Abstract
Failure of antiretroviral therapy (ART) in HIV-1 infection is a critical issue for the physicians treating HIV patients. The major cause of drug failure is the development of resistance mutations in reverse transcriptase (RT) and/or protease (PR) genes. Mutations associated with drug resistance decrease drug effectiveness. This study was conducted to assess drug resistance profile of the entire PR gene in 90 HIV-1 patients consisting of 23 ART non-responsive, 32 ART responsive and 35 drug naive patients. It was observed that the majority of the sequences (94.4%) belonged to subtype C and (5.5%) to subtype A1. The ART non-responsive and responsive patients were treated with either first line of ART regimen (two NRTI and one NNRTI) or second line of ART regimen that included additional one protease inhibitor (PI). All the patients in each group except one responsive patient had various minor resistance mutations. Thus, drug failures in ART non-responsive patients may not always be due to drug resistance mutations instead other factors may also be responsible for drug failures such as non-compliance, suboptimal dose or drug interaction. The presence of minor drug resistance mutations in drug naive patients is suggestive of transmitted resistance mutations.
Keywords: HIV-1, ART, Drug resistance, Protease inhibitors, Protease gene, Drug naive patients
Introduction
Globally out of 36.9 million individuals living with HIV, only 21.7 million are reported to have access to ART according to WHO report [48]. In India, of 2.3 million people having HIV-1, 1.65 million people are reported to have registered for ART treatment [33]. The persistent efforts of controlling HIV-1 by therapeutic interventions have resulted in the development of anti-retroviral drug resistance too, creating a demand for formulation of newer drugs. To lessen the development of drug resistance, therefore, a quest for administration of ART regimen as a cocktail popularly known as HAART (Highly Active Antiretroviral Therapy) instead of monotherapy was recommended. The HAART significantly reduced the morbidity and mortality of HIV-1 infected subjects [30]. After initial success of HAART, the drug resistance still persisted and then simply it was referred to as anti-retroviral therapy (ART). Gradually, it was realized that ART failure was widespread and became a major cause of concern in ART clinics. The major cause of drug failure though remains the development of drug resistance mutations in the virus but there are various other factors as well such as poor drug absorption, noncompliance, drug interaction etc. [25]. Patients who fail the first-line regimen subsequently are prescribed the second line regimen, which essentially includes protease inhibitors in combination therapy. In India, the preferred choice of protease inhibitor (PI) in second line regimen includes Atazanavir/ritonavir (ATV/r), the alternative being Lopinavir/ritonavir (LPV/r) [31].
The effectiveness of all currently prescribed HIV antiretroviral drugs is limited by the emergence of drug resistant variants, which frequently show extensive cross-resistance within each drug class [28]. Drug resistant mutations in the polymerase gene severely impede ART regimen in HIV [35]. Several lines of evidence on emerging trends on drug resistance are from North America and Europe where HIV-1 subtype B is prevalent. Contrary to Africa and South-East Asia including India, where majority of the world’s HIV-infected individuals’ harbouring subtype C live, there is a dearth of data on HIV-induced drug resistance and pattern. Published data suggest that the currently available protease and reverse transcriptase inhibitors are active against non-B viruses as they are against subtype B viruses and thus the emergence of drug resistance has worldwide implications for all HIV-1 subtypes [16].
Majority of the literature and information on the drug resistance in HIV-1 reverse transcriptase comes from different region of India on first line drug regimen failure of antiretroviral therapy and drug naive patients [2, 38]. Very limited studies are available from India and Indian subcontinent on protease drug resistance mutations. Some of the studies on PR gene from India and sub-Saharan African countries, South Africa, Djibouti and Nigeria reported no major resistance mutations among patients failing first-line antiretroviral therapy [3, 7, 8, 21, 38]. In contrast, a few other reports from India as well as sub-saharan African countries have shown both major and minor mutations, L10I, K20R, D30N, E35D, M36L/I, M46I, L63P, V82A, N88D, L90M and I93L in PR gene with drug failure [2, 7, 17, 21, 24]. Similar data has been reported in drug naive patients, some of them showing no PR mutation while others showing both major and minor mutations (L10I, K20R, I13V, D30N, M36I, K45R, L63P, H69K, A71V, V82A and I93L) [4, 13, 20, 39, 42, 43]. The current study focused on a comparative assessment of the presence of drug resistance mutations in three groups of HIV-infected subjects; those receiving ART but were non-responsive, those responsive to ART and drug naive patients. The drug resistance profile was determined according to Stanford HIV Resistance Data Base [6], Rega v8.02 Algorithm [37], IAS-USA mutation list [12] and ANRS_09/2012 Algorithm [10].
Materials and methods
Patient population and sample collection
Blood samples were collected from 109 HIV patients attending anti-retroviral therapy clinic at Department of Medicine, Lok Nayak Jai Prakash Narayan Hospital, New Delhi during 2013–2016. Based on ART treatment, 32 of them were ART non-responsive, 40 were ART responsive while 37 of them never received ART and thus were defined as drug naive. A standardized questionnaire was also prepared to collect demographic data. The study was approved by ethical committee of Jamia Millia Islamia and written consent was obtained from all the patients. Based on clinical history as specified by WHO guidelines, the patients were categorized as ART-non-responsive, ART-responsive and drug naive. The criteria for ART failure (non-responsive) or ART susceptibility (responsive) was selected as recommended by WHO; i.e., patients with new or recurrent WHO stage 4 conditions indicating immunological failure resulting from 50% fall of CD4 count from on-treatment peak value and persistent plasma viral load levels above 5000 copies ml−1 were classified as non-responsive while the patients with plasma viral load levels below 5000 copies ml−1 were classified as responsive. Patients who had never been administered any HIV drug were classified as HIV-naive [47].
ART treatment of patients was based on national recommendations by NACO [32]. Decisions to treat and switch to second-line drug regimen in the patients were mainly guided by clinical, immunological and virological failure of ART, as defined by WHO. The First-line regimen included two nucleoside analogues (NRTI) and one non-nucleoside reverse transcriptase inhibitor (NNRTI). Second-line regimen included several combinations (i) one NRTI, one NNRTI with one protease inhibitor (PI), (ii) Two NRTI with one PI, (iii) two NRTI with two PI.
Whole blood of 5–7 ml was drawn in K3 EDTA vacutainer (Becton Dickinson, USA). Peripheral Blood Mononuclear Cells (PBMCs) were isolated from the blood samples using standard Histopaque-1077 (Sigma-Aldrich, St Louis, USA). The absolute CD4 Cell count was done at intervals of 6 months by FACS Calibur (Beckton Dickinson, USA) and viral loads were enumerated using AMPLICOR HIV-1 Monitor test, version 1.5 (Roche Molecular Systems Inc., Branchburg, NJ, USA) as per manufacturer’s instructions.
Genotyping of HIV-1 protease region of pol gene
The genotyping of drug resistance mutations was carried-out by extraction of proviral DNA from the frozen PBMCs through Qiagen DNeasy kit (Qiagen GmbH, Germany), according to the manufacturer’s protocol. The DNA was finally stored at − 20 °C until the time of testing. The two sets, an outer and an inner set of primers were used to amplify the PR gene using nested PCR. The first amplification was done to amplify an approximately 454-bp fragment using an outer primer set consisting of forward primer PR-F1 (5′-ACCAGAGCCAACAGCCCCACCA-3′) and reverse primer PR-R1 (5′-CTTTTGGGCCATCCATTCCTGGC-3′). A subsequent second round of PCR amplification was performed using inner set of primers, consisting of forward primer PR-F2 (5′-GAAGCAGGAGCCGATAGACAAGG-3′) and reverse primer PR-R2 (5′-CTTTTG GGCCATCCATTCCTGGC-3′) to amplify a 394-bp fragment on a Gene Amp PCR system SC-200 (Kyratec, Australia). The PCR cycling conditions for outer PCR were 95 °C for 5 min, followed by 35 cycles at 95 °C for 15 s, 65 °C for 30 s, and 72 °C for 45 s with final extension at 72 °C for 7 min and for inner PCR were 95 °C for 5 min, followed by 25 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 1 min with final extension at 72 °C for 10 min. The amplified PCR products were run on 1.5% agarose gel electrophoresis with ethidium bromide and visualized under gel documentation system Gel doc 2000 (BioRad, USA).
All the PCR products were sequenced commercially by the Sanger dideoxy method (GCC Biotech, Calcutta, India; Xcelris, Bangalore, India, BGI, Beijing, China). Briefly, automated nucleotide sequencing of purified PCR products was done using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Sequencing was performed on both the strands using forward and reverse primers of the gene. Sequence analysis was done using Clustal X version 2.0.10 (http://ebi.ac.uk/pub/software/clustalw2/2.0.10/) and carried-out from both strands of the gene while HXB2 was used as a reference sequence strain.
Drug resistance analysis
Drug resistance analysis was carried-out using the Calibrated Population resistance (CPR) tool employing the Stanford Surveillance Drug Resistance Mutation (SDRM) (http://hivdb.stanford.edu) [6], REGA algorithm version 8.02 (v8.02;http://www.Kuleuven.ac.be.rega/cev/links) [37] IAS-USA mutation list [12] and, National Agency for AIDS Research (ANRS) (ANRS_09/2012 Algorithm) (http://www.hivfrenchresistance.org/) [10] for quality assessment and drug-resistance mutations interpretation.
Clade typing and phylogenetic analysis of full length sequence of protease gene
The subtyping of HIV-1 PR sequences was done using REGA, a tool of HIV sequence database (v2.0; http://dbpartners.stanford.edu/RegaSubtyping/) [36]. The phylogenetic tree (neighbor-joining) was constructed through PHYLIP package [9]. The bootstrapping was done with SEQBOOT (replications set as 1000) followed by DNADIST, NEIGHBOR and CONSENSE with the F84 model, transition: transversion ratio of 1.5 with empirical base frequencies. The bootstrap values were placed on nodes. The bootstrap replicates indicate the number of times the partition of the species into the two sets which are separated by that branch occurred among the trees, out of 1000 trees. The M-group consensus was used as “outgroup” to determine ancestral states and roots of the “ingroups”. The tree was rooted with the “outgroup”. The tree was visualized using Tree-view (version 1.6.6; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Statistical analysis
Descriptive statistical analyses were performed on non-responsive, responsive and naive HIV-1 groups with respect to different demographic variables. All the baseline and virological/immunological properties were summarized as frequency for categorical variables, mean ± standard deviation or median {Interquartile range (IQR)} for quantitative variables using IBM SPSS software (Version 20;http://www-01.ibm.com/software/analytics/spss/).
Results
Clinical parameters of patient population
Among the 109 HIV-1 seropositive samples, genotyping results were obtained from 90 samples (23 non-responsive, 32 responsive and 35 drug naive) of the participants. All the sequences had intact open reading frame (ORF). A total of nineteen samples failed PCR amplification, nine from non-responsive, eight from responsive and two from drug naive groups. The male to female patient ratio was 3.4:1 with a male preponderance of 84 (77.06%) to 25 females (22.93%), their ages ranged from 20 to 70 years although majority of patients (40.36%) were between 30 and 40 years in each of the three groups. Most of the patients reported were married (77.98%). Based on WHO guidelines, majority (44, 40.36%) of the patients were diagnosed with clinical stage II followed by a total of 34 (31.19%) patients with clinical stage III, 19 (17.43%) patients with clinical stage IV and 12 (11%) patients with clinical stage I. The vast majority of them (89.9%) acquired HIV-1 through heterosexual transmission followed by Intravenous Drug Users (IDU, 5.5%) and blood transfusion (BT, 4.5). A detailed account of these parameters along with the drug regimen at the time of treatment is presented in Table 1. First line of ART regimen was administered to 23 (71.8%) of nonresponsive patients and 34 (85%) to responsive patients. The second line of ART regimen was administered to 9 (28.1%) nonresponsive and 6 (15%) responsive patients (Table 1). The mean value of HIV-1 viral load for non-responsive group was log10 4.60 copies/ml, with a median value of 4.33 log10copies/ml [IQR 3.98–5.19 log10 copies/ml]. In responsive patients, the mean viral load was log101.56 copies/ml with a median value of 1.30 log10 copies/ml [IQR 1.30–1.69 log10 copies/ml]. The CD4 cell count among non-responsive patients ranged from 167 to 351 cells/mm3 with the median value, 257 cells/mm3 and among responsive patients ranged from 323 to 585 cells/mm3 with the median value, 437 cells/mm3.
Table 1.
Clinical parameter and treatment history of 109 HIV-1infected nonresponsive, responsive and drug naive patients
| Parameters | Nonresponsive | Responsive | Drug Naive |
|---|---|---|---|
| Number of patients | 32 | 40 | 37 |
| Sex (male/female) | 27/5 (84.3%/15.6%) | 27/13 (67.5%/ 32.5%) | 30/7(81%/18.9%) |
| Sex ratio (male: female) | 5.4:1 | 2.07:1 | 4.2:1 |
| Age (in years) | |||
| 20 + to ≤ 30 | 4 (12.5%) | 5 (12.5%) | 14 (37.8%) |
| 30 + to ≤ 40 | 13 (40.6%) | 15 (37.5%) | 16 (43.2%) |
| 40 + to ≤ 50 | 9 (28.1%) | 6 (15%) | 4 (10.8%) |
| 50 + to ≤ 60 | 5 (15.6%) | 10 (25%) | 2 (5.4%) |
| 60 + to ≤ 70 | 1 (3.1%) | 4 (10%) | 1 (2.7%) |
| Mean age ± SD* (in yrs) | 40.03 ± 10.13 | 43.02 ± 11.45 | 35.37 ± 9.94 |
| Median age (IQR)** | 36.5 (33–45.5) | 39.5 (35.5–53) | 32 (29.5–40) |
| Marital status | |||
| Married | 23 (71.8%) | 34 (85%) | 28 (75.6%) |
| Unmarried | 7 (21.8%) | 5 (12.5%) | 8 (21.6%) |
| Widow | 2 (6.2%) | 1 (2.5%) | 1 (2.7%) |
| WHO clinical stage | |||
| I | 2 (6.2%) | 6 (15%) | 4 (10.8%) |
| II | 13 (40.6%) | 16 (40%) | 15 (40.5%) |
| III | 9 (28.1%) | 11 (27.5%) | 14 (37.8%) |
| IV | 8(25%) | 7 (17.5%) | 4 (10.8%) |
| Mode of transmission | |||
| Heterosexual | 29 (90.6%) | 35 (87.5%) | 34 (91.8%) |
| Blood Transfusion | 1 (3.1%) | 2 (5%) | 2 (5.4%) |
| Intravenous Drug User | 2 (6.2%) | 3(7.5%) | 1 (2.7%) |
| ART Drug Regimen | |||
| First-line ART regimen | 23 (71.8%) | 34 (85%) | – |
| Second-line ART regimen | 9 (28.1%) | 6 (15%) | – |
| CD4 Count (cells/cu.mm.) | |||
| Median (IQR)) ** | 257 (167–351) | 437 (323–585) | – |
| HIV-1 RNA (log10copies/ml) | |||
| Mean ± S.D.* | 4.60 ± 0.84 | 1.56 ± 0.45 | – |
| Median (IQR) ** | 4.33(3.98–5.19) | 1.30 (1.30–1.69) | – |
*SD denotes Standard Deviation; **IQR denotes Interquartile Range
Analysis of drug resistance mutations
Analysis of various mutations in the study population revealed that the minor resistance mutations against Atazanavir and Tripanavir were present in 100% of non-responsive and drug naive patients while against Nelfinavir it was found to be 91.3% and 94.2% and against Lopinavir, it was found to be 73.9% and 65.71%, respectively. In ART-responsive patients the minor resistance mutations against Atazanavir (ATV), Tripanavir (TPV), Nelfinavir (NFV) and Lopinavir (LPV) were found to be 93.75%, 96.87%, 78.12% and 87.5%, respectively.
In patients non-responsive to ART, two major mutations D30N (1/23, 4.35%) and M46I (1/23, 4.35%) were found in one of the patients who was treated with first line of drug regimen. No major mutation, however, was observed in responsive and drug naive patients. The minor mutations, however, were found in all the three groups of the patients. In non-responsive patients, five minor mutations at positions 36, 63, 69, 89, and 93 singly or in combination were present in all patients as shown in parentheses with percentage; H69K (23/23, 100%), I93L (22/23, 95.65%), M36I and L89M (19/23, 82.6%), L63P (14/23, 60.87%) and K20R (5/23, 21.74%). In addition, less commonly found mutations that included G16E (1/23, 4.35%), M36V (1/23, 4.35%), M36L (3/23, 13.04%), D60E (4/23, 17.39%), I64V and I64L (1/23, 4.35%), T74S (2/23, 8.70%) and V82I (1/23, 4.35%) were also observed. In responsive patients, most frequent minor resistance mutations included H69K (31/32, 96.88%), I93L (30/32, 93.75%), M36I and L89M (28/32, 87.50%), L63P (25/32, 78.13%) and K20R (8/32, 25%), while some other minor mutations such as M36L, D60E, I62V, T74S, T74A, V77I and V82I were present in one patient each and I64M in four of the patients (4/32, 12.5%) (Table 2). In drug naive patients too, minor mutations were found singly or in combination in any one of the patients as given in parentheses with percentage; H69K (35/35, 100%), M36I (33/35, 94.28%), I93L (30/35, 85.7%), L89M (27/35, 77%), L63P (17/35, 48.5%) and K20R (10/35, 28.5%). Other minor mutations that are reported less commonly were also found including G16E (4/35, 11.4%), I64L (4/35, 11.4%), I64M (5/35, 14.2%), T74S (4/35, 11.4%), V82I (5/35, 14.2%) and M36V and D60E one patient each (Table 2). It is worth to note that among all non-responsive and responsive patient only 26% (6/23) and 18.75% (6/32) patients respectively received protease inhibitors; Atazanavir (ATV)/Ritonavir (RTV) or Lopinavir (LPV) but no major mutation was found against them.
Table 2.
Major and Minor amino-acid mutations associated with resistance to protease inhibitors in nonresponsive, responsive and ART naive patients (90)
| S. No | Mutations | Number of nonresponsive patients (Mutational Frequency in percentage, n = 23) | Number of responsive patients (Mutational Frequency in percentage, n = 32) | Number of naive patients (Mutational Frequency in percentage, n = 35) | Resistance profile a |
|---|---|---|---|---|---|
| Major mutations | |||||
| 1 | D30N | 1 (4.35) | 0 (0) | 0 (0) | NFV (R)1,2,3 |
| 2 | M46I | 1 (4.35) | 0 (0) | 0 (0) |
IDV(R)4, ATV(L)1, IDV(L)1, LPV(L)1 NFV(I)1, SQV(L)3, FPV(L)1 |
| Minor mutations | |||||
| 1 | L10I | 0 (0) | 1 (3) | 0 (0) | ATV(L)2, IDV(L)2, LPV(L)2, NFV(L)2, SQV(L)2 |
| 2 | G16E | 1 (4.35) | 0 (0) | 4 (11.4) | ATV(L)2 |
| 3 | K20R | 5 (21.74) | 8 (25) | 10 (28.5) | ATV(L)2,3, IDV(L)2,3, LPV(L)2,3, NFV(L)3, SQV(L)3, FPV(L)3 |
| 4 | E35D | 7 (30.0) | 12 (37.5) | 10 (28.5) | - |
| 5 | M36I | 19 (82.6) | 28 (87.5) | 33 (94.28) | ATV(L)2, IDV(L)2, NFV(L)2, TPV(L)2 |
| 6 | M36V | 1 (4.35) | 0 (0) | 2 (5.7) | ATV(L)2, TPV(L)2 |
| 7 | M36L | 3 (13.04) | 1 (3) | 0 (0) | ATV(L)2, TPV(L)2 |
| 8 | D60E | 4 (17.39) | 1 (3) | 2 (5.7) | ATV(L)2 |
| 9 | I62V | 0 (0) | 1 (3) | 0 (0) | IDV(L)3, NFV(L)3, SQV(L)2,3 |
| 10 | L63P | 14 (60.87) | 25 (78.1) | 17 (48.5) | LPV(L)2 |
| 11 | L63V | 3 (13) | 1 (3) | 0 (0) | – |
| 12 | L63S | 1 (4.35) | 0 (0) | 4 (11.4) | – |
| 13 | L63A | 2 (8.7) | 2 (6) | 6 (17.1) | – |
| 14 | L63T | 2 (8.7) | 0 (0) | 4 (11.4) | – |
| 15 | I64V | 1 (4.35) | 0 (0) | 0 (0) | ATV(L)2,NFV(L)3, LPV(L),3 |
| 16 | I64L | 1 (4.35) | 0 (0) | 4 (11.4) | ATV(L)2, |
| 17 | I64M | 0 (0) | 4 (12.5) | 5 (14.2) | - |
| 18 | H69K | 23 (100) | 31 (97) | 35 (100) | TPV(L)2 |
| 19 | T74S | 2 (8.70) | 1 (3) | 4 (11.4) | ATV(L)3,IDV(L)3,NFV(I)1,SQV(L)3 |
| 20 | T74A | 0 (0) | 1 (3) | 0 (0) | ATV(L)3, NFV(L)3, SQV(L)3 |
| 21 | V77I | 0 (0) | 1 (3) | 0 (0) | IDV(L)2, NFV(L)2, SQV(L)2 |
| 22 | V82I | 1 (4.35) | 1 (3) | 5 (14.2) | ATV(L)2 |
| 23 | L89M | 18 (78.26) | 28 (87.5) | 27 (77) | TPV(L)2 |
| 24 | I93L | 22 (95.65) | 30 (93.7) | 30 (85.7) | ATV(L)2 |
Capital letter L, I, R in parenthesis denotes low level resistance, intermediate resistance, highly resistance respectively
a Resistance was analyzed using the 1 Stanford HIV Resistance Database (HIVDBv 6.2), 2 IAS-USA mutation list, 3 Rega v8.02 Algorithm and 4ANRS_09/2012 Algorithm
n denotes total number of patients
Drug listed in the table: NFV-Nelfinavir, IDV- Indinavir, ATV- Atazanavir, LPV- lopinavir, SQV- Saquinavir,
FPV-Fosamprenavir, TPV-Tipranavir
HIV-1 subtype analysis by phylogenetic tree construction
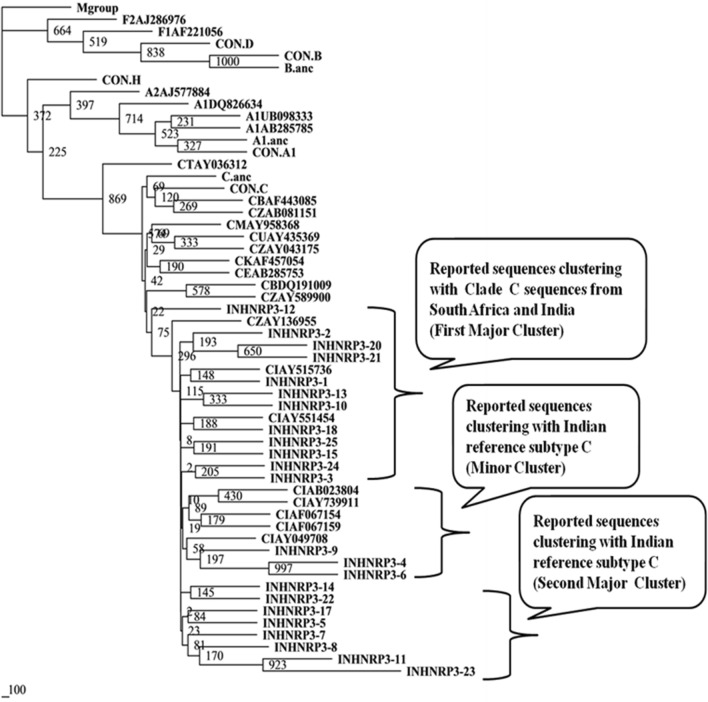
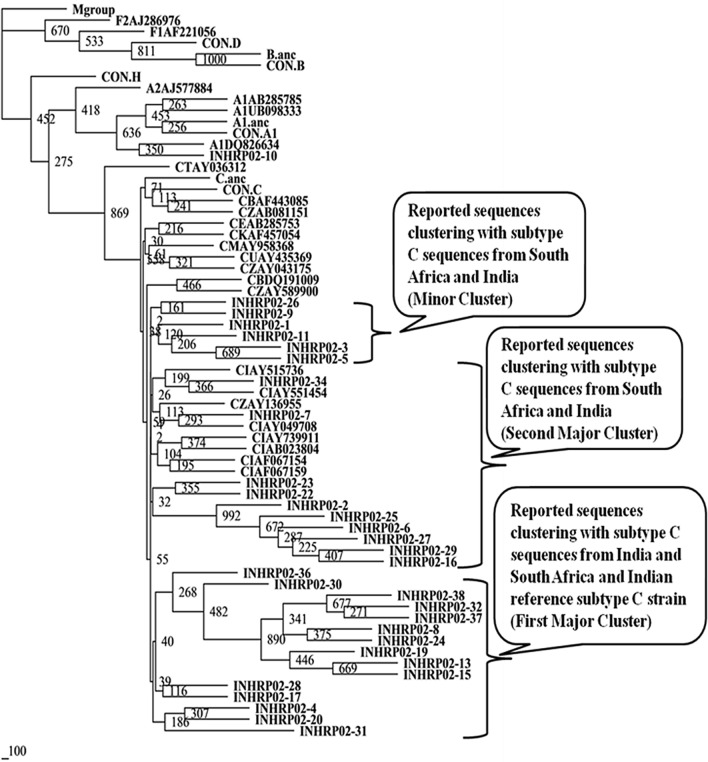
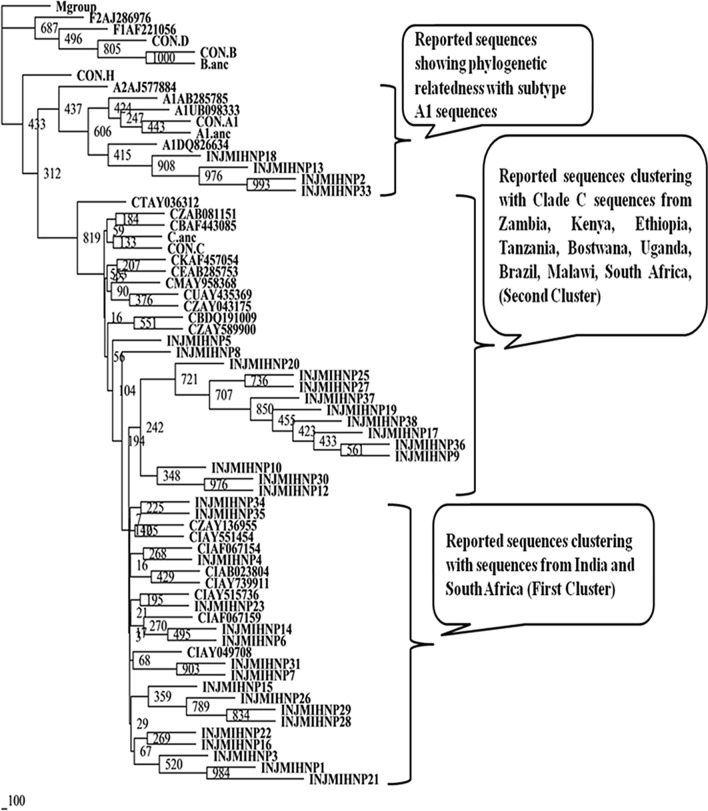
Nucleotide sequences of protease gene from 90 patients were analyzed using REGA HIV-1 subtyping tool v2.0 and phylogenetic trees were constructed utilizing earlier reported sequences from different countries (Figs. 1, 2 and 3) [22]. It was observed that 85 (94.44%) of the HIV-1 sequences were subtype C. Non-clade C sequences were found in 5 (5.55%) individuals and belonged to subtype A1. It is worthy to note that all the five subtype A1 sequences were from four drug naive patient (INJMIHNP-2, INJMIHNP-13, INJMIHNP-18 and INJMIHNP-33) and one-responsive patient (INHRP02-10) population. Subtype analysis of the 23 protease sequences from non-responsive group revealed two major clusters and one minor cluster of clade C virus. One cluster (n = 12, 52.17%) was closer to sequences from South Africa and India (IN), the second cluster (n = 8, 34.78%), and the minor cluster (n = 3, 13.04%) were closer to Indian reference subtype C (Fig. 1). Similarly, the responsive protease sequences showed two major clusters and one minor cluster. The first major cluster (n = 15, 46.87%) was more closer to subtype C sequences from India and South Africa and Indian reference Subtype C strain (CINAB023804), the second major cluster (n = 10, 31.25%)) showed evolutionary distance from subtype C sequences from India and South Africa and Indian reference Subtype C strain (CINAB023804) and the minor cluster (n = 6, 18.75%) also showed distant phylogenetic relatedness to previously reported subtype C sequences from India and South Africa and Indian reference subtype C (Fig. 2). Sequences from naive patients revealed two major clusters (n = 17, 48.57%), one being closer to both South African (CZAY136955) and Indian sequences (CINAY515736, CINAB023804 etc.) while second cluster (n = 14, 40%) was closer to clade C sequences from Zambia, Ethiopia, Bostwana, Uganda, Brazil, Kenya, Tanzania, Malawi and South Africa (Fig. 3).
Fig. 1.
Phylogenetic relationship based on protease gene sequences (non-responsive group) of HIV-1 with other reported subtypes and consensus sequences from other clades. Consensus sequences and other sequences included in the tree are obtained from Los Alamos HIV Sequence Database. The clusters are indicated in the callouts. The bootstrapping was done with SEQBOOT (replications set as 1000). The scale bar indicates nucleotide substitution. Our study populations have been indicated as INHNRP3
Fig. 2.
Phylogenetic relationship based on protease gene sequences (responsive group) of HIV-1 with other reported subtypes and consensus sequences from other clades. Consensus sequences and other sequences included in the tree are obtained from Los Alamos HIV Sequence Database. The clusters are indicated in the callouts. The bootstrapping was done with SEQBOOT (replications set as 1000). The scale bar indicates nucleotide substitution. Our study populations have been indicated as INHRP02
Fig. 3.
Phylogenetic relationship based on protease gene sequences (drug naive group) of HIV-1 with other reported subtypes and consensus sequences from other clades. Consensus sequences and other sequences included in the tree are obtained from Los Alamos HIV Sequence Database. The clusters are indicated in the callouts. The bootstrapping was done with SEQBOOT (replications set as 1000). The scale bar indicates nucleotide substitution. Our study populations have been indicated as INJMIHNP
Discussion
The anti-HIV drugs have sufficiently delayed the progression of AIDS in HIV patients by prolonging their life and improving the quality of life. However, their effectiveness has been severely inhibited by the emergence of drug-resistant and cross-resistant mutants that renders AIDS with no definitive cure. The HIV drug resistance (HIVDR) remains a major concern worldwide especially in low- and middle income countries including India due to the acquisition and transmission of HIV-1 drug resistance strains. Further, failure to ARV therapy due to HIV-1 drug resistant viruses increases significant risk of morbidity and mortality among HIV patients, particularly in developing countries where choice for ART are limited. Majority of data on HIVDR comes from developed countries because of their robust HIV surveillance and treatment practices [12]. In India, however, no consolidated data on HIVDR is available except a few sporadic studies [7, 15, 38].
The current study was carried out to know comparative genetic evaluation of the drug resistance mutations in patients who show failure or success to ART and those who never had received HIV drugs and thus classified as HIV drug-naive. Across the patient population in each group except one non-responsive patient none of the patients had major mutation. This non-responsive patient harboured two major PI mutations, D30N and M46I in protease gene. The patient was given first line of ART drug regimen (2NRTI plus NNRTI) and did not receive any second line drug regimen (Protease inhibitor). Interestingly, six non-responsive patients (3 of them shown in Table 1 could not be amplified) though received second line drug regimen that included one protease inhibitor, did not show any major mutation suggesting some other factor to be responsible for drug failure. Several studies from India and sub-Saharan African countries, South Africa, Djibouti and Nigeria reported similar findings with no PR major resistance mutations among patients failing antiretroviral therapy [3, 7, 8, 21, 38]. However, a study from northern India on non-responsive patients has reported that patients receiving first line drug regimen showed PI drug resistance mutations in 12 out of 128 patients (about 10.9%) which is higher than the current study [43]. In a study from Zambia, Sue et al. reported the frequency of PI major mutations to be around 5% in non-responsive patients on first line of drug regimen similar as found in our study [40]. In ART-responsive patients and drug naive patients no major PI mutation was observed in this study, which is similar to the reports with some earlier studies from elsewhere [8, 13, 14]. A few other studies on drug naive patients, however, reported major PI mutations with varied frequency from 1.4 to 0% [1, 2, 20, 43].
The major PI drug resistance mutations in non-responsive patients, D30N causes resistance only to NFV but no cross resistance to other PIs [41]. The M46I confers high resistance to IDV (according to ANRS Algorithm) [10] while low levels of resistance to ATV, FPV, IDV, and LPV and intermediate resistance to NFV as per Stanford HIV DR Database [6]. The D30N and M46I mutations in non-responsive patient in our study are suggestive of transmitted resistance as no PI inhibitor was prescribed to the patient.
All the patients irrespective of categories exhibited various minor mutations that are reported to be associated with low level resistance to PIs. The non-responsive and drug naive patients showed 100% minor resistance mutations to PI, which is similar to other studies on non-responsive and drug naive patients from India [1, 2, 4, 14]. The higher frequency of minor mutations ranging from 12.5 to to 91% in non-responsive patients has been reported from various other countries too [8, 19, 21]. In patients responsive to ART, 97% of the sequences had minor mutations.
The published data suggests that the selection of secondary/minor protease mutations helps in repairing the enzymatic function to rescue viral fitness [5]. Thus large number of minor mutations presents in this study population irrespective of their being non-responsive, responsive or drug naive may have compensatory role to maintain viral fitness. High prevalence of minor mutations in drug naive patients in our study also suggests increased frequency of transmitted mutations.
Most common amino-acid variants were found to be located at seven polymorphic positions including codon positions 10, 20, 36, 63, 71, 77 and 93. The polymorphism found at amino-acid positions 20, 36, 63 and 93 is reported to be the most polymorphic protease position [49]. The amino-acid variants on these positions contribute to drug resistance in combinatorial form while a single mutation does not cause the drug resistance by itself. Some of these minor mutations tend to compensate the decrease in catalytic efficiency of the protease caused by some other mutations thus helping in maintaining fitness of the virus [5, 26]. The minor mutations at positions 36, 63, 69, 89, and 93 were observed in more than 84% of HIV sequences across all the three groups. The mutation M36I/V/L was observed in 95.65%, 90% and 100% of patients belonging to non-responsive, responsive and drug naive patient groups respectively. These mutations on above positions are reported to be present in 13% of subtype B population but more than 80% in subtype C, which supports our data on subtype C [34]. The common pattern of minor mutations M36I, H69K, and L89M are associated with reduced susceptibility and diminished virological response to TPV [29]. In the current study, the mutation pattern M36I, H69K and L89M was observed in 14 (60.86%) non-responsive, 25 (78.15%) responsive and 26 (74.28%) drug naive patients.
The another pattern of mutations M36I, K20R, and I93L have been reported to putatively occur more frequently in HIV-1 subtype C than subtype B [29]. They are among the accumulated mutations associated with reduced virologic response or failure to atazanavir/ritonavir containing regimen [11, 12]. The K20R mutation was found in 5 (22%) non-responsive, 8 (25%) responsive and 10 (28.5%) drug naive patients. The I93L was found in 22 (96%) non-responsive, 30 (93.75%) responsive and 30 (85.7%) drug naive patients. The IAS-USA mutation list documented that I93L is associated with low levels of resistance against ATV only [12] while K20R is associated with low level resistance to multiple protease inhibitors such as ATV, IDV and LPV according to IAS-USA mutation list and Rega v8.02 Algorithm [12, 37].
The L63P (leucine-to-proline) in protease gene is a polymorphic position that strongly enhances viral replicative fitness of HIV-1 under pressure of ART [44]. The L63P mutation is reported to be highly prevalent in Indian PR sequences and elsewhere [21, 45] and in this study too, this mutations was present in 14 (60.87%) of the non-responsive, 25 (78.1%) of the responsive and 17 (48.57%) drug naive sequences. Another well-recognized mutation E35D, which was present both in non-responsive as well as responsive patients, is reported to affect the conformational equilibrium between the closed and semi-open conformations of the free protease as well as significant reduction in free energy binding of the protease to its substrate or Amprenavir. It has also been shown that the E35D mutation reduces interaction with the HLA B44 molecule impeding cellular immune response [23]. It is thus postulated to favour escape from the immune system in addition to conferring drug resistance [27]. The other minor mutations across all the population in the study included D60E, I62V, I64V/L/M, T74A, V77I, and V82I with varied frequency from 3 to 17.3%, which are associated with low resistance to one or more PIs inhibitors, i.e. atazanavir, saquinavir, lopinavir, nelfinavir, and indinavir. The mutation T74S, which confers intermediate level resistance to nelfinavir (Stanford HIV Resistance Database) [6] if present with K14R, it slightly increases resistance to IDV too. The combination of T74S and K14R was found in one non-responsive patient in the present study [46].
In Indian subcontinent, subtype C is most prevalent and represents more than 80% of HIV-1 infections. In our study, majority of the sequences obtained from the patients, except five, were characterized as subtype C (94.4%) as revealed by analysis of protease gene sequences using REGA, (HIV-1 sub typing tool) [36] and phylogenetic tree construction [9]. Similar findings have been reported in other studies as well [2, 38, 39, 43]. The pattern of clustering shows evolutionary relatedness among the sequences. However, in all the trees of PR gene sequences, certain sequences of the study dataset clustered together to form Indian subclade. This finding along with similar data reported previously supports the fact that HIV-1 subtype C sequences have subclusters [18].
In conclusion, the current study found major mutations (D30N and M46I) in one of the non-responsive patients with the possibility of being acquired through transmitted resistance mutation as the patient did not receive drug treatment regimen with PI inhibitor. The study also is indicative of involvement of various other factors too such as poor drug compliance, unfavourable drug interactions, insufficient drug absorption or change in drug metabolism in non-responsive patients. Presence of high frequency of minor drug resistance mutations irrespective of patient group whether non-responsive, responsive or drug naive may confer low level drug resistance in addition to compensatory role in maintaining viral fitness. Certain polymorphic sites in the protease gene strongly enhance viral replicative fitness under pressure of ART. Thus, determination of such polymorphic sites may enable better understanding of survival of the virus in the presence of ART and the subsequent drug failure. It may also be construed clearly that pattern of mutations play crucial role in drug resistance rather than individual mutations. Thus identification of pattern of mutations and studying further their role in drug resistance may facilitate development of better treatment regimens.
Acknowledgements
We sincerely thank the Council of Scientific and Industrial Research (CSIR), New Delhi, Government of India,for providing financial support to conduct this study (vide grant number 27(0236)/10/EMR-II).
Funding
This research study was supported by Council of Scientific and Industrial Research (CSIR), New Delhi, Government of India (vide grant number 27(0236)/10/EMR-II).
Data availability
The Gene Bank accession numbers of PR nucleotide sequences of non-responsive, responsive, and naive patients groups from this study are KF689039 to KF689073, KF689074 to KF689105 and KF689106 to KF689128.
Declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical approval and consent to participate
All patients were included with their written informed consent. Based on clinical history as specified by WHO guidelines (2010) and the study was approved by the institutional ethics committee of Jamia Millia Islamia, New Delhi.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arora SK, Gupta S, Toor JS, Singla A. Drug resistance-associated genotypic alterations in the pol gene of HIV type 1 isolates in ART-naive individuals in North India. AIDS Res Hum Retroviruses. 2008;24:125–130. doi: 10.1089/aid.2007.0156. [DOI] [PubMed] [Google Scholar]
- 2.Balakrishnan P, Kumarasamy N, Kantor R, et al. HIV type 1 genotypic variation in an antiretroviral treatment-naive population in southern India. AIDS Res Hum Retroviruses. 2005;21:301–305. doi: 10.1089/aid.2005.21.301. [DOI] [PubMed] [Google Scholar]
- 3.Chaplin B, Eisen G, Idoko J, et al. Impact of HIV type 1 subtype on drug resistance mutations in Nigerian patients failing first-line therapy. AIDS Res Hum Retroviruses. 2011;27:71–80. doi: 10.1089/aid.2010.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturbhuj DN, Hingankar NK, Srikantiah P, et al. Transmitted HIV drug resistance among HIV-infected voluntary counseling and testing centers (VCTC) clients in Mumbai, India. AIDS Res Hum Retroviruses. 2010;26:927–932. doi: 10.1089/aid.2010.0032. [DOI] [PubMed] [Google Scholar]
- 5.Condra JH, Schleif WA, Blahy OM, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;37:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 6.Stanford Drug Resistance Database. http://hivdb.stanford.edu Accessed 14 February 2014
- 7.Deshpande A, Jauvin V, Magnin N, et al. Resistance mutations in subtype C HIV type 1 isolates from Indian patients of Mumbai receiving NRTIs plus NNRTIs and experiencing a treatment failure: resistance to AR. AIDS Res Hum Retroviruses. 2007;23:335–340. doi: 10.1089/aid.2006.0183. [DOI] [PubMed] [Google Scholar]
- 8.ElmiAbar A, Jlizi A, Darar HY, Kacem MA, Slim A. HIV-1 drug resistance genotyping from antiretroviral therapy (ART) naive and first-line treatment failures in Djiboutian patients. Diagn Pathol. 2012;7:138. doi: 10.1186/1746-1596-7-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J (1993) PHYLIP package (v3.69). http://evolution.genetics.washington.edu/phylip/getme.html. Accessed 07 July 2014
- 10.French ANRS (National Agency for AIDS Research) AC11 resistance group (2012) HIV-1 genotypic drug resistance interpretations algorithms. http://www.hivfrenchresistance.org/hivfrenchres.pdf. Accessed 13 September 2014
- 11.Johnson VA, Brun-Vézinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1:2007. Top HIV Med. 2007;15:119–125. [PubMed] [Google Scholar]
- 12.Johnson VA, Calvez V, Gunthard HF, et al. Update of the drug resistance mutations in HIV-1. Top Antivir Med. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 13.Kandathil AJ, Kannangai R, Abraham OC, et al. The frequency of HIV-I drug resistance mutations among treatment-naive individuals at a tertiary care centre in south India. Int J STD AIDS. 2009;20:522–526. doi: 10.1258/ijsa.2008.008403. [DOI] [PubMed] [Google Scholar]
- 14.Kandathil AJ, Kannangai R, Abraham OC, Sudarsanam TD, Pulimood SA, Sridharan G. Genotypic resistance profile of HIV-1 protease gene: a preliminary report from Vellore, south India. Indian J Med Microbiol. 2008;26:151–154. doi: 10.4103/0255-0857.40530. [DOI] [PubMed] [Google Scholar]
- 15.Kandathil AJ, Kannangai R, Varghese R, et al. Drug resistant mutations detected by genotypic drug resistance testing in patients failing therapy in clade c HIV-1 infected individuals from India. Indian J Med Microbiol. 2009;27:231–236. doi: 10.4103/0255-0857.53205. [DOI] [PubMed] [Google Scholar]
- 16.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. Plos Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantor R, Zijenah LS, Shafer RW, et al. HIV-1 subtype C reverse transcriptase and protease genotypes in Zimbabwean patients failing antiretroviral therapy. AIDS Res Hum Retroviruses. 2002;18:1407–1413. doi: 10.1089/088922202320935483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar M, Jain SK, Pasha ST, Chattopadhaya D, Lal S, Rai A. Genomic diversity in the regulatory nef gene sequences in Indian isolates of HIV type 1: emergence of a distinct subclade and predicted implications. AIDS Res Hum Retroviruses. 2006;22:1206–1219. doi: 10.1089/aid.2006.22.1206. [DOI] [PubMed] [Google Scholar]
- 19.Kyeyune F, Gibson RM, Nankya I, et al. Low-frequency drug resistance in HIV-infected Ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob Agents Chemother. 2016;60:3380–3397. doi: 10.1128/AAC.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lall M, Gupta RM, Sen S, et al. Profile of primary resistance in HIV-1-infected treatment-naive individuals from Western India. AIDS Res Hum Retroviruses. 2008;24:987–990. doi: 10.1089/aid.2008.0079. [DOI] [PubMed] [Google Scholar]
- 21.Levison JH, Orrel C, Gallien S, et al. Virologic failure of protease inhibitor-based second-line antiretroviral therapy without resistance in a large HIV treatment program in South Africa. PLoS ONE. 2012;7:e32144. doi: 10.1371/journal.pone.0032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Los Alamos HIV Database. https://www.hiv.lanl.gov/components/sequence/HIV/search/search.html. Accessed July 2018
- 23.Manosuthi W, Butler DM, Pérez-Santiago J, et al. (2010) Protease polymorphisms in HIV-1 subtype CRF01_AE represent selection by antiretroviral therapy and host immune pressure. AIDS. 2010;24:411–416. doi: 10.1097/QAD.0b013e3283350eef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marconi VC, Sunpath H, Lu Z, et al. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008;46:1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz M (2000) Resistance, fitness, adherence, and potency: mapping the paths to virologic failure. JAMA 283:250–251.https ://doi.org/10.1001/jama.283.2.250 [DOI] [PubMed]
- 26.Martinez-Picado J, Savara AV, Sutton L, D'Aquila RT. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/JVI.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiselbach H, Horn AH, Harrer T, Sticht H. Insights into amprenavir resistance in E35D HIV-1 protease mutation from molecular dynamics and binding free-energy calculations. J Mol Model. 2007;13:297–304. doi: 10.1007/s00894-006-0121-3. [DOI] [PubMed] [Google Scholar]
- 28.Miller V, Larder BA (2001) Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir Ther 6:25–44. [PubMed]
- 29.Miri L, Ouladlahsen A, Kettani A, Bensghir R, Marhoum El filali K, Wakrim L. Characterization of protease resistance-associated mutations in HIV type 1 drug-naive patients following the increasing prevalence of the CRF02_AG strain in Morocco. AIDS Res Hum Retroviruses. 2012;28:571–577. doi: 10.1089/aid.2011.0225. [DOI] [PubMed] [Google Scholar]
- 30.Mocroft A, Vella S, Benfield TL et al (1998)Changing patterns of mortality across Europe in patients infected with HIV-1. EuroSIDA Study Group. Lancet 352:1725–1730.10.1016/s0140-6736(98)03201-2 [DOI] [PubMed]
- 31.National AIDS Control Organisation, Ministry of health and Family Welfare (2013) Antiretroviral therapy guidelines for HIV-infected adults and Adolescents. http://naco.gov.in/sites/default/files/Antiretroviral%20Therapy%20Guidelines%20for%20HIV-Infected%20Adults%20and%20Adolescents%20May%202013%281%29_0.pdf. Accessed 5 April 2019
- 32.National AIDS Control Organisation, Ministry of health and Family Welfare, Government of India (2008) Draft Guidelines for national roll-out of second line ART. National Guidelines on second line ART for adults and adolescents. http://www.nacoonline.org/. Accessed 12 March 2011
- 33.National AIDS Control Organization, Ministry of health and Family Welfare (2014) Annual Report 2013–2014. http://naco.gov.in/sites/default/files/NACO_English%202013-14.pdf. Accessed 12 June 2014
- 34.Ode H, Matsuyama S, Hata M, Neya S, Kakizawa J, Sugiura W, Hoshino T. Computational characterization of structural role of the non-active site mutation M36I of human immunodeficiency virus type 1 protease. J Mol Biol. 2007;370:598–607. doi: 10.1016/j.jmb.2007.04.081. [DOI] [PubMed] [Google Scholar]
- 35.Perrin L, Telenti A. HIV treatment failure: testing for HIV resistance in clinical practice. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 36.REGA HIV-1 Subtyping Tool v2.0, University of Leuven. http://dbpartners.stanford.edu/RegaSubtyping/html/ subtypinghiv.html. Accessed 13 March 2014
- 37.REGA algorithm v8.02, University of Leuven(2013). http://www.kuleuven.ac.be/rega/cev/links) Accessed 14 February 2014
- 38.Saini S, Bhalla P, Gautam H, Baveja UK, Pasha ST, Dewan R. Resistance-associated mutations in HIV-1 among patients failing first-line antiretroviral therapy. J Int Assoc Physicians AIDS Care (Chic) 2012;11:203–209. doi: 10.1177/1545109711421217. [DOI] [PubMed] [Google Scholar]
- 39.Sen S, Tripathy SP, Patil AA, et al. High prevalence of human immunodeficiency virus type 1 drug resistance mutations in antiretroviral treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007;23:1303–1308. doi: 10.1089/aid.2007.0090. [DOI] [PubMed] [Google Scholar]
- 40.Seu L, Mulenga LB, Siwingwa M, et al. Characterization of HIV drug resistance mutations among patients failing first-line antiretroviral therapy from a tertiary referral center in Lusaka, Zambia. J Med Virol. 2015;87:1149–1157. doi: 10.1002/jmv.24162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev. 2002;15:247–277. doi: 10.1128/cmr.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha S, Ahmad H, Shekhar RC, et al. Prevalence of HIV drug resistance mutations in HIV type 1 isolates in antiretroviral therapy naive population from Northern India. AIDS Res Treat. 2012 doi: 10.1155/2012/905823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinha S, Shekhar RC, Ahmad H, et al. Prevalence of HIV drug resistance mutation in the northern Indian population after failure of the first line antiretroviral therapy. Curr HIV Res. 2012;10:532–538. doi: 10.2174/157016212802429785. [DOI] [PubMed] [Google Scholar]
- 44.Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother. 2003;51:229–240. doi: 10.1093/jac/dkg079. [DOI] [PubMed] [Google Scholar]
- 45.Sune C, Brennan L, Stover DR, Klimkait T. Effect of polymorphisms on the replicative capacity of protease inhibitor-resistant HIV-1 variants under drug pressure. Clin Microbiol Infect. 2004;10:119–126. doi: 10.1111/j.1469-0691.2004.00832.x. [DOI] [PubMed] [Google Scholar]
- 46.Velazquez-Campoy A, Todd MJ, Vega S, Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci USA. 2001;98:6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization (2010) Antiretroviral Therapy for HIV Infection in Adults and Adolescents, Recommendations for a public Health Approach. http://apps.who.int/iris/bitstream/10665/44379/1/9789241599764_eng.pdf. Accessed 11 February 2011 [PubMed]
- 48.World Health Organization (2018) WHO HIV/AIDS Fact Sheet. https://www.who.int/en/news-room/fact-sheets/detail/hiv-aids/. Accessed 19 July 2018
- 49.Yahi N, Tamalet C, Tourrès C, et al. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J Clin Microbiol. 1999;37:4099–4106. doi: 10.1128/JCM.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Gene Bank accession numbers of PR nucleotide sequences of non-responsive, responsive, and naive patients groups from this study are KF689039 to KF689073, KF689074 to KF689105 and KF689106 to KF689128.