Abstract
In fission yeast as well as in higher eukaryotic organisms, entry into mitosis is delayed in cells containing damaged or unreplicated DNA. This is accomplished in part by maintaining the Cdc25 phosphatase in a phosphorylated form that binds 14-3-3 proteins. In this study, we generated a mutant of fission yeast Cdc25 that is severely impaired in its ability to bind 14-3-3 proteins. Loss of both the DNA damage and replication checkpoints was observed in fission yeast cells expressing the 14-3-3 binding mutant. These findings indicate that 14-3-3 binding to Cdc25 is required for fission yeast cells to arrest their cell cycle in response to DNA damage and replication blocks. Furthermore, the 14-3-3 binding mutant localized almost exclusively to the nucleus, unlike wild-type Cdc25, which localized to both the cytoplasm and the nucleus. Nuclear accumulation of wild-type Cdc25 was observed when fission yeast cells were treated with leptomycin B, indicating that Cdc25 is actively exported from the nucleus. Nuclear exclusion of wild-type Cdc25 was observed upon overproduction of Rad 24, one of the two fission yeast 14-3-3 proteins, indicating that one function of Rad 24 is to keep Cdc25 out of the nucleus. In support of this conclusion, Rad 24 overproduction did not alter the nuclear location of the 14-3-3 binding mutant. These results indicate that 14-3-3 binding contributes to the nuclear exclusion of Cdc25 and that the nuclear exclusion of Cdc25 is required for a normal checkpoint response to both damaged and unreplicated DNA.
The cell division cycle is a series of temporally regulated events, with each event being dependent upon the proper execution of the preceding event. Checkpoints maintain the order of events in the cell cycle by preventing cell cycle progression at inappropriate times (18). The DNA replication checkpoint monitors S-phase completion and prevents mitosis in its absence, and the G2/M DNA damage checkpoint monitors genome integrity and delays the onset of mitosis when damage is detected. In most organisms the DNA replication and damage checkpoints prevent entry into mitosis by maintaining Cdc2 in its tyrosine-phosphorylated (inhibited) form (4, 21, 50, 51). In higher eukaryotic organisms, the Wee1 and Myt1 protein kinases negatively regulate mitosis by phosphorylating Cdc2. Wee1 phosphorylates Cdc2 exclusively on tyrosine 15, whereas Myt1 is capable of phosphorylating Cdc2 on both threonine 14 and tyrosine 15 (8, 19, 20, 30, 34, 35, 37, 38, 46, 47, 59). In the fission yeast, Schizosaccharomyces pombe, the Wee1 and Mik1 tyrosine kinases negatively regulate mitosis by phosphorylating Cdc2 on Tyr 15 (13, 27, 44, 45). Cdc25 is a universally conserved protein phosphatase that promotes mitotic entry by dephosphorylating Cdc2 on Tyr 15/Thr 14, thereby activating Cdc2 (11, 17, 28, 53, 55). A major effort in cell cycle research has been to elucidate how checkpoint pathways interface with the cell cycle machinery to maintain Cdc2 in its inhibited form and hence to prevent entry into mitosis.
In all eukaryotic organisms, phosphoinositide 3-like kinases contribute an important if not essential checkpoint function. In fission yeast, the phosphoinositide 3-like kinase SpRad3 is a critical component of both the DNA replication and damage checkpoints. Downstream of SpRad3 are two structurally distinct protein kinases, SpChk1 and SpCds1 (2, 39, 56, 57). SpChk1 becomes hyperphosphorylated in a Rad3-dependent manner in response to DNA damage (57), and SpCds1 becomes hyperphosphorylated in S-phase in a Rad3-dependent manner in response to both DNA damage and replication blocks (29, 32). Phosphorylation increases the intrinsic protein kinase activity of SpCds1 (29, 32). chk1− cells fail to arrest their cell cycle in the presence of DNA damage and subsequently die (2, 56), whereas cds1− cells do arrest their cell cycle when treated with hydroxyurea (HU) (39). HU is an inhibitor of ribonucleotide reductase, and cells treated with HU become depleted for deoxyribonucleotides and arrest DNA synthesis. The HU arrest in cds1− cells is dependent upon chk1+, which is normally dispensable in this pathway (6, 29, 62). cds1− cells are still sensitive to HU treatment, indicating that Cds1 is required for recovery from replication blocks, and Chk1 is unable to complement this essential function (39). Mammalian counterparts of Chk1 and Cds1 have recently been reported (5, 9, 14, 33, 52).
Cdc25, Mik1, and Wee1 have all been implicated as targets of the G2/M checkpoint pathways. Mik1 protein levels increase in a Cds1-dependent manner when DNA synthesis is inhibited by HU (6). In addition, loss of Mik1 exacerbates the checkpoint defect observed in cells expressing a phosphorylation mutant of Cdc25 that is partially defective in 14-3-3 binding (62). The SpWee1 tyrosine kinase (42) and the SpCdc25 protein phosphatase (15, 16, 62) are potential substrates of the SpChk1 protein kinase. In the case of SpCdc25, phosphorylation by Chk1 enables 14-3-3 binding (62). The Xenopus laevis and human Chk1 protein kinases also phosphorylate Cdc25 on 14-3-3 binding sites (25, 52). Disruption of the interactions between Cdc25 and 14-3-3 compromises G2/M checkpoints in humans (49), in fission yeast (62), and in Xenopus laevis (26). Studies conducted with several organisms indicate that G2/M checkpoints operate to maintain Cdc25 in a 14-3-3-bound form rather than to induce 14-3-3 binding (26, 31, 49, 62). In fission yeast, 14-3-3 binding keeps Cdc25 out of the nucleus, and the nuclear exclusion of Cdc25 requires Chk1 function in the presence of DNA damage (31). Chk1 also binds directly to 14-3-3 proteins in response to DNA damage, but the significance of this interaction awaits further study (10).
In a previous study, we demonstrated that SpCds1 and SpChk1 phosphorylate Cdc25 on equivalent sites in vitro (62). Three of the phosphorylation sites were identified as S99, S192, S359. Substitution of alanine for each of the three serines reduced but did not eliminate the binding of 14-3-3 to SpCdc25. In addition, fission yeast cells induced to express a triple mutant of Cdc25 containing alanine in place of serine at each of the three phosphorylation sites were partially impaired in their ability to arrest cell cycle progression in response to HU treatment. We concluded that Cds1 and Chk1 regulate Cdc25/14-3-3 interactions as part of the checkpoint response to replication blocks. However, it was recently reported that fission yeast cells lacking both Cds1 and Chk1 still contain Cdc25 bound to 14-3-3 proteins both before and after DNA damage (10). The authors interpreted their findings to indicate that the cellular response to DNA damage in fission yeast does not involve phosphorylation of Cdc25 by Cds1 and Chk1 in the regulation of Cdc25/14-3-3 interactions. Because several protein kinases have been implicated in regulating Cdc25/14-3-3 interactions, we believed that the best way to address the importance of 14-3-3/Cdc25 interactions to G2/M checkpoint control in fission yeast was to generate a mutant of Cdc25 that no longer bound 14-3-3. In this study, we generated a mutant of Cdc25 that is severely impaired in its ability to bind to 14-3-3 proteins. We then monitored the ability of fission yeast cells expressing this mutant to arrest cell cycle progression in the presence of unreplicated or damaged DNA. In addition, we monitored the intracellular location of wild-type Cdc25 as well as phosphorylation site mutants of Cdc25. Results demonstrate that in fission yeast, the cellular response to DNA damage and replication blocks does require Cdc25/14-3-3 interactions and that one function of 14-3-3 binding is to keep Cdc25 out of the nucleus.
MATERIALS AND METHODS
General procedures and reagents.
Growth of yeast strains and plasmid transformations were done by standard methods (36). Recombinant baculoviruses were generated with the BAC-TO-BAC Baculovirus Expression System (GIBCO BRL). Glutathione S-transferase (GST)-Cdc25 and mutants were detected with affinity-purified GST antibody. Rad 24 and insect cell 14-3-3 proteins were detected with 14-3-3 (K-19) antibodies (Santa Cruz). HA-tagged Rad 24 was detected with 12CA5 (Boehringer Mannheim). Bound primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Cappel) and an enhanced chemiluminescence substrate detection system (Amersham Life Sciences, Inc.). Microcystin was purchased from Calbiochem. LLnL (N-acetyl-Leu-Leu-norleucinal), leupeptin, aprotinin, phenylmethylsulfonyl fluoride, DAPI (4′,6-diamidino-2-phenylindole), HU, glutathione (GSH)-agarose beads, protein A-Sepharose, and Calcofluor were purchased from Sigma Chemical Co. Methyl methanesulfonate (MMS) was purchased from Acros.
Yeast strains.
All strains are derived from 972h− and 975h+. Full genotypes are as follows: TE282 (cdc25-22 leu1-32 h+), TE938 (cdc25-22 mik1::ura4+ ade6− leu1-32 ura4-D18 h−) (62), and TE236 (leu1-32 ura4-D18 h−) (22).
Plasmids used for bacterial expression studies.
The complete coding sequence of fission yeast Cdc25 (596 amino acids; Swiss-Prot accession no. P06652) was cloned into pGEX2TN(EcoNI-HpaI) (62) as an NdeI-SmaI fragment to generate pGEX2TN(HpaI)-Cdc25Sp. PCR was used to generate the deletion and point mutants described below. All mutations were verified by sequencing.
(i) pGEX2TN(HpaI)-Cdc25(1–184).
PCR primers GGAATTCCATATGGATTCTCCGCTTTCTTCACTT (NdeI site underlined) and TCCCCCGGGTCAACGAAGGTAGGAATCAAAAGA (SmaI site underlined) were used to amplify nucleotides encoding the first 184 amino acids of Cdc25. The PCR product was digested with NdeI and SmaI and then subcloned into pGEX2TN(EcoNI-HpaI).
(ii) pGEX2TN(HpaI)-Cdc25(150–306).
PCR primers GGAATTCCATATGATTGCTTACAGCTTTAAGCCT (NdeI site underlined) and CTAGTCTAGATTAAAATCTTCTAAGTGTAGAGAGGGAATGCATGGCAGTAGGAGA ATCATT (XbaI site underlined) were used to amplify nucleotides encoding amino acids 150 to 596 of Cdc25. The PCR product was digested with NdeI and BglII and subcloned into the NdeI-BglII site of pGEX2TN(HpaI)-Cdc25Sp to generate pGEX2TN(HpaI)-Cdc25(150–596). pGEX2TN(HpaI)-Cdc25(150–596) was digested with HindIII, blunt ended, and recircularized to generate pGEX2TN(HpaI)-Cdc25(150–306).
(iii) pGEX2TN(HpaI)-Cdc25(315–596).
PCR primers CGGATCCCATATGTTACCCAAGATAAATAAA (NdeI site underlined) and CTAGTCTAGATTAAAATCTTCTAAGTGTAGAGAGGGAATGCATGGCAGTAGGAGAAT CATT (XbaI site underlined) were used to amplify nucleotides encoding the COOH terminus of Cdc25. The PCR product was digested with NdeI and XbaI and then subcloned into pGEX2TN(HpaI)-Cdc25Sp.
(iv) pGEX2TN(HpaI)-Cdc25(3A).
pGEX2TN(HpaI)-Cdc25(3A) encodes Cdc25 with alanines substituted for S99, S192, and S359.
(v) pGEX2TN(HpaI)-Cdc25(9A).
pGEX2TN(HpaI)-Cdc25(9A) encodes Cdc25 with alanines substituted for S99, S148, S178, S192, S204, S206, S234, S359, and T226.
Oligonucleotides used to generate Cdc25 phosphorylation site mutants.
Oligonucleotides used to generate Cdc25 phosphorylation site mutants are as follows (mutations are underlined): S148A (CGTGTTTCCGCCACTATTGCTTAC and GATAGCAATAGTGGCGGAAACACG), S99A (CGATCTCTTGCTTGTACTGTAGAA andTTCTACAGTACAAGCAAGAGATCG), S178A (TCGAGTTCTGCTTTTGATTCC and GGATACAAAAGCAGAACTCGA), S192A (CTCACGTTCTCGAGCATCAGGCAACGC and GCGTTGCCTGATGCTCGAGAACGTGAG), S204A (TTGCGATCCAGAGCGAGTTCCTCATAT and ATATGAGGAACTCGCTCTGGATCGCAA), S206A (ATCCAGATCGAGTGCCTCATATTCCAT and ATGGAATATGAGGCACTCGATCTGGAT), S204A and S206A (TCCAGAGCGAGTGCCTCATAT and ATATGAGGCACTCGCTCTGGA), T226A (CGCCATTTGGCTTATGCCTTA and TAAGGCATAAGCCAAATGGCG), S234A (CGTACCTGTGCTCAGTCGAGCAAC and GTTGCTCGACTGAGCACAGGTACG), S359A (TCGTACCCAAGCCATGTTTCTCAA and TTGAGAAACATGGCTTGGGTACGA), T561A (ACGAAACGCTGCTTTTATGC and GCATAAAAGCAGCGTTTCGT), and S567A (CGTACTAAAGCTTATACTTTT and AAAAGTATAAGCTTTAGTACG).
Plasmids used for fission yeast studies.
pREP81-cdc25+ was made by subcloning the NdeI-SmaI fragment of Cdc25Sp from pGEX2TN(HpaI)-Cdc25Sp into pREP81 (a gift of T. Enoch). pREP81-cdc25(3A) and pREP81-cdc25(9A) were made by replacing the NdeI-BglII fragment of pREP81-cdc25+ with the NdeI-BglII fragment of pGEX2TN(HpaI)-Cdc25(3A) and pGEX2TN(HpaI)-Cdc25(9A), respectively. pREP41x-GFP was made by amplifying the green fluorescence protein (GFP) coding sequence from pVT-102U-GFP (54) and cloning the PCR product into the SmaI site of pREP41X. pVT-102U-GFP contains two mutations (F64L and S65T) to enhance the GFP signal, and our PCR primers were designed to eliminate the NdeI site within the GFP sequence: 5′-end primer (ATGAGTAAAGGAGAAGAACTT), 3′-end primer (GGGAAGTGACATATGACCTCCACCTTTGTATAGTTCATCCATGCC) (NdeI site underlined); NdeI 5′-end primer (CTGTTTCATGTGATCTGGGTA), and NdeI 3′-end primer (TACCCAGATCACATGAAACAG). pREP41x-GFP-WT, pREP41x-GFP-3A, and pREP41x-GFP-9A were made by subcloning the NdeI-SmaI fragments of Cdc25Sp from pGEX2TN(HpaI)-Cdc25Sp, pGEX2TN(HpaI)-Cdc25(3A), and pGEX2TN(HpaI)-Cdc25(9A), respectively, into pREP41x-GFP. HA-Rad24 was amplified from pBB131-Rad24 (62) with the 5′-end primer CCATGGCTTACCCATACGATGTTCCAGATTACGCTGCAGGAATGTCTACTACTTCTCGTGAAGAT (NcoI site underlined) and the 3′-end primer GGAATTCCTATCTATGCGTCGCCTTGG (EcoRI site underlined). The PCR product was blunt ended and cloned into the SmaI site of pUC19 to generate pUC19-HA-Rad24. pUC19-HA-Rad24 was digested with NcoI and EcoRI, and HA-Rad24 was cloned into pBB131 to generate pBB131-HA-Rad24. To make pREP2-HA-rad24 and pREP82-HA-rad24, HA-Rad24 was amplified from pBB131-HA-Rad24 with the 5′-end primer CAGGGAATTCCATATGGCTTACCCATACGATGTTCCAGAT (NdeI site underlined) and the 3′-end primer GATCCCCCGGGCTATCTATGCGTCGCCTTGG (SmaI site underlined), and the PCR product was digested with NdeI and SmaI and cloned into pREP2 and pREP82, respectively. Mutations in the putative nuclear export sequence (NES) of Rad24 were made by PCR with the following two mutagenic primers: TCTACTTTAGCCATGCAATTGGCGCGTGAC and GTCACGCGCCAATTGCATGGCTAAAGTAGA (mutations underlined) to generate pREP82-HA-rad24(I222A, L226A).
pREP3x-GST-cdc25(WT), pREP3x-GST-cdc25(3A), and pREP3x-GST-cdc25(9A) were made by subcloning the HpaI-SmaI fragments from pGEX2TN(HpaI)-Cdc25(Sp), pGEX2TN(HpaI)-Cdc25(3A), and pGEX2TN(HpaI)-Cdc25(9A), respectively, into the SmaI site of pREP3X (gift of T. Enoch).
Plasmids used for insect cell expression studies.
pFASTBAC1-GST-Cdc25(WT) and pFASTBAC1-GST-Cdc25(9A) were made by subcloning the HpaI-SmaI fragment encoding GST-Cdc25Sp and GST-Cdc25(9A) from pGEX2TN(HpaI)-Cdc25Sp and pGEX2TN(HpaI)-Cdc25(9A), respectively, into the StuI site of pFASTBAC1 (GIBCO BRL).
Identification of residues of Cdc25 phosphorylated by Cds1 in vitro.
To identify phosphorylation sites, full-length and truncated forms (amino acids 1 to 184, 150 to 306, and 315 to 596) of Cdc25Sp were purified from bacteria as GST fusion proteins. Kinase assays were performed in vitro with purified Cds1 as described previously (62). Trypsin digestion, reverse-phase high-pressure liquid chromatography (HPLC), and Edman degradation of selected HPLC fractions were performed to identify potential phosphorylation sites (49, 62). Alanines were substituted for potential phosphorylation sites and experiments were repeated to confirm the loss or movement of an HPLC fraction.
14-3-3-binding assays.
The in vitro binding studies with phosphorylated Cdc25 and recombinant Rad 24 were performed essentially as described previously (62). Briefly, 1 to 2 μg of GST-Cdc25 was purified from JM109-expressing cells by using agarose beads, phosphorylated with 50 ng of GST-Cds1 at room temperature for 30 min, washed, and then incubated at room temperature for 15 min with 100 μl of binding buffer containing BL21 lysates with 200 to 600 ng of Rad24 or Rad24(I222A, L226A). GSH beads were washed three times with LiCl buffer (50 mM Tris [pH 8.0], 0.5 M LiCl). To monitor Cdc25/14-3-3 interactions in insect cells, Sf9 cells were infected with recombinant baculovirus encoding GST-Cdc25-WT or GST-Cdc25-9A. Two days after infection, cells were lysed in NETN buffer (43) and GST-fusion proteins were purified on GSH-agarose beads. Precipitates were washed with LiCl buffer and were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Coprecipitation of insect cell 14-3-3 proteins was determined by Western blotting. For binding studies conducted with fission yeast, TE236 was sequentially transformed with either pREP82-HA-rad24 or pREP82-HA-rad24(I222A, L226A) along with pREP3x-GST-cdc25(WT), pREP3x-GST-cdc25(3A), or pREP3x-GST-cdc25(9A). Cells were grown for ∼ 20 h at 30°C in medium lacking thiamine, pelleted, and washed once with lysis buffer (50 mM Tris [pH 7.4], 1 mM EDTA, 0.2 M NaCl, 50 mM NaF, 0.1% Nonidet P-40, 10% glycerol, 2 mM dithiothreitol, 1 μM microcystin, 100 μM LLnL, 20 μM leupeptin, 0.15 U of aprotinin per ml, 2 mM phenylmethylsulfonyl fluoride). The cells were resuspended in 30 μl of lysis buffer, 0.6 g of glass beads was added, and the cells were broken by shaking in a Fast-Prep machine (Bio 101, Inc.). Cell lysates were recovered by two washes with 0.5 ml of lysis buffer each followed by centrifugation. Supernatants from the two washes were combined and clarified again by centrifugation. Approximately 80 μg of total cellular protein was reserved for SDS-PAGE, 1 to 2 mg of protein was incubated with GSH-agarose beads, and 0.5 to 1 mg of protein was immunoprecipitated with 12CA5 antibody prebound to protein A-Sepharose. After incubation at 4°C for 60 to 80 min, the beads were washed four times with 0.5 ml of lysis buffer. Proteins were resolved by SDS-PAGE and analyzed by Western blotting. GST-Cdc25 was detected with GST antibody, and HA-Rad24 was detected with 12CA5.
Checkpoint studies.
Yeast cells were grown in Edinburgh minimal medium (Bio 101, Inc.) with appropriate selections (36). The DNA replication checkpoint was analyzed as described previously (12, 62). Briefly, TE282 and TE938 transformed with pREP81-cdc25+, pREP81-cdc25(3A), or pREP81-cdc25(9A) were grown at 30 to 32°C for about 40 h. The cells were diluted to an absorbance at 595 nm of 0.2, and HU was added to a final concentration of 12 mM. For analysis of cuts, cells were grown in HU for the indicated times and then fixed and stained with DAPI. At least 100 cells were counted for each data point. To determine the septation index of control cells or cells treated with MMS (0.1%), the cells were harvested at the indicated time points, fixed with methanol, and stained with Calcofluor and DAPI as described previously (12). Septa were detected as bright lines in cells stained with Calcofluor and DAPI.
Localization of GFP-Cdc25 proteins in fission yeast.
TE236 cells containing pREP41x-GFP, pREP41x-GFP-WT, pREP41x-GFP-3A, and pREP41x-GFP-9A alone or in combination with pREP2-HA-rad24 were grown for 16 to 20 h at 30°C in the absence of thiamine. Living cells were observed and photographed with an Olympus microscope equipped with a digital camera. In some cases, leptomycin B was added to a final concentration of 20 ng/ml or MMS was added to a final concentration of 0.03%. The cells were observed after 1 to 3 h of incubation.
RESULTS
The Cds1 and Chk1 protein kinases phosphorylate Cdc25 on equivalent sites in vitro to create 14-3-3 binding sites (62). Serines 99, 192, and 359 were previously identified as three of the sites phosphorylated by Cds1 and Chk1 (62). A triple mutant of Cdc25, containing alanine in place of serine at each of the three phosphorylation sites (Cdc25-3A), is impaired in its ability to bind 14-3-3 proteins relative to wild-type Cdc25. Furthermore, fission yeast cells expressing Cdc25-3A are partially compromised in their ability to arrest cell cycle progression in response to replication blocks (62). Fission yeast cells expressing a single substitution of alanine in place of serine 99 are also partially impaired in their ability to arrest cell cycle progression in response to HU treatment (15). These findings indicate that 14-3-3 binding to Cdc25 is an important component of the DNA replication checkpoint response. We wished to generate a mutant of Cdc25 that was incapable of binding to 14-3-3 proteins in order to reexamine the role of Cdc25/14-3-3 interactions in the DNA replication checkpoint and to determine whether Cdc25/14-3-3 interactions are also important in the DNA damage checkpoint. The approach we took was to identify all of the sites in Cdc25 that are phosphorylated by Cds1 in vitro and to substitute alanines for each phosphorylated residue. Various mutated forms of Cdc25 were then generated and tested for their ability to bind to 14-3-3 proteins. One mutant, denoted Cdc25-9A, was found to be severely impaired in 14-3-3 binding and was therefore used throughout this study (see below).
Identification of 12 residues of Cdc25 phosphorylated by Cds1 in vitro.
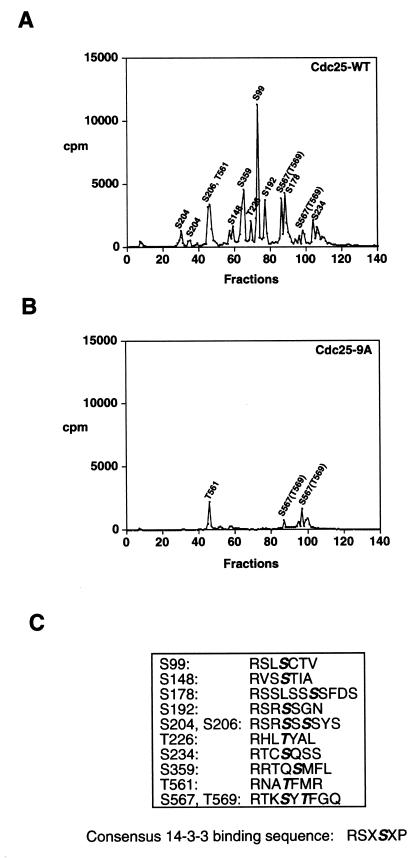
Kinase assays were performed to identify all of the residues of Cdc25 that were phosphorylated by Cds1 in vitro. Cds1 was purified as a GST fusion protein from baculovirus-infected insect cells as described previously (62). Cds1 kinase assays were then performed in vitro in the presence of bacterially purified Cdc25 (both full length and various subdomains). Radiolabeled Cdc25 was subjected to trypsin proteolysis, and phosphopeptides were analyzed by reverse-phase HPLC (Fig. 1A). Selected fractions were then subjected to Edman degradation (see Materials and Methods). Several phosphorylation sites were identified by this approach. Proteins containing mutations substituting alanine for serine/threonine at identified phosphorylation sites were made, and the experiments were repeated to confirm the sequencing data. This approach enabled us to identify nine new phosphorylation sites in addition to the three identified previously (Fig. 1A and C). These new sites include S148, S178, S204, S206, T226, S234, T561, S567, and T569. In this study, we characterized a mutant of Cdc25 (denoted Cdc25-9A) which contains alanine in place of either serine or threonine at positions 99, 148, 178, 192, 204, 206, 226, 234, and 359. As seen in Fig. 1B, Cdc25-9A was poorly phosphorylated by Cds1 in vitro and was defective in 14-3-3 binding (see below).
FIG. 1.
Identification of 12 residues of Cdc25 phosphorylated by Cds1 in vitro. (A and B) Cdc25-WT and Cdc25-9A were purified as GST fusion proteins, and kinase assays were performed in vitro in the presence of the Cds1 protein kinase. Radiolabeled GST-Cdc25-WT (A) and GST-Cdc25-9A (B) were digested with trypsin, and the tryptic peptides were resolved by reverse-phase HPLC. Column fractions were collected and monitored for the presence of radioactivity. Identified residues are shown above the fraction in which they elute. Peptides containing phosphorylated S234 and S567/T569 elute in more than one fraction. (C) Phosphorylation sites (bold and italics) and neighboring residues. The consensus sequence for 14-3-3 binding as defined by Muslin et al. (40) is indicated.
Cdc25-9A protein containing nine phosphorylation site mutations is defective in 14-3-3 binding.
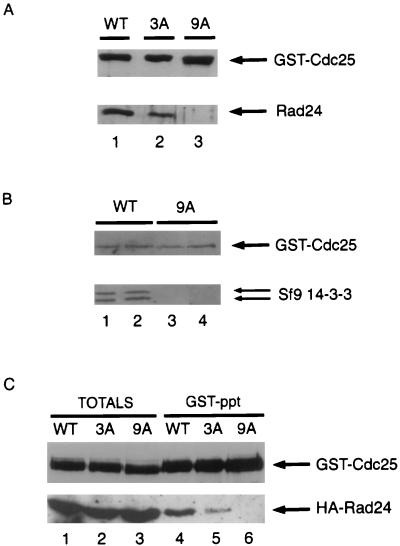
Three assays were used to monitor the ability of 14-3-3 proteins to bind to wild-type and mutant forms of Cdc25. In the first assay, Cds1 was used to phosphorylate wild-type and mutant forms of Cdc25 in vitro. Phosphorylated forms of Cdc25 were then incubated with soluble Rad 24 (14-3-3) protein. The binding of 14-3-3 to Cdc25 was monitored by immunoblotting (Fig. 2A). Rad 24 binding is not detected in the absence of Cdc25 phosphorylation (62). As reported previously (62), phosphorylation of Cdc25 by Cds1 facilitated 14-3-3 binding in vitro (lane 1) and mutation of serines 99, 192, and 359 resulted in reduced 14-3-3 binding relative to wild-type Cdc25 (lane 2). The Cdc25-9A protein containing nine phosphorylation site mutations was severely impaired in its ability to bind 14-3-3 proteins in this assay (lane 3). In a second assay, recombinant baculoviruses encoding GST-Cdc25 and GST-Cdc25-9A were generated and used to infect insect cells. Complex formation with endogenous insect cell 14-3-3 proteins was monitored by immunoblotting (Fig. 2B). As seen in Fig. 2B, a complex between GST-Cdc25 and 14-3-3 could readily be detected in insect cells (lanes 1 and 2). In contrast, 14-3-3 did not detectably associate with the Cdc25-9A protein (lanes 3 and 4). In the third assay, Cdc25-WT, Cdc25-3A, and Cdc25-9A were individually coexpressed as GST fusion proteins in fission yeast along with HA-tagged Rad 24 (Fig. 2C). Coprecipitation of Rad 24 with wild-type Cdc25 was observed by using GSH-agarose to isolate GST-Cdc25-WT followed by immunoblotting with antibody specific for the HA tag (lane 4). Significantly less Rad 24 was observed to coprecipitate with Cdc25-3A (lane 5), and interactions between Cdc25-9A and Rad 24 were not observed in this assay (lane 6). We analyzed several combinations of mutations in SpCdc25 and never found a single mutation that completely eliminated 14-3-3 binding (unpublished result). This indicates that fission yeast Cdc25 has more than one 14-3-3 binding site, but how many of the nine residues mutated in Cdc25-9A actually contribute to 14-3-3 binding is not known.
FIG. 2.
Association between Cdc25 and 14-3-3 proteins in vitro and in vivo. (A) Cdc25-WT (lane 1), Cdc25-3A (lane 2), and Cdc25-9A (lane 3) were purified as GST fusion proteins, and kinase assays were performed in vitro in the presence of the Cds1 protein kinase. Binding assays were then performed in the presence of purified Rad 24, and the reaction products were subjected to SDS-PAGE. Association of Rad 24 with Cdc25 was monitored by immunoblotting for Cdc25 (top) and Rad 24 (bottom). (B) Lysates were prepared from Sf9 insect cells infected with recombinant baculovirus encoding either GST-Cdc25-WT (WT; lanes 1 and 2) or GST-Cdc25-9A (lanes 3 and 4). GSH-agarose was used to precipitate the Cdc25 protein from either 400 μg (lanes 1 and 3) or 600 μg (lanes 2 and 4) of total cellular lysate. Precipitates were resolved by SDS- PAGE and immunoblotted for Cdc25 (top) and endogenous insect cell 14-3-3 proteins (bottom). (C) TE236 cells (leu1-32 ura4-D18 h−) cotransformed with pREP82-HA-rad24 and pREP3x-GST-cdc25(WT), pREP3x-GST-cdc25(3A), or pREP3x-GST-cdc25(9A) were grown in thiamine-free medium for ∼20 h. Lysates were prepared and analyzed directly by SDS-PAGE (lanes 1 to 3) or were incubated with GSH-agarose beads (lanes 4 to 6) prior to SDS-PAGE. Coprecipitation of Cdc25 with Rad 24 was analyzed by Western blotting. Lanes 1 to 3 represent 5% of the input lysate used in the precipitation assays. The experiments were all performed at least three times.
DNA replication and damage checkpoints are defective in fission yeast cells expressing Cdc25-9A.
To investigate the role of phosphorylation on 14-3-3 binding sites in the DNA replication checkpoint, wild-type cdc25+, a cdc25 mutant containing alanine substitutions at positions 99, 192, and 359 (cdc25-3A), and a cdc25 mutant containing alanine substitutions at positions 99, 148, 178, 192, 204, 206, 226, 234, and 359 (cdc25-9A) were introduced into TE282, a fission yeast strain bearing a temperature-sensitive allele of cdc25. Expression of the exogenous wild-type and mutant cdc25 was controlled by a weakened nmt1+ promoter on the REP81-based vector (3). cdc25+, cdc25-3A, and cdc25-9A were expressed to similar levels and fully rescued the cell cycle arrest of cdc25-22 at the restrictive temperature, as indicated by the absence of elongated cells (data not shown). Neither cdc25+, cdc25-3A, nor cdc25-9A significantly advanced mitosis as judged by the absence of “wee” cells in the cultures, indicating that Cdc25 and the mutant proteins were not overexpressed to a level that perturbed mitotic timing (data not shown).
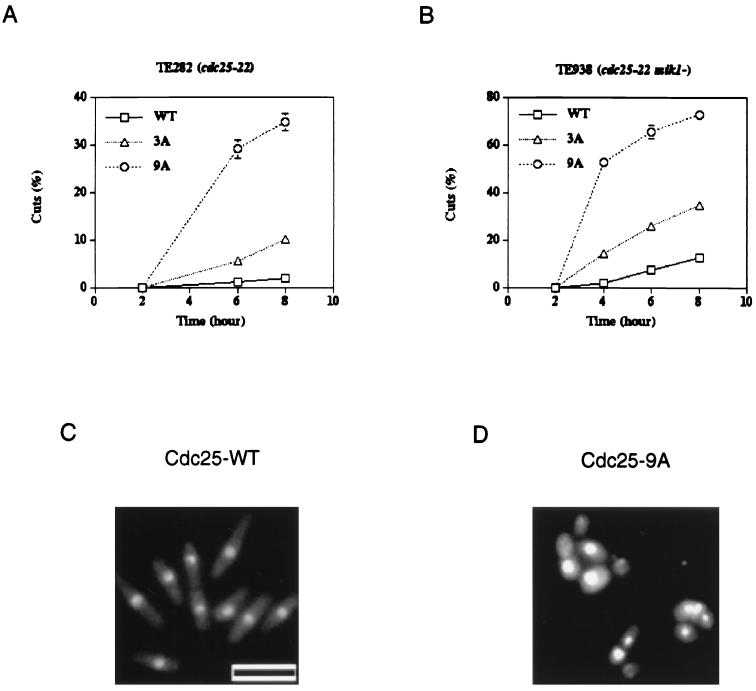
To test the integrity of the checkpoint response to unreplicated DNA, each strain was incubated in HU and the formation of cuts was monitored (Fig. 3). Cuts are indicative of cells undergoing cytokinesis without nuclear division and are scored by counting cells with a single nucleus located to one side of the division plate or cells with nuclei bisected by the division plate (Fig. 3D). As reported previously (62), cells expressing wild-type Cdc25 had an intact checkpoint response whereas cells expressing Cdc25-3A had a partially compromised checkpoint response (Fig. 3A). Interestingly, cells expressing Cdc25-9A showed a significant defect in their response to HU, with 35% of the cells exhibiting a cut phenotype by 8 h (Fig. 3A). To test the contribution of the Mik1 tyrosine kinase to the checkpoint response, Cdc25, Cdc25-3A, and Cdc25-9A were each expressed in TE938 (cdc25-22 mik1−). Each strain was incubated in HU and analyzed for cuts as described above (Fig. 3B to D). Cells expressing wild-type Cdc25 had a largely intact checkpoint response, as indicated by the presence of elongated uninucleate cells (Fig. 3C). Cells expressing Cdc25-9A were severely impaired in their ability to arrest cell cycle progression in the presence of unreplicated DNA (Fig. 3D), with 75% of the cells exhibiting cuts by 8 h (Fig. 3B). Cells expressing Cdc25-3A had a phenotype intermediate between those observed for Cdc25-WT and Cdc25-9A (Fig. 3A and B) (62).
FIG. 3.
Mutation of canonical 14-3-3 binding sites disrupts the checkpoint response to unreplicated DNA. (A and B) cdc25-22 (TE282) (A) and cdc25-22 mik1− (TE938) (B) cells were transformed with pREP81-cdc25+ (WT), pREP81-cdc25(3A) (3A), or pREP81-cdc25(9A) (9A). Cells were treated with HU and analyzed as described previously (62). At the indicated times, cells were fixed and stained with DAPI and the percentage of cut cells was determined by counting at least 100 cells. Five to seven independent transformants were assayed for each data point. (C and D) Photographs of HU-treated cdc25+ mik1− cells (C) and cdc25-9A mik1− cells (D) at 6 h. Bar, 10 μm.
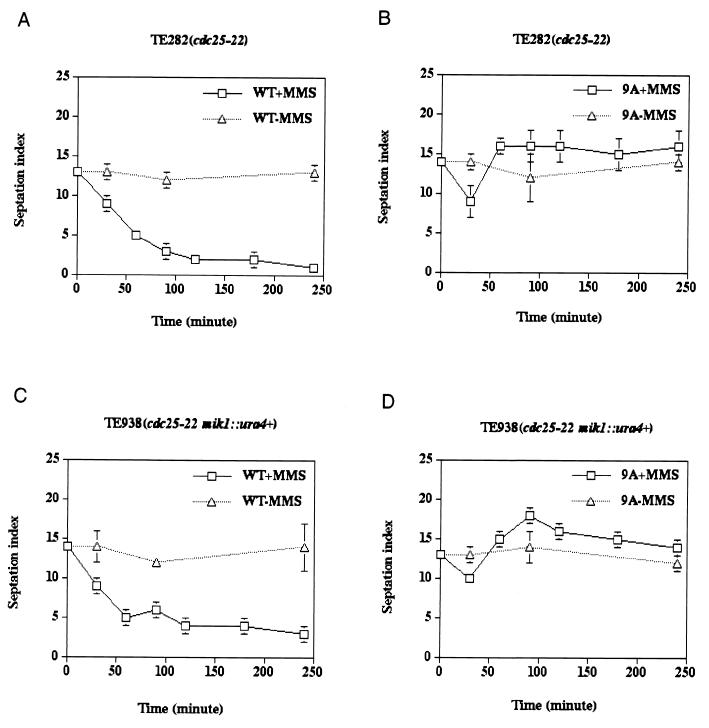
We next examined the integrity of the DNA damage checkpoint response in cells expressing either Cdc25-WT or the Cdc25-9A mutant protein (Fig. 4). When exponentially growing cultures of cells (TE282 and TE938) expressing Cdc25-WT were treated with MMS to induce DNA damage, cell cycle arrest was observed. Arrest was characterized by a rapid drop in the septation index in treated cells compared with untreated cells and simultaneous elongation and accumulation of uninucleate cells (Fig. 4A and C). This was in contrast to cells expressing Cdc25-9A, where a transient drop in the septation index was observed 30 min after MMS treatment and was followed by a rise and stabilization from 60 to 240 min (Fig. 4B and D). Thus, cells expressing Cdc25-9A continued to divide in the presence of damaged DNA. Furthermore, cells expressing Cdc25-9A were more sensitive to killing by MMS than were cells expressing Cdc25-WT (data not shown).
FIG. 4.
Mutation of canonical 14-3-3 binding sites disrupts the checkpoint response to damaged DNA. cdc25-22 (TE282) (A and B) and cdc25-22 mik1− (TE938) (C and D) cells were transformed with pREP81-cdc25+ (WT) or pREP81-cdc25(9A) (9A). Cells were either untreated (−MMS) or incubated with 0.1% MMS (+MMS). At the indicated times, samples were harvested and processed for Calcofluor and DAPI staining. The septation index [(number of septated cells/total number of cells) × 100] was determined. Three independent transformants were assayed for each data point.
Intracellular localization of wild-type and phosphorylation site mutants of Cdc25.
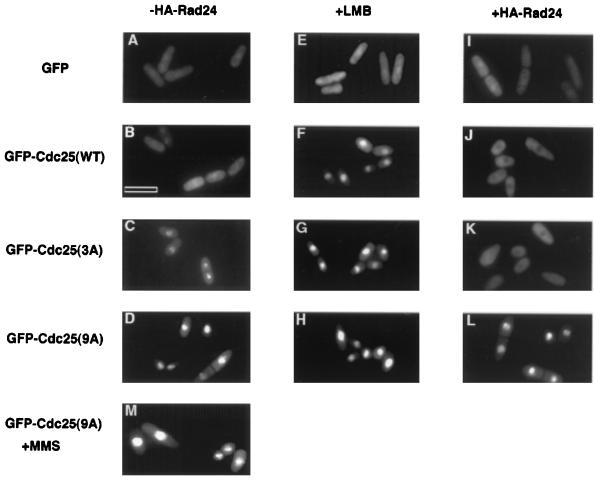
It has recently been reported that the two fission yeast 14-3-3 proteins (Rad 24 and Rad 25) function to keep Cdc25 out of the nucleus throughout interphase and in response to checkpoint activation (31). We therefore examined the subcellular localization of Cdc25-WT and the two phosphorylation site mutants (Cdc25-3A and Cdc25-9A) under a variety of conditions (Fig. 5). Wild-type and mutant Cdc25 proteins tagged with GFP were expressed, and live fission yeast cells were visualized by microscopy. GFP-Cdc25 and its mutants were able to rescue cdc25-22, demonstrating that the GFP tag did not affect Cdc25 function (data not shown). GFP alone was found to be both cytoplasmic and nuclear (Fig. 5A). GFP-tagged Cdc25-WT also localized to both the cytoplasm and the nucleus (Fig. 5B). Nuclear accumulation of Cdc25-WT (Fig. 5F) but not GFP (Fig. 5E) was observed upon treatment of cells with leptomycin B (LMB), an inhibitor of the export factor exportin 1 (Crm1) (23, 41). This indicates that wild-type Cdc25 is actively exported from the nucleus in a Crm1-dependent fashion. Overexpression of Rad 24 resulted in the nuclear exclusion of Cdc25-WT (Fig. 5J), and this effect was reversed by treatment of cells with LMB (data not shown). GFP localization was not altered upon overproduction of Rad 24 (Fig. 5I). These results support a role for 14-3-3 binding in keeping Cdc25 out of the nucleus. A more pronounced nuclear localization was observed for GFP-Cdc25-3A (Fig. 5C) compared with wild-type Cdc25, and overproduction of Rad 24 resulted in an enhanced nuclear exclusion (Fig. 5K). As was observed for Cdc25-WT LMB treatment resulted in the nuclear accumulation of Cdc25-3A (Fig. 5G). Strikingly, GFP-Cdc25-9A exhibited preferential nuclear localization (Fig. 5D), and neither LMB treatment (Fig. 5H), overproduction of Rad 24 (Fig. 5L), nor MMS treatment (Fig. 5M) substantially altered the localization of Cdc25-9A.
FIG. 5.
Localization of Cdc25 and phosphorylation site mutants in fission yeast. TE236 (leu1-32 ura4-D18 h−) transformed with pRep41x-GFP or pREP41x-GFP-cdc25(WT), pREP41x-GFP-cdc25(3A), or pREP41x-GFP-cdc25(9A), in the absence (A to H and M) or in the presence (I to L) of pREP2-HA-rad24 were grown in thiamine-free medium for ∼ 20 h. In some cases, cultures were incubated for 1 to 2 h with 20 ng of LMB per ml prior to analysis (E to H). In one case, cells were treated with 0.03% MMS and photographed 2 h later (M). Live cells were observed by using a conventional fluorescence microscope. Bar, 10 μm.
Mutations in putative NES of Rad 24 impairs binding to Cdc25.
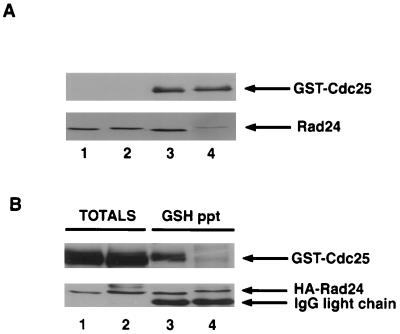
It has been proposed that Rad 24 contains an NES that contributes an “attachable” NES to mediate the nuclear export of Cdc25 (31). The sequence comprising the putative NES consists of the sequences STLIMQLLRDNLTLW (amino acids 219 to 233), and this sequence is conserved in Rad 25 as well as all of the mammalian 14-3-3 homologs. A mutant of Rad 24 containing alanine in place of isoleucine 222 and leucine 226 is more nuclear than wild-type Rad 24 and fails to deplete Cdc25 from the nucleus upon irradiation (31). These results demonstrate that I222 and L226 are required for Rad 24 function. However, the question remains whether these residues actually comprise an authentic NES, given that this sequence regulates the interactions between 14-3-3 and its binding partners. For example, mutations within this region of 14-3-3ζ disrupt the interactions of 14-3-3ζ with Raf1, Cbl and c-Bcr (58). Thus, it is possible that mutation of I222 and L226 in Rad 24 actually reduces the affinity of Rad 24 for Cdc25 rather than disrupting an NES function. To test this hypothesis, we generated a mutant of Rad 24 containing alanine in place of I222 and L226 and tested its ability to bind to Cdc25 in vitro (Fig. 6A) and in fission yeast (Fig. 6B). As seen in Fig. 6, Rad 24(I222A, L226A) showed a slight reduction in electrophoretic mobility on SDS gels compared with wild-type Rad 24. More strikingly, Rad 24(I222A, L226A) appeared severely impaired in its ability to bind to Cdc25 both in vitro and in vivo, indicating that mutation of residues I222 and L226 perturbs stable interactions between Cdc25 and 14-3-3.
FIG. 6.
Rad 24(I222A, L226A) is impaired in its ability to bind to Cdc25. (A) GST-Cdc25 was purified on GSH beads, phosphorylated by Cds1 in vitro, and incubated with ∼200 ng of Rad 24 or Rad 24(I222A, L226A). Precipitates were washed and subjected to SDS-PAGE. Association of Rad 24 with Cdc25 was monitored by immunoblotting for Cdc25 (top) and Rad 24 (bottom). Lanes: 1 and 2, 50 ng of Rad24 and Rad24(I222A, L226A) as input controls, respectively; 3 and 4, binding of phosphorylated GST-Cdc25 to Rad24 and Rad24(I222A, L226A), respectively. (B) TE236 cells (leu1-32 ura4-D18 h−) cotransformed with pREP3x-GST-cdc25(WT) and either pREP82-HA-rad24 (lanes 1 and 3) or pREP82-HA-rad24(I222A, L226A) (lanes 2 and 4) were induced for ∼ 20 h. Lysates were prepared and analyzed directly by SDS-PAGE (lanes 1 and 2) or were incubated with anti-HA antibody bound to protein A beads (lanes 3 and 4) prior to SDS-PAGE. Coimmunoprecipitation of Cdc25 with Rad 24 was analyzed by Western blotting. Lanes 1 and 2 represent 10% of the input lysate used in the precipitation assays. IgG, immunoglobulin G.
DISCUSSION
In this study, we generated a mutant of SpCdc25 that is severely impaired in its ability to bind to the fission yeast 14-3-3 proteins (Rad 24 and Rad 25). When expressed in fission yeast, this mutant Cdc25 protein localized almost exclusively to the nucleus, in contrast to wild-type Cdc25, which localized to both the cytoplasm and the nucleus. Inhibition of Crm1-mediated nuclear export resulted in the nuclear accumulation of wild-type Cdc25, indicating that wild-type Cdc25 normally shuttles between the nucleus and the cytoplasm. Overproduction of Rad 24 caused wild-type Cdc25 to localize exclusively to the cytoplasm, whereas nuclear localization of the 14-3-3 binding mutant was not altered upon Rad 24 overproduction. Finally, cells expressing the 14-3-3 binding mutant exhibited defective G2/M checkpoint responses. Taken together, these results suggest that 14-3-3 binding regulates the intracellular compartmentalization of Cdc25 and establish that 14-3-3 binding to Cdc25 is required for fission yeast cells to arrest cell cycle progression if damaged or incompletely replicated DNA is detected.
To identify residues in Cdc25 that mediate 14-3-3 binding upon phosphorylation, we first identified residues in Cdc25 that are phosphorylated by the Cds1 protein kinase in vitro. We previously demonstrated that both the Cds1 and Chk1 protein kinases phosphorylate Cdc25 in vitro at 14-3-3 binding sites but identified only three of the sites at that time (62). Cds1 was used in this study because it is more active than Chk1 when overproduced in insect cells. In total, we identified 12 phosphorylation sites and then characterized a mutant of Cdc25 with 9 of these residues mutated to alanine (denoted Cdc25-9A) because this mutant was severely impaired in its ability to bind 14-3-3 proteins. Importantly, Cdc25-9A fully complemented the cdc25-22 temperature-sensitive allele at the nonpermissive temperature, demonstrating that the nine mutations do not grossly affect the function of Cdc25. Human and Xenopus Cdc25C each have a single phosphorylation site that mediates 14-3-3 binding, and in both cases mutation of this residue ablates 14-3-3 binding (26, 49). In contrast, fission yeast Cdc25 has several phosphorylation sites, and it is not known how many actually contribute to 14-3-3 binding. The 14-3-3 binding sites in human and Xenopus Cdc25C match well with the consensus 14-3-3 binding sequence (40, 60), while those in fission yeast Cdc25 match less well (Fig. 1C). Throughout the course of this study, we analyzed several combinations of mutations in SpCdc25 and never found a single mutation that completely eliminated 14-3-3 binding. This indicates that fission yeast Cdc25 has more than one 14-3-3 binding site. Given that 14-3-3 is a dimer with the capability of binding two phosphorylation sites simultaneously (60), it is possible that in fission yeast, two lower-affinity residues function together to facilitate high-affinity 14-3-3 binding.
By tagging wild-type and mutant forms of Cdc25 with GFP, we observed the subcellular localization of Cdc25 in living cells (Fig. 5). In most cases, GFP-Cdc25 was observed to be evenly distributed throughout the cell. However, some cells showed enhanced nuclear fluorescence while others exhibited a more pronounced cytoplasmic fluorescence. These results are consistent with those reported by Lopez-Girona et al. (31). Nuclear accumulation of Cdc25-WT was observed when cells were treated with LMB. This indicates that Cdc25 normally shuttles between the nucleus and the cytoplasm. Cdc25-WT was excluded from the nucleus upon Rad 24 overproduction, indicating that Rad 24 binding contributes to the nuclear exclusion of Cdc25. Further evidence in favor of 14-3-3 proteins regulating the nuclear exclusion of Cdc25 comes from the analysis of the two Cdc25 mutants. Cdc25-3A and Cdc25-9A were distinguishable in terms of 14-3-3 binding, nuclear localization, and responses to Rad 24 overproduction. The Cdc25-9A mutant, which was severely impaired in 14-3-3 binding, localized to the nucleus, and neither Rad 24 overproduction nor DNA damage affected its nuclear location. The Cdc25-3A mutant, which was partially defective in 14-3-3 binding, showed more nuclear localization than Cdc25-WT but less than Cdc25-9A did, and, unlike Cdc25-9A, Cdc25-3A was partially excluded from the nucleus upon Rad 24 overproduction. Given that 14-3-3 is competent to bind Cdc25, the simplest interpretation of our data is that 14-3-3 binding either reduces the rate of nuclear import of Cdc25 or enhances its rate of nuclear export.
Rad 24 reportedly contains an NES, and it has been proposed that Rad 24 contributes an “attachable” NES to mediate the nuclear export of Cdc25 (31). The sequence comprising the putative NES consists of the sequence STLIMQLLRDNLT LW (amino acids 219 to 233). A mutant of Rad 24 containing alanine in place of isoleucine 222 and leucine 226 [Rad 24(I222A, L226A)] is reported to be more nuclear than wild-type Rad 24 and to be unable to deplete Cdc25 from the nucleus upon irradiation (31). We confirmed that Rad 24(I222A, L226A) was more nuclear than wild-type Rad 24 and that overexpression of wild-type Rad 24 but not Rad24(I222A, L226A) resulted in the nuclear exclusion of Cdc25 (Fig. 5 and data not shown). Furthermore, cell elongation was noted upon overexpression of wild-type but not mutant Rad24 (data not shown). These results are consistent with the NES hypothesis, which states that 14-3-3 contributes an NES to export Cdc25 from the nucleus. However, we found that Rad 24(I222A, L226A) was severely impaired in its ability to bind to Cdc25 both in vitro and in vivo (Fig. 6). Furthermore, fluorescence polarization experiments with a fluorescein-labelled phospho-Raf peptide as ligand demonstrated that Rad24(I222A, L226A) had a Kd at least 30 times higher than wild-type Rad24 (data not shown). Thus, the phenotypes described for the Rad24(I222A, L226A) mutant by Lopez-Girona et al. (31) could easily be accounted for by the failure of the Rad 24 mutant to efficiently bind to Cdc25 or possibly to other target proteins. Interestingly, Xenopus Cdc25 contains its own NES, and in this case 14-3-3 binding appears to regulate the rate of nuclear import of Cdc25 as opposed to its nuclear export (24, 61). Thus, it is possible that 14-3-3 binding perturbs the nuclear import of fission yeast Cdc25 rather than actually promoting its nuclear export by providing an attachable NES or that the same sequence in Rad 24 has dual functions.
Upon completion of DNA synthesis or after recovery from DNA damage, Cdc25 must accumulate in the nucleus to promote mitotic entry. This could be accomplished by either reduced phosphorylation or enhanced dephosphorylation of Cdc25 followed by loss of 14-3-3 binding. Differential subcellular localization provides a level of regulation that potentially explains how phosphorylation of Cdc25 and subsequent 14-3-3 binding function to prevent premature activation of Cdc2 in response to G2/M checkpoint activation. However, in the absence of checkpoint activation, fission yeast cells expressing Cdc25-9A were indistinguishable from those expressing Cdc25-WT, even though Cdc25-9A was constitutively nuclear. Given that fission yeast Cdc2 has been reported to be nuclear (1, 7), these results indicate that nuclear colocalization of Cdc25 with Cdc2 is not sufficient to advance cells into mitosis from S phase during a normal cell cycle. Thus, the importance of 14-3-3/Cdc25 interactions in coupling S-phase completion to M-phase entry during a normal cell cycle is unclear due to the apparent lack of any phenotype in cells expressing Cdc25-9A.
In an earlier study, we demonstrated that reduced 14-3-3 binding to Cdc25 coupled with a deletion in mik1+ partially compromised the DNA replication checkpoint. Here we show that a more complete disruption of Cdc25/14-3-3 interactions coupled with mik1+ deletion results in an almost complete bypass of the DNA replication checkpoint. We also establish a role for 14-3-3/Cdc25 interactions in the DNA damage checkpoint, since cells expressing the 9A mutant of Cdc25, which is severely impaired in 14-3-3 binding, do not arrest cell cycle progression following DNA damage. Thus, in fission yeast, 14-3-3 binding to Cdc25 is required for cell cycle arrest in response to both DNA damage and replication blocks.
Studies conducted with a wide variety of organisms indicate that G2/M checkpoints operate to maintain Cdc25 in a 14-3-3-bound form rather than to induce 14-3-3 binding. In humans, Cdc25C is bound to 14-3-3 proteins throughout interphase (49). A similar situation exists in Xenopus (that is, Cdc25 is stoichiometrically bound to 14-3-3 proteins throughout interphase), and so it is unclear how checkpoints could further stimulate the binding of 14-3-3 (26). That checkpoints operate to maintain rather than induce 14-3-3 binding to Cdc25 is also supported by studies with fission yeast. In fission yeast, activation of the DNA replication checkpoint with HU did not alter the phosphorylation state of Cdc25, suggesting that the DNA replication checkpoint operates to maintain Cdc25 in a phosphorylated state to ensure 14-3-3 binding (62). In addition, the amount of 14-3-3 bound to Cdc25 has not been observed to change during G2/M checkpoint responses in fission yeast (10, 31).
In humans there are at least three protein kinases that potentially regulate the interactions between Cdc25 and 14-3-3 proteins. These kinases include C-TAK1 (43, 48), Chk1 (5, 52), and Cds1/Chk2 (5, 9, 33). Elimination of Chk1 in either Xenopus or fission yeast does not detectably alter the levels of 14-3-3 bound to Cdc25 (10, 25, 31). A recently published study reported that fission yeast cells lacking both Cds1 and Chk1 still contain Cdc25 bound to 14-3-3 proteins both before and after DNA damage (10). The authors interpreted their findings to indicate that neither Chk1 nor Cds1 function is required for Cdc25/14-3-3 interactions either before or after DNA damage. Given what we know about the interactions between the human and fission yeast Cdc25 proteins with 14-3-3, it is perhaps not unexpected that loss of Cds1 and Chk1 could affect checkpoint function without noticeably affecting overall 14-3-3/Cdc25 interactions. As indicated above, at least three kinases potentially regulate Cdc25/14-3-3 interactions in humans. C-TAK1 resides in the cytoplasm, whereas Chk1 and Cds1/Chk2 are in the nucleus. Thus, elimination of any two of the kinases, as was done by Chen et al. (10), would not be expected to eliminate the interactions between Cdc25 and 14-3-3 in human cells. Furthermore, the studies of Lopez-Girona et al. (31) and the findings reported here indicate that the major role of 14-3-3 binding in the case of fission yeast is to keep Cdc25 out of the nucleus. Thus, elimination of two nuclear checkpoint kinases (Chk1 and Cds1) could abrogate the checkpoint response by allowing Cdc25 to accumulate in the nucleus without affecting 14-3-3 binding to the bulk population of Cdc25 remaining in the cytoplasm. Furthermore, Lopez-Girona et al. (31) showed that DNA damage does not cause Cdc25 to be excluded from the nucleus in the absence of Chk1, indicating that Chk1 does indeed regulate Cdc25 function in fission yeast.
In summary, our data support the hypothesis that both the DNA replication and the DNA damage checkpoints use similar mechanisms to arrest the cell cycle in fission yeast. The mechanism involves maintaining Cdc25 in a phosphorylated form that binds 14-3-3 proteins. 14-3-3 binding keeps Cdc25 out of the nucleus, away from its substrate, either by inhibiting nuclear import or by indirectly facilitating nuclear export. The net effect is to prevent Cdc25 from activating Cdc2 and thereby to delay entry into mitosis. At this time, Cds1 and Chk1 remain viable candidate kinases for mediating Cdc25/14-3-3 interactions during a G2/M checkpoint response.
ACKNOWLEDGMENTS
We thank M. Yoshida for providing leptomycin B and S. Wente for the use of her microscope. We thank S. Heximer and K. Blumer for providing pVT-102U-GFP and K. Chrispell Forbes and T. Enoch for providing plasmids, strains, and methods. We thank C. Nguyen and A. Shaw for assistance in the fluorescence polarization studies. We thank K. Blumer, T. Muslin, A. Shaw, and members of our laboratory for helpful suggestions and comments.
This work was supported by the NIH. H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alfa C E, Ducommun B, Beach D, Hyams J S. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature. 1990;347:680–682. doi: 10.1038/347680a0. [DOI] [PubMed] [Google Scholar]
- 2.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or the thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 4.Blasina A, Paegle E S, McGowan C H. The role of inhibitory phosphorylation of Cdc2 following DNA replication block and radiation-induced damage in human cells. Mol Biol Cell. 1997;8:1013–1023. doi: 10.1091/mbc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1998;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 6.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 7.Booher R, Alfa E, Hyams J, Beach D. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 8.Booher R N, Holman P S, Fattaey A. Human Myt1 is a cell cycle regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272:22300–22306. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 9.Brown A, Lee C-H, Schwarz J K, Mitiku N, Griffith D, Piwnica-Worms H, Chung J H. A human Cds1-related kinase that functions downstream of ATM in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Liu T-H, Walworth N C. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 1999;13:675–685. doi: 10.1101/gad.13.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunphy W G, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 12.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 13.Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- 14.Flaggs G, Plug A W, Dunks K M, Mundt K E, Ford J C, Quiggle M R E, Taylor E M, Westphal C H, Ashley T, Hoekstra M F, Carr A M. ATM-dependent interactions of a mammalian Chk1 homolog with meiotic chromosomes. Curr Biol. 1997;7:977–986. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- 15.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 17.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 18.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 19.Honda R, Ohba Y, Yasuda H. The cell cycle regulator, human p50wee1, is a tyrosine kinase and not a serine/tyrosine kinase. Biochem Biophys Res Commun. 1992;186:1333–1338. doi: 10.1016/s0006-291x(05)81552-9. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi M, Nagata A, Jinno S, Suto K, Okayama H. Wee1+-like gene in human cells. Nature. 1991;353:80–83. doi: 10.1038/353080a0. [DOI] [PubMed] [Google Scholar]
- 21.Jin P, Gu Y, Morgan D O. Role of inhibitory Cdc2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostrub C F, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 24.Kumagai A, Dunphy W G. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumagai A, Guo Z, Emami K, Wang S X, Dunphy W G. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumagai A, Yakowec P S, Dunphy W G. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M S, Enoch T, Piwnica-Worms H. Mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J Biol Chem. 1994;269:30530–30537. [PubMed] [Google Scholar]
- 28.Lee M S, Ogg S, Xu M, Parker L L, Donoghue D J, Maller J L, Piwnica-Worms H. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol Biol Cell. 1992;3:73–84. doi: 10.1091/mbc.3.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsay H D, Griffiths D J F, Edwards R, Murray J M, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Stanton J J, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 32.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka S, Huang M, Elledge S J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 34.McGowan C H, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr 15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGowan C H, Russell P. Cell cycle regulation of human WEE1. EMBO J. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 37.Mueller P R, Coleman T R, Dunphy W G. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller P R, Coleman T R, Kumagai A, Dunphy W G. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 39.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 40.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 41.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade or crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;9:6320–6324. [PubMed] [Google Scholar]
- 42.O’Connell M J, Raleigh J M, Verkade H M, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogg S, Gabrielli B, Piwnica-Worms H. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J Biol Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 44.Parker L L, Atherton-Fessler S, Lee M S, Ogg S, Falk F L, Swenson K I, Piwnica-Worms H. Cyclin promotes the tyrosine phosphorylation of p34cdc2 in a wee1+ dependent manner. EMBO J. 1991;10:1255–1263. doi: 10.1002/j.1460-2075.1991.tb08067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker L L, Atherton-Fessler S, Piwnica-Worms H. p107wee1 is a dual specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc Natl Acad Sci USA. 1992;89:2917–2921. doi: 10.1073/pnas.89.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker L L, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human wee1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 47.Parker L L, Sylvestre P J, Byrnes III M J, Liu F, Piwnica-Worms H. Identification of a 95-kDa WEE1-like tyrosine kinase in HeLa cells. Proc Natl Acad Sci USA. 1995;92:9638–9642. doi: 10.1073/pnas.92.21.9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng C-Y, Graves P R, Ogg S, Thoma R S, Byrnes M J, Wu Z, Stephenson M, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 49.Peng C-Y, Graves P R, Thoma R S, Wu Z, Shaw A, Piwnica-Worms H. Mitotic- and G2-checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine 216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 50.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 51.Rhind N, Russell P. Tyrosine phosphorylation of Cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:3782–3787. doi: 10.1128/mcb.18.7.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation via Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 53.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci USA. 1993;90:3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasa S P, Bernstein L S, Blumer K J, Linder M E. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:5584–5589. doi: 10.1073/pnas.95.10.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strausfeld U, Labbe J C, Fesquet D, Cavadore J C, Picard A, Sadhu K, Russell P, Doree M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human cdc25 protein. Nature. 1991;351:242–245. doi: 10.1038/351242a0. [DOI] [PubMed] [Google Scholar]
- 56.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 57.Walworth N C, Bernards R. rad-dependent responses of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Zhang L, Liddington R, Fu H. Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3ζ disrupts its interactions with Raf-1 kinase. J Biol Chem. 1998;273:16297–16304. doi: 10.1074/jbc.273.26.16297. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aikten A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 61.Yang J, Wonkler K, Yoshida M, Kornbluth S. Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint in fission yeast requires Cdc25p phosphorylation by Cds1p or Chk1p. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]