Abstract
Purpose
The aim of this study was to establish and evaluate a fully automatic deep learning system for the diagnosis of COVID-19 using thoracic computed tomography (CT).
Materials and methods
In this retrospective study, a novel hybrid model (MTU-COVNet) was developed to extract visual features from volumetric thoracic CT scans for the detection of COVID-19. The collected dataset consisted of 3210 CT scans from 953 patients. Of the total 3210 scans in the final dataset, 1327 (41%) were obtained from the COVID-19 group, 929 (29%) from the CAP group, and 954 (30%) from the Normal CT group. Diagnostic performance was assessed with the area under the receiver operating characteristic (ROC) curve, sensitivity, and specificity.
Results
The proposed approach with the optimized features from concatenated layers reached an overall accuracy of 97.7% for the CT-MTU dataset. The rest of the total performance metrics, such as; specificity, sensitivity, precision, F1 score, and Matthew Correlation Coefficient were 98.8%, 97.6%, 97.8%, 97.7%, and 96.5%, respectively. This model showed high diagnostic performance in detecting COVID-19 pneumonia (specificity: 98.0% and sensitivity: 98.2%) and CAP (specificity: 99.1% and sensitivity: 97.1%). The areas under the ROC curves for COVID-19 and CAP were 0.997 and 0.996, respectively.
Conclusion
A deep learning–based AI system built on the CT imaging can detect COVID-19 pneumonia with high diagnostic efficiency and distinguish it from CAP and normal CT. AI applications can have beneficial effects in the fight against COVID-19.
Keywords: COVID-19, Pneumonia, Artificial intelligence (AI), Deep learning, Computed tomography (CT)
1. Introduction
In the city of Wuhan in China, a pneumonia outbreak that developed due to a novel coronavirus was detected in December 2019. The outbreak could not be taken under control and it spread all around the world, resulting in a pandemic. The novel coronavirus was defined as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease that resulted from the virus was defined as coronavirus disease 2019 (COVID-19) by the World Health Organization.1 As of March 5, 2021, approximately 116 million COVID-19 cases and higher than 2.5 million deaths were reported all around the world.2
Community-acquired pneumonia (CAP) is a pulmonary parenchymal infection acquired outside a healthcare setting. Despite bacterial infections are responsible for the majority of CAP, viral infections also are not uncommon.3, 4. The clinical symptoms of CAP and COVID-19 associated pneumonia are generally similar.5 The test used as standard to confirm the diagnosis of COVID-19 is reverse transcription polymerase chain reaction (RT-PCR). However, RT-PCR sensitivity may not be high enough for early diagnosis and treatment of patients. Several studies have shown that thoracic computed tomography (TCT), which can be easily accessed in many hospitals, is a useful test that can be used in the diagnosis of COVID-19 pneumonia under current pandemic conditions. In addition, RT-PCR gives false negative results while abnormalities compatible with COVID-19 pneumonia may be observed in TCT when the viral load is insufficient. TCT has been shown to have a higher sensitivity in the diagnosis of COVID-19 compared to RT-PCR samples. Therefore, TCT may play a very important role in the early detection and treatment of COVID-19 pneumonia.6, 7, 8
Artificial intelligence (AI) is one of the most recent topics in medical imaging and has led to major changes in diagnostic imaging systems. The development of deep learning methods and especially the use of convolutional neural networks (CNNs) have led to significant performance gains compared to classical machine learning techniques. AI applications are currently used in areas such as evaluation of lung nodules, detection of diffuse lung diseases, diagnosis of tuberculosis and pneumonia in thoracic imaging.9 Recently, a number of articles have reported that deep learning techniques have promising results in increasing pulmonary nodule detection sensitivity and predicting malignancy.10, 11 In addition, AI applications were used to differentiate viral pneumonia from bacterial pneumonia on chest radiography in pediatric patients prior to the COVID-19 pandemic and successful results were obtained.12 Recently, the efficacy of AI applications in the diagnosis of COVID-19 pneumonia has been evaluated in some recent studies and has been shown to have high diagnostic performance in this field.13, 14., 15., 16., 17.
In this study, a private dataset (CT-MTU) of TCT images that can be used as an instance of imminent deep learning applications in the field of medical has been utilized. This retrospectively planned study aims to evaluate the effectiveness and diagnostic performance of artificial intelligence applications in the diagnosis of COVID-19 pneumonia using TCT images. CAP and normal TCT images were also included in the study to test the robustness of the model.
2. Materials and methods
2.1. Patients
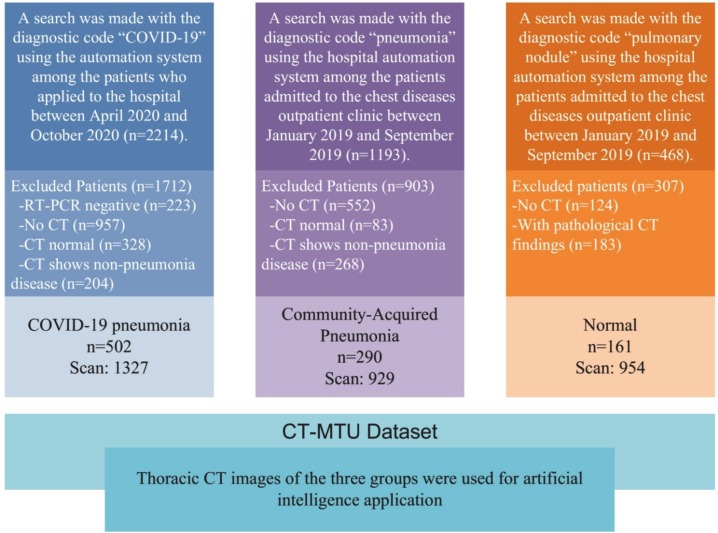
Patients who applied to Malatya Turgut Ozal University Training and Research Hospital and met the study criteria were included in this retrospectively planned study. This study was conducted in a tertiary university hospital, the primary admission center for patients with COVID-19 in the region. This study was conducted in accordance with the Declaration of Helsinki and ethics committee approval was obtained from Fırat University Ethics Committee for the study (ethics committee number/date: 416900/08.10.2020). A search was made with the diagnostic code “COVID-19” among the patients who applied to the hospital between April 01, 2020 and October 01, 2020 using the hospital automation system for the COVID-19 group. Patients with negative RT-PCR results, no or normal TCT, and signs of non-pneumonia disease on TCT were excluded from the study. In conclusion, a total of 502 COVID-19 pneumonia patients with confirmed positive RT-PCR results and TCT images for SARS-COV-2 were included in the study. A search was made with the diagnostic code “pneumonia” using the hospital automation system among the patients who applied to the chest diseases outpatient clinic between January 01, 2019 and September 01, 2019 for the CAP group. A total of 290 CAP patients were included in the study after exclusion of patients with no or normal TCT and signs of non-pneumonia disease on TCT. A search was made with the diagnostic code “pulmonary nodule” using the hospital automation system among the patients who applied to the chest diseases outpatient clinic between January 01, 2019 and September 01, 2019 to determine the Normal control group. After excluding patients with no CT and pathological CT findings, a total of 161 patients with normal TBT were included in the study as the Normal group. The Normal control group consisted of patients with completely normal CT findings. Patients with any pathology on CT including pulmonary nodules were excluded from this group. The demographic characteristics of the three groups is given in Table 1 .
Table 1.
Demographic characteristics of the three groups.
| COVID-19 | CAP | Normal | p | |
|---|---|---|---|---|
| Patients, n(%) | 502 (52.7) | 290 (30.4) | 161 (16.9) | |
| Exams, n(%) | 1327 (41) | 929 (29) | 954 (30) | |
| Age, yrs | ||||
| Mean | 66.3 ± 15.1 | 66.8 ± 16.7 | 42.3 ± 18.6 | <0.001 |
| <40 | 31 (6.2) | 24 (8.3) | 87 (54) | |
| 40–65 | 182 (36.3) | 88 (30.3) | 51 (31.7) | |
| >65 | 289 (57.6) | 178 (61.4) | 23 (14.3) | |
| Sex, male/female | 289/213 | 174/116 | 81/80 | >0.05 |
As a result, the final cohort was formed as 502 COVID-19, 290 CAP, and 161 Normal. A total of 3210 TCT scans were selected from 953 patients included in the study. Of the total 3210 scans in the final dataset, 1327 (41%) were obtained from the COVID-19 group, 929 (29%) from the CAP group, and 954 (30%) from the Normal CT group. The diagram including the design of the study is provided in Fig. 1 .
Fig. 1.
Schematic view of study design.
2.2. CT protocol
All the images were obtained via a 16-slice CT device (Philips Medical Systems, Shenyang, China) with patients at the end of inspiration and in the supine position. The axial images were obtained craniocaudally and included the body parts from the thoracic inlet to the diaphragm. No contrast media was used during the scans. The scans were performed with the following technical parameters: 120 kV, 250 mA, 0.625 slice thickness, 512 × 512 matrix. The reconstructed images were also obtained and used in the current study.
2.3. CT analysis
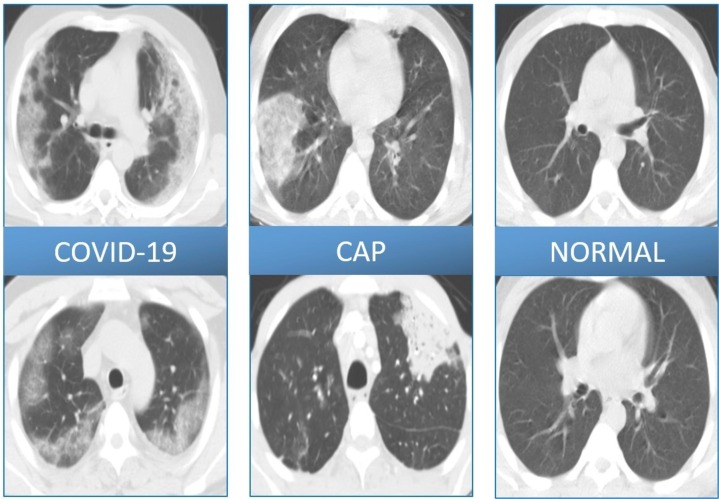
CT images were analyzed by a radiologist and three pulmonologist with at least 8 years of experience (E.I., A.A.G., N.K.B. and M.S. with 15, 11, 10 and 8 years of experiences in chest imaging interpretation, respectively). In CT, the lung was segmented manually to remove extrapulmonary sites. The entire dataset was pre-processed by adjusting the CT window width and lung window level. Lesion sections in COVID-19 or CAP patients were manually labeled and used as the reference standard to train the deep neural network. The representative images from CT-MTU dataset is demonstrated in Fig. 2 .
Fig. 2.
Representative images from CT-MTU dataset.
2.4. MTU-COVNet: deep CNN architecture for classifying CT images
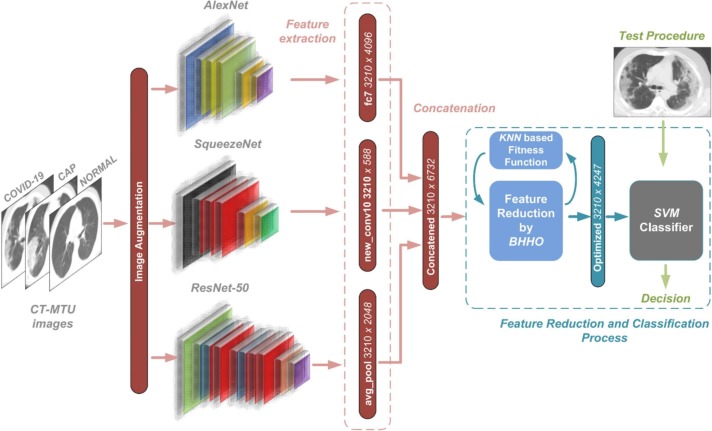
In this section, the evaluation of the proposed method is presented. The proposed approach includes the feature reduction with the Binary Harris Hawk Optimization (BHHO) method to execute the classification process on CT-MTU dataset by concatenating the deeper layers of pre-trained CNN models AlexNet, ResNet-50 and SqueezeNet. The Support Vector Machine Classifier (SVMC) was employed to label the test TCT images with their putative features and compare the performance of the classification via some evaluation metrics. Three pre-trained deep network models, ResNet-50, AlexNet, and SqueezeNet are preferred due to ease of use, training time consumption, and structural simplicity.18., 19, 20. The input images were resized to 224 × 224 to be compatible with ResNet-50, 227 × 227 for AlexNet and SqueezeNet models. The number of the data was expanded by applying image augmentation methods as online throughout the learning period. The imbalance problem in the number of images for the classes may not be suitable for building a robust image classifier. Besides, image augmentation methods have been used to eliminate the overfitting problems in the literature. The image data augmentation library of MATLAB was used to perform the augmentation. The augmented image data store applies a combination of multiple transformations, such as; random rotation (in the range [−1010] degrees), vertically and horizontally reflection, and shear (in the range of [−0.7 0.7] horizontal and vertical) to the training data. The classification process proposed in this study consists of three stages: feature extraction using pre-trained deep network structures, reducing the size of the obtained feature vectors by the BHHO method, and finally determining the labels of the CT images by utilizing the SVMC. The deep CNN features were extracted from fc7, avg_pool and new_conv10 activations from AlexNet, ResNet-50 and SqueezeNet, respectively. At first, four different feature sets were constructed by the output of the fc7, avg_pool and new_conv10 layers, such as AlexNet (fc7 3210 × 4096), ResNet-50 (avg_pool 3210 × 2048), SqueezeNet (new_conv10 3210 × 588) and AlexNet (fc7) + ResNet-50 (avg_pool) + SqueezeNet (new_conv10). Consequently, 3210 × 6732 dimensional concatenated feature vectors were obtained. Since the concatenated deep features contain many avoidable components in their structure, the necessity of feature selection methods arose. The feature selection method aims to obtain the best features that define the target class during the classification process by removing unrelated characteristics. In this way, classification quality is improved, and temporal problems are eliminated.30 In this study, the BHHO algorithm was used to achieve lower computation costs and higher accuracy performance by reducing feature sizes. This technique is a metaheuristic optimization algorithm that is the preferred wrapper method for feature selection.21 The size of the total feature vector decreased from 3210 × 6732 to 3210 × 4247 after the optimization process. All the feature sets were normalized according to the zero mean. The SVMC was used in the classification stage with 70% training and 30% testing data partition. SVMC aims to find the most appropriate separator plane that classifies the dataset as much as possible by determining the situation where the distance between the two classes is the greatest.22., 23., 24 The impact of the proposed method on its accuracy and evaluation metrics are discussed with the computational efficiency. All the experiments were performed in a MATLAB environment running on a PC with AMD Ryzen 52,600 3.4GHz CPU, 64 GB memory, and 12 GB NVIDIA GeForce RTX 2080 TI GPU. The proposed approach is demonstrated in Fig. 3 .
Fig. 3.
Illustration of the proposed approach.
2.5. Statistical analysis
The evaluation metrics such as accuracy, sensitivity, specificity, precision, F1 score, and Matthew Correlation Coefficient (MCC) were statistically computed from the confusion matrix to evaluate the quantitative performance of the proposed method. The confusion matrix consists of rows and columns for predicted classes and actual classes, respectively. The diagonal cells correspond to observations classified correctly, while other cells correspond to observations classified incorrectly. The number of observations and the percentage of the total number of observations are given in each cell. The rightmost column of the chart shows the percentages of all samples estimated to belong to each class that were classified correctly (precision metrics) and incorrectly (false discovery rate metrics). The row showing the percentages of all samples is at the bottom of the matrix for each class classified correctly (true positive rate metrics) and incorrectly (false negative rate metrics). The cell in the bottom right corner of the matrix shows the overall accuracy. Also, one of the most widely used metrics to evaluate the performance of machine learning algorithms is the Receiver Operating Characteristic (ROC) curve, which provides the true-positive rate as a function of the false-positive rate. Statistical analyses were performed using MATLAB packages. The selected evaluation metrics are defined as:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
3. Results
3.1. Patient characteristics
There was no statistical difference between the groups in terms of gender (p = 0.13, X2:4.07). The ages of the patients in the Normal group were statistically significantly lower compared to the other two groups (p < 0.001 in both groups) whereas there was no difference between COVID-19 and CAP patients in terms of age (p > 0.05) (Table 1).
3.2. Performance of MTU-COVNet
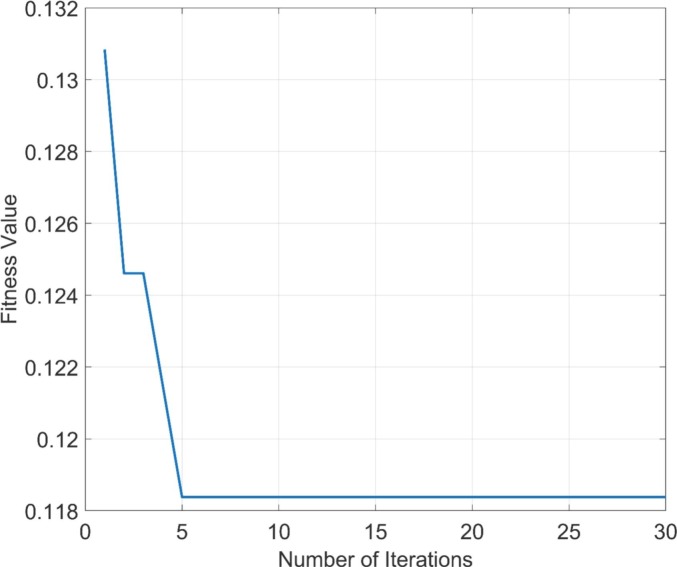
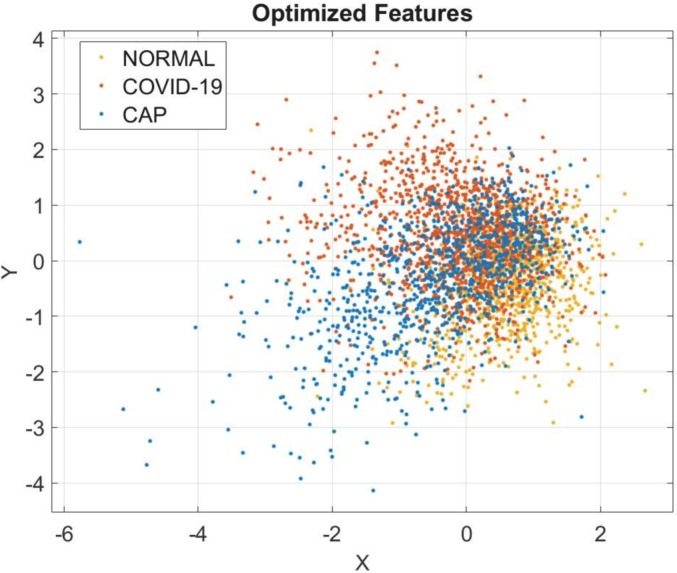
In this subsection, a performance comparison of the pre-trained models and the proposed architecture is given. Fig. 4 illustrates the convergence curve of BHHO methods for the most characteristic features from the concatenated deep features. The size of the total feature vector decreased from 3210 × 6732 to 3210 × 4247 after optimization, while the total accuracy value increased to 97.7%. The distribution of patients in the 3210 × 4247 dimensional feature space given in Fig. 5 . Dimension reduction in the concatenated feature vector was completed by taking feedback from the KNN (K-Nearest Neighbors) objective function. Thus, a feature set with the highest accuracy could be identified.
Fig. 4.
The convergence curve of the BHHO method.
Fig. 5.
The scatter plot of the optimized features.
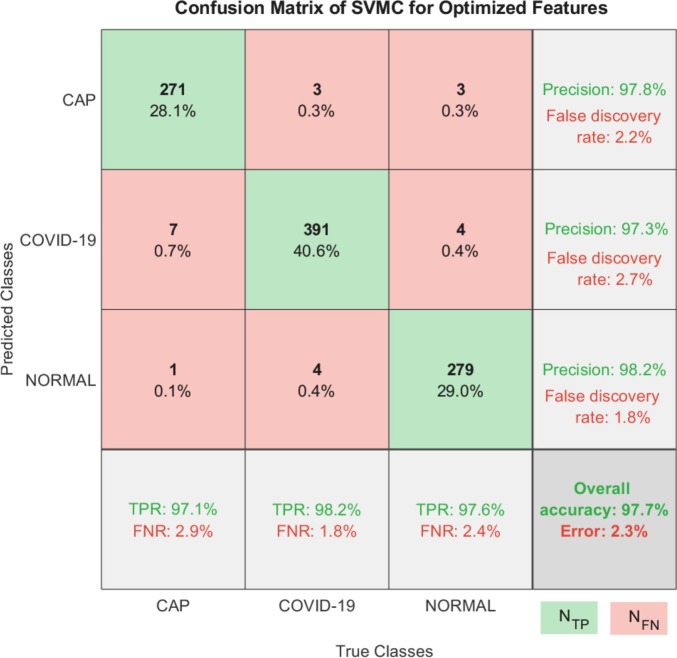
Fig. 6 demonstrates the multi-class confusion matrix of the SVMC that was fed by the optimized feature set. According to the confusion matrix, there are 22 misclassified samples among 963 test samples. The most misclassified samples were found to be in the CAP class, with 8 samples. The detailed classification results of the SVMC for all feature sets from selected layers were given in Table 2 . Here, we can see a remarkable performance enhancement in all classes and overall criteria.
Fig. 6.
The multi-class confusion matrix of the SVMC with optimized features. (TPR: True positive rate, FNR: False negative rate, NTP: Observations classified correctly, NFN: Observations classified incorrectly. *The percentage of the total number of observations are given in each cell.)
Table 2.
The performance comparison of the MTU-COVNet and the other CNN for each class.
| Deep CNN architectures | Class | Accuracy | Specificity | Sensitivity | Precision | F1 | MCC |
|---|---|---|---|---|---|---|---|
| SqueezeNet (new_conv10 layer) | CAP | 82.37% | 96.50% | 82.37% | 90.51% | 86.25% | 81.21% |
| COVID-19 | 88.44% | 89.73% | 88.44% | 85.85% | 87.13% | 77.86% | |
| NORMAL | 91.64% | 94.53% | 91.64% | 87.67% | 89.61% | 85.10% | |
| AlexNet (fc7 layer) | CAP | 89.57% | 97.08% | 89.57% | 92.57% | 91.04% | 87.51% |
| COVID-19 | 91.48% | 93.79% | 91.48% | 91.25% | 91.36% | 85.24% | |
| NORMAL | 93.71% | 96.16% | 93.71% | 91.16% | 92.41% | 89.16% | |
| ResNet-50 (avg_pool layer) | CAP | 91.37% | 97.81% | 91.37% | 94.42% | 92.87% | 90.07% |
| COVID-19 | 94.22% | 95.22% | 94.22% | 93.28% | 93.75% | 89.31% | |
| NORMAL | 96.52% | 97.78% | 96.52% | 94.86% | 95.68% | 93.83% | |
| Concataned | CAP | 94.24% | 98.54% | 94.24% | 96.32% | 95.27% | 93.39% |
| COVID-19 | 96.48% | 97.70% | 96.48% | 96.73% | 96.60% | 94.22% | |
| NORMAL | 97.21% | 97.78% | 97.21% | 94.90% | 96.04% | 94.35% | |
| Optimized | CAP | 97.13% | 99.12% | 97.13% | 97.83% | 97.48% | 96.46% |
| COVID-19 | 98.24% | 98.05% | 98.24% | 97.26% | 97.75% | 96.16% | |
| NORMAL | 97.55% | 99.26% | 97.55% | 98.24% | 97.89% | 97.01% |
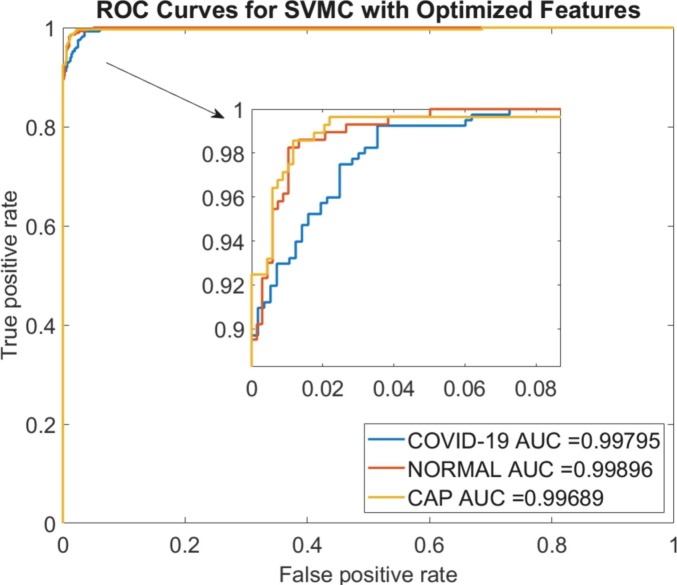
COVID-19 class achieved the best accuracy result, whereas the CAP and Normal classes have the performance of 97.13% and 97.55%, respectively. The obtained results show that the Normal class had higher values of specificity, precision, F1 score, and MCC as 99.26%, 98.24%, 97.89%, and 97.01%, respectively. The ROC curves of the predicted classes with AUC values were given in Fig. 7 . The area of the ROC curve was obtained as 0.997, 0.999, and 0.996 for COVID-19, Normal, and CAP classes, respectively.
Fig. 7.
The ROC curves of all classes for optimized features.
The general classification scores of the SVMC is given in Table 3 . The results in the table indicate that, the best total metric values for the CT-MTU dataset were observed in the optimized network with 4247 features. The model with optimized features from concatenated layers reached an overall accuracy of 97.7% for the CT-MTU dataset. The rest of the total performance metrics specificity, sensitivity, precision, F1 score, and MCC were 98.8%, 97.6%, 97.8%, 97.7%, and 96.5%, respectively.
Table 3.
The general classification scores of the SVMC for CT-MTU dataset.
| Net | The general classification scores of the SVMC |
|||||
|---|---|---|---|---|---|---|
| Acc. | Spec. | Sens. | Prec. | F1 | MCC | |
| SqueezeNet | 87.64% | 93.59% | 87.48% | 88.01% | 87.66% | 81.39% |
| AlexNet | 91.59% | 95.68% | 91.58% | 91.66% | 91.61% | 87.31% |
| ResNet-50 | 94.08% | 96.94% | 94.03% | 94.19% | 94.10% | 91.07% |
| Concatenated | 96.05% | 98.01% | 95.98% | 95.98% | 95.97% | 93.99% |
| Optimized | 97.72% | 98.81% | 97.64% | 97.78% | 97.71% | 96.54% |
The classification scores of the SVMC for CT-MTU dataset for optimized features are given in Table 3, on the last line.
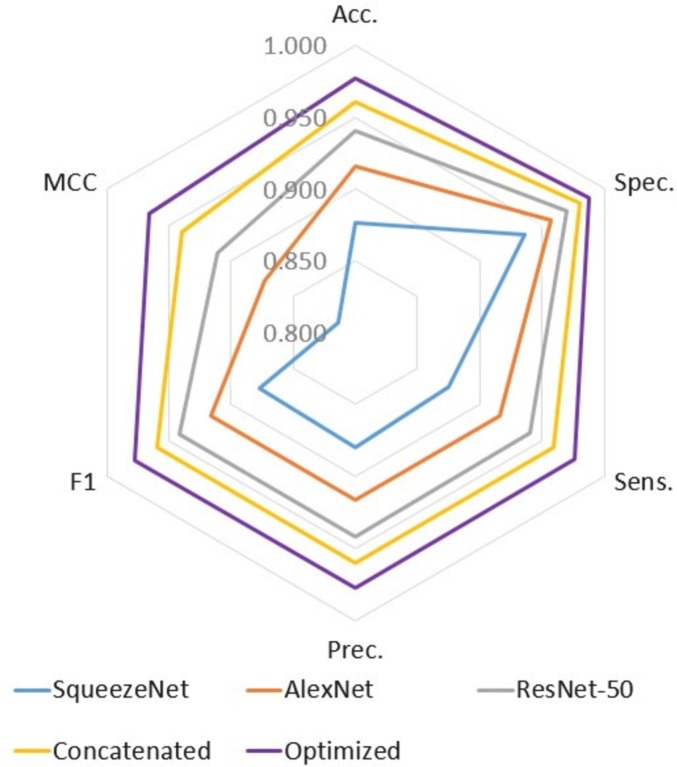
The radar chart in Fig. 8 , plotted according to the data in Table 3, depicts a comparison of performances of the proposed method and other features set. The proposed method has ability to provide an improvement in MCC. Accuracy and F1 score computed on confusion matrices have been among the most popular metrics in classification tasks. These evaluation metrics can lead to inflated, overly optimistic results in imbalance dataset. An effective solution to the class imbalance issue comes from the highest MCC. The proposed method provided 3%, 5%, 9% and 15% enhancements over Concatenated, ResNet-50, AlexNet and SqueezeNet, respectively.
Fig. 8.
Radar chart of the performance in terms of all feature sets.
4. Discussion
In this study, we proposed a novel hybrid methodology for diagnosing the COVID-19 pneumonia with optimized features from multi pre-trained net. The feature selection method based on the metaheuristic optimization algorithm was used to achieve lower computation costs and higher accuracy performance by reducing the feature size. An example was illustrated where the feature selection with BHHO technique can provide novel insight into medical research and test the model with the CT images of COVID-19, CAP and Normal situations. We found in an independent test dataset that this model showed high diagnostic performance in detecting COVID-19 pneumonia and CAP (diagnostic accuracy: 98.2% and 97.1, respectively). In addition, we determined the areas under the ROC curves for the COVID-19, CAP, and Normal groups as 0.997, 0.996, and 0.999, respectively.
The number of COVID-19 cases and disease mortality are rapidly increasing all over the world despite the measures taken.2 COVID-19 often involves the lower respiratory tract and leads to pneumonia. CAP and COVID-19-associated pneumonia can be clinically confused. However, it is crucial to distinguish COVID-19 pneumonia from CAP and isolate these patients as it may cause significant public health problems in the current pandemic conditions. In addition, early recognition and hospitalization of severe forms of COVID-19 patients are vital due to different treatment approaches and high mortality rates.5., 25 RT-PCR is considered the reference standard for the diagnosis of COVID-19. However, it has been reported that TCT can be used as a reliable and fast approach to COVID-19 screening.6, 7, 8 Typical CT results observed in COVID-19 pneumonia are bilateral, peripheral, scattered ground glass opacities and consolidations.26 It is important that radiologists are familiar with the typical CT features associated with this new infection and the imaging criteria for an alternative diagnosis, given the important role of TCT in the diagnosis of COVID-19. However, one of the most important problems in the pandemic process is the lack of sufficient number of radiologists to evaluate TBT images. In addition, these patients have to be followed not only by chest disease specialists who are familiar with lung radiology but also by many physicians from different branches during the pandemic. Artificial intelligence applications have become very important for the diagnosis of COVID-19 in order to accelerate the diagnosis of the disease and to support clinicians in the pandemic conditions where a limited number of chest diseases doctors cannot handle the current patient burden.
The efficacy of AI applications in differentiating COVID-19 pneumonia from other pneumonia in TCT sections has also been analyzed in some studies so far. Diagnostic yield was found to be between 85% and 99% in these studies.27 One of the first studies on this subject was conducted by Li et al..13 Li et al. developed a CT-based artificial intelligence software to detect patients with COVID-19 pneumonia and found the sensitivity and specificity of this software for the diagnosis of COVID-19 pneumonia to be 90% and 96%, respectively. Data including 400 COVID-19, 1396 community-acquired pneumonia and 1173 non-pneumonia (normal or non-pneumonia disease) patients were trained on the network in this study. The network was tested and evaluated with data from 68 patients with COVID-19, 155 patients with community-acquired pneumonia, and 130 patients with non-pneumonia. Diagnostic efficiency was found to be high in this study, which has a very large data set. 924 COVID-19 and 342 other pneumonia patients created an artificial intelligence software using an educational dataset including CT scans in a study by Wang et al..15 They used two data sets for testing in this study: the first data set included 102 COVID-19 cases and 124 other pneumonia cases whereas the other data set included 92 COVID-19 cases and 69 other pneumonia cases. The sensitivity and specificity for the diagnosis of COVID-19 pneumonia were found to be 79% and 81%, respectively, in this study. Similarly, Zheng et al.16 developed an artificial intelligence software using a total of 540 patients, 313 of whom had COVID-19 pneumonia. The AI program evaluated 540 patients by dividing them into a test dataset according to training/validation datasets and different time periods, and the sensitivity and specificity of the test for COVID-19 pneumonia were found to be 90% and 91%, respectively, in this study. TBT has proven to be a very sensitive technique in the diagnosis of COVID-19 pneumonia, but its specificity is relatively low. Although it is easy to distinguish COVID-19 pneumonia from bacterial pneumonia with CT, it may be difficult to differentiate, especially from viral interstitial pneumonias.6, 28. In our study, all pneumonia cases including interstitial pneumonias were included in the community-acquired pneumonia group. Therefore, in our study, the effectiveness of artificial intelligence in distinguishing between CAP subgroups and COVID-19 pneumonia was not examined. Cordoba et al.29 evaluated whether artificial intelligence application is useful for differentiating COVID-19 pneumonia from non-COVID-19 interstitial pneumonia. In this study which included 115 patients, it was determined that a machine learning model based on whole lung HRCT radiomic footprint could be useful in differentiating COVID-19 pneumonia from non-COVID-19 interstitial pneumonia.
This study by the authors reveals that the proposed approach that one conducted with the private dataset obtained from local patients appears to have a higher diagnostic performance in the diagnosis of COVID-19 than the studies in the literature. The CAP and normal CT images were also included in the study to test the robustness of the model. One of the main reasons for this achievement is the large-scale images in our dataset. A combination of layers from pre-trained deep networks is used to achieve more complex and detailed features. In addition, the best features that define the target class were obtained in the classification process by removing the unrelated features with the feature selection method. The untruthful accuracy improvement brought about by imbalanced data distribution has been resolved by eliminating the temporal problems in features. In this way, classification quality is improved. Accordingly, the diagnostic accuracy, sensitivity, and specificity of AI administration in the detection of COVID-19 pneumonia were 98.2, 98.0, and 98.2, respectively.
Although the proposed structure yields worthy performance compared to others, it has several limitations. It can take advantage of a larger cohort size in different locations to see how different patient groups impact our model's success. Besides, the prognosis of the disease supported by clinical findings may affect the outcomes of the decision mechanism. Different CT slices belonging to a patient were used under the same class. Some of these images should be extracted into a separate file, and it could be beneficial to use the model in the testing phase. Any of these factors could have influenced the performance measures in our study.
In conclusion, the results of this study showed that MTU-COVNet, an up-to-date network, distinguishes COVID-19 pneumonia from CAP and normal CT with high diagnostic efficiency in TBT images. AI applications can have beneficial effects in the fight against COVID-19 by accelerating the diagnostic process, increasing diagnostic efficiency, and reducing the workload of doctors working especially on the front lines. Real-time applications of AI will be on the agenda to help doctors detect COVID-19 infection in the near future.
CRediT authorship contribution statement
İn E.: Conceptualization, Methodology, Investigation, Resources, Formal analysis, Writing - Original Draft Kavuran G.: Conceptualization, Methodology, Data Curation, Software, Formal analysis, Writing - Original Draft Geçkil AA: Resources, Methodology, Writing - Original Draft Berber NK: Resources, Methodology Şahin M: Resources, Methodology. All authors read and approved the final manuscript.
Availability of data
The data that support the findings of this study are available from the corresponding author on reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Batah S.S., Fabro A.T. Pulmonary pathology of ARDS in COVID-19: a pathological review for clinicians. Respir Med. 2021;176 doi: 10.1016/j.rmed.2020.106239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University (JHU) Johns Hopkins Univ; 2020. Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) [Google Scholar]
- 3.Metlay J.P., Waterer G.W. Update in adult community-acquired pneumonia: key points from the new American Thoracic Society/Infectious Diseases Society of America 2019 guideline. Curr Opin Pulm Med. 2020;26:203–207. doi: 10.1097/MCP.0000000000000671. [DOI] [PubMed] [Google Scholar]
- 4.Olson G., Davis A.M. Diagnosis and treatment of adults with community-acquired pneumonia. JAMA. 2020;323:885. doi: 10.1001/jama.2019.21118. [DOI] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zu Z.Y., Di Jiang M., Xu P.P., Chen W., Ni Q.Q., Lu G.M., et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chassagnon G., Vakalopoulou M., Paragios N., Revel M.-P. Artificial intelligence applications for thoracic imaging. Eur J Radiol. 2020;123 doi: 10.1016/j.ejrad.2019.108774. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y., Feng X., Chi W., Li Z., Duan W., Liu H., et al. Deep learning aided decision support for pulmonary nodules diagnosing: a review. J Thorac Dis. 2018;10:S867–S875. doi: 10.21037/jtd.2018.02.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanc D., Racine V., Khalil A., Deloche M., Broyelle J.-A., Hammouamri I., et al. Artificial intelligence solution to classify pulmonary nodules on CT. Diagn Interv Imaging. 2020;101:803–810. doi: 10.1016/j.diii.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Rajaraman S., Candemir S., Kim I., Thoma G., Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl Sci. 2018;8:1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X. 2020. Development and evaluation of an AI System for COVID-19 diagnosis. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J. 2020. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. 2020;56:2000775. doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H. 2020. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. [DOI] [Google Scholar]
- 18.Antonellis G., Gavras A.G., Panagiotou M., Kutter B.L., Guerrini G., Sander A.C., et al. Shake table test of large-scale bridge columns supported on rocking shallow foundations. J Geotech Geoenviron Eng. 2015;141 doi: 10.1061/(ASCE)GT.1943-5606.0001284. [DOI] [Google Scholar]
- 19.He K., Zhang X., Ren S., Sun J. 2016 IEEE conf. comput. vis. pattern recognit. IEEE; 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 20.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. 2016. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and< 0.5 MB model size. [Google Scholar]
- 21.Too Abdullah, Saad Mohd. A new quadratic binary Harris hawk optimization for feature selection. Electronics. 2019;8:1130. doi: 10.3390/electronics8101130. [DOI] [Google Scholar]
- 22.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995 doi: 10.1023/A:1022627411411. [DOI] [Google Scholar]
- 23.Vapnik V. The support vector method of function estimation. Nonlinear model. Springer US; Boston, MA: 1998. pp. 55–85. [DOI] [Google Scholar]
- 24.Burges C.J.C. A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov. 1998;2:121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 25.Tian J., Xu Q., Liu S., Mao L., Wang M., Hou X. Comparison of clinical characteristics between coronavirus disease 2019 pneumonia and community-acquired pneumonia. Curr Med Res Opin. 2020;36:1747–1752. doi: 10.1080/03007995.2020.1830050. [DOI] [PubMed] [Google Scholar]
- 26.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozsahin I., Sekeroglu B., Musa M.S., Mustapha M.T., Uzun Ozsahin D. Review on diagnosis of COVID-19 from chest CT images using artificial intelligence. Comput Math Methods Med. 2020;2020:1–10. doi: 10.1155/2020/9756518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng M.Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Wang H., et al. Imaging profile of the COVID-19 infection radiologic findings and literature review. Radiol Cardiothorac Imaging. 2020;13(1) doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardobi N., Benetti G., Cardano G., Arena C., Micheletto C., Cavedon C., et al. CT radiomic models to distinguish COVID-19 pneumonia from other interstitial pneumonias. Radiol Med. 2021;126(8):1037–1043. doi: 10.1007/s11547-021-01370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavuran G. SEM-Net: Deep features selections with Binary Particle Swarm Optimization Method for classification of scanning electron microscope images. Materials Today Communications. 2021;27 doi: 10.1016/j.mtcomm.2021.102198. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.