have coelomycetous asexual morphs [4,14,15,16], while Neooccultibambusa forms chlamydospores in culture or has as hyphomycetous asexual morphs [6,9,17].
Occultibambusaceae is a small family with 18 species [18]. To date, this family has been reported from China, Italy, Japan, and Thailand [4,5,6,7,8,9,10,11,12,13,14,15,16,17,19]. With the exception of species of Neooccultibambusa and Brunneofusispora, most species of Occultibambusaceae are saprobes on dead bamboo [4,8,10,12,13]. Neooccultibambusa has been found on a wide variety of hosts such as Ammophila sp., Pandanus sp. and Tectona grandis [6,9,11,17]. Brunneofusispora was reported on dead wood and Clematis sp. in terrestrial habitats and decaying wood submerged in freshwater habitats [5,7,15,16].
Occultibambusa is typified by O. bambusae D.Q. Dai & K.D. Hyde and is characterized by solitary or gregarious, raised, immersed, subglobose to conical, dark brown, uni-loculate, coriaceous ascostromata with black, papillate ostioles, bitunicate, fissitunicate, eight-spored, broadly cylindrical to clavate asci, and fusiform, pale brown to brown, 1–(3)-septate ascospores, with or without a sheath [4,8,10,12,13]. Eight species are accommodated in Occultibambusa [18]. However, only O. fusispora Phookamsak, D.Q. Dai & K.D. Hyde has a known coelomycetous asexual morph and is characterized by multi-loculate, eustromatic, immersed, solitary to gregarious, globose to subglobose, black conidiomata with long papillate necks and enteroblastic, phialidic, determinate, cylindrical to ampulliform, hyaline, smooth, aseptate conidiogenous cells bearing oblong to cylindrical, hyaline, aseptate, guttulate, smooth-walled conidia [4].
Seriascoma is typified by S. didymosporum Phookamsak, D.Q. Dai, S.C. Karunarathana & K.D. Hyde. Seriascoma didymosporum and S. yunnanense Rathnayaka & K.D. Hyde are accommodated in the genus [1,12,18]. Seriascoma is characterized by solitary or gregarious, erumpent, subglobose or elongated, uni- to multi-loculate, coriaceous ascostromata, immersed under a clypeus, bitunicate, fissitunicate, eight-spored, clavate asci with short to long furcate pedicels and 1–3-seriate, fusiform, asymmetric, 1-septate, hyaline ascospores with or without a sheath [4,12,13]. The asexual morph of this genus has only been reported in S. didymosporum and is characterized by eustromatic, solitary to gregarious, semi-immersed to erumpent, conical, black, uni- to multi-loculate conidiomata and enteroblastic, phialidic, determinate, cylindrical to ampulliform, hyaline, aseptate, smooth-walled conidiogenous cells bearing oblong, hyaline, aseptate, smooth-walled conidia [4].
During our studies on bambusicolous fungi in southwest China and Thailand, three new fungal strains belonging to Occultibambusaceae were collected and isolated from Yunnan Province in China. This study introduces two novel species in Occultibambusa and Seriascoma based on multi-locus phylogenetic analyses and morphological characteristics. In addition, Occultibambusa kunmingensis C.X. Liu, H. Zhang & K.D. Hyde is reported from a terrestrial environment for the first time.
2. Materials and Methods
2.1. Collection, Examination, Isolation and Preservation
Dead bamboo branches and culms were collected from Mengla County, Xishuangbanna Dai Autonomous Prefecture, Yunnan Province, China in January 2019 and Honghe County, Honghe Hani and Yi Autonomous Prefecture, Yunnan Province, China, in October 2020. Samples were stored in plastic Ziploc bags and taken to the laboratory at Kunming Institute of Botany, CAS, Kunming, Yunnan Province, China for observation and examination following the method described by Senanayake et al. [20]. Fungal fruiting bodies on host substrates were visualized under a Motic SMZ 140 series dissecting stereoscope and photographed by digital camera. Vertical sections of ascostromata and conidiomata and other micro-morphological characteristics (e.g., peridium, pseudoparaphyses, asci, ascospores, conidiogenous cells and conidia) were observed and captured with a Nikon ECLIPSE Ni compound microscope connected with a Canon EOS 600D digital camera. The Tarosoft (R) Image FrameWork version 0.9.7 program was used to measure the size (10–20 measurements of each structure) of fungal characteristics. Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA) was used to edit and combine photographic plates. Ex-type living culture of Occultibambusa fusispora (MFLUCC 11-0127) was also loaned from Mae Fah Luang University Culture Collection, Chiang Rai, Thailand (MFLUCC). It was aseptically sub-cultured in a laminar flow and incubated at room temperature (20–25 °C) for sequencing. Specimens of new taxa and new collections obtained for this study have been deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), Yunnan, China and the Herbarium Mycologicum Academiae Sinicae (HMAS), Beijing, China. Living cultures have been deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and Kunming Institute of Botany Culture Collection, Kunming, China (KUMCC). Facesoffungi and Index Fungorum numbers have been registered for the newly described taxa [21,22]. New species have been established based on the guidelines of Jeewon and Hyde [23].
2.2. DNA Extraction, PCR Amplification and Sequencing
Genomic DNA of new fungal isolates and a loaned strain of Occultibambusa fusispora (MFLUCC 11-0127) was extracted from fresh mycelia using Biospin Fungus Genomic DNA extraction kit (BioFlux®, Hangzhou, China) following the manufacturer’s instructions. DNA amplification was performed by polymerase chain reaction (PCR). Five primer pairs viz. ITS5/ITS4 [24], LR0R/LR5 [25], NS1/NS4 [24], EF1-983F/EF1-2218R [26] and fRPB2-5F/fRPB2-7cR [27] were used to amplify the fragments of the internal transcribed spacers (ITS1-5.8S-ITS2), the 28S large subunit rDNA (LSU), the 18S small subunit rDNA (SSU), the translation elongation factor 1-alpha (TEF1-α), and the partial RNA polymerase second largest subunit (RPB2), respectively. PCR was carried out based on 25 µL total volume per reaction, containing 2 µL of fungal genomic DNA, 1 µL of each forward and reverse primer, 12.5 µL of 2 × Power Taq PCR Master Mix (a mixture of EasyTaqTM DNA Polymerase, dNTPs, and optimized buffer; Beijing BioTeke Corporation, China) and 8.5 µL of sterilized double-distilled water (ddH2O). The PCR thermal cycle profiles for ITS, LSU, SSU, and TEF1-α gene was processed under the following conditions: an initial denaturation at 94 °C for 3 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min, and finally kept at 4 °C. We followed the PCR thermal cycle profiles for RPB2 gene in Jiang et al. [28]. Final PCR products were sent to TsingKe Biological Technology (Beijing) Co., Ltd., China for PCR purification and sequencing. The Sanger dideoxy sequencing method was used for the new strains. The quality of sequences was checked by both manual and FinchTV v. 1.4.0 (http://www.geospiza.com/Products/finchtv.shtml (accessed on 5 April 2021)).
2.3. Alignment and Phylogenetic Analyses
The nucleotide BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 April 2021)) was applied to discover taxa closely related to our three new isolates (KUMCC 21-0019, KUMCC 21-0020 and KUMCC 21-0021). Similarity indices from the BLAST search indicated that KUMCC 21-0019 and KUMCC 21-0020 belong to Occultibambusa (Occultibambusaceae) and KUMCC 21-0021 belongs to Seriascoma (Occultibambusaceae). Therefore, to reveal accurate phylogenetic placements of our three new isolates, multi-gene phylogeny of Occultibambusaceae and the closely related family Nigrogranaceae (Pleosporales, Dothideomycetes) were done based on maximum-likelihood and Bayesian inference methods. DNA sequences of representative taxa in Occultibambusaceae and Nigrogranaceae are shown in Table 1. Sequence alignments and phylogenetic analyses were carried out following methods described by Dissanayake et al. [29]. Preliminarily individual DNA sequence matrixes were aligned via the online platform, MAFFT v. 7.475 [30]. Aligned sequence datasets were trimmed by TrimAl v. 1.3 via the web server phylemon 2 (http://phylemon.bioinfo.cipf.es/utilities.html (accessed on 20 April 2021)) and then improved where necessary using BioEdit v. 6.0.7 [31], i.e., complementing the missing bases at the start and end of the consensus sequence. Individual gene datasets were analyzed by maximum likelihood criteria in order to compare the congruence of tree topologies.
Table 1.
Taxa names, strain numbers, and GenBank accession numbers of taxa used for the present phylogenetic analyses.
| Taxa Names | Strain Numbers | Origin | Substrate/Host | GenBank Accession Numbers | Refs. | ||||
|---|---|---|---|---|---|---|---|---|---|
| LSU | SSU | TEF1-α | RPB2 | ITS | |||||
| Brunneofusispora clematidis | MFLUCC 17-2070 | Chiang Rai, Thailand | Dead stems of Clematis subumbellata | MT214570 | NG_070658 | MT394629 | MT394692 | MT310615 | [7] |
| Brunneofusispora hyalina | MFLUCC 21-0008 | Chiang Mai, Thailand | Decaying wood | MW287234 | MW485613 | MW512606 | MW512609 | MW260330 | [16] |
| Brunneofusispora sinensis | KUMCC 17-0030 | Yunnan, China | Dead wood | MH393557 | MH393556 | MH395329 | / | MH393558 | [5] |
| Brunneofusispora sinensis | MFLUCC 20-0016 | Yunnan, China | Dead branches of Magnolia denudata | MT159624 | MT159636 | MT159607 | MT159613 | MT159630 | [15] |
| Brunneofusispora sp. | X135 | China | Ageratina adenophora | / | / | / | / | MK304223 | [16] |
| Massarina rubi | CBS 691.95 | Austria | Ulmus glabra | FJ795453 | GU456301 | / | FJ795470 | / | Unknown |
| Massarina rubi | MUT 4323 | Italy | Rhizomes of Posidonia oceanica | KF636772 | / | / | / | KF636766 | Unpublished |
| Massarina rubi | MUT 4887 | Italy | Flabellia petiolata | KP671721 | KT587318 | / | / | KR014359 | Unpublished |
| Massarina sp. | MUT 4860 | Italy | Flabellia petiolata | KP671730 | KT587325 | / | / | KR014362 | Unpublished |
| Neooccultibambusa chiangraiensis | MFLUCC 12-0559 | Chiang Rai, Thailand | Dead twigs of Tectona grandis | KU764699 | NG_061230 | KU872761 | / | NR_154238 | [6] |
| Neooccultibambusa jonesii | MFLUCC 16-0643 | Italy | Dead and stems of Ammophila arenaria | NG_059741 | NG_062422 | / | / | / | [9] |
| Neooccultibambusa pandanicola | KUMCC 17-0179 | Yunnan, China | Dead leaves of Pandanus utilis | MG298940 | MG298942 | MG298943 | MG298944 | MG298941 | [17] |
| Neooccultibambusa thailandensis | MFLUCC 16-0274 | Prachuap Khiri Khan, Thailand | Dead leaf of Pandanus sp. | MH260308 | MH260348 | MH412780 | MH412758 | MH275074 | [11] |
| Nigrograna mackinnonii | E5202H | Ecuador | Dead stems of Guazuma ulmifolia | KJ605422 | JX264155 | JX264154 | JX264156 | JX264157 | Unpublished |
| Nigrograna obliqua | MRP | Austria | Ribes uva-crispa | KX650561 | / | KX650532 | KX650581 | KX650561 | [37] |
| Nigrograna obliqua | BW4 | Austria | A twig of Sambucus racemosa | KX650557 | / | KX650529 | / | KX650557 | [37] |
| Occultibambusa aquatica | MFLUCC 11-0006 | Chiang Rai, Thailand | Bamboo | KX698110 | KX698112 | / | / | / | [8] |
| Occultibambusa bambusae | MFLUCC 11-0394 | Chiang Mai, Thailand | Dead culms of bamboo | KU863113 | KU872117 | KU940194 | KU940171 | KU940124 | [4] |
| Occultibambusa bambusae | MFLUCC 13-0855 | Chiang Rai, Thailand | Dead culms of bamboo | KU863112 | KU872116 | KU940193 | KU940170 | KU940123 | [4] |
| Occultibambusa chiangraiensis | MFLUCC 16-0380 | Chiang Rai, Thailand | Dead stems of Bambusoideae sp. | KX655546 | NG_062421 | KX655561 | KX655566 | / | [8] |
| Occultibambusa fusispora | MFLUCC 11-0127 | Chiang Rai, Thailand | Dead branches of bamboo | NG_059669 | / | KU940195 | KU940172 | NR_154340 | [4] |
| Occultibambusa fusispora | MFLUCC 11-0127II | Chiang Rai, Thailand | Dead branches of bamboo | MZ329032 | MZ329028 | MZ325466 | MZ325469 | MZ329036 | This study |
| Occultibambusa hongheensis | KUMCC 21-0020 | Yunnan, China | Dead branches of bamboo | MZ329033 | MZ329029 | MZ325467 | / | MZ329037 | This study |
| Occultibambusa jonesii | GZCC 16-0117 | Guizhou, China | Dead culms of bamboo | NG_066381 | NG_065104 | KY814756 | KY814758 | / | [10] |
| Occultibambusa kunmingensis | KUN-HKAS 102151 | Yunnan, China | Decaying bam | MN913733 | MT864342 | MT954407 | MT878453 | MT627716 | [13] |
| Occultibambusa kunmingensis | KUMCC 21-0019 | Yunnan, China | Submerged bamboo | MZ329034 | MZ329030 | / | / | MZ329038 | This study |
| Occultibambusa maolanensis | GZCC 16-0116 | Guizhou, China | Dead culms of bamboo | KY628323 | KY628325 | KY814757 | KY814759 | / | [10] |
| Occultibambusa pustula | MFLUCC 11-0502 | Chiang Rai, Thailand | Dead culm of bamboo | KU863115 | NG_062419 | / | / | NR_154341 | [4] |
| Ohleria modesta | MGC | Spain | Branches of Chamaecytisus proliferus | KX650562 | / | KX650533 | KX650582 | KX650562 | [37] |
| Ohleria modesta | OM | Spain | Branches of Chamaecytisus proliferus | KX650563 | KX650513 | KX650534 | KX650583 | KX650563 | [37] |
| Seriascoma bambusae | KUMCC 21-0021 | Yunnan, China | Dead culms of bamboo | MZ329035 | MZ329031 | MZ325468 | MZ325470 | MZ329039 | This study |
| Seriascoma didymosporum | MFLUCC 11-0179 | Chiang Rai, Thailand | Dead culms of bamboo | NG_059670 | KU872119 | KU940196 | KU940173 | NR_154433 | [4] |
| Seriascoma didymosporum | MFLUCC 11-0194 | Chiang Rai, Thailand | Dead culms of bamboo | KU863117 | KU872120 | KU940197 | KU940174 | KU940128 | [4] |
| Seriascoma sp. | KUMCC 21-0007 | Yunnan, China | Dead branches of bamboo | MW981347 | MZ325471 | MZ325472 | MZ325473 | MW981351 | [38] |
| Seriascoma yunnanense | MFLU 19-0690 | Yunnan, China | Dead branches of bamboo | NG_068303 | MN174694 | MN381858 | MN210324 | / | [12] |
| Versicolorisporium triseptatum | HHUF 28815 | Honshu, Japan | Dead culms of Pleioblastus chino | NG_042318 | NG_060995 | / | / | NR_119392 | [14] |
The ex-type strains are in bold. Abbreviations: GZCC: Guizhou Culture Collection, Guizhou, China; HHUF: Herbarium of Hirosaki University, Japan; KUMCC: Kunming Institute of Botany Culture Collection, Kunming, China; KUN-HKAS: Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica, Yunnan, China; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUT: Mycotheca Universitatis Taurinensis, Torino, Italy.
Maximum-likelihood (ML) analysis was performed via the online portal CIPRES Science Gateway v. 3.3 [32], with RAxML-HPC v.8 on XSEDE (8.2.12) tool, using default settings but following the adjustments: the GAMMA nucleotide substitution model and 1000 rapid bootstrap replicates. The evolutionary model of nucleotide substitution for Bayesian inference (BI) analysis was selected independently for each locus using MrModeltest 2.3 [33]. GTR+I+G was the best-fit for LSU, TEF1-α, and RPB2 loci under the Akaike Information Criterion (AIC), while the GTR+G substitution model was the best-fit for the ITS locus and HKY+I+G was the best-fit for the SSU locus. BI analysis was performed via MrBayes v. 3.1.2 [34]. Markov chain Monte Carlo sampling (MCMC) was used to determine posterior probabilities (PP) [35,36]. Six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled every 100th generation. The 0.15 “temperature” value was set in MCMC heated chain. All sampled topologies beneath the asymptote (20%) were discarded as part of a burn-in procedure and the remaining 8000 trees were used for calculating posterior probabilities (PP) in the 50% majority rule consensus tree (when split frequency lower than 0.01).
The tree topologies generated in this study were visualized on FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 April 2021)). The phylogram was edited and redrawn by using Microsoft Office PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA) and converted to tiff file on Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA). New sequences generated from the present study were deposited in GenBank (Table 1). The final alignment and phylogram were submitted to TreeBASE (submission ID: 28553, https://www.treebase.org/ (accessed on 20 July 2021)).
3. Results
3.1. Phylogenetic Analyses
The combined LSU, SSU, TEF1-α, RPB2 and ITS sequence matrix comprises 36 strains of representative species in Occultibambusaceae, the closely related family Nigrogranaceae, and Ohleria modesta (MGC and OM) as the outgroup. The dataset consists of 4308 total characters, including gaps (LSU: 1–832 bp, SSU: 833–1855 bp, TEF1-α: 1856–2791 bp, RPB2: 2792–3855 bp, ITS: 3856–4308 bp). The best scoring ML tree was selected to represent the phylogenetic relationships of two new taxa and a new record taxon with other representative taxa in Occultibambusaceae (Figure 1), with the final ML optimization likelihood value of −20,955.880345 (ln). All free model parameters were estimated by GTRGAMMA model, with 1331 distinct alignment patterns and 26.65% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.246035, C = 0.250866, G = 0.268453, T = 0.234646, with substitution rates AC = 2.237705, AG = 4.680757, AT = 1.669097, CG = 1.580519, CT = 10.758320, GT = 1.000000. The gamma distribution shape parameter alpha = 0.169891 and the Tree-Length = 1.401029. The final average standard deviation of split frequencies at the end of total MCMC generations was calculated as 0.003559 in BI analysis.
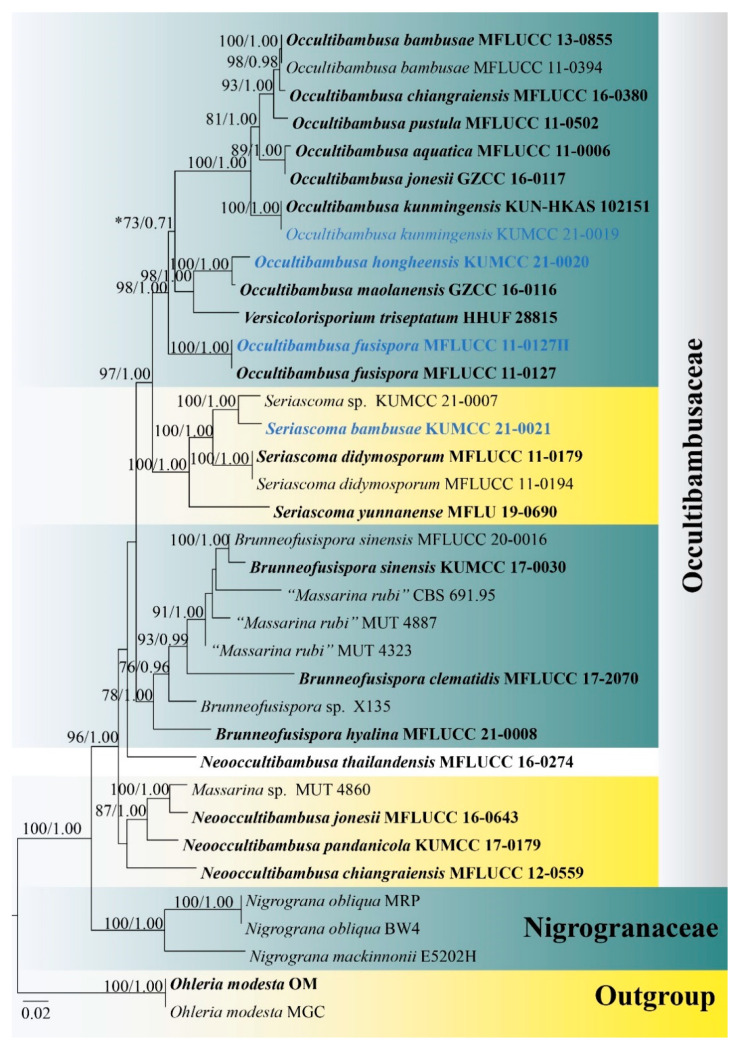
Figure 1.
RAxML tree based on LSU, SSU, TEF1-α, RPB2, and ITS sequence matrix representing the phylogenetic relationships of taxa in Occultibambusaceae. The tree is rooted to Ohleria modesta (MGC and OM). Bootstrap support values for ML equal to or greater than 70% and the Bayesian posterior probabilities equal to or higher than 0.95 PP are indicated above the nodes as ML/PP. Ex-type strains are in bold and the new species and new record are indicated in blue. * These values 73/0.71 are indicated on the node to discuss the separation of Occultibambusa taxa.
Tree topologies generated based on ML and BI analyses were similar in the present study and the ML phylogenetic tree is shown in Figure 1. All genera in Occultibambusaceae formed well-resolved clades, except for Versicolorisporium, which clustered within Occultibambusa. Neooccultibambusa thailandensis formed an independent lineage separated from other Neooccultibambusa species. Multi-locus phylogeny demonstrated that the new isolates (KUMCC 21-0019, KUMCC 21-0020 and KUMCC 21-0021) belong to Occultibambusaceae. KUMCC 21-0019 and KUMCC 21-0020 clustered within the Occultibambusa clade, and KUMCC 21-0021 grouped with the other Seriascoma species. KUMCC 21-0020 is sister to O. maolanensis with high statistical support (100% ML, 1.00 PP). Thus, Occultibambusa hongheensis sp. nov. (KUMCC 21-0020) is hereby introduced. The strain KUMCC 21-0019 shared the same branch length with the type strain of O. kunmingensis (KUN-HKAS 102151) with high statistical support (100% ML, 1.00 PP). Therefore, the new strain KUMCC 21-0019 is identified as O. kunmingensis, whereas O. fusispora (MFLUCC 11-0127II) was re-sequenced from the ex-type living culture and the newly generated sequences were found to be consistent with O. fusispora (MFLUCC 11-0127), clarifying the correctness of phylogenetic placement of O. fusispora as basal to Occultibambusa. Strain KUMCC 21-0021 formed a distinct subclade with Seriascoma sp. (KUMCC 21-0007) with high statistical support (100% ML, 1.00 PP). Hence, Seriascoma bambusae (KUMCC 21-0021) is introduced as a new species.
3.2. Taxonomy
3.2.1. Occultibambusa hongheensis H.B. Jiang, K.D. Hyde & Phookamsak, sp. nov.
Index Fungorum number: IF558429; Facesoffungi number: FoF 09884; Figure 2
Figure 2.
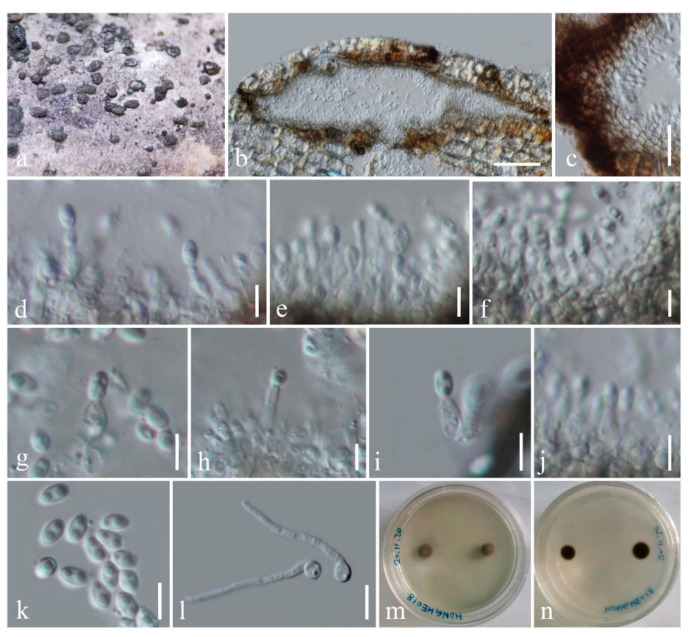
Occultibambusa hongheensis (HMAS 249944, holotype). (a,b) Appearance of ascostromata on the host; (c) Periphysate ostiole; (d,e) Vertical sections of ascostroma; (f) Peridium arranged in textura angularis, with palisade-like cells at the side; (g) Pseudoparaphyses; (h–k) Asci; (l–o) Ascospores [(o) Ascospore with mucilaginous sheath stained by Indian ink]. Scale bars: (d,e) = 100 μm; (c,f) = 50 μm; (j) = 30 μm; (g–i,k) = 20 μm; (l–o) = 15 μm.
Etymology: The specific epithet “hongheensis” refers to the location, Honghe, Yunnan Province of China, where the new species was collected.
Holotype: HMAS 249944
Saprobic on dead branches of bamboo. Sexual morph: Ascostromata 180–340 μm high, 400–550 μm diam., solitary or gregarious, immersed under host cortex, ampulliform, conical to subglobose, flattened at the base, uni- to bi-loculate, black, coriaceous, with 80–125 μm broad, central, periphysate ostiole. Peridium 40–130 μm thick, of unequal thickness, thin at the base, thick at sides, composed of several layers of pseudoparenchymatous cells of textura angularis, with palisade-like cells on the sides, outer layers consisting of dark brown pseudoparenchymatous cells, fused with host tissues, paler towards the inner layers. Hamathecium dense, composed of 1–2 μm wide, septate, branched, anastomosed, cellular pseudoparaphyses. Asci (78–)80–130(–137) × (18–)19–23(–25) μm (x = 107.5 × 21.5 μm, n = 20), eight-spored, bitunicate, fissitunicate, cylindric-clavate to clavate, with a short pedicel, apically rounded with a distinct ocular chamber. Ascospores (25–)27–30 × (5.5–)8–9(–10) μm (x = 28.8 × 8.4 μm, n = 20), partially overlapping 2-seriate, fusiform, 1-septate, slightly constricted at the septum, asymmetrical, upper cell broader and longer than the lower cell, straight to somewhat curved, hyaline when young and becoming pale brown when mature, smooth-walled, with guttules, surrounded by a broad mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes produced from both cells of ascospores. Colonies were grown on PDA, reaching 30 mm after four weeks at room temperature (10–20 °C), under normal light conditions, colonies on PDA cottony, circular, raised, dense, pale grey to dark grey from above and below. Mycelium superficial to immersed in media, with branched, septate, smooth hyphae.
Material examined: China, Yunnan Province, Honghe Autonomous Prefecture, Honghe County, on the roadside (23°16′32.26″ N, 102°25′30.37″ E, altitude 1544.29 m), on dead branches of bamboo in a terrestrial environment, 28 October 2020, H.B. Jiang, HONGHE012 (HMAS 249944, holotype), ex-type living culture, KUMCC 21-0020.
Notes: An ITS nucleotide blast search found the new isolate to be closely related to Versicolorisporium triseptatum HHUF 28815 (89.19% similarity), Neooccultibambusa thailandensis MFLUCC 16-0274 (88.27% similarity), and Massarina sp. MUT 4860 (87.65% similarity), while LSU and TEF1-α nucleotide blast searches indicated that this new isolate belongs to Occultibambusa. Occultibambusa hongheensis is most similar to O. maolanensis but differs in having pale brown ascospores with a broad mucilaginous sheath, longer asci (78–137 μm vs. 66–94 μm) [10], and smaller ascostromata (400–550 μm diam. vs. 544–600 µm diam.) [10]. Based on multi-locus phylogenetic analyses, O. hongheensis is sister to O. maolanensis with high statistical support (100% ML, 1.00 PP; Figure 1). There are 14 base pair (1.54%; not including gaps) differences between O. hongheensis and O. maolanensis in comparing a total of 910 nucleotides across the TEF1-α region.
3.2.2. Occultibambusa kunmingensis C.X. Liu, H. Zhang & K.D. Hyde in Dong et al., Fungal Diversity 105: 471
Index Fungorum number: IF557930; Facesoffungi number: FoF09272; Figure 3
Figure 3.
Occultibambusa kunmingensis (KUN-HKAS 112011). (a) Ascostromata on a dead bamboo branch; (b) Vertical section of ascostroma with ostiole; (c) Peridium; (d–g) Asci [(g) Asci with pseudoparaphyses]; (h–k) Ascospores; (l) Germinating ascospore. Scale bars: (b) = 200 μm; (c,g) = 30 μm; (d–f,l) = 20 μm; (h–k) = 10 μm.
Holotype: HKAS 102151
Saprobic on dead branches of bamboo, visible as raised, navicular black spots on the host. Sexual morph: Ascostromata 170–220 μm high, 350–550 μm diam., solitary, scattered or gregarious (in-group, 2–3 ascomata), immersed under host’s cortex, raised to superficial, ampulliform, flattened at the base, uni-loculate, dark brown to black, coriaceous, with a short, central, minutely papillate ostiole protruding the host. Peridium 30–120 μm thick, of unequal thickness, thin at the base, thicker at the sides, composed of several layers of brown pseudoparenchymatous cells, fused with host tissues, arranged in a textura angularis, with palisade-like cells at the sides. Hamathecium dense, composed of 2.4–3 μm wide, septate, branched, cellular pseudoparaphyses. Asci (76–)83–106(–115) × (11–)12–14(–15) μm (x = 95 × 13.2 μm, n = 20), eight-spored, bitunicate, fissitunicate, cylindric-clavate to clavate, with a short pedicel or subsessile, apically rounded with a narrow, well-developed ocular chamber. Ascospores (30–)34–36(–37.5) × (4.5–)5–6 μm (x = 35.7 × 5.6 μm, n = 20), overlapping 1–2-seriate, or twisted, brown to dark brown, fusiform, with acute ends,1–(3)-septate, occasionally the upper cell larger and longer than the lower cell, straight to slightly curved, with 1–2 large guttules in each cell, lacking a mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes produced from both ends of ascospore. Colonies growing slowly on PDA, reaching 20 mm in three weeks at room temperature under normal light conditions. Cottony, circular, raised, dark brown from above and below. Mycelium superficial to immersed in media, with branched, septate, smooth hyphae.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, near Bubeng Field Station-Xishuangbanna Station for Tropical Rain Forest Ecosystem Studies, on dead branches of bamboo in the terrestrial environment, 25 January 2019, H.B. Jiang & R. Phookamsak, BN009 (KUN-HKAS 112011; HMAS 249943), living culture, KUMCC 21-0019.
Known host and habitats: bamboo, freshwater, and terrestrial ([13], this study).
Known distribution: Yunnan, China ([13], this study).
Notes: Our collection is morphologically similar to Occultibambusa kunmingensis. Based on nucleotide comparisons of ITS, LSU, and SSU pairwise [23], the new isolate has consistent base pairs in comparison to the type strain of O. kunmingensis. Thus, we identify the new collection as O. kunmingensis. Occultibambusa kunmingensis was reported as a saprobe on decaying bamboo submerged in freshwater habitats in Yunnan, China [13] and it has never been reported from terrestrial habitats. Thus, we report this species as a saprobe on bamboo in terrestrial habitat for the first time, suggesting that this species can live in both terrestrial and/or aquatic environments. Alternatively, the freshwater records may have resulted from bamboo recently falling in water.
3.2.3. Seriascoma bambusae H.B. Jiang, K.D. Hyde & Phookamsak, sp. nov.
Index Fungorum number: IF558430; Facesoffungi number: FoF 09885; Figure 4
Figure 4.
Seriascoma bambusae (KUN-HKAS 112014, holotype). (a) Conidiomata on surface of dead bamboo culms; (b) Vertical section of conidioma; (c) Wall of conidioma; (d–j) Conidiogenous cells bearing conidia; (k) Conidia; (l) Germinating conidia; (m,n) Culture from above and reverse. Scale bars: (b) = 50 μm; (c) = 20 μm; (l) = 10 μm; (d–k) = 5 μm.
Etymology: The specific epithet “bambusae” refers to the host, bamboo, on which the new species was collected.
Holotype: KUN-HKAS 112014
Saprobic on dead culms of bamboo. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 170–380 μm diam., 110–150 μm high, solitary to gregarious, immersed under the host’s cortex, raised, becoming superficial, dull, black, elongate-conical to lenticular or dome-shaped, uni- to bi-loculate, glabrous. Locules 95–220 μm diam., 35–140 μm high, clustered, dark brown to black, subglobose. Peridium 10–35 μm thick, thin- to thick-walled, of unequal thickness, thick at the sides, thin at the base, composed of host and fungal tissue, with several layers of dark brown to black, pseudoparenchymatous cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 5.6–7.2 × 1.6–3.5 μm (x = 6.4 × 2.5 μm, n = 20), enteroblastic, phialidic, determinate, discrete, cylindrical to ampulliform, hyaline, aseptate, smooth-walled. Conidia 3.5–4 × 2–2.3 μm (x = 3.8 × 2.2 μm, n = 20), subglobose to ellipsoidal, hyaline, 2-guttulate, aseptate, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies were growing slowly on PDA, reaching 5 mm in one week at room temperature (10–20 °C), under normal light conditions, colonies cottony, circular, raised, greyish to dark brown from above and below. Mycelium superficial or immersed in media, with branched, septate, smooth hyphae.
Material examined: China, Yunnan Province, Honghe Autonomous Prefecture, Honghe County, on roadside (23°11′40.61″ N, 102°23′6.73″ E, altitude 2012.36 m), on dead culms of bamboo in terrestrial environment, 28 October 2020, H.B. Jiang, HONGHE018 (KUN-HKAS 112014, holotype) Ibid. (HMAS 249945, isotype), ex-type living culture, KUMCC 21-0021.
Notes: Seriascoma bambusae is typical of the asexual morph of Seriascoma in having immersed, eustromatic conidiomata and enteroblastic, phialidic, cylindrical to ampulliform, hyaline, aseptate conidiogenous cells bearing hyaline conidia. Seriascoma bambusae is most similar to Seriascoma sp. (KUMCC 21-0007) in having multi-loculate conidiomata [38], while S. didymosporum has uni-loculate conidiomata. However, S. bambusae can be distinguished from Seriascoma sp. (KUMCC 21-0007) in having smaller conidiomata (170–380 μm diam. vs. 320–510 μm diam.) and smaller, subglobose conidia (3.5–4 × 2–2.3 μm vs. 4.5–5 × 2–2.4 μm) [38]. Pairwise nucleotide comparison of ITS and TEF1-α sequence data also showed that S. bambusae differs from Seriascoma sp. (KUMCC 21-0007) in 22/ 502 bp (4.38%) and 26/ 928 bp (2.80%), respectively.
4. Discussion
Species of Occultibambusa have been discovered in both freshwater and terrestrial habitats (Table 2). Presently, all Occultibambusa species have been reported as saprobes on dead bamboo, indicating that the host preference of the genus is restricted to bamboo. Occultibambusa has currently been reported from China and Thailand (Table 2). More than 1500 bamboo species are distributed worldwide [39], especially in subtropical and tropical regions [40] Therefore, there is a high potential to discover more new species of the genus from bamboos in other regions [41]. Most species in Occultibambusa have similar morphology, but they can be distinguished by dimensions of ascostromata, asci and ascospores and color of ascospores (Table 2). In addition, significant phylogenetic distances of ITS, TEF1-α, and RPB2 can also be used.
Table 2.
Synopsis of morphological characteristics of Occultibambusa.
| Species Name | Sexual Morph | Origin | Host | Habitat | References | ||
|---|---|---|---|---|---|---|---|
| Ascostromata | Asci | Ascospores | |||||
| Occultibambusa aquatica | 180–280 × 100–250 μm, subglobose, brown to dark brown, papillate ostiole | 73–86 × 9–13 μm, clavate, with a short furcate pedicel | 19–25 × 3.5–6.5 μm, 2-seriate, narrow fusiform with acute ends, 1-septate, not constricted at the septum, brownish, with sheath | Chiang Rai, Thailand | Submerged bamboo | Freshwater | [8] |
| O. bambusae | 400–550 × 150–200 μm, subglobose, dark brown to black, papillate ostiole | (50–)60–80(−90) × (9.5–)11.5–14.5(−15) μm, broadly cylindrical, with a short furcate pedicel | (22–)23.5–27.5 × 4.5–7 μm, 2–3-seriate, slightly broad fusiform, 1-septate, not constricted at the septum, dark brown, with sheath | Chiang Rai, Thailand | Dead bamboo | Terrestrial | [4] |
| O. chiangraiensis | 352–520 × 195–295 μm, depressed globose to subglobose, brown to light brown, ostiole with a slit-like opening | 47–92 × 12–16 μm, clavate-oblong, with a short pedicel | 16–24 × 5–7 μm, 2-seriate, pale brown to reddish brown, fusiform, tapering towards the ends, (1–)3-septate, not constricted at the septa, without any mucilaginous sheaths and appendages | Chiang Rai, Thailand | Dead stem of Bambusoideae sp. | Terrestrial | [8] |
| O. fusispora | 240–275 × 135–185 μm, conical with wedged sides, brown to dark brown, papillate ostiole | (60–)65–90(−110) × (11–)12–14(−15)(−16) μm, clavate to cylindric-clavate, with a short furcate pedicel | (20–)22–25(−26) × 5–6(−6.5) μm, 2-seriate, fusiform with acute ends, light brown, 1–(2–3)-septate, not constricted at the septa, without any mucilaginous sheaths and appendages | Chiang Rai, Thailand | Dead bamboo | Terrestrial | [4] |
| O. hongheensis | 400–550 × 180–340 µm, ampulliform, conical to subglobose, black, ostiolate | (78–)80–130(–137) × (18–)19–23(–25) μm, cylindrical to clavate, with a short pedicel | (25–)27–30 × (5.5–)8–9(–10) μm, 2-seriate, inequilateral-fusiform, pale brown, 1-septate, slightly constricted at the septum, with a broad mucilaginous sheath | Yunnan, China | Dead bamboo | Terrestrial | This study |
| O. jonesii | 200–260 × 196–236 µm, subglobose, dark brown, papillate ostiole | (65–)75–89(–105) × 13.5–19 µm, broadly cylindrical to clavate, with a short pedicel | 27–33.5 × 5.5–6.5 µm, 1–3-seriate, inequilateral-fusiform, brown to grayish, 1-septate, constricted at the septum, without any mucilaginous sheaths and appendages | Guizhou, China | Dead bamboo | Terrestrial | [10] |
| O. kunmingensis | 220–260 × 110–150 μm, ellipsoidal, black, ostiolate | 110–140(–160) × 13–16.5 μm, cylindric-clavate, with a short to long pedicel | 32–40 × 5–6.5 μm, 3–4-seriate, fusiform, brown, 1-septate, constricted at the septum, without any mucilaginous sheaths and appendages | Yunnan, China | Submerged bamboo | Freshwater | [13] |
| 350–550 × 170–220 μm, ampulliform, dark brown to black, minutely papillate ostiole | (76.4–)83–106(–115) × (11–)12–14(–15) μm, cylindric-clavate to clavate, with a short pedicel or subsessile | (30–)34–36(–37.5) ×(4.5–)5–6 μm, 1–2-seriate, fusiform, brown to dark brown, 1–(3)-septate, slightly constricted at the septum, lacking a gelatinous sheath | Yunnan, China | Dead bamboo | Terrestrial | This study | |
| O. maolanensis | 544–600 µm diam., subglobose to slightly conical, dark brown, papillate ostiole | (66–)77–85(–94) × 17–20(–24) µm, broadly cylindrical to clavate, with a short pedicel | 25–31 × 8–10 µm, 2–4-seriate, inequilateral-fusiform, light brown, 1-septate, slightly constricted at the septum, without any mucilaginous sheaths and appendages | Guizhou, China | Dead bamboo | Terrestrial | [10] |
| O. pustula | 200–300 ×150–200 μm, conical, black, ostiolate | 80–105 × 8–12 μm, cylindrical, with a short furcate pedicel | 22–25 × 5–5.5 μm, 2–3-seriate, slightly broad-fusiform, hyaline to pale brown, 1-septate, not constricted at the septum, with sheath | Chiang Rai, Thailand | Dead bamboo | Terrestrial | [4] |
| 320–350 ×190–220 μm, ellipsoidal, black, papillate ostiole | (60–)78–125 × 12.5–15.5 μm, mostly broadly clavate or sometimes narrowly clavate, with a short or long pedicel | 22–29 × 6–8 μm, 1–2-seriate, fusiform, pale brown and 1-septate when young, dark brown and 3-septate when mature, constricted at the septa, without sheath | Yunnan, China | Submerged wood | Freshwater | [13] | |
The phylogenetic placement of Occultibambusa fusispora is unstable in several previous publications. Occultibambusa fusispora was separated from all Occultibambusa species and Versicolorisporium triseptatum in Dong et al. [13] and Wanasinghe et al. [15], while Phukhamsakda et al. [7] showed that Occultibambusa fusispora clustered with O. maolanensis and Versicolorisporium triseptatum. Therefore, in order to resolve this issue, we re-sequenced the ex-type living culture of Occultibambusa fusispora. Previously, Dai et al. [4] did not sequence the SSU region of this species, while we sequenced SSU, ITS, LSU, TEF1-α, and RPB2 regions. In our phylogeny, the newly generated sequences of O. fusispora (MFLUCC 11-0127II) are consistent with MFLUCC 11-0127 (100% ML, 1.00 PP; Figure 1) and separated well from all Occultibambusa species and Versicolorisporium triseptatum with high statistical support (98% ML, 1.00 PP; Figure 1).
Occultibambusa fusispora matches the typical morphology of sexual morph of Occultibambusa; however, it cannot be compared with asexual morphs of other Occultibambusa species because O. fusispora is the only species of this genus known in its holomorph, as the asexual morph was induced on bamboo pieces in vitro. In addition, our phylogeny showed O. fusispora is basal to Occultibambusa and Versicolorisporium clade. Therefore, in order to give a more reliable explanation for the placement of Occultibambusa fusispora, further studies on Occultibambusa species had better be focused on the induction of asexual morph sporulation in vitro. Induction of asexual morph sporulation in vitro can be performed by following the method described in Phookamsak et al. [42].
In the present study, Occultibambusa maolanensis and O. hongheensis clustered with Versicolorisporium triseptatum and were separated from the main Occultibambusa clade with low statistical support (73% ML, 0.71 PP; Figure 1). In addition, the nucleotide BLAST search of SSU sequence of V. triseptatum indicated that V. triseptatum has consistent base pairs with O. maolanensis. The phylogenetic position of O. maolanensis and V. triseptatum concurs with the studies of Dong et al. [13] and Wanasinghe et al. [15]. Occultibambusa maolanensis and O. hongheensis cannot be compared with Versicolorisporium triseptatum as they are known from different morphs. Occultibambusa maolanensis and O. hongheensis have the typical morphology of the sexual morph of Occultibambusa. The asexual morph of Occultibambusa is very different from Versicolorisporium. Therefore, the congeneric status of Occultibambusa and Versicolorisporium is pending further studies.
Versicolorisporium is a poorly known coelomycetous genus with V. triseptatum collected in Japan on dead culms of Pleioblastus chino and Sasamorpha borealis (bamboo) [14]. Fresh collections and sequencing of Versicolorisporium are needed in order to solve its confusing phylogenetic placement.
Serisacoma is presently known as saprobic on bamboo and dead and decaying wood in the terrestrial or freshwater habitats distributed in China and Thailand [4,12,13,38]. The genus accommodates only three species, suggesting that more taxa await discovery [41]. The sexual morphs of Seriascoma can be distinguished based on dimensions of ascostromata and ascospores, and the number of locules. The asexual morphs of Seriascoma can be distinguished based on dimensions of conidiomata and conidia, the number of locules, and the shape of conidia (Table 3).
Table 3.
Synopsis of morphological characteristics of Seriascoma.
| Species Name | Sexual Morph | Asexual Morph | References | ||||
|---|---|---|---|---|---|---|---|
| Ascostromata | Asci | Ascospores | Conidiomata | Conidiogenous Cells | Conidia | ||
| Seriascoma bambusae | N/A | N/A | N/A | 170–380 μm diam., 110–150 μm high, uni- to bi-loculate | 5.6–7.2 × 1.6–3.5 μm | 3.5–4 × 2–2.3 μm, subglobose to ellipsoidal | This study |
| S. didymosporum | 1000–1900 μm diam., 150–320 μm high, multi-loculate | (56–)60–75(−80) × 8–11(−13) μm | 11–12(−14.5) × 3–4 μm, clavate to fusiform, with upper cell shorter and wider than lower cell | 250–470 μm diam., 110–170 μm high, uni-loculate | 4–7(−8) × 1.5–3 μm | 4–5.5 × 1.5–2 μm, oblong, with rounded to obtuse ends | [4] |
| 200–250 μm diam., 120–170 μm high, uni-loculate | 70–95 × 9–11 μm | 10.5–14.5 × 3.5–5 μm, clavate to fusiform, with upper cell shorter and wider than lower cell | N/A | N/A | N/A | [13] | |
| S. yunnanense | 275–400 μm diam., 175–205 μm high, uni-loculate | 44–83 × 10–20 μm | 22–30 × 5–7.2 μm, slightly broad and fusiform, with upper cell larger than lower cell, surrounded by a gelatinous sheath | N/A | N/A | N/A | [12] |
Acknowledgments
We acknowledge the Biology Experimental Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the facilities of their molecular laboratory. Shaun Pennycook at Manaaki Whenua Landcare Research is thanked for help in the Latin naming of the new fungal species. Austin Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing. Hong-Bo Jiang would like to thank Mae Fah Luang University for his PhD scholarship. Chiang Mai University is thanked for partially supporting this research work.
Author Contributions
Conceptualization, H.-B.J., R.P., K.D.H. and S.C.K.; data curation, H.-B.J.; formal analysis, H.-B.J. and R.P.; funding acquisition, R.P., K.D.H., P.E.M., J.-C.X., J.K. and S.C.K.; methodology, H.-B.J. and R.P.; project administration, S.C.K.; supervision, R.P., K.D.H., P.E.M., J.-C.X., P.K., J.K. and S.C.K.; writing—original draft, H.-B.J. and R.P.; writing—review and editing, R.P., K.D.H., P.E.M., J.-C.X., P.K., J.K. and S.C.K. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the National Science Foundation of China (NSFC) under projects 31851110759 and 31850410489 (grant no. Y81I982211), “The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species” (grant no. DBG6080013), “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub-region” (grant no. RDG6130001), CAS President’s International Fellowship Initiative (PIFI) young staff (grant no. 2020FYC0002 and Y9215811Q1), Key Research Project Agroforestry Systems for Restoration and Bio-industry Technology Development (grant no. 2017YFC0505101), Ministry of Sciences and Technology of China (grant no. 2017YFC0505100), and Chiang Mai University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study are deposited in GenBank (Table 1). The finalized alignment and tree were submitted to TreeBASE (submission ID: 28553, https://www.treebase.org/ (accessed on 20 July 2021)). Specimens of new taxa and new collections obtained for this study have been deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), Yunnan, China and the Herbarium Mycologicum Academiae Sinicae (HMAS), Beijing, China. Living cultures have been deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and Kunming Institute of Botany Culture Collection, Kunming, China (KUMCC).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Boonmee S., Lücking R., Bhat J.D., Liu N., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 2.Wijayawardene N.N., Hyde K.D., Al-Ani L.K.T., Tedersoo L., Haelewaters D., Rajeshkumar K.C., Zhao R.L., Aptroot A., Leontyev D.V., Saxena R.K., et al. Outline of Fungi and fungus-like taxa. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- 3.Pem D., Hongsanan S., Doilom M., Tibpromma S., Wanasinghe D.N., Dong W., Liu N.G., Phookamsak R., Phillips A.J.L., Jeewon R., et al. https://www.dothideomycetes.org: An online taxonomic resource for the classification, identification, and nomenclature of Dothideomycetes. Asian J. Mycol. 2019;2:287–297. doi: 10.5943/ajom/2/1/19. [DOI] [Google Scholar]
- 4.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 5.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B.G., Maharachchikumbura S.S.N., Raspe’ O., Karunarathna S.K., Wanasinghe D.N., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 6.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.K., Bhat D.J., Taylor J.E., Bahkali A.H., McKenzie E.H.C., Hyde K.D. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2017;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 7.Phukhamsakda C., McKenzie E.H.C., Phillips A.J.L., Jones E.B.G., Bhat D.J., Stadler M., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 8.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H.C., Jones E.B.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q., et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 9.Jayasiri S.C., Hyde K.D., Jeewon R., Bhat D.J., Camporesi E., Kang J.C. Neooccultibambusajonesii, a novel taxon within Occultibambusaceae. Mycosphere. 2016;7:1458–1472. doi: 10.5943/mycosphere/7/9/17. [DOI] [Google Scholar]
- 10.Zhang J.F., Liu J.K., Hyde K.D., Yang W., Liu Z.Y. Fungi from Asian Karst formations II. Two new species of Occultibambusa (Occultibambusaceae, Dothideomycetes) from karst landforms of China. Mycosphere. 2017;8:550–559. doi: 10.5943/mycosphere/8/4/4. [DOI] [Google Scholar]
- 11.Tibpromma S., Hyde K.D., McKenzie E.H.C., Bhat D.J., Phillips A.J.L., Wanasinghe D.N., Samarakoon M.C., Jayawardena R.S., Dissanayake A.J., Tennakoon D.S., et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018;93:1–160. doi: 10.1007/s13225-018-0408-6. [DOI] [Google Scholar]
- 12.Rathnayaka A.R., Dayarathne M.C., Maharachchikumbura S.S.N., Liu J.K., Tennakoon D.S., Hyde K.D. Introducing Seriascomayunnanense sp. nov. (Occultibambusaceae, Pleosporales) based on evidence from morphology and phylogeny. Asian J. Mycol. 2019;2:245–253. doi: 10.5943/ajom/2/1/15. [DOI] [Google Scholar]
- 13.Dong W., Wang B., Hyde K.D., McKenzie E.H.C., Raja H.A., Tanaka K., Abdel-Wahab M.A., Abdel-Aziz F.A., Doilom M., Phookamsak R., et al. Freshwater Dothideomycetes. Fungal Divers. 2020;105:319–575. doi: 10.1007/s13225-020-00463-5. [DOI] [Google Scholar]
- 14.Hatakeyama S., Tanaka K., Harada Y. Bambusicolous fungi in Japan (7): A new coelomycetous genus, Versicolorisporium. Mycoscience. 2008;49:211–214. doi: 10.1007/S10267-008-0409-5. [DOI] [Google Scholar]
- 15.Wanasinghe D.N., Wijayawardene N.N., Xu J.C., Cheewangkoon R., Mortimer P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China) PLoS ONE. 2020;15:e0235855. doi: 10.1371/journal.pone.0235855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calabon M.S., Jones E.B.G., Boonmee S., Doilom M., Lumyong S., Hyde K.D. Five novel freshwater ascomycetes indicate high undiscovered diversity in lotic habitats in Thailand. J. Fungi. 2021;7:117. doi: 10.3390/jof7020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde K.D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K.W.T., Dayarathne M.C., de Silva N.I., Dissanayake A.J., Ekanayaka A.H., et al. Mycosphere notes 169–224. Mycosphere. 2018;9:271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- 18.Species Fungorum. [(accessed on 30 January 2021)]. Available online: http://www.speciesfungorum.org/Names/Names.asp.
- 19.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E.B.G., Liu N.G., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 20.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Hurdeal V.G., Pem D., Dissanayake L.S., Wijesinghe S.N., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 21.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat D.J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I., et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 22.Index Fungorum. [(accessed on 30 January 2021)]. Available online: http://www.indexfungorum.org/Names/Names.asp.
- 23.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 24.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Volume 18. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 25.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehner S. Primers for Elongation Factor 1-alpha (EF1-alpha) 2001. [(accessed on 30 December 2020)]. Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf.
- 27.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H.B., Hyde K.D., Jayawardena R.S., Doilom M., Xu J.C., Phookamsak R. Taxonomic and phylogenetic characterizations reveal two new species and two new records of Roussoella (Roussoellaceae, Pleosporales) from Yunnan, China. Mycol. Prog. 2019;18:577–591. doi: 10.1007/s11557-019-01471-9. [DOI] [Google Scholar]
- 29.Dissanayake A.J., Bhunjun C.S., Maharachchikumbura S.S.N., Liu J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere. 2020;11:2652–2676. doi: 10.5943/mycosphere/11/1/18. [DOI] [Google Scholar]
- 30.Katoh K., Rozewicki J., Yamada K.D. Mafft online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall T. Bioedit Version 6.0.7. 2004. [(accessed on 1 March 2021)]. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html.
- 32.Miller M.A., Pfeiffer W., Schwartz T. Creating the cipres science gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; New York, NY, USA: IEEE; 2010. pp. 1–8. [DOI] [Google Scholar]
- 33.Nylander J.A.A. MrModeltest2 v. 2.3 (Program for Selecting DNA Substitution Models Using PAUP*) Evolutionary Biology Centre; Uppsala, Sweden: 2008. [Google Scholar]
- 34.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 35.Rannala B., Yang Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 36.Zhaxybayeva O., Gogarten J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. Genomics. 2002;3:1–15. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaklitsch W.M., Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016;85:35–64. doi: 10.1016/j.simyco.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boonmee S., Wanasinghe D.N., Calabon M.S., Huanraluek N., Chandrasiri S.K.U., Jones E.B.G., Rossi W., Leonardi M., Singh S.K., Rana S., et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021 doi: 10.1007/s13225-021-00489-3. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai D.Q., Tang L.Z., Wang H.B. A review of bambusicolous ascomycetes. In: Abdul Khalil H.P.S., editor. Bamboo: Current and Future Prospects. IntechOpen; London, UK: 2018. pp. 165–183. [DOI] [Google Scholar]
- 40.Hyde K.D., Zhou D.Q., Dalisay T. Bambusicolous fungi: A review. Fungal Divers. 2002;9:1–14. [Google Scholar]
- 41.Hyde K.D., Jeewon R., Chen Y.J., Bhunjun C.S., Calabon M.S., Jiang H.B., Lin C.G., Norphanphoun C., Sysouphanthong P., Pem D., et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020;103:219–271. doi: 10.1007/s13225-020-00458-2. [DOI] [Google Scholar]
- 42.Phookamsak R., Norphanphoun C., Tanaka K., Dai D.Q., Luo Z.L., Liu J.K., Su H.Y., Bhat D.J., Bahkali A.H., Mortimer P.E., et al. Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 2015;74:143–197. doi: 10.1007/s13225-015-0352-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences generated in this study are deposited in GenBank (Table 1). The finalized alignment and tree were submitted to TreeBASE (submission ID: 28553, https://www.treebase.org/ (accessed on 20 July 2021)). Specimens of new taxa and new collections obtained for this study have been deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), Yunnan, China and the Herbarium Mycologicum Academiae Sinicae (HMAS), Beijing, China. Living cultures have been deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and Kunming Institute of Botany Culture Collection, Kunming, China (KUMCC).