Abstract
The mycotoxin fumonisin (FB) has become a major problem in maize products in southeastern Asia. Fumonisin can affect the health of humans and many animals. Fumonisin contamination can be reduced by detoxifying microbial enzyme. Screening of 95 potent natural sources resulted in 5.3% of samples yielding a total of five bacterial isolates that were a promising solution, reducing approximately 10.0–30.0% of fumonisin B1 (FB1). Serratia marcescens, one of the dominant degrading bacteria, was identified with Gram staining, 16S rRNA gene, and MALDI-TOF/TOF MS. Cell-free extract showed the highest fumonisin reduction rates, 30.3% in solution and 37.0% in maize. Crude proteins from bacterial cells were analyzed with a label-free quantification technique. The results showed that hydrolase enzymes and transferase enzymes that can cooperate in the fumonisin degradation process were highly expressed in comparison to their levels in a control. These studies have shown that S. marcescens 329-2 is a new potential bacterium for FB1 reduction, and the production of FB1-reducing enzymes should be further explored.
Keywords: fumonisin, mycotoxin reduction, Serratia marcescens
1. Introduction
Mycotoxins, which are secondary metabolites produced by fungi [1], cause serious problems to animal and human health. Plant pathogenic fungi are one of the fungal groups causing crop health problems. This damage has a direct effect on agricultural production and the economy [2]. Mycotoxins are accumulated during fungal colonization of plants by fungi before harvest. Fungi such as Fusarium graminearum, F. verticillioides, F. proliferatum, and sometimes Aspergillus flavus are present before harvest. Another group of fungi can occur after harvest, as reported for so-called storage fungi such as Penicillium verrucosum and A. flavus [3,4]
Fumonisins are mycotoxins that are mainly produced by F. verticillioides (Sacc.) Nirenberg (previously F. moniliforme, Sheldon) and F. proliferatum (Matsush.) Nirenberg [5]. To date, 28 structurally related fumonisin analogs have been identified. Three of the fumonisins B1, B2, and B3 occur abundantly [6]. Fumonisin B1 (FB1) is a highly toxic fumonisin analog that causes equine leukoencephalomalacia in horses, hepatocarcinogenesis in rats, and pulmonary edema in swine [7]. Maize products are primarily contaminated with fumonisins [8], which are related to starburst symptoms in maize [9,10]. In the Biomin world mycotoxin survey 2020, it was reported that fumonisins contaminated various commodities, especially maize. The contaminated maize was found at high levels of 96% in Asia, 70% in North America, and 71% in Europe. Some positive samples were reported to contain maximum concentrations of fumonisins at 30,872 ppb, 66,588 ppb, and 13,902 ppb, respectively [11].
Physical, chemical, and biological principles are used to develop strategies to eliminate fumonisin contamination in food and feed. Even so, physical, and chemical approaches have certain drawbacks in terms of costly instrumentation and nutritional losses. Biological detoxification using an enzyme technology is a promising strategy. Enzymes can reduce mycotoxin toxicity by transforming mycotoxins to less toxic metabolites. In some cases, the use of an enzyme can provide a practical approach to the nutrition in feed [12,13,14,15].
The first report of fumonisin microbial detoxification was given by Duvick et al. [16]. Microbes were isolated from moldy maize kernels and stalk tissue. They can grow with FB1 as their sole carbon source. They were Gram-negative bacteria identified as Exophiala spinifera and Rhinocladiella atrovirens. In 1999, Blackwell et al. [17] reported that the fungal species E. spinifera produces soluble extracellular esterase and can transform FB1 to hydrolyzed FB1, the amino polyol AP1, and free tricarballylic acid. The hydrolyzed FB1 has been demonstrated to have a greatly reduced toxicity compared to FB1 [18]. Benedetti et al. [19] reported that the bacterial strain NCB 1492, isolated from soil samples using an enrichment culture technique, can degrade FB1 as the sole carbon and nitrogen source in phosphate buffer. Sequences identified using 16S rDNA analysis were related to Delftia/Comamonas. Heinl et al. [20] investigated two genes involved in fumonisin degradation from Sphingopyxis sp. MTA144. The deesterification of FB1 to hydrolyzed FB1 was catalyzed by recombinant carboxylesterase in the same manner as the deamination of hydrolyzed FB1 in the presence of pyruvate and pyridoxal phosphate. In 2016, Masching et al. [21] noted that a commercial FUMzyme feed supplement that contains the fumonisin carboxylesterase FumD prevented changes in the sphinganine-to-sphingosin (Sa/So) ratio in turkeys and pigs. Hence, only a few microorganisms and enzymes have been successful in reducing FB1. The objectives of this study were to screen for fumonisin-degrading bacterial strains in natural sources and determine the proteomic profile of bacterial intracellular enzymes with a regard to FB1 reduction.
2. Results
2.1. Acclimatization and Isolation of Potential Fumonisin-Degrading Bacteria
Screening of bacteria for fumonisin degradation was performed through an acclimatization process. After acclimatization of natural sources such as maize, rice, soil, and fermented fluid with crude fumonisins, the degradation efficiency was determined. From the potential samples from 95 natural sources, we found five samples from which FB1-degrading bacteria were isolated. The FB1-degrading bacteria from natural sources were in approximately 5.3% of the natural source samples. Four were found in maize samples (isolates S2, 302-2, 329-2, and 412), and one was from fermented fluid (isolate P1) (Table 1). All bacteria were purified and collected for further study.
Table 1.
Number of samples collected from various natural sources.
| Natural Source | Number of Screened Samples |
Potential Degrading Samples |
|---|---|---|
| Maize | 37 | 4 |
| Rice | 12 | 0 |
| Soil | 8 | 0 |
| Fermented fluid | 38 | 1 |
| Total | 95 | 5 |
2.2. Fumonisin B1 Removal Activity by a Selected Bacterial Isolate and Determination of the Active Components
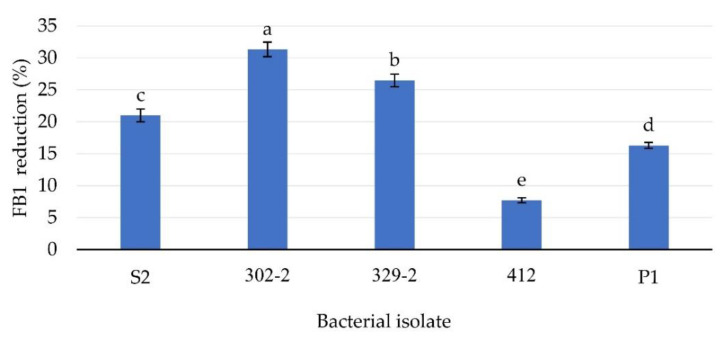
Each selected bacterial isolate was tested using PBS containing FB1 at 5 ppm and the FB1 degrading activities were observed. The reduction rates of these isolates ranged from 7.72% to 31.34% after 24 h of incubation. The highest FB1 reduction rate (31.34%) was exhibited by bacterial isolate 302-2, followed by isolate 329-2 at 26.48% (Figure 1).
Figure 1.
Percentage of FB1 reduction by all isolates after 24 h of incubation with the FB1 standard at 5 ppm. Different lowercase letters above the columns represent significant differences by ANOVA (p < 0.05), n = 5.
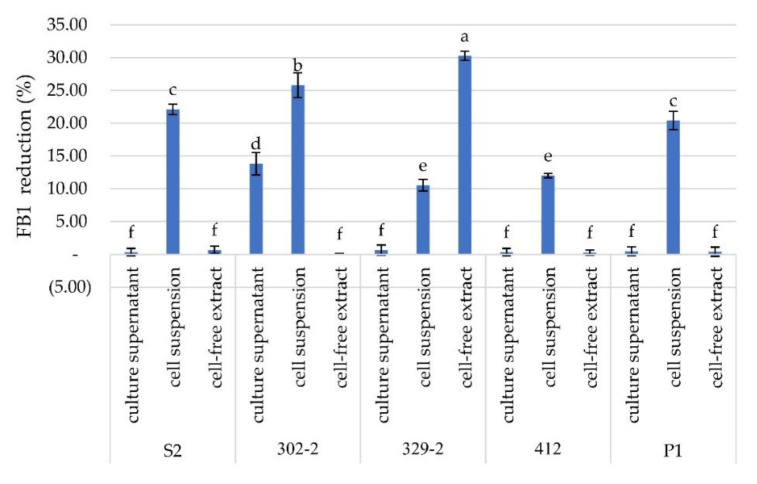
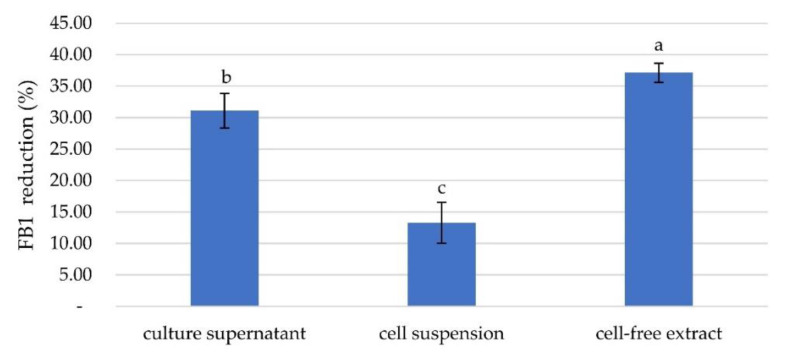
For further investigation, three bacterial preparations, the cell suspension, culture supernatant, and cell-free extract, were prepared from the five bacterial isolates. The efficiency of FB1 degradation was determined for each portion. The percent reduction ranged from 0–30.29%. The highest reduction occurred with treatment by isolate 329-2 cell-free extract, which resulted in a reduction rate of 30.29%, followed by reductions of 25.80% using the cell suspension of 302-2 and 22.13% using the cell suspension of S2. The reduction rate of 302-2 culture supernatant was 13.82%, while those of cell suspensions of 412 and 329-2 were 12.02% and 10.55%, respectively. Culture supernatants and cell-free extract of bacterial isolates S2, 412, and P1 showed no reduction; moreover, the culture supernatant of isolate 329-2 was effective. Each portion showed a different reduction rate. This result might be due to the active protein involved in FB1 reduction, which was contained in different bacterial fractions. (Figure 2). An in situ study showed that bacterial isolate 329-2 was the most capable of FB1 degradation. Then, further study of cell-free bacterial isolate 329-2 extract was performed in ground maize. The cell-free extract of 329-2 had the highest reduction rate at 37.00%, followed by the culture supernatant at 31.30% and by the cell suspension at 13.40% (Figure 3).
Figure 2.
Percentage of FB1 reduction by culture supernatants, cell suspensions, and cell-free extracts in solution after 24 h of incubation with FB1 standard at 5 ppm. Different lowercase letters above the columns represent significant differences by ANOVA (p < 0.05), n = 5.
Figure 3.
Percentage of FB1 reduction by the culture supernatant, cell suspension, and cell-free extract in ground maize after 24 h of incubation with the FB1 standard at 5 ppm. Different lowercase letters above the columns represent significant differences by ANOVA (p < 0.05), n = 5.
2.3. Bacterial Identification
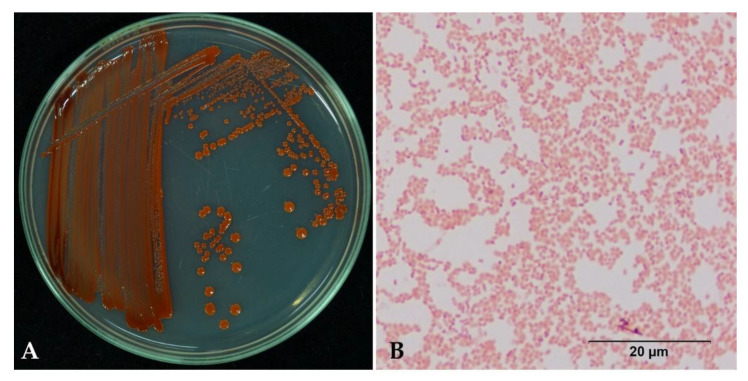
Bacterial isolate 329-2 was isolated from maize and formed red pigmentation on round colonies and an entire margin on NGA after 48 h of incubation under aerobic conditions. The isolate was a rod-shaped and Gram-negative bacterium (Figure 4).
Figure 4.
Microscopic and macroscopic examination of Serratia marcescens 329-2. (A) The colony morphology of 329-2 on nutrient glucose agar. (B) Rod-shaped cells observed by microscope.
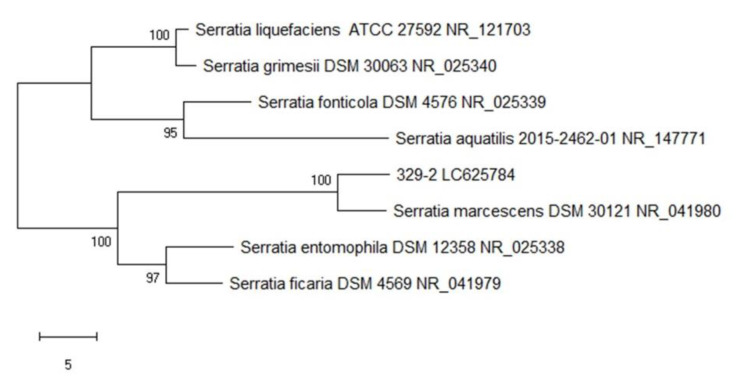
The relationships of isolate 329-2 and other closely related bacterial species are shown in Figure 5. Isolate 329-2 appears to be closely related to Serratia marcescens. Furthermore, a BLAST search at the NCBI indicated that the 16S rDNA sequence of isolate 329-2 was most similar to that of Serratia marcescens DSM 30121 (accession number: NR_041980). The 16S rDNA sequence of isolate 329-2 has been deposited in the GenBank database under accession number LC625784.
Figure 5.
Phylogenetic tree based on 16S rRNA gene sequences of isolate 329-2 and related taxa.
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/TOF MS) showed that isolate 329-2 belonged to S. marcescens, and the highest score (2.361) was for S. marcescens DSM 12481 (Table 2).
Table 2.
MALDI-TOF/TOF MS analysis of Serratia marcescens 329-2.
| Rank | Quality | Matched Pattern | Score |
|---|---|---|---|
| 1 | +++ | Serratia marcescens DSM 12481 DSM | 2.361 |
| 2 | +++ | Serratia marcescens DSM 12485 DSM | 2.345 |
| 3 | +++ | Serratia marcescens 13103_1 CHB | 2.319 |
| 4 | +++ | Serratia marcescens subsp. marcescens DSM 30121T DSM | 2.306 |
| 5 | ++ | Serratia marcescens subsp. sakuensis CIP 107489T HAM | 2.240 |
| 6 | ++ | Serratia ureilytica DSM 16952T DSM | 2.155 |
| 7 | ++ | Serratia marcescens DSM 30122 DSM | 2.147 |
| 8 | ++ | Serratia marcescens (PX) 24086109 MLD | 2.062 |
| 9 | ++ | Serratia marcescens DSM 12483 DSM | 2.039 |
| 10 | + | Serratia entomophila DSM 12358T DSM | 1.942 |
2.4. Proteins Expression during Fumonisin B1 Reduction by S. marcescens 329-2
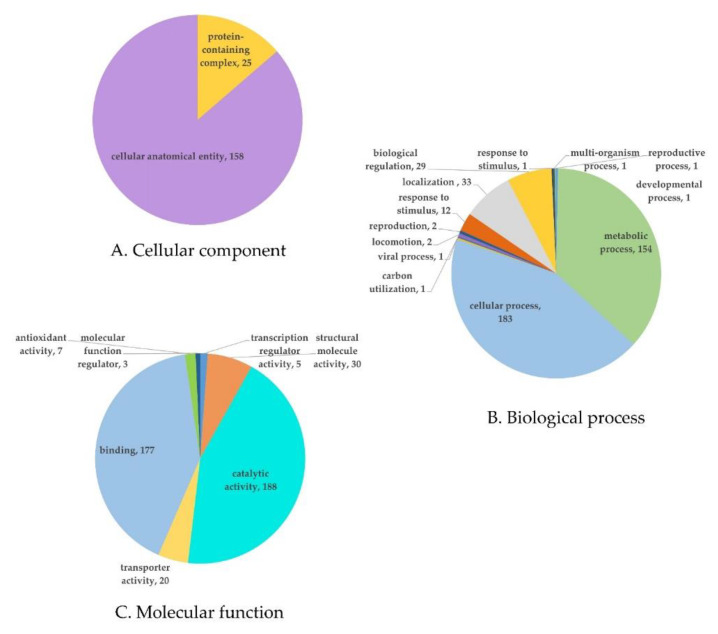
The protein expression results identified 461 differentially expressed proteins with p < 0.05, and of these proteins (Supplementary Tables S1 and S2), 159 showed upregulated expression and 25 showed downregulated expression in the treatment group. To evaluate the expression of protein functions, we annotated these proteins based on the gene ontology (GO) databases. The identified level 2 GO terms (related to cellular components, molecular functions, and biological processes) associated with the differentially expressed proteins are shown in Figure 6. Gene annotation of the expressed proteins showed their relationship to cellular components for 183 proteins, biological processes for 421 proteins, and molecular functions for 430 proteins. Major concerns exist regarding the biological process in which they are activated, and the proteins involved in cellular process (183) and metabolic process (154). In the category of molecular function, the majority of proteins were related to catalytic activity (188 proteins) and protein binding (177 proteins) (Figure 6).
Figure 6.
Gene ontology (GO) classifications of the differentially expressed proteins during fumonisin degradation by S. marcescens 329-2.
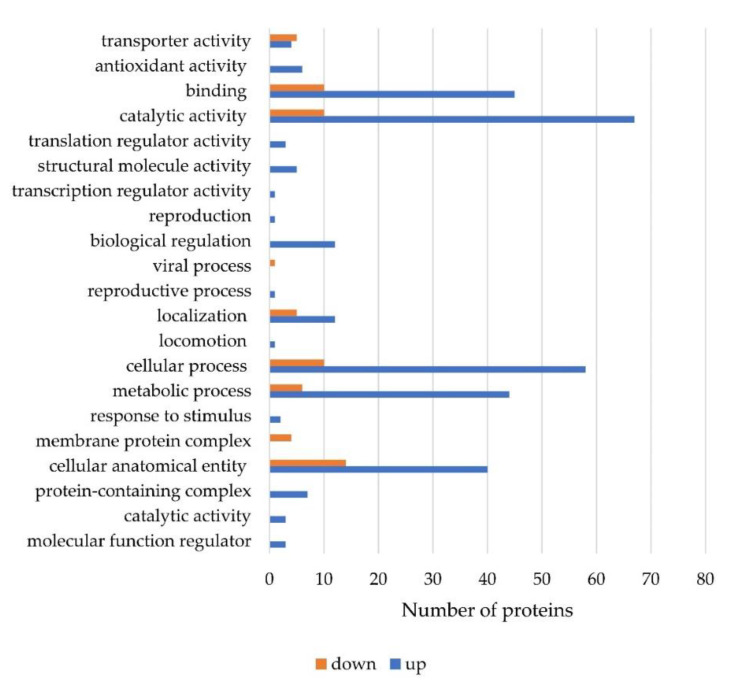
The highly matching proteins upregulated in FB1 degradation were categorized into catalytic activity, binding protein function, and metabolic process under in molecular function (Figure 7). The protein expression data are shown in Table 3 and Table 4, with the fold change values in expression relative to the noninduced protein levels from S. marcescens. The upregulated proteins in Table 3 indicate the proteins that were highly related to FB1 degradation by S. marcescens. The details of each protein were compared within the UniProt database. This indicates the function related to the protein degradation of FB1. In relation to FB1, proteins involved in the cell catalysis process include hydrolase proteins and aminotransferase proteins. The upregulated hydrolase function was evidenced by entries A0A6N3ZRH0 (fumarylacetoacetate hydrolase family protein, 4.91), A0A656VL53 (alpha/beta hydrolase, 4.90), A0A6I4GZS8 (hydrolase, 3.80), and A0A6M5I193 (MBL fold metallo-hydrolase, 3.64), and the transferase enzymes that can activate chemical groups in FB1 included A0A6H1E4N5 (Acetylornithine/succinyl-diaminopimelate aminotransferase, 4.1), Q6MXC8 (methyl transferase, 3.94), A0A656V5R8 (5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, 3.46), and A0A0U6KIH4 (GTP cyclohydrolase, 3.34). Other proteins included the isomerase A0A0G8B4P9 (peptidyl-prolyl cis-trans isomerase, 3.58), the protein synthesis protein A0A6M5HVT2 (4-hydroxy-tetrahydrodipicolinate synthase, 5.33), and the cell metabolism protein V5YV29 (maltodextrin-binding protein, 6.89). ABC transporter, cellular process, transcription, nucleic acid proteins were also induced, which means that cellular processes were also activated for the degradation response.
Figure 7.
Comparison of protein upregulation and downregulation during fumonisin degradation by S. marcescens 329-2.
Table 3.
Identification of the upregulated proteins (>3-fold change) in FB1-treated S. marcescens 329-2 compared with the control group.
| Entry | Protein Names | Gene Names | Fold Change |
|---|---|---|---|
| A0A6G9UZ48 | ABC transporter substrate-binding protein | HCG50_10660 | 8.20 |
| A0A6N0D898 | Porin OmpC | ompC | 7.94 |
| A0A3E2ENK9 | Amino acid ABC transporter substrate-binding protein | gltI | 7.12 |
| V5YV29 | Maltodextrin-binding protein | malE | 6.89 |
| A0A080V044 | Universal stress protein | uspA | 5.42 |
| A0A6M5HVT2 | 4-hydroxy-tetrahydrodipicolinate synthase | dapA | 5.33 |
| A0A6G9UQE2 | ABC transporter substrate-binding protein | HCG50_08880 | 5.33 |
| A0A1Q4NZ53 | Superoxide dismutase | BHU62_14220 | 4.99 |
| A0A6G9UU24 | Phosphate-binding protein PstS | pstS | 4.98 |
| A0A6N3ZRH0 | Fumarylacetoacetate hydrolase family protein | G3M84_1332 | 4.91 |
| A0A656VL53 | Alpha/beta hydrolase | AB868_03825 | 4.90 |
| A0A6M5HYD0 | Phenylacetate-CoA oxygenase/reductase subunit PaaK | paaK | 4.78 |
| A0A3E2EF40 | Malate dehydrogenase | mdh | 4.68 |
| A0A6N0D0Q5 | Neutral metalloproteinase | F0335_18215 | 4.49 |
| A0A086FBX8 | Transcription termination/antitermination protein NusG | nusG | 4.39 |
| A0A2V4FJ05 | Peptide deformylase | def | 4.35 |
| V5YU98 | Extracellular solute-binding protein | E4655_11925 | 4.27 |
| A0A5Q8BY15 | YtfJ family protein | EGJ31_19890 | 4.20 |
| A0A1Q5WAZ4 | Oligopeptide ABC transporter substrate-binding protein OppA | A8A12_03045 | 4.18 |
| A0A6H1E4N5 | Acetylornithine/succinyldiaminopimelate aminotransferase | argD | 4.10 |
| A0A6N3ZYZ9 | 2,3-diphosphoglycerate-dependent phosphoglycerate mutase | gpmA | 4.06 |
| A0A0P0Q8S3 | ABC transporter substrate-binding protein | AR325_02675 | 4.06 |
| A0A6N0CVJ7 | Superoxide dismutase | sodB | 4.03 |
| A0A5Q8C0J8 | Organic hydroperoxide resistance protein | EGJ31_14360 | 4.02 |
| V5YUS0 | Periplasmic serine endoprotease DegP-like | degQ | 4.01 |
| Q6MXC8 | Methyltransferase | SMR0272 | 3.94 |
| A0A2S4XAJ8 | Surface composition regulator | glgS | 3.89 |
| A0A1Q5WEW3 | Antibiotic biosynthesis monooxygenase | A8A12_06980 | 3.87 |
| A0A6I4GZS8 | Hydrolase | GMA22_24835 | 3.80 |
| A0A221FKL4 | UPF0234 protein BVG93_01845 | BVG93_01845 | 3.75 |
| A0A6H3S2C0 | ATP-dependent protease subunit HslV | hslV | 3.71 |
| A0A6M5I193 | MBL fold metallo-hydrolase | HMI62_20840 | 3.64 |
| A0A6N0DB57 | Protein deglycase HchA | hchA | 3.62 |
| A0A2V4G7I4 | Amino acid ABC transporter substrate-binding protein | glnH | 3.62 |
| A0A5C7CH16 | VOC family protein | FOT62_15570 | 3.58 |
| A0A0G8B4P9 | Peptidyl-prolyl cis-trans isomerase | fkpA | 3.58 |
| A0A656VU86 | Long-chain fatty acid transport protein | AB868_00683 | 3.55 |
| A0A2S4 × 857 | Histidine ABC transporter substrate-binding protein HisJ | hisJ | 3.47 |
| A0A656V5R8 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | metE | 3.46 |
| A0A0G8BFE1 | Cystine ABC transporter substrate-binding protein | tcyJ | 3.43 |
| A0A1C3HIZ5 | Aconitate hydratase B | acnB | 3.35 |
| A0A0U6KIH4 | GTP cyclohydrolase 1 | folE | 3.34 |
| A0A6N0CW48 | Branched-chain amino acid ABC transporter substrate-binding protein | F0335_15805 | 3.32 |
| A0A6N3ZXZ8 | ABC transporter substrate-binding protein | G3M84_09620 | 3.32 |
| A0A6G8TTH4 | Autoinducer 2-binding protein LsrB | G5643_21680 | 3.29 |
| A0A656VPU2 | Uncharacterized protein | AB868_00798 | 3.26 |
| A0A1C3HHX7 | Nitrogen regulatory protein P-II | glnB | 3.23 |
| A0A1Q5WH71 | Thiol:disulfide interchange protein | dsbA | 3.22 |
| A0A0G8B466 | 2-dehydro-3-deoxygluconokinase | AR325_02155 | 3.22 |
| A0A0M5K334 | Transaldolase | tal | 3.18 |
| A0A080UWJ0 | Peptidyl-prolyl cis-trans isomerase | fklB | 3.15 |
| A0A6N0D450 | Two-component system response regulator BaeR | baeR | 3.13 |
| A0A6N0CZA0 | Glucose-6-phosphate isomerase | pgi | 3.10 |
| A0A0F6KTS7 | 2-dehydro-3-deoxy-phosphogluconate aldolase | eda | 3.10 |
| V5YUY6 | Stringent starvation protein A | sspA | 3.09 |
| A0A6M5HTX8 | Uncharacterized protein | HMI62_14785 | 3.08 |
| A0A1Q4NZT3 | DUF1471 domain-containing protein | BHU62_12635 | 3.03 |
| A0A086FJA0 | 50S ribosomal protein L9 | rplI | 3.02 |
Table 4.
Identification of the downregulated proteins (<0.50-fold change) in FB1-treated S. marcescens 329-2 compared with the control group.
| Entry | Protein Names | Gene Names | Fold Change |
|---|---|---|---|
| A0A514I8F1 | DUF3251 domain-containing protein | FG174_04965 | 0.50 |
| A0A086FBJ7 | Outer membrane protein assembly factor BamD | bamD | 0.48 |
| A0A6G8TR73 | Outer membrane lipoprotein RcsF | rcsF | 0.48 |
| A0A080UZC4 | Tol-Pal system protein TolR | tolR | 0.48 |
| A0A2V4H3Z0 | Ribose-phosphate pyrophosphokinase | prs | 0.47 |
| V5YU02 | Divisome-associated lipoprotein YraP | yraP | 0.47 |
| A0A0M4S0F9 | Glutamine synthetase | glnA | 0.47 |
| A0A379ZG33 | Phage shock protein A | pspA | 0.47 |
| Q5J5B8 | Outer membrane protein assembly factor BamC | nlpbsm | 0.46 |
| A0A379ZA39 | Outer membrane protein slp | slp | 0.46 |
| A0A080V8M2 | Heat shock chaperone IbpB | ibpB | 0.46 |
| A0A6I4HK57 | Outer membrane protein assembly factor BamB | bamB | 0.46 |
| A0A0P0QBU0 | Glycoprotein-polysaccharide metabolism protein | A8A12_17600 | 0.46 |
| A0A6I6ZSG9 | ATP synthase subunit alpha | atpA | 0.46 |
| A0A6N0CYE2 | Phosphate acetyltransferase | pta | 0.45 |
| A0A6N0CUX7 | Glycine-tRNA ligase subunit beta | glyS | 0.43 |
| A0A1Q5WD75 | NAD(P)H dehydrogenase (quinone) | wrbA | 0.42 |
| A0A6I6ZPJ4 | Terminase | GV243_19590 | 0.40 |
| A0A3E2EPF1 | Outer membrane | ompA | 0.39 |
| A0A1Q4NYR4 | ATP synthase gamma chain | atpG | 0.38 |
| A0A6M5IGG0 | SPOR domain-containing protein | HMI62_24000 | 0.35 |
| A0A1Q4P2F3 | Fructose-1,6-bisphosphatase class 1 | fbp | 0.34 |
| A0A6H3SAN4 | Glyceraldehyde-3-phosphate dehydrogenase | gapA | 0.34 |
| A0A0M3UJK8 | ATP synthase subunit beta | atpD | 0.30 |
| A0A6G8TKN9 | Uncharacterized protein | Uncharacterized protein | 0.21 |
Proteins that were downregulated (Table 4), which means that the proteins that were highly present in the control group, were mostly related to cellular anatomical entities and cellular processes and included proteins such as ATP synthase, glyceraldehyde-3-phosphate dehydrogenase, fructose-1,6-bisphosphatase class, and outer membrane proteins.
3. Discussion
FB1 is one of the most important mycotoxins produced by several species of Fusarium, mainly F. verticillioides or F. proliferatum, which frequently occur in maize kernels and affect grain quality [22,23]. FB1 is a potential natural contaminating toxin. The fumonisin-contaminated products increase yearly with global warming [24]. The degradation of fumonisins is a concern because it causes contamination of feed products. To decrease the severity of contamination with fumonisin-producing fungi in the field before harvest, biocontrol agents against fumonisin-producing fungi have been studied, such as lactic acid bacteria from corn silage [25], Pediococcus pentosaceus (L006) isolated from maize leaves [26], and Lactobacillus plantarum MYS6, a probiotic bacterium [27]. In an early report, Becker et al. [28] treated 50 to 1000 µM fumonisin with human intestinal bacteria. The results showed no effect on fumonisin decrease, bacterial growth, or metabolic substances. From our study, only 5.3% of natural tested samples (n = 95) showed FB1 degradation with the acclimatized method. Most of the potential samples were from maize and fermented fluid. Within this study, five bacterial isolates caused reductions of 8 to 32%. Although fumonisins are a highly important mycotoxin in maize samples worldwide, only some microorganisms, such as Exophiala spinifera isolate 2141.10, Rhinocladiella atrovirens and the Gram-negative bacterium 2412.1 isolated from maize [16,17], the Delftia/Comamonas group isolated from soil [19], and the microbial consortium SAAS79 isolated from spent mushroom compost [12], have been reported to effectively degrade FB1 at the post-harvest stage. In this study, the cell-free extract showed the highest fumonisin reducing rate, at 40% in solution and 30% in maize. The FB1 reduction factor may be a crude enzyme from inside bacterial cells [12,29].
In this study, we identified a bacterial isolate based on Gram staining, the 16S rRNA gene, and MALDI-TOF MS. Regarding the macroscopic characteristics, bacterial isolate 329-2 formed red pigment, round colonies, and an entire margin. The bacterium was Gram-negative, straight rod–shaped and 0.6–0.8 µM in diameter, similar to S. marcescens [30,31].
The 16S rRNA gene, used to investigate bacterial phylogeny and taxonomy, is the most frequent housekeeping genetic marker and is more reliable than other genes for various reasons. First, the 16S rRNA gene is present in all bacteria, typically as part of a multigene family or operon. Second, the 16S rRNA gene is a more accurate measure over time and is not altered, implying that random sequence changes are a more accurate measure of time. Finally, the 16S rRNA gene is large enough for informatics purposes [32,33,34]. The strain here whose 16S rRNA gene was sequenced was identified as a S. marcescens strain closely related to S. marcescens DSM 30121 (accession number: NR_041980). The 16S rRNA gene was also successfully used for Serratia species identification [35,36].
MALDI-TOF MS is an efficient high-throughput technology for identifying and evaluating proteins [37,38]. MALDI-TOF MS was successful for the identification of proteins from whole bacterial cells from various sources [39,40,41] and from S. marcescens [42,43]. In this study, the MALDI-TOF/TOF MS results showed isolate 329-2 belonging to S. marcescens. The highest score for matching was 2.361 for S. marcescens DSM 12481, indicating highly probable species identification [44]. Based on Gram staining, 16S rRNA gene sequence analysis, and MALDI-TOF MS, we conclude that bacterial isolate 329-2 was S. marcescens.
S. marcescens has been reported to be a potential biocontrol agent for plant pathogens causing several diseases such as damping-off disease in cyclamen caused by Rhizoctonia solani [45], damping-off disease in cucumber caused by Phytophthora capsici [46], and blast disease caused by Pyricularia oryzae in rice [47]. Moreover, S. marcescens has been demonstrated to be a plant growth-promoting agent inducing systemic resistance in cucumber against Fusarium wilt disease caused by F. oxysporum [48] and it can induce systemic resistance, enhanced salinity tolerance, and inhibit F. graminearum infection in wheat [49]. Guo et al. [50] reported that S. marcescens inhibited the germination of F. proliferatum and suppressed fumonisins accumulation in an in vitro study. However, the use of S. marcescens as a biocontrol agent was still concerned about a human opportunistic pathogen. In our study, we focused on intracellular enzymes without bacterial living cells. Red-pigmented S. marcescens 329-2 was used for a potential new enzyme from bacterial sources. It was not similar to the strain described by Carbonell et al. [51], as the non-pigmented strain of S. marcescens was mostly a human opportunistic pathogen.
To understand the proteins related to the degradation process, bacterial cells were analyzed with label-free techniques. Label-free MS-based quantitative proteomic analysis was attempted to further characterize fumonisin degradation. Many abundant proteins identified and quantified in the degradation process were upregulated, as shown in Table 3, or downregulated, as shown in Table 4. The major points of interest were indicated in a previous report by Blackwell, Gilliam, Savard, Miller and Duvick [17] on a soluble extracellular esterase from E. spinifera isolate 2141.10, which transformed fumonisin B1 to the amino polyol AP1 and free tricarballylic acid. Moreover, two genes encoding a carboxylesterases (fumD) and aminotransferases (fumI) for fumonisin degradation by esterification and hydrolysis were described from the bacterium Sphingopyxis sp. [20,52].
The label-free quantification data showed upregulated esterase and transferase proteins. Potential proteins associated with fumonisin degradation, including hydrolases and transferases, were examined. Potential proteins associated with fumonisin degradation included alpha/beta hydrolase (A0A656VL53) and acetylornithine/succinyldiaminopimelate aminotransferase (A0A6H1E4N5), with fold changes of 4.90 and 4.10, respectively.
Alpha/beta hydrolases are hydrolase families. Hydrolases are a group of enzymes that act as biochemical catalysts. A hydrolase is an enzyme that catalyzes hydrolysis of C-O, C-N, C-C, and phosphoric anhydride bonds. The enzymes use H2O to break a chemical bond, which typically results in the degradation of a larger molecule into smaller molecules. Hydrolases are classified as EC 3 enzymes. One common example of hydrolase enzymes is esterases, which include enzymes such as lipases, phosphatases, glycosidases, peptidases, and nucleosidases [53,54]. Montella et al. [55] reported that esterases hydrolyze ester bonds, which are present in a wide range of insecticides, including fumonisin. Fumonisin B1 esterase (EC 3.1.1.87) [20]. Later, they named the gene encoding carboxylesterase activity fumD. We assume that alpha/beta hydrolase may be involved in FB1 conversion to HPB1 and tricarboxylic acids.
Transferases, an enzyme class, can transfer various chemical groups from one compound to another. The enzymes work with functional groups such as the amino group (-NH2) (transferred from amino acids to keto groups in the case of transaminase), phosphate, methyl (-CH3), and sulfur-containing groups. The enzymes may react with one end of fumonisin (-CHNH2CH3). One of the enzymes identified was acetylornithine/succinyldiaminopimelate aminotransferase, which is related to the -CHNH2CH3 end of the fumonisin structure.
The results revealed that the degradation activity of S. marcescens was related to upregulated hydrolase and transferase enzymes. The specific enzymes of S. marcescens can decrease FB1 abundance, but they may show different specificities for the structure of fumonisin than the carboxylesterase and aminotransferase from Sphingopyxis sp. that have been marketed as FUMzyme.
4. Conclusions
In this study, we isolated and identified the bacterium S. marcescens 329-2 from maize with high FB1 reduction activity. This is the first report of S. marcescens reducing FB1. The FB1 reduction by the cell-free extract of S. marcescens 329-2 was more effective than that by the culture supernatant and cell suspension in FB1 solution and maize. We also found hydrolase and transferase with upregulated expression in bacterial cells, which may indicate potential for the development of FB1-reducing enzymes.
5. Materials and Methods
5.1. Acclimatization and Isolation of Potential Fumonisin-Degrading Bacteria
5.1.1. Acclimatization of Bacteria from Natural Resources
Natural sources such as maize (starburst symptom and fumonisin-contaminated samples), rice (bakanae disease samples), soil, and fermented fluid were collected randomly from the continuous production area in Thailand. Five grams or milliliters of sample was added to 50 mL of nutrient glucose broth (NGB, 3 g of beef extract, 5 g of peptone, and 5 g of glucose) and incubated for 24 h followed by transfer of 1 mL into 20 mL of NGB with crude fumonisins (total fumonisins B1, B2, and B3) at 3 ppm, which refer to average level of positive sample of maize contaminated fumonisins in Asia [11]. The crude fumonisins were prepared by the inoculation of ground maize with F. verticillioides (fumonisins-producing strain). After 45 d, the crude fumonisins were extracted with 70% methanol and filtrated samples were diluted 1:20 with distilled water before taking into ELISA assay (AgraQuant® Total Fumonisin Assay, Romer Lab®, Singapore), range 0.25–5.0 ppm with LOD = 0.20 ppm and LOQ = 0.25 ppm. Two hundred microliters of conjugation solution were mixed with 100 μL of each standard or sample. One hundred microliters of the mixture were transferred into the reaction wells and incubated for 10 min. The reaction wells were washed 5 times with distilled water and added with 100 μL of substrate solution. After 5 min of incubation at RT, one hundred microliters of stop solution were added into the mixture. Absorbance was measured by a microplate reader using a 450 nm filter (Tecan, Hombrechtikon, Switzerland). The data was interpreted by the AQ FUM form provided by the company.
5.1.2. Isolation of Potential Fumonisin-Degrading Bacteria
The acclimatized samples were incubated at room temperature on shaker in the dark for 15 d. One hundred microliters of bacterial suspension was prepared by using agar spread on nutrient glucose agar (NGA) plates. The plates were incubated with alternating periods of 12 h darkness/light at 25 ± 2 °C for 24 h. A single colony of the withstand bacteria was cross streaked on NGA three times. The pure culture was used for analysis of reduced FB1 activity.
5.2. Fumonisin B1 Removal Activity by a Selected Bacterial Isolate
Bacteria were identified based on the removal of FB1 as described by Niderkorn, Morgavi, Pujos, Tissandier and Boudra [25] with some modifications. Overnight culture of a bacterial pellet was adjusted to 0.2 OD using PBS, pH 7 (phosphate buffer solution; 8 g of NaCl, 0.2 g of KCl, 0.2 g of KH2PO4, and 1.44 g of Na2HPO4). One hundred microliters of bacterial suspension was mixed with FB1 solution (Biopure, Tulln, Austria) to final concentration of 5 ppm in a 1000 µL reaction. The concentration was set at 5 ppm according to the European Union maximum limits for fumonisins B1 and B2 established in the complementary and complete feeding stuffs for pigs, horses, rabbits, and pet animals [56]. The high FB1 concentration was set to strengthen the screening of potential selected bacterial isolates in a short period of 24 h. A positive control containing only FB1 in PBS and a negative control containing only a bacterial suspension in PBS were used. All tubes were incubated at 37 °C in the dark for 24 h. After incubation, the samples were determined the FB1 concentration with ELISA assay (AgraQuant® Total Fumonisin Assay, Romer Lab®, Singapore), range 0.25–5.0 ppm with LOD = 0.20 ppm and LOQ = 0.25 ppm, followed topic 5.1.1. The rate of FB1 degradation was calculated using the following formula: (concentration of FB1 control-concentration of FB1 residual)/concentration of FB1 control × 100%.
5.3. Determination of the Active Components
The bacterial culture was separated into culture supernatant, cell suspension, and cell-free extract fractions. All components were used to screen for fumonisin-degrading activity according to Wang et al. [57]. The culture supernatant and cell suspension were obtained by centrifugation at 10,000 rpm and 4 °C for 15 min. Then, the culture supernatant was filtered through 0.22 μm sterile cellulose acetate filters. The cell pellet was washed twice with PBS, pH 7, before being suspended again in the same buffer. Cell-free extract was prepared by disintegrating (5 s on/off) the cell suspension using a sonicator for 30 min in an ice bath. After that, the cell debris was removed by centrifugation at 13,000 rpm and 4 °C for 15 min. To obtain cell-free extract, the supernatant was filtered through 0.22 μm sterile cellulose acetate filters.
FB1 removal activity was determined as follows: 250 µL of FB1 (10 µg/mL) was mixed with 250 µL of the culture supernatant, cell suspension, or cell extract. All reaction systems were placed on a rotary shaker for 24 h.
To determine the potential of active components in a realistic matrix, we used 20 g of 5 ppm spiked ground maize in a 50 mL laboratory bottle. Then, five hundred microliters of the culture supernatant, cell suspension, or cell-free extract was dropped in the middle of the ground maize and incubated for 24 h at RT.
Each sample was extracted according to the ELISA kit instructions. The result was compared to that of the controls without active components.
5.4. Bacterial Identification
Gram staining, 16S rRNA gene sequence analysis, and MALDI-TOF MS were used for bacterial identification. One loop of an overnight culture was smeared on a slide for Gram staining following the method reported by Davies et al. [58]. The GeneJET Genomic DNA Purification Kit (Thermo Scientific, Vilnius, Lithuania) was used to extract genomic DNA according to the manufacturer’s instructions. The 16S rRNA gene was amplified by polymerase chain reaction (PCR) using the primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) [59,60]. PCR amplification was performed by a T Gradient (Biometra, Goettingen, Germany). Amplification conditions consisted of pre-denaturation at 94 °C for 3 min followed by 35 cycles of 94 °C for 1 min, 56 °C for 30 s, and 72 °C for 1 min, with final extension at 72 °C for 10 min. PCR products were confirmed using agarose gel electrophoresis (1X agarose in TBE buffer). The PCR products were purified and sequenced by Sanger sequencing (Apical Scientific, Selangor, Malaysia). Nucleotide sequence comparisons were performed using the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/BLAST/, accessed date 23 March 2021). Similar 16S rDNA sequences were downloaded from GenBank and manually reviewed, after which all the sequences were aligned, and a phylogenetic tree was constructed using neighbor joining by MEGA X [61].
Bacteria were transferred to NGA and incubated at room temperature for 24 h before analysis. MALDI-TOF MS was performed by the Salaya Central Instrument Facility, Mahidol University, Nakhon Pathom, using an Autoflex MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The standard Bruker interpretive criteria were applied as follows: unreliable identification (score 0.000–1.699); probable genus identification (score 1.700–1.999); secure genus and probable species identification (score 2.000–2.299); and highly probable species identification (score 2.300–3.000) [44].
5.5. Proteins Expression during Fumonisin B1 Reduction by S. marcescens 329-2
A bacterial pellet was harvested after 3 days of incubation on NGB, washed with PBS and centrifuged at 10,000 rpm for 15 min. The bacterial pellet was incubated in 3 ppm FB1 solution, and a control was inoculated in PBS without FB1. The test condition was set for 7 days to ensure the protein expression during FB1 reduction.
The cell pellet was trypsin-digested following protocol. The cell pellet was washed twice in 100 µL of PBS. The cell pellet was resuspended twice in a lysis buffer (10% sodium deoxycholate (Tokyo Chemical Industry, Tokyo, Japan), 10 mM Tris (2-carboxyethyl) phosphine hydrochloride, (Sigma-Aldrich, St. Louis, MO, USA), 40 mM 2-chloroacetamide (Sigma-Aldrich, St. Louis, MO, USA), and 50 mM phosphate buffer, pH 8.0), boiled at 95 °C for 10 min, and sonicated for 15 min. Cell debris was pelleted by centrifugation at 10,000 rpm for 5 min, and the clarified lysate was transferred into a new tube. The lysate was diluted 1:10 for trypsin digestion using Trypsin Gold, mass spectrometry grade (Promega, Madison, WIUSA) at an enzyme/substrate ratio of 1:50, and digestion was performed overnight at 37 °C. The digested sample was acidified to a final concentration of trifluoroacetic acid (Sigma-Aldrich, St. Louis, MO, USA) at 0.5%, and sodium deoxycholate was extracted by adding an equal volume of ethyl acetate and vigorous shaking. The organic phase was removed after centrifugation at 10,000 rpm for 5 min. The aqueous solution was transferred to a new tube and submitted to lyophilization. Label-free quantification and data analysis were performed by the Salaya Central Instrument Facility, Mahidol University, Nakhon Pathom, with NanoLC (Ultimate 3000, Thermo Scientific, Vienna, Austria) using an Acclaim PepMap RSLC C18 column (75 µm × 150 mm, Thermo Scientific, Vienna, Austria). The mobile phases were 2% (v/v) acetonitrile with 0.1% (v/v) formic acid (phase A) and 80% (v/v) acetonitrile with 0.1% (v/v) formic acid (phase B). The linear gradient elution was as follows: 0–5 min, 3% B; 5–45 min, 3–45% B; 45–50 min, 90% B; and 50–60 min, 3% B. The masses of the peptides were determined using a Sciex Triple TOF 6600+ instrument (AB Sciex, Framingham, MA, USA).
Acknowledgments
We would like to thank member of Physiology of Plant Disease Laboratory, Department of Plant Pathology, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand for kindly support. This research is supported by graduate study development scholarship from the National Research Council of Thailand as of 2020 fiscal year and the Postharvest Technology Innovation Center, Ministry of Higher Education, Science, Research and Innovation, Bangkok, Thailand.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13090638/s1, Table S1: Upregulation proteins in FB1-treated S. marcescens 329-2 compared with the control group, Table S2: Downregulation proteins in FB1-treated S. marcescens 329-2 compared with the control group.
Author Contributions
Conceptualization, P.K., C.R. and R.P.; methodology, P.K., C.R. and R.P.; formal analysis, P.K., C.R. and R.P.; investigation, C.R.; writing—original draft preparation, P.K., C.R. and R.P.; writing—review and editing, P.K., C.R. and R.P.; visualization, P.K., C.R. and R.P.; supervision, C.R. and R.P.; project administration, C.R.; funding acquisition, C.R. and R.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by graduate study development scholarship from the National Research Council of Thailand as of 2020 fiscal year and Postharvest Technology Innovation Center, Ministry of Higher Education, Science, Research and Innovation, Bangkok, Thailand.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request, please contact the contributing authors.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
S. marcescens showed the potential to degrade FB1 with hydrolase and transferase activity.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shibamoto T., Bjeldanes L.F. In: Chapter 6-Fungal Toxins Occurring in Foods. Taylor S.L., editor. Academic Press; San Diego, CA, USA: 1993. pp. 97–116. [Google Scholar]
- 2.Pohland A.E. Mycotoxins in review. Food Addit. Contam. 1993;10:17–28. doi: 10.1080/02652039309374126. [DOI] [PubMed] [Google Scholar]
- 3.Ayalew A. Mycotoxins and surface and internal fungi of maize from Ethiopia. Afr. J. Food Agric. Nutr. Dev. 2010;10 doi: 10.4314/ajfand.v10i9.62890. [DOI] [Google Scholar]
- 4.Tola M., Kebede B., Yildiz F. Occurrence, importance and control of mycotoxins: A review. Cogent Food Agric. 2016;2:1191103. doi: 10.1080/23311932.2016.1191103. [DOI] [Google Scholar]
- 5.Leslie J.F., Summerell B.A. The Fusarium Laboratory Manual. Blackwell Publishing; Hoboken, NJ, USA: 2006. [Google Scholar]
- 6.EFSA Risks for animal health related to the presence of fumonisins, their modified forms and hidden forms in feed. EFSA J. 2018;16:e05242. doi: 10.2903/j.efsa.2018.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rheeder J.P., Marasas W.F.O., Vismer H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002;68:2101–2105. doi: 10.1128/AEM.68.5.2101-2105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sydenham E.W., Shephard G.S., Thiel P.G., Marasas W.F.O., Stockenstrom S. Fumonisin contamination of commercial corn-based human foodstuffs. J. Agric. Food Chem. 1991;39:2014–2018. doi: 10.1021/jf00011a028. [DOI] [Google Scholar]
- 9.Presello D.A., Botta G., Iglesias J., Eyhérabide G.H. Effect of disease severity on yield and grain fumonisin concentration of maize hybrids inoculated with Fusarium verticillioides. Crop Prot. 2008;27:572–576. doi: 10.1016/j.cropro.2007.08.015. [DOI] [Google Scholar]
- 10.Munkvold G.P., Desjardins A.E. Fumonisins in maize: Can we reduce their occurrence? Plant Dis. 1997;81:556–565. doi: 10.1094/PDIS.1997.81.6.556. [DOI] [PubMed] [Google Scholar]
- 11.Biomin BIOMIN World Mycotoxin Survey 2020. [(accessed on 15 May 2021)]. Available online: https://www.biomin.net/downloads/2020-biomin-world-mycotoxin-survey-report/#c41851.
- 12.Zhao Z., Zhang Y., Gong A., Liu N., Chen S., Zhao X., Li X., Chen L., Zhou C., Wang J. Biodegradation of mycotoxin fumonisin B1 by a novel bacterial consortium SAAS79. Appl. Microbiol. Biotechnol. 2019;103:7129–7140. doi: 10.1007/s00253-019-09979-6. [DOI] [PubMed] [Google Scholar]
- 13.Lyagin I., Efremenko E. Enzymes for detoxification of various mycotoxins: Origins and mechanisms of catalytic action. Molecules. 2019;24:2362. doi: 10.3390/molecules24132362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., Hassan Y.I., Lepp D., Shao S., Zhou T. Strategies and methodologies for developing microbial detoxification systems to mitigate mycotoxins. Toxins. 2017;9:130. doi: 10.3390/toxins9040130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loi M., Fanelli F., Liuzzi V.C., Logrieco A.F., Mulè G. Mycotoxin biotransformation by native and commercial enzymes: Present and future perspectives. Toxins. 2017;9:111. doi: 10.3390/toxins9040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvick J., Rood T., Maddox J., Gilliam J. Detoxification of mycotoxins in planta as a strategy for improving grain quality and disease resistance: Identification of fumonisin-degrading microbes from maize. In: Kohmoto K., Yoder O.C., editors. Molecular Genetics of Host-Specific Toxins in Plant Disease: Proceedings of the 3rd Tottori International Symposium on Host-Specific Toxins, Daisen, Tottori, Japan, August 24–29, 1997. Springer; Dordrecht, The Netherlands: 1998. pp. 369–381. [Google Scholar]
- 17.Blackwell B.A., Gilliam J., Savard M., Miller J., Duvick J. Oxidative deamination of hydrolyzed fumonisin B1 (AP1) by cultures of Exophiala spinifera. Nat. Toxins. 1999;7:31–38. doi: 10.1002/(SICI)1522-7189(199902)7:1<31::AID-NT36>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Collins T.F., Sprando R.L., Black T.N., Olejnik N., Eppley R.M., Shackelford M.E., Howard P.C., Rorie J.I., Bryant M., Ruggles D.I. Effects of aminopentol on in utero development in rats. Food Chem. Toxicol. 2006;44:161–169. doi: 10.1016/j.fct.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Benedetti R., Nazzi F., Locci R., Firrao G. Degradation of fumonisin B1 by a bacterial strain isolated from soil. Biodegradation. 2006;17:31–38. doi: 10.1007/s10532-005-2797-y. [DOI] [PubMed] [Google Scholar]
- 20.Heinl S., Hartinger D., Thamhesl M., Vekiru E., Krska R., Schatzmayr G., Moll W.-D., Grabherr R. Degradation of fumonisin B1 by the consecutive action of two bacterial enzymes. J. Biotechnol. 2010;145:120–129. doi: 10.1016/j.jbiotec.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Masching S., Naehrer K., Schwartz-Zimmermann H.E., Sarandan M., Schaumberger S., Dohnal I., Nagl V., Schatzmayr D. Gastrointestinal degradation of fumonisin B(1) by carboxylesterase FumD prevents fumonisin induced alteration of sphingolipid metabolism in turkey and swine. Toxins. 2016;8:84. doi: 10.3390/toxins8030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logrieco A., Bottalico A., Mulé G., Moretti A., Perrone G. Epidemiology of toxigenic fungi and their associated mycotoxins for some mediterranean crops. Eur. J. Plant Pathol. 2003;109:645–667. doi: 10.1023/A:1026033021542. [DOI] [Google Scholar]
- 23.Voss K.A., Riley R.T. Fumonisin toxicity and mechanism of action: Overview and current perspectives. Food Saf. 2013;1:49–69. doi: 10.14252/foodsafetyfscj.2013006. [DOI] [Google Scholar]
- 24.Medina Á., González-Jartín J.M., Sainz M.J. Impact of global warming on mycotoxins. Curr. Opin. Food Sci. 2017;18:76–81. doi: 10.1016/j.cofs.2017.11.009. [DOI] [Google Scholar]
- 25.Niderkorn V., Morgavi D.P., Pujos E., Tissandier A., Boudra H. Screening of fermentative bacteria for their ability to bind and biotransform deoxynivalenol, zearalenone and fumonisins in an in vitro simulated corn silage model. Food Addit. Contam. 2007;24:406–415. doi: 10.1080/02652030601101110. [DOI] [PubMed] [Google Scholar]
- 26.Dalie D.K., Deschamps A.M., Atanasova-Penichon V., Richard-Forget F. Potential of Pediococcus pentosaceus (L006) isolated from maize leaf to suppress fumonisin-producing fungal growth. J. Food Prot. 2010;73:1129–1137. doi: 10.4315/0362-028X-73.6.1129. [DOI] [PubMed] [Google Scholar]
- 27.Deepthi B.V., Poornachandra Rao K., Chennapa G., Naik M.K., Chandrashekara K.T., Sreenivasa M.Y. Antifungal attributes of Lactobacillus plantarum MYS6 against fumonisin producing Fusarium proliferatum associated with poultry feeds. PLoS ONE. 2016;11:e0155122. doi: 10.1371/journal.pone.0155122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker B., Bresch H., Schillinger U., Thiel P.G. The effect of fumonisin B1 on the growth of bacteria. World J. Microbiol. Biotechnol. 1997;13:539–543. doi: 10.1023/A:1018513308847. [DOI] [Google Scholar]
- 29.Wang Y., Zhao C., Zhang D., Zhao M., Zheng D., Lyu Y., Cheng W., Guo P., Cui Z. Effective degradation of aflatoxin B(1) using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017;224:166–173. doi: 10.1016/j.biortech.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 30.John G.H., Noel R.K., Peter H.A.S., James T.S., Stanly T.W. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins; Baltimore, MD, USA: 1994. [Google Scholar]
- 31.Li B., Yu R., Liu B., Tang Q., Zhang G., Wang Y., Xie G., Sun G. Characterization and comparison of Serratia marcescens isolated from edible cactus and from silkworm for virulence potential and chitosan susceptibility. Braz. J. Microbiol. 2011;42:96–104. doi: 10.1590/S1517-83822011000100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L.-Q., Liu Z.-Q., Zheng Y.-G., Shen Y.-C. Identification and characterization of Serratia marcescens ZJB-09104, a nitrile-converting bacterium. World J. Microbiol. Biotechnol. 2010;26:817–823. doi: 10.1007/s11274-009-0238-5. [DOI] [Google Scholar]
- 33.Patel J.B. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 2001;6:313–321. doi: 10.2165/00066982-200106040-00012. [DOI] [PubMed] [Google Scholar]
- 34.Woese C.R. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey A.K., Chaudhary P., Singh S.B., Arora A., Kumar K., Chaudhry S., Nain L. Deciphering the traits associated with PAH degradation by a novel Serratia marcesencs L-11 strain. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2012;47:755–765. doi: 10.1080/10934529.2012.660108. [DOI] [PubMed] [Google Scholar]
- 36.Stancu M.M. Highly solvent tolerance in Serratia marcescens IBBPo15. Braz. Arch. Biol. Technol. 2016;59 doi: 10.1590/1678-4324-2016160268. [DOI] [Google Scholar]
- 37.Neville S.A., LeCordier A., Ziochos H., Chater M.J., Gosbell I.B., Maley M.W., van Hal S.J. Utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 2011;49:2980–2984. doi: 10.1128/JCM.00431-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timperio A.M., Gorrasi S., Zolla L., Fenice M. Evaluation of MALDI-TOF mass spectrometry and MALDI biotyper in comparison to 16S rDNA sequencing for the identification of bacteria isolated from Arctic sea water. PLoS ONE. 2017;12:e0181860. doi: 10.1371/journal.pone.0181860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauget M., Valot B., Bertrand X., Hocquet D. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 2017;25:447–455. doi: 10.1016/j.tim.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Li Y., Shan M., Zhu Z., Mao X., Yan M., Chen Y., Zhu Q., Li H., Gu B. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect. Dis. 2019;19:941. doi: 10.1186/s12879-019-4584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dingle T.C., Butler-Wu S.M. Maldi-tof mass spectrometry for microorganism identification. Clin. Lab. Med. 2013;33:589–609. doi: 10.1016/j.cll.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Rödel J., Mellmann A., Stein C., Alexi M., Kipp F., Edel B., Dawczynski K., Brandt C., Seidel L., Pfister W., et al. Use of MALDI-TOF mass spectrometry to detect nosocomial outbreaks of Serratia marcescens and Citrobacter freundii. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:581–591. doi: 10.1007/s10096-018-03462-2. [DOI] [PubMed] [Google Scholar]
- 43.Othman M.A., El-Zamik F.I., Hegazy M.I., Salama A.S.A. Isolation and identification of egyptian strains of Serratia marcescens producing antibacterial and antioxidant prodigiosin pigment. Zagazig J. Agric. Res. 2019;46:1573–1582. doi: 10.21608/zjar.2019.48175. [DOI] [Google Scholar]
- 44.Ng L.S.Y., Sim J.H.C., Eng L.C., Menon S., Tan T.Y. Comparison of phenotypic methods and matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for the identification of aero-tolerant Actinomyces spp. isolated from soft-tissue infections. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1749–1752. doi: 10.1007/s10096-011-1496-3. [DOI] [PubMed] [Google Scholar]
- 45.Someya N., Kataoka N., Komagata T., Hirayae K., Hibi T., Akutsu K. Biological control of cyclamen soilborne diseases by Serratia marcescens strain B2. Plant Dis. 2000;84:334–340. doi: 10.1094/PDIS.2000.84.3.334. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto H., Sato M., Sato Z., Isaka M. Biocontrol of Phytophthora capsici by Serratia marcescens F-1-1 and analysis of biocontrol mechanisms using transposon-insertion mutants. Jpn. J. Phytopathol. 1998;64:287–293. doi: 10.3186/jjphytopath.64.287. [DOI] [Google Scholar]
- 47.Jaiganesh V., Eswaran A., Balabaskar P., Kannan C. Antagonistic activity of Serratia marcescens against Pyricularia oryzae. Not. Bot. Horti Agrobot. Cluj. Napoca. 2007;35 doi: 10.15835/nbha352219. [DOI] [Google Scholar]
- 48.Press C.M., Loper J.E., Kloepper J.W. Role of iron in rhizobacteria-mediated induced systemic resistance of cucumber. Phytopathology. 2001;91:593–598. doi: 10.1094/PHYTO.2001.91.6.593. [DOI] [PubMed] [Google Scholar]
- 49.Singh R.P., Jha P.N. The multifarious PGPR Serratia marcescens CDP-13 augments induced systemic resistance and enhanced salinity tolerance of wheat (Triticum aestivum L.) PLoS ONE. 2016;11:e0155026. doi: 10.1371/journal.pone.0155026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Z., Zhang X., Wu J., Yu J., Xu M., Chen D., Zhang Z., Li X., Chi Y., Wan S. In vitro inhibitory effect of the bacterium Serratia marcescens on Fusarium proliferatum growth and fumonisins production. Biol. Control. 2020;143:104188. doi: 10.1016/j.biocontrol.2020.104188. [DOI] [Google Scholar]
- 51.Carbonell G.V., Della Colleta H.H.M., Yano T., Darini A.L.C., Levy C.E., Fonseca B.A.L. Clinical relevance and virulence factors of pigmented Serratia marcescens. FEMS Immunol. Med. Microbiol. 2000;28:143–149. doi: 10.1111/j.1574-695X.2000.tb01469.x. [DOI] [PubMed] [Google Scholar]
- 52.Hartinger D., Heinl S., Schwartz H.E., Grabherr R., Schatzmayr G., Haltrich D., Moll W.-D. Enhancement of solubility in Escherichia coli and purification of an aminotransferase from Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B(1) Microb. Cell Factories. 2010;9:62. doi: 10.1186/1475-2859-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devlin T.M. Textbook of Biochemistry. John Wiley; New York, NY, USA: 2002. [Google Scholar]
- 54.McKee T., McKee J.R. Biochemistry. McGraw-Hill; Boston, MA, USA: 2003. [Google Scholar]
- 55.Montella I.R., Schama R., Valle D. The classification of esterases: An important gene family involved in insecticide resistance-A review. Mem. Inst. Oswaldo Cruz. 2012;107:437–449. doi: 10.1590/S0074-02762012000400001. [DOI] [PubMed] [Google Scholar]
- 56.European Commission Commission Recommendation of 17 August 2006 on the presence of deoxynivalenol, zearalenone, ochratoxin A, T-2 and HT-2 and fumonisins in products intended for animal feeding (2006/576/EC) Off. J. Eur. Union. 2006;L229:7–9. [Google Scholar]
- 57.Wang J.Q., Yang F., Yang P.L., Liu J., Lv Z.H. Microbial reduction of zearalenone by a new isolated Lysinibacillus sp. ZJ-2016-1. World Mycotoxin J. 2018;11:571–578. doi: 10.3920/WMJ2017.2264. [DOI] [Google Scholar]
- 58.Davies J.A., Anderson G.K., Beveridge T.J., Clark H.C. Chemical mechanism of the gram stain and synthesis of a new electron-opaque marker for electron microscopy which replaces the iodine mordant of the stain. J. Bacteriol. 1983;156:837–845. doi: 10.1128/jb.156.2.837-845.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons, Inc.; New York, NY, USA: 1991. pp. 115–175. [Google Scholar]
- 60.Turner S., Pryer K.M., Miao V.P., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request, please contact the contributing authors.