Abstract
Human transcription factor IIF (TFIIF) is an α2β2 heterotetramer of RNA polymerase II-associating 74 (RAP74) and RAP30 subunits. Mutagenic analysis shows that the N-terminal region of RAP74 between L155 (leucine at codon 155) and M177 is important for initiation. Mutants in this region have reduced activity in transcription, but none are inactive. Single amino acid substitutions at hydrophobic residues L155, W164, I176, and M177 have similar activity to RAP74(1–158), from which all but three amino acids of this region are deleted. Residual activity can be explained because each of these mutants forms a complex with RAP30 and recruits RNA polymerase II into the preinitiation complex. Mutants are defective for formation of the first phosphodiester bond from the adenovirus major late promoter but do not appear to have an additional significant defect in promoter escape. Negative DNA supercoiling partially compensates for the defects of TFIIF mutants in initiation, indicating that TFIIF may help to untwist the DNA helix for initiation.
Accurate initiation from human pre-mRNA promoters requires the cooperation of general transcription factors and RNA polymerase II (reviewed in references 15, 31, and 38). For promoters containing a TATA box, an ordered in vitro pathway for assembly of an active transcription complex has been defined. TATA-binding protein (TBP) binds to the TATAAA sequence. Transcription factor IIB (TFIIB) can then associate with TBP and promoter DNA to form a TBP-TFIIB-promoter complex. TFIIF escorts RNA polymerase II to the promoter. TFIIE recruits TFIIH, which is a large protein complex that includes two subunits that are DNA helicases. The helicase activities of TFIIH are believed to open the DNA helix for initiation. In addition to its helicase module, TFIIH also includes a subassembly that contains a kinase-cyclin pair of subunits that phosphorylates the carboxy terminal domain of the largest subunit of RNA polymerase II.
Human TFIIF is an α2β2 heterotetramer of RAP74 and RAP30 subunits (RAP for RNA polymerase II-associating protein) (3, 8, 27, 50). Both subunits of TFIIF participate in stable recruitment of RNA polymerase II to the promoter (4, 9, 25), and both are necessary for accurate initiation from linear DNA templates in vitro (24, 28, 44). From negatively supercoiled DNA templates, the requirement for TFIIH and for ATP hydrolysis can be bypassed for initiation from many promoters (13, 33, 34, 45, 46). In these transcription systems, generally, TBP, TFIIB, and RNA polymerase II are minimally required to detect accurate initiation. TFIIE may be dispensable for transcription or it may be stimulatory (19, 33, 34). TFIIE is thought to have multiple roles in initiation. TFIIE recruits TFIIH, but TFIIE also makes contacts to TFIIF and TBP (30, 54), and TFIIE may have a role in DNA untwisting (19). Like TFIIE, TFIIF may be either stimulatory or, in some cases, required for transcription from supercoiled templates. Both TFIIF subunits contribute to transcription from supercoiled templates, although in some cases the RAP30 subunit may be more important than RAP74 (13, 46). Transcription systems have also been developed in which the region surrounding the initiation site cannot form base pairs because of sequence noncomplementarity. Partially mismatched templates are called “bubble” templates. The bubble template system minimally requires TBP, TFIIB, and RNA polymerase II for activity, but TFIIF strongly stimulates accurate initiation (32). Apparently, TFIIF contributes to a step in the initiation pathway that is distinct from formation of the open complex. For instance, TFIIF might help orient the +1 base of the template strand relative to the RNA polymerase II active site for initiation. Both RAP30 and RAP74 contribute to accurate initiation, and experiments with supercoiled templates indicate that both TFIIE and TFIIF may stimulate initiation by helping to untwist the DNA helix (reference 19 and the present study).
Isomerization is a complex progression of conformational changes in DNA and proteins that accompanies transcription initiation (5). From studies of the mechanism of initiation of Escherichia coli RNA polymerase, which is a homolog of eukaryotic RNA polymerase II, untwisting of the DNA helix appears to occur prior to strand separation in the formation of the fully open complex. At reduced temperature (i.e., 15°C), the E. coli RNA polymerase holoenzyme can form a complex on the promoter in which the DNA is topologically unwound, but the DNA strands are not yet separated (5, 6, 36). This intermediate is designated closed complex II. Prior to formation of the open complex, the promoter DNA appears to be wrapped around RNA polymerase, and the degree of DNA untwisting is equivalent to that observed in the open complex (1, 11). So helix untwisting and extensive promoter isomerization appear to be a prerequisite for open complex formation (5, 6).
TFIIF was recently shown to have a fundamental role in isomerization of the RNA polymerase II preinitiation complex (10, 37). In these studies, site-specific photo-cross-linking probes were placed beside a 32P radiolabel at many positions throughout the adenovirus major late promoter. Complexes were assembled with TBP, TFIIB, RNA polymerase II, TFIIF, and TFIIE, in either the presence or the absence of TFIIH, and the positions of particular factors were localized by the transfer of radiolabel from DNA to protein. Photo-cross-linking experiments done either in the absence of RAP74 or in the presence of RAP74 mutants indicated that TFIIF induces promoter DNA to wrap tightly around RNA polymerase II and the general factors (37). TFIIF containing the deletion mutant RAP74(1–172), although able to support a fully assembled preinitiation complex, did not support the tightly wrapped DNA structure. RAP74(1–205) and more complete versions of RAP74, however, did support this more active conformation. The region of RAP74 between amino acids 172 and 205, therefore, appears to have a critical function in complex isomerization, which involves tight wrapping of promoter DNA around RNA polymerase II and the general factors. By analogy to transcription by E. coli RNA polymerase, the TBP-TFIIB-RNA polymerase II-TFIIF-TFIIE complex might resemble closed complex II, forming a structure in which the DNA is wrapped around the complex and substantially untwisted but the DNA strands are not yet separated (5, 37). In this report, we analyzed a complete set of substitution mutants constructed in human RAP74 between amino acids 138 and 215. We suggest that the region of RAP74 between amino acids L155 and M177 may stimulate untwisting of the DNA helix for initiation.
MATERIALS AND METHODS
RAP74 mutagenesis and reconstitution of recombinant TFIIF.
RAP74 deletion mutants were constructed by PCR or as previously described (48). Most of the amino acid substitution mutants and the Δ170–177 mutant were constructed by using the Stratagene QuickChange Mutagenesis kit. A few mutants were constructed with the Promega Altered-Site II system.
RAP74 mutants and the RAP30 construct used in this study have a C-terminal six-histidine tag (49, 50). Recombinant proteins were isolated by chromatography on Qiagen Ni-nitrilotriacetic acid resin in buffer containing 4 M urea, as described earlier (49, 50). RAP30 and RAP74 were combined in equimolar amounts and dialyzed into buffer lacking urea to reconstitute the TFIIF complex (50).
In vitro transcription from linear DNA templates.
An extract derived from the nuclei of human HeLa cells was used as the source of transcription factors (41). TFIIF was removed from the extract by immunoprecipitation with anti-RAP74 and anti-RAP30 antibodies, as described previously (7, 28). Activity was reconstituted by addition of human recombinant TFIIF or a TFIIF mutant. The template for transcription was plasmid pML carrying the adenovirus major late promoter from position −258 to +196. The template was digested with endonuclease SmaI at position +217 relative to the transcription start site. Recombinant TFIIF, extract, and DNA template were combined for 60 min at 30°C. Transcription was initiated with 100 μM concentrations of ATP, CTP, and GTP and 5 μCi (0.25 μM) of [α-32P]UTP for 1 min. Results were very similar for wild-type (wt) TFIIF and mutants when reactions were initiated with ATP, CTP, and UTP (data not shown), suggesting that the pulse protocol with all four nucleoside triphosphates (NTPs) primarily reflects initiation events rather than elongation efficiencies. Furthermore, by using the 1-min pulse-labeling protocol with all four NTPs, it was previously shown that wt RAP74 and deletion mutants such as RAP74(1–172) supported synthesis of short transcripts of comparable lengths (28). After the pulse-labeling, reactions were chased for 30 min with 1 mM ATP, CTP, GTP, and UTP in the presence of 0.25% Sarkosyl. Addition of Sarkosyl dissociates pausing and termination factors from RNA polymerase II, such as N-TEF (negative transcription elongation factor) (29), factor 2 (52), NELF (negative elongation factor) (53), and DSIF (DRB-sensitivity inducing factor; DSIF subunits are homologs of yeast Spt4 and Spt5) (47). With the HeLa extract system, the addition of Sarkosyl approximately doubles the yield of runoff transcripts and allows efficient conversion of short transcripts to the runoff position (28). At some template positions Sarkosyl may also induce transcriptional pausing (16). Elongation for 30 min in the presence of Sarkosyl is sufficient to complete all previously initiated chains (16).
In vitro transcription with supercoiled DNA templates.
To determine the relationship between template supercoiling and TFIIF function, a defined in vitro system was established with recombinant human general transcription factors and highly purified calf RNA polymerase II (13, 33, 34, 45). Human recombinant TBPc (amino acids 155 to 335; the highly conserved C-terminal repeats of TBP) was the kind gift of Z. Sean Juo (22). Human recombinant TFIIB (14), TFIIA (42), and TFIIE (35) were produced by using expression clones kindly provided by Danny Reinberg. The production of TFIIF and TFIIF mutants was as described above. RNA polymerase II was isolated from calf thymus (18). The template for transcription was the supercoiled plasmid pML(C2AT)Δ71, which contains the adenovirus major late promoter from position −71 to +10 fused to a 406-nucleotide G-free cassette (the G-free cassette transcript is 416 nucleotides) (40). The buffer for in vitro transcription contained 12 mM HEPES (pH 7.9), 60 mM KCl, 12% glycerol, 6 mM MgCl2, 1 mg of bovine serum albumin per ml, 0.12 mM EDTA, 0.12 mM EGTA, and 1.2 mM dithiothreitol. Reaction mixtures (20 μl) contained 200 ng of supercoiled template, 0.3 pmol of RNA polymerase II, 0.8 pmol of TBPc, 1 pmol of TFIIA (except for the experiment shown in Fig. 9), 1 pmol of TFIIB, 1 pmol of TFIIE, and 1 pmol of TFIIF. An amount of 0.5 pmol of TFIIF supported maximal activity in this assay (data not shown). After 20 min of preincubation, 600 μM ATP and CTP and 25 μM [α-32P]UTP (5 μCi per reaction) were added. For the reactions shown in Fig. 8A, the reaction was stopped at various times. For the reaction shown in Fig. 8B, 0.05% Sarkosyl was added to the reactions at various times to inhibit further initiation, and elongation was continued for an additional 30 min to complete all previously initiated chains. Transcripts were isolated and analyzed as previously described (28).
FIG. 9.
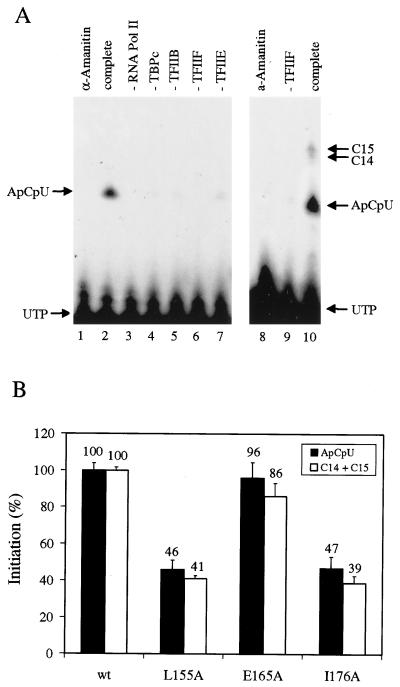
RAP74 mutants are defective for formation of the first phosphodiester bond from the adenovirus major late promoter. (A) Abortive and productive initiation assays. Abortive transcription reactions in the presence of 500 μM ApC plus 0.25 μM [α-32P]UTP (left panel) and productive transcription reactions in the presence of 500 μM ApC plus 0.25 μM [α-32P]UTP plus 500 μM CTP (right panel) are shown. The complete system (lanes 1, 2, 8, and 10) included TBPc, TFIIB, RNA polymerase II, TFIIF, and TFIIE. Omission of any one reaction component (as indicated) severely reduced or eliminated transcription. Abortive and productive initiation were completely sensitive to 1 μg of α-amanitin per ml (lanes 1 and 8). (B) Abortive (black bars) and productive (white bars) initiation assays with TFIIF mutants. Error bars indicate the standard deviations. Samples were done in triplicate.
FIG. 8.
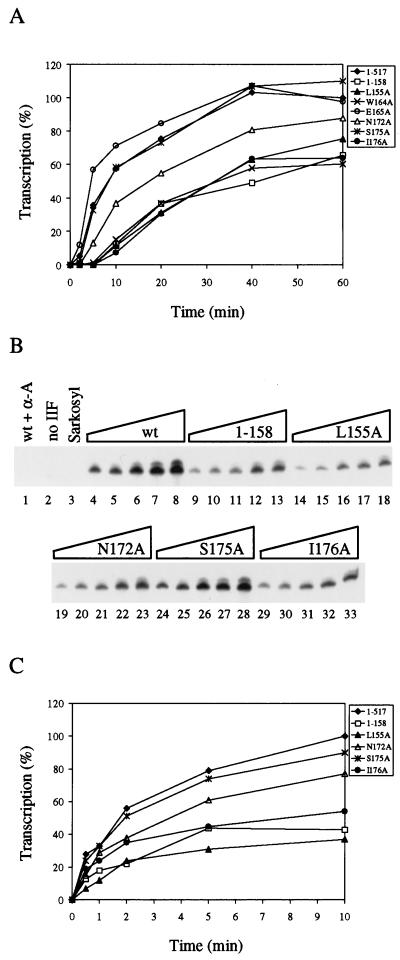
Negative DNA supercoiling partially rescues the activity of defective RAP74 mutants. (A) Reactions were reconstituted with wt TFIIF or mutants, and the accumulation of accurate transcripts was determined as a function of time (2, 5, 10, 20, 40, or 60 min). Reactions were done as in Fig. 7B, substituting TFIIF mutants for wt TFIIF. Transcription from the G-free cassette was quantitated by using a phosphorimager. (B) Defective RAP74 mutants support slow accumulation of transcripts. Reactions were done as in Fig. 7B, except that at the time indicated (0.5, 1, 2, 5, or 10 min) 0.05% Sarkosyl was added to the reaction to block further initiation, and elongation was continued for an additional 30 min to complete all previously initiated chains. The reaction in lane 1 was a 10-min time point in the presence of wt TFIIF and 1 μg of α-amanitin per ml. (C) Phosphorimager quantitation of the data shown in panel B.
For the abortive and productive initiation assays shown in Fig. 9, TFIIA was omitted from the reactions. Transcription was initiated with 500 μM ApC dinucleotide and 5 μCi of [α-32P]UTP (0.25 μM), in the absence or presence of 500 μM CTP, as noted. Abortive initiation reactions were stopped after 30 min. For productive initiation assays, the incubation continued for 30 min and was then chased for 10 min with addition of 1 mM UTP and 0.5 mM CTP. Reactions were stopped by addition of 4 μl of 200 mM EDTA, 1% sodium dodecyl sulfate (SDS), 10% glycerol, and 0.025% bromophenol blue. Samples were heated at 90°C for 2 min and electrophoresed in a 23% polyacrylamide gel (40:1 [wt/wt] acrylamide to methylene-bisacrylamide) containing 50% (wt/vol) urea.
Electrophoretic mobility shift assay.
The electrophoretic mobility shift experiment was done as previously described (28, 48). The DNA probe was the adenovirus major late promoter from position −53 to +14 relative to the transcription start site at +1. The 15-μl reaction mixtures contained recombinant yeast TBP (0.6 pmol), human TFIIB (0.6 pmol), calf RNA polymerase II (0.3 pmol), and human TFIIF or TFIIF mutant (1.0 pmol, except as noted). Reactions were electrophoresed in a 4% polyacrylamide gel and analyzed by autoradiography.
RESULTS
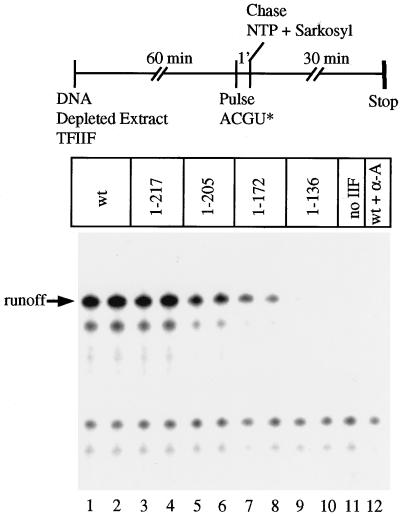
Previous work indicated that the N-terminal domain of RAP74 is very important for accurate initiation of transcription (28). To test the initiation activities of RAP74 mutants, an extract derived from HeLa cell nuclei was depleted of TFIIF by immunoprecipitation with anti-RAP74 and anti-RAP30 antibodies. To restore initiation activity, the TFIIF heterotetramer was reconstituted from recombinant RAP74 and RAP30 subunits and added back to the TFIIF-depleted extract (Fig. 1). The template for transcription was the adenovirus major late promoter treated with endonuclease SmaI, which in this plasmid has a cleavage site at position +217 downstream of the +1 position. Transcription was initiated with a 1-min pulse of 100 μM ATP, CTP, and GTP and 0.25 μM [α-32P]UTP. Sarkosyl was added to 0.25%, and elongation was continued for 30 min with 1 mM of each NTP. In this protocol, Sarkosyl dissociates elongation and termination factors from RNA polymerase II. Without Sarkosyl treatment, ca. 50% of the initiated chains terminate before the +217 position (29). Sarkosyl can also induce transcriptional pausing (16), which accounts for the α-amanitin-resistant transcripts that are shorter than the +217 runoff in Fig. 1. When Sarkosyl is added followed by a 30-min elongation time, short transcripts are efficiently extended to the runoff position. In this assay, RAP74(1–217) was almost as active as wt RAP74. RAP74(1–205) and RAP74(1–172) had progressively lower activities, and RAP74(1–136) was inactive.
FIG. 1.
The region of RAP74 between amino acids 136 and 217 is critical for accurate initiation from the adenovirus major late promoter. An extract derived from HeLa cell nuclei was depleted of TFIIF by immunoprecipitation with anti-RAP74 and anti-RAP30 antibodies. Template DNA, depleted extract, and a TFIIF sample (10 pmol) were combined and incubated for 60 min. Then, 100 μM ATP, CTP, and GTP and 0.25 μM [α-32P]UTP were added for 1 min. The reaction was chased for 30 min with 0.25% Sarkosyl and 1 mM ATP, CTP, GTP, and UTP. TFIIF complexes assembled with the indicated RAP74 mutant were tested in duplicate, as indicated. RAP74(1–136) does not form a stable complex with RAP30 so, for the reactions shown in lanes 9 and 10, subunits were added separately. The reaction in lane 12 contained wt TFIIF and 1 μg of α-amanitin per ml.
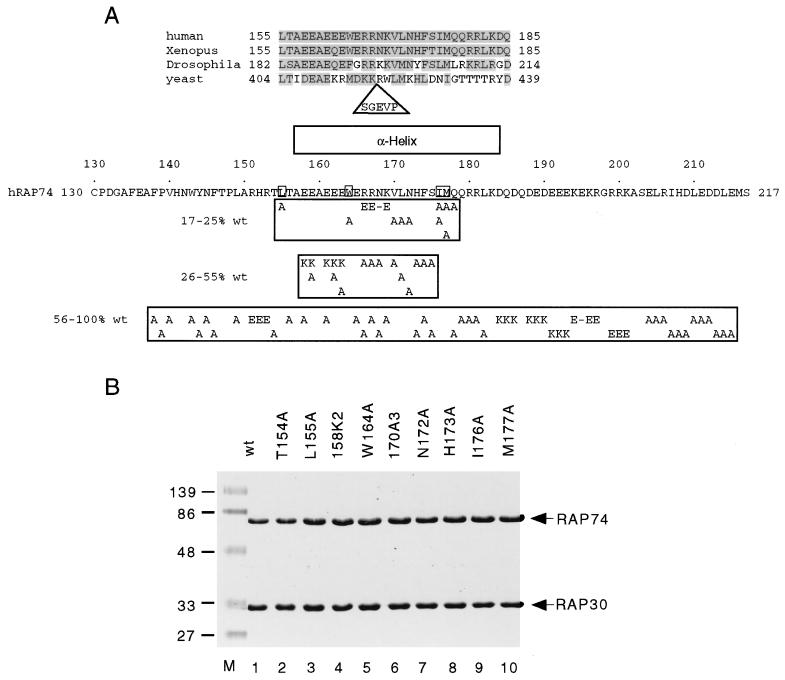
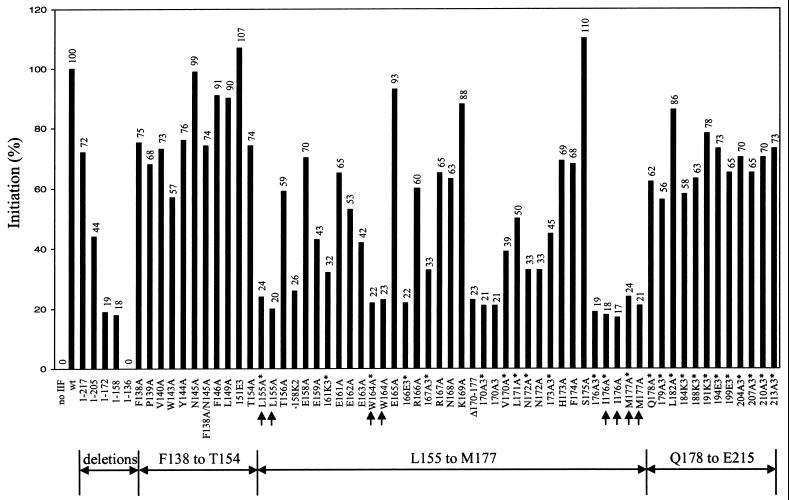
To determine which amino acid side chains in the region from codons 136 to 217 are most important for RAP74 function, a large set of site-directed mutants was constructed (Fig. 2). In Fig. 2A an alignment of human RAP74 with homologs from Xenopus laevis, Drosophila melanogaster, and Saccharomyces cerevisiae is shown. Secondary structure prediction analysis (39) indicates that amino acids 157 to 183 of human RAP74 may constitute an α-helix, and a similar structure is likely to be preserved in RAP74 homologs. Single, double, and triple amino acid substitutions are indicated beneath the sequence and categorized according to their activities in initiation (Fig. 3). Single substitution mutants are named according to the RAP74 amino acid that is substituted: L155A, W163A, etc. Multiple substitution mutants are named for the position of the first substituted amino acid, as in 161K3*, which is a triple-charge reversal mutant including the substitutions E161K, E162K, and E163K. The asterisk indicates that the mutation was constructed in RAP74(1–217) rather than in wt RAP74(1–517). A combination of alanine substitutions and charge reversal mutations was constructed based on the idea that these changes might alter activity without inducing long-range changes in protein conformation. Mutant RAP74 subunits were produced in E. coli, purified, and reconstituted with RAP30 in vitro to form TFIIF complexes. A polyacrylamide gel containing a representative panel of TFIIF mutants is shown in Fig. 2B.
FIG. 2.
Mutants in the RAP74 subunit of human TFIIF. (A) The region of human RAP74 between amino acids 130 and 217 is shown. At the top of the figure, a critical region of RAP74 homologues from human (2, 7), Xenopus (12), Drosophila (23), and yeast (S. cerevisiae) (17, 43) sources is aligned. The yeast sequence contains a 5-amino-acid insertion, as indicated. PHD analysis indicates that a segment of this sequence is α-helical (indicated by an open bar) (39). Substituted amino acids are indicated beneath the human sequence, and the most critical amino acids are boxed in the sequence. Each cluster of letters positioned beneath the sequence indicates a single, double, or triple amino acid substitution mutant, categorized according to its activity in initiation (Fig. 3). (B) TFIIF mutants were reconstituted from recombinant subunits produced in E. coli. Representative TFIIF samples (5 μg) were electrophoresed in a 12% polyacrylamide gel in the presence of SDS. The gel was stained with Coomassie blue.
FIG. 3.
Activities of RAP74 mutants in accurate initiation from the adenovirus major late promoter. The method for transcription assays was the same as that used in Fig. 1. The activity of wt RAP74 is reported as 100%. Reported values are the average of two determinations, which never varied by more than 15% from the average. An asterisk indicates that a mutant was constructed in RAP74(1–217) rather than in wt RAP74(1–517). The consistency of determinations is indicated by the similarity of results with particular amino acid substitutions constructed in both RAP74(1–217) and RAP74(1–517). Arrows indicate the single amino acid substitution mutants with the most severe defects.
The activities of RAP74 mutants in initiation from the adenovirus major late promoter are shown in Fig. 3. All of the TFIIF mutants were added at a saturating concentration (see Fig. 5), so reported values represent the maximum activity for each mutant. None of the substitution mutants between amino acids F138 and T154 or between Q178 and E215 demonstrated a severe defect in initiation, although some of these changes caused moderate defects. Mutants between Q178 to E215 were constructed in RAP74(1–217), so their activities should be compared to RAP74(1–217) rather than to wt RAP74. Sequences between amino acids 138 to 154 and 178 to 215, therefore, are not critical for initiation. Sequences between L155 and M177, however, were found to be very important for initiation from this promoter. Single amino acid substitutions that have the greatest defects are found at hydrophobic residues, L155, W164, I176, and M177. The activities of alanine substitution mutants at these positions are similar to the activities of the internal deletion mutant RAP74(Δ170–177) and the C-terminal truncations RAP74(1–172) and RAP74(1–158), from which significant portions of the critical region between L155 and M177 are deleted. Inactivation of the L155 to M177 region of RAP74 by amino acid substitution or deletion resulted in 17 to 23% wt activity. Some amino acid substitutions in the L155 to M177 region have an intermediate effect on transcription, including T156A, 158K2, E158A, E159A, 161K3*, E161A, E162A, E163A, R166A, 167A3*, R167A, N168A, V170A, L171A, N172A, 173A3*, H173A, and F174A. Although 158K2 has only 26% wt activity, substitution mutants E158A and E159A have only an intermediate defect. Also, mutants such as N172A are shown in this and other assay systems to have a consistently higher activity than the most affected mutants (see Fig. 8). Mutations that have a large or intermediate defect in transcription cluster within or immediately adjacent to the predicted α-helix between positions 157 and 183 (Fig. 2).
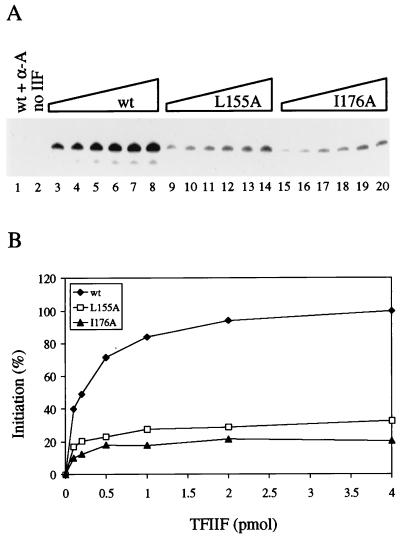
FIG. 5.
TFIIF mutants have a similar affinity for transcription complexes compared to wt TFIIF but a lower transcriptional activity. (A) Transcriptional activity from the adenovirus major late promoter as a function of TFIIF concentration. Lanes 3 to 8, 9 to 14, and 15 to 20 contain 0.1, 0.2, 0.5, 1.0, 2.0, and 4.0 pmol of the indicated TFIIF sample, respectively. Lane 1 contained 4.0 pmol of wt TFIIF and 1 μg of α-amanitin per ml. Lane 2 contained no TFIIF. (B) Phosphorimager quantitation of the data shown in panel A.
Mutants with the most pronounced transcriptional defects have been analyzed by gel permeation chromatography. This analysis indicates that RAP74 mutants form α2β2 heterotetramers with RAP30 when assembled into the TFIIF complex (data not shown), as does wt RAP74 (3, 8, 27, 50). By several criteria, these mutants have a very similar affinity for transcription complexes compared to wt TFIIF (Fig. 4 to 6).
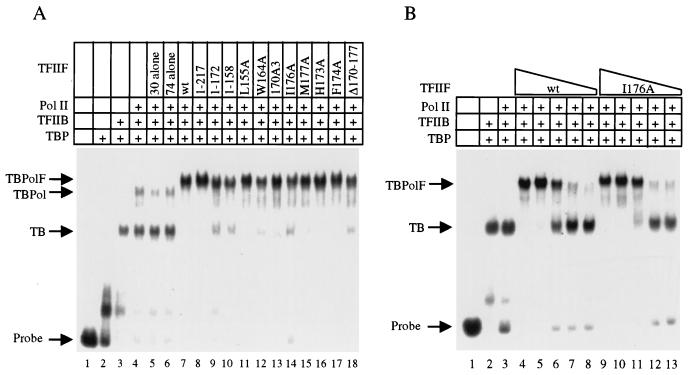
FIG. 4.
TFIIF mutants support efficient preinitiation complex assembly. (A) TFIIF mutants recruit RNA polymerase II to a complex of TBP and TFIIB formed at the adenovirus major late promoter. Reactions contain yeast TBP (T; 0.6 pmol), human TFIIB (B; 0.6 pmol), human TFIIF (F; 1.0 pmol), and calf RNA polymerase II (Pol II or Pol; 0.3 pmol), as indicated. (B) The defective I176A mutant supports preinitiation complex formation as efficiently as wt TFIIF. The protocol was the same as that in panel A except that lanes 4 to 8 and lanes 9 to 13 contained 2, 1.5, 1, 0.5, and 0.25 pmol of the indicated TFIIF sample.
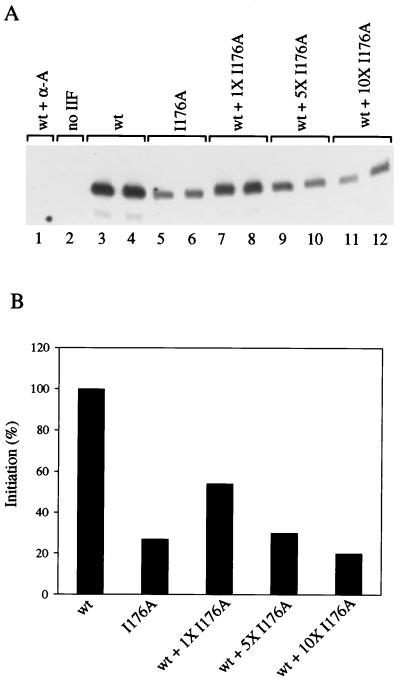
FIG. 6.
The transcriptionally defective I176A mutant competes with wt TFIIF for the formation of transcription complexes. (A) Competition assay. Lanes 1, 3, 4, and 7 to 12 contained 2 pmol of wt TFIIF. Lanes 5 to 8 contained 2 pmol of I176A mutant, lanes 9 to 10 contained 10 pmol of I176A mutant, and lanes 11 and 12 contained 20 pmol of I176A mutant. No inhibition or “squelching” of accurate transcription was observed with the addition of 20 pmol of a TFIIF sample in these reactions (data not shown), so the decrease in signal observed in lanes 7 to 12 is due to competition. No TFIIF was added to the reaction in lane 2. The reaction in lane 1 contained 2 pmol of wt TFIIF and 1 μg of α-amanitin per ml. Accurate transcription assays were done as in Fig. 1. (B) Phosphorimager quantitation of the data in panel A.
Electrophoretic mobility shift experiments were done to measure the capacity of TFIIF mutants to assemble a TBP-TFIIB-RNA polymerase II-TFIIF complex on the adenovirus major late promoter (Fig. 4). A TBP-TFIIB complex forms efficiently on and around the strong TATAAA box of the promoter (Fig. 4A, lane 3). RNA polymerase II binds weakly to the TBP-TFIIB complex (lane 4) and, at the protein concentrations used in these experiments, the addition of either the RAP30 or the RAP74 subunit of TFIIF alone was not sufficient to improve RNA polymerase II recruitment (lanes 5 and 6). Addition of the TFIIF complex, however, induced quantitative formation of the TBP-TFIIB-RNA polymerase II-TFIIF complex (lane 7). Even the most defective RAP74 mutants appeared to recruit RNA polymerase II as efficiently as did wt TFIIF (compare lane 7 to lanes 8 to 18). The I176A mutant has 17% wt activity in transcription from a linear DNA template (Fig. 3), but when titrated from a limiting to a saturating concentration in the mobility shift experiment, I176A recruited RNA polymerase II to the promoter with the same efficiency as wt TFIIF (Fig. 4B; compare lanes 4 to 8 with lanes 9 to 13).
To extend the argument that TFIIF mutants efficiently form transcription complexes, the concentration of wt TFIIF necessary to support accurate transcription was compared to that of the strongly affected mutants L155A and I176A (Fig. 5). If TFIIF mutants had a defect in complex assembly, transcriptional activity would be expected to saturate at a higher protein concentration for the mutants than for wt TFIIF. However, no clear difference in the concentration requirement for maximal activity in initiation was observed (Fig. 5), indicating that mutants and wt TFIIF have similar affinities for transcription complexes. From similar titration experiments, we found that at least 20 pmol of TFIIF could be added to these reactions without causing inhibition of the transcription reaction, for instance, by “squelching.” These experiments demonstrate that 10 pmol of wt or mutant TFIIF was a saturating amount in the transcription assay, so low activities of mutants reported in Fig. 3 cannot be attributed to reduced affinity for transcription complexes. TFIIF mutants appear to be properly folded and to support transcription complex assembly, but these mutants have specific defects in initiation.
Another indication that TFIIF mutants assemble transcription complexes normally comes from functional competition between the defective I176A mutant and wt TFIIF (Fig. 6). Addition of a one- or a fivefold molar excess of the I176A mutant over wt TFIIF to a TFIIF-depleted HeLa extract progressively reduced accurate transcription (Fig. 6A, compare lanes 3 and 4 with lanes 7 to 10). A 10-fold molar excess of the mutant reduced transcription to the level supported by I176A alone (compare lanes 5 and 6 with lanes 11 and 12). Gel data is quantitated in Fig. 6B. Because addition of 20 pmol of wt or mutant TFIIF to these reactions supported the same activity as the addition of 2 pmol of the same TFIIF sample (data not shown), there was no evidence for “squelching” of transcription by adding an excess of TFIIF. The inhibition observed in lanes 7 to 12, therefore, appears to be attributable to competition rather than “squelching” or nonspecific blocking of the transcription reaction.
To further characterize the activities of TFIIF mutants in transcription, a defined system was established that requires a negatively supercoiled DNA template (Fig. 7). Purified calf thymus RNA polymerase II was combined with human recombinant TFIIA, TFIIB, TFIIF, and TFIIE. The template for transcription was a supercoiled plasmid DNA containing the adenovirus major late promoter fused to a G-free cassette. Accurate initiation was indicated by synthesis of the G-free cassette transcript. Because the template was negatively supercoiled, TFIIH was not required to open the DNA strands for initiation (13, 33, 34, 45). The recombinant human general transcription factors and calf RNA polymerase II used in this assay are shown in Fig. 7A, and the assay is validated in Fig. 7B. Accurate initiation is completely dependent on RNA polymerase II, TBP, TFIIB, and TFIIF. TFIIA is not required for initiation. TFIIE is strongly stimulatory but not essential, a finding in agreement with a previous report (19). The reaction is completely inhibited by 1 μg of α-amanitin per ml, as expected. Accurate transcription appears to be completely dependent on negative supercoiling of the DNA template because activity is abolished by relaxation of the template with E. coli DNA topoisomerase I or by linearization with a restriction endonuclease (data not shown). The faster mobility gel band appears to result from transcriptional pause site about 30 bases from the end of the G-free cassette, because this shorter RNA appears to be a precursor to the full-length cassette transcript (data not shown). Both the RAP30 and RAP74 subunits of TFIIF were required for detectable levels of transcription in this assay (data not shown).
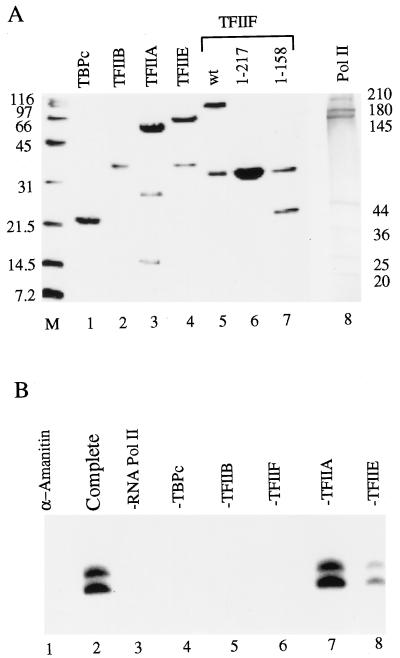
FIG. 7.
Recombinant human TFIIA, TBPc, TFIIB, TFIIF, TFIIE, and calf RNA polymerase II support accurate initiation from a supercoiled adenovirus major late promoter template. (A) Transcription factors used in this experiment were analyzed by SDS-polyacrylamide gel electrophoresis. The 15% polyacrylamide gel shown in the left panel was stained with Coomassie blue. RAP74(1–217) and RAP30 comigrated in the gel (lane 6). The 4 to 15% gradient polyacrylamide gel of the calf RNA polymerase II sample (right panel) was stained with silver nitrate. (B) General transcription factor requirements for initiation from a supercoiled template. TFIIA, TBPc, TFIIB, TFIIF, TFIIE, and calf RNA polymerase II were incubated for 20 min with a supercoiled plasmid containing the adenovirus major late promoter linked to a G-free cassette. Then, 600 μM ATP and CTP and 25 μM [α-32P]UTP were added, and transcription was continued for 30 min. “Complete” indicates that all reaction components were included (lane 2). Individual reaction components were omitted from the reaction, as indicated (lanes 3 to 8). The reaction in lane 1 contained the complete system in the presence of 1 μg of α-amanitin per ml. The more rapidly migrating transcript band results from transcriptional pausing about 30 nucleotides before the end of the 416 nucleotide G-free cassette (data not shown).
The capacity of RAP74 mutants to support accurate transcription from the supercoiled template was analyzed by observing the accumulation of G-free cassette transcripts as a function of time (Fig. 8). A saturating amount of TFIIF was added to each reaction (data not shown). Defective mutants such as L155A, W164A, and I176A and mutant 1–158 accumulated transcripts slowly compared to wt TFIIF. The S175A and E165A mutants, which were not defective in transcription from a linear template (Fig. 3), accumulated full-length transcripts from the supercoiled template at a similar rate compared to wt RAP74. N172A, which has low to moderate activity in the extract system (Fig. 3), has moderate activity in the defined transcription system. Because the defects of TFIIF mutants were apparent in both the defined and the extract system, the most important contacts of TFIIF for initiation are likely to be with components of the core transcription apparatus: TBP, TFIIB, RNA polymerase II, TFIIE, and/or promoter DNA. In this assay, defective mutants appear to initiate transcription inefficiently, but these mutants may also have defects in promoter escape and elongation. From other experiments, we have determined that RAP74 mutants that are defective in initiation are also defective in stimulating elongation by RNA polymerase II (reference 28 and data not shown).
To confirm that RAP74 mutants are slow to initiate, the assay was modified to recover all productively initiated RNA chains formed as a function of time (Fig. 8B and C). This was accomplished by using the anionic detergent Sarkosyl. In this assay, Sarkosyl can have two effects. First, Sarkosyl inhibits initiation by RNA polymerase II. This effect is demonstrated because, when Sarkosyl was added prior to NTP substrates, no accurate transcription was observed (Fig. 8B, lane 3). Second, Sarkosyl may eliminate the effects of elongation factors, so the addition of the detergent and the subsequent 30-min elongation time are likely to eliminate any effects of different TFIIF mutants on elongation. The results of this experiment support the conclusion that the most defective RAP74 mutants initiate inefficiently. As expected, mutants L155A, I176A, and 1–158 had the slowest initiation rates. N172A had intermediate activity, and S175A had the same activity as wt RAP74.
Productive transcription depends on initiation and promoter escape. To determine whether the defect of RAP74 mutants is in the formation of the first phosphodiester bond or the formation of the first stable transcript, these processes were analyzed (Fig. 9). A supercoiled DNA template containing the adenovirus major late promoter was combined with general transcription factors TBPc, TFIIB, RNA polymerase II, TFIIF, and TFIIE. To monitor abortive initiation, transcription was initiated with ApC and [α-32P]UTP. In this case, only the first phosphodiester bond could be formed, resulting in the synthesis of ApCpU (Fig. 9A, left panel). The trinucleotide cannot be retained stably in the RNA polymerase II active site, so multiple abortive products can be formed from each transcription complex. Synthesis of the ApCpU product was completely sensitive to 1 μg of α-amanitin per ml and appeared to be completely dependent on RNA polymerase II and TFIIF. Synthesis of ApCpU was strongly dependent on addition of TBPc, TFIIB, and TFIIE. Abortive initiation also required DNA supercoiling, because relaxed circular DNA templates and linear templates did not support transcription with these reaction components (data not shown). To measure productive initiation, transcription was initiated with ApC, [α-32P]UTP, and CTP (Fig. 9A, right panel). In this case, the 15-mer 5′-ACUCUCUUCCCCUCC-3′ (C15) was synthesized from the adenovirus major late promoter. Because ATP was omitted from the reaction, elongation was stalled at C14 and C15, prior to addition of A16. C14 and C15 transcripts have escaped the promoter, because these short RNAs can be quantitatively chased to the end of the G-free cassette (data not shown).
Abortive transcription continued even when productive transcription was possible, because synthesis of ApCpU was detected when CTP was included in the reaction (lane 10). Because TFIIH is absent from the system, the open complex is not formed by the natural mechanism and may be only transiently maintained. If the open complex is unstable, as we suggest, this may explain why abortive transcription is so prevalent in the supercoiled template system. The ApCpU trinucleotide signal was quantitated by a phosphorimager to be about sixfold more intense than that for C14 and C15. The trinucleotide has a fivefold lower specific activity than the C14 and C15 transcripts, so ca. 30 ApCpU trinucleotides were synthesized for each C14 or C15 transcript.
When TFIIF mutants were tested in abortive and productive initiation assays, their primary defect appeared to be in formation of the first phosphodiester bond (Fig. 9B). The strongly defective mutants L155A and I176A were found to be equally defective for abortive and productive initiation compared to wt TFIIF. The E165A mutant was used in this experiment as a control because it carries a substitution within the L155 to M177 region of RAP74, but this mutant does not appear to have a defect in transcription (Fig. 3 and 8A). L155A and I176A are defective for formation of the first phosphodiester bond from the adenovirus major late promoter. These mutants have essentially the same defect relative to wt TFIIF in production of C14 and C15 transcripts that they have in formation of the first phosphodiester bond. The defect in C14 and C15 synthesis, therefore, appears to result primarily from the prior defect in initiation.
Comparing transcription from linear and supercoiled templates, negative DNA supercoiling appeared to partially rescue the defect of TFIIF mutants (Table 1). When we used a supercoiled template, at the 60-min time point in Fig. 8A, strongly defective mutants demonstrated 60 to 65% wt activity. By contrast, with a linear DNA template and an extract transcription system, strongly defective TFIIF mutants demonstrated only 17 to 23% wt activity (Fig. 3). The higher relative activity supported by these mutants from the supercoiled template than from the linear template indicates that negative DNA supercoiling may partially compensate for the normal function of RAP74 in initiation.
TABLE 1.
A negatively supercoiled DNA template partially compensates for the defects of TFIIF mutants in initiationa
| TFIIF | Linear DNA (% wt) | Supercoiled DNA (% wt) |
|---|---|---|
| wt | 100 | 100 |
| S175A | 110 | 107 |
| E165A | 93 | 98 |
| L155A | 20 | 65 |
| W164A | 23 | 60 |
| N172A | 33 | 88 |
| I176A | 17 | 64 |
DISCUSSION
The region of RAP74 between L155 and M177 enhances accurate initiation from the adenovirus major late promoter by facilitating formation of the first phosphodiester bond (Fig. 9). Alanine substitutions at hydrophobic residues L155, W164, I176, and M177 have a similar defect in initiation to a deletion from 1 to 158, from which most of the L155-to-M177 region is removed. These mutants, however, form normal complexes with the RAP30 subunit and enter transcription complexes with similar affinity compared to wt TFIIF. Even the most defective mutants in the L155-to-M177 region bring RNA polymerase II into a complex with TBP and TFIIB, formed at the adenovirus major late promoter (Fig. 4A). The transcriptionally defective mutant I176A recruits RNA polymerase II to the promoter with the same efficiency as wt TFIIF (Fig. 4B). Defective mutants L155A and I176A were found to saturate a transcription reaction at a similar concentration to wt TFIIF (Fig. 5). Furthermore, TFIIF containing the defective I176A mutant competed with wt TFIIF for assembly of transcription complexes (Fig. 6). There is no indication, therefore, that mutants in the critical L155-to-M177 region of RAP74 have a defect in RAP30 binding or in transcription complex assembly.
The L155-to-M177 region of RAP74 is conserved in evolution and is likely to be α-helical (Fig. 2). The distribution of strongly affected mutants, however, indicates that, if this region is within an α-helix, at least two surfaces of this structure may contribute to function. Notably, W164 would appear on the opposite face of a helix from L155, I176, and M177. In the future, a molecular structure of TFIIF will be useful in the full interpretation of the results from mutagenic studies.
Because the L155A, W164A, and I176A mutants are partially defective in a defined transcription system containing only TBP, TFIIB, RNA polymerase II, TFIIF, TFIIE, and promoter DNA (Fig. 8 and 9), the most important contacts of TFIIF in initiation are likely to be with these general factors and/or DNA. These TFIIF mutants have very similar defects in elongation stimulation to their defects in initiation (27a). Because the elongation assay was done in a system containing only RNA polymerase II, TFIIF, template DNA, and nascent RNA, the most important contacts of TFIIF are likely to be with RNA polymerase II and DNA. Contacts with RNA cannot be relevant in initiation complexes that contain no RNA. If we assume that the L155-to-M177 region of RAP74 is not involved in RAP30 binding (see above), this region may contact RNA polymerase II and/or DNA.
From protein affinity chromatography experiments, it was shown that the region between amino acids 172 and 205 is important for binding of RAP74 to itself (37). The critical mutations identified in the present study, therefore, might lie within a dimerization region that is important in maintaining the TFIIF heterotetramer. Estimates of the native molecular weights of TFIIF samples by gel permeation chromatography, however, indicated that even the most severely affected mutants in this region, including Δ170–177 and I176A, form heterotetramers with RAP30 (data not shown). The region of RAP74 between L155 and M177 may be involved in dimerization, but this region also may contribute to another important contact with RNA polymerase II and/or template DNA, as discussed above. Because the TFIIF heterotetramer appears to be maintained in strongly defective RAP74 mutants, the L155-to-M177 region does not appear to be essential for dimerization.
Site-specific DNA-protein photo-cross-linking studies have recently demonstrated that the region of RAP74 between amino acids 172 and 205 is critically important to form a tight wrap of adenovirus major late promoter DNA around a preinitiation complex containing TBP, TFIIB, RNA polymerase II, TFIIF, and TFIIE (37). Significant DNA bending around RNA polymerase II was inferred from photo-cross-linking studies of the TBP-TFIIB-RNA polymerase II-TFIIF complex (26), although the degree of DNA wrapping appeared to be more extensive in a complex that also contained TFIIE (37). In studies with the complex containing TFIIE, the deletion mutant RAP74(1–172) was found to assemble into the preinitiation complex but failed to support a tightly wrapped structure and failed to support formation of many specific photo-cross-links to general transcription factors and RNA polymerase II within the core promoter (10, 37). RAP74(1–205), on the other hand, appeared to support DNA wrapping, apparently with the same efficiency as wt RAP74. RAP74(1–205), however, is not as active in transcription as wt RAP74 (Fig. 1 and 3), indicating that a tight DNA wrap may be necessary but not sufficient to fully isomerize the preinitiation complex. The interpretation of the photo-cross-linking data was that the region of RAP74 between amino acids 172 and 205 is critically important for forming the tight DNA wrap around RNA polymerase II and for conformational isomerization of the preinitiation complex (37).
Although these regions of RAP74 only partially overlap, the critical region that we identify between amino acids L155 and M177 is likely to correspond to the region previously mapped by deletion mutagenesis between amino acids 172 and 205. RAP74 deletion mutants 1–158 and 1–172 support the same activity in transcription (Fig. 3). As judged by transcriptional activity, therefore, the deletion interval between amino acids 172 and 205 cannot be distinguished from the interval between amino acids 158 and 205. The region from amino acids 158 to 205 corresponds closely to the L155-to-M177 region mapped in the present study. A comparison of the activity of substitution and deletion mutants in initiation further demonstrates this conclusion. Single amino acid substitution mutants L155A, W164A, I176A, and M177A have indistinguishable activity in transcription compared to deletion mutants 1–158 and 1–172 (Fig. 3). Each of these substitutions and deletions, therefore, appear to eliminate the activity of this region of RAP74. The L155-to-M177 region is predicted to overlap with an extended α-helix in the RAP74 structure (Fig. 2). The deletion from 1 to 172 would be expected to damage this helical structure, and this mutation removes critical residues I176 and M177. The deletion from 1 to 158 removes the region predicted to be helical, along with critical residues W164, I176, and M177. Single alanine substitutions at L155, W164, I176, and M177 are predicted to maintain the helical structure but may eliminate RAP74 dimer contacts or contacts to RNA polymerase II and/or to DNA.
A deletion endpoint that lies just beyond a functional protein region may affect the activity of the nearby region. An example of this point is the 1–205 mutant, which does not appear to be missing any critical amino acids and yet has a lower activity than the 204A3*, 207A3*, 210A3*, and 213A3* substitution mutants (Fig. 3). Presumably, the reduced activity of mutant 1–205 does not result from deletion of any particular amino acid side chain between amino acids 205 and 217 but rather results from a partial disruption of the proximal L155-to-M177 region. The deletion from 1 to 205 is of further interest, because a comparison of its behavior in transcription assays and in photo-cross-linking studies indicates that transcription assays may be more reliable than the current photo-cross-linking techniques for identifying defects in RAP74. Mutant 1–205 is clearly defective in initiation, although it has no apparent defect when analyzed by photo-cross-linking.
The only RAP74 mutant in this collection that was completely inactive for initiation was mutant 1–136 (Fig. 3). This mutant fails to form a stable complex with the RAP30 subunit and fails to enter transcription complexes (28). It may be that a primary function of the region of RAP74 between amino acids 1 and 158 is to orient the RAP30 subunits of TFIIF. The region between L155 and M177 is not required for association with the RAP30 subunit, and this region does not appear to be important for recruitment of RNA polymerase II to the promoter. The L155-to-M177 region appears to have a distinct function in initiation.
A comparison of the activities of TFIIF mutants in transcription assays utilizing linear versus supercoiled templates may be revealing for TFIIF function. Initiation from linear DNA templates requires ATP hydrolysis and the general transcription factor TFIIH, which includes two subunits with ATP-dependent DNA helicase activities. Negatively supercoiled DNA templates allow initiation in the absence of TFIIH, presumably because untwisting of the DNA template by supercoiling allows the remaining general factors to open the helix for initiation. The exposure of single-stranded thymidines to potassium permanganate has been detected for promoters opened by TFIIH and ATP hydrolysis (20, 21, 51), but detection of the open complex formed in the absence of TFIIH with a supercoiled template has not been described. Without the normal ATP-driven mechanism for promoter opening, the supercoiled template may support only transient strand separation for initiation. TFIIE stimulates transcription from supercoiled templates and, in the system we have established, both the RAP74 and RAP30 subunits of TFIIF are essential for transcription from the supercoiled template. DNA supercoiling appears to partially compensate for RAP74 mutants in the L155-to-M177 region, because, from a supercoiled template, initiation activity in the presence of strongly defective mutants is from 60 to 65% wt as opposed to 17 to 23% wt from a linear DNA template (Table 1). The more-severe defect of RAP74 mutants in initiation from a linear template is also observed in the elongation activities of these mutants from a linear DNA template (27a). Based on these observations, we suggest that the L155-to-M177 region of RAP74 may have an activity in helix untwisting that stimulates both initiation and elongation.
Isomerization of the RNA polymerase II preinitiation complex has been hypothesized to involve sharp bending of DNA at the TATA box of the adenovirus major late promoter, sharp bending of DNA through the RNA polymerase II active site (5, 26, 37), and tight wrapping of DNA around RNA polymerase II and the general transcription factors (37). Constraining the DNA between two bends by the tight wrap was hypothesized to untwist the DNA helix for initiation (5, 37). Both TFIIF and TFIIE appear to contribute to DNA wrapping (37) and perhaps helix untwisting (reference 19 and the present study).
In E. coli, the transcriptional intermediate designated closed complex II appears to be topologically altered by −1.7 helical turns, the same untwisting that was observed for the open complex (1, 11). This topological change in DNA is likely to represent both helix untwisting and DNA wrapping. Assuming that a similar situation occurs in the TBP-TFIIB-RNA polymerase II-TFIIF-TFIIE complex and assuming that approximately 10 bp are untwisted, with an increase in the distance between stacked bases of ca. 1.5 Å per untwisted bp, the DNA length between the TATAAA box and the transcriptional start site would increase by 15 Å. Such a change in helix twist would alter the position and might alter the face of the helix on which the initiating template base is oriented relative to the RNA polymerase II active site. We propose that the functions of TFIIE and TFIIF in initiation are, in part, to untwist the DNA helix to orient the +1 base with the RNA polymerase II catalytic center and/or to present the untwisted helix as a substrate for the helicases of TFIIH for strand opening.
ACKNOWLEDGMENTS
We thank Danny Reinberg, Z. Sean Juo, and Michelle Sawadogo for clones and Z. Sean Juo for generously providing a sample of human TBPc. We gratefully acknowledge Augen A. Pioszak for construction of some of the RAP74 mutants used in this work.
This research was supported by a grant from the American Cancer Society. Additional support was provided by the Research Excellence Fund of Michigan State University and the Michigan State University Agricultural Experiment Station.
REFERENCES
- 1.Amouyal M, Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- 2.Aso T, Vasavada H A, Kawaguchi T, Germino F J, Ganguly S, Kitajima S, Weissman S M, Yasukochi Y. Characterization of cDNA for the large subunit of the transcription initiation factor TFIIF. Nature. 1992;355:461–464. doi: 10.1038/355461a0. [DOI] [PubMed] [Google Scholar]
- 3.Conaway J W, Conaway R C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 4.Conaway R C, Garrett K P, Hanley J P, Conaway J W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors alpha and beta gamma promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulombe B, Burton Z F. DNA bending and wrapping around RNA polymerase: a revolutionary model to describe transcriptional mechanisms. Microbiol Mol Biol Rev. 1998;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deHaseth P L, Zupancic M L, Record M T., Jr RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkelstein A, Kostrub C F, Li J, Chavez D P, Wang B Q, Fang S M, Greenblatt J, Burton Z F. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature. 1992;355:464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- 8.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 9.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamper H B, Hearst J E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- 12.Gong D W, Hasegawa S, Wada K, Roeder R G, Nakatani Y, Horikoshi M. Elucidation of three putative structural subdomains by comparison of primary structure of Xenopus and human RAP74. Nucleic Acids Res. 1992;20:6736. doi: 10.1093/nar/20.24.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 14.Ha I, Lane W S, Reinberg D. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 15.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley D K, Roeder R G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 17.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 18.Hodo H G, Blatti S P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977;16:2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 19.Holstege F C, Tantin D, Carey M, van der Vliet P C, Timmers H T. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege F C, van der Vliet P C, Timmers H T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Smale S T, Gralla J D. A common ATP requirement for open complex formation and transcription at promoters containing initiator or TATA elements. J Biol Chem. 1993;268:6535–6540. [PubMed] [Google Scholar]
- 22.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 23.Kephart D D, Price M P, Burton Z F, Finkelstein A, Greenblatt J, Price D H. Cloning of a Drosophila cDNA with sequence similarity to human transcription factor RAP74. Nucleic Acids Res. 1993;21:1319. doi: 10.1093/nar/21.5.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kephart D D, Wang B Q, Burton Z F, Price D H. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 25.Killeen M, Coulombe B, Greenblatt J. Recombinant TBP, transcription factor IIB, and RAP30 are sufficient for promoter recognition by mammalian RNA polymerase II. J Biol Chem. 1992;267:9463–9466. [PubMed] [Google Scholar]
- 26.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitajima S, Tanaka Y, Kawaguchi T, Nagaoka T, Weissman S M, Yasukochi Y. A heteromeric transcription factor required for mammalian RNA polymerase II. Nucleic Acids Res. 1990;18:4843–4849. doi: 10.1093/nar/18.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Lei, L., and Z. F. Burton. Submitted for publication.
- 28.Lei L, Ren D, Finkelstein A, Burton Z F. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall N F, Price D H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxon M E, Goodrich J A, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 31.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 32.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 33.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 34.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 35.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Structure and functional properties of human general transcription factor IIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 36.Polyakov A, Severinova E, Darst S A. Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 37.Robert F, Douziech M, Forget D, Egly J-M, Greenblatt J, Burton Z F, Coulombe B. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 39.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 40.Sawadogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 42.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 43.Sun Z W, Hampsey M. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan S, Aso T, Conaway R C, Conaway J W. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 45.Timmers H T. Transcription initiation by RNA polymerase II does not require hydrolysis of the beta-gamma phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 47.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B Q, Burton Z F. Functional domains of human RAP74 including a masked polymerase binding domain. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 49.Wang B Q, Kostrub C F, Finkelstein A, Burton Z F. Production of human RAP30 and RAP74 in bacterial cells. Protein Expr Purif. 1993;4:207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- 50.Wang B Q, Lei L, Burton Z F. Importance of codon preference for production of human RAP74 and reconstitution of the RAP30/74 complex. Protein Expr Purif. 1994;5:476–485. doi: 10.1006/prep.1994.1067. [DOI] [PubMed] [Google Scholar]
- 51.Wang W, Carey M, Gralla J D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 52.Xie Z, Price D. Drosophila Factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J Biol Chem. 1997;272:31902–31907. doi: 10.1074/jbc.272.50.31902. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 54.Yokomori K, Verrijzer C P, Tjian R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci USA. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]