Abstract
The objective of this study was to compare Reverse Hybridisation Assay with conventional sequencing for determination of Hepatitis C Virus Genotype and Subtypes. Anti-HCV antibody was determined followed by HCV RNA extraction which was used for (1) viral load determination (2) qualitative real-time PCR RHA for genotyping and (3) conventional sequencing. Compared to conventional sequencing, accuracy of RHA results was 96.55% for determination of genotype (κ = 0.93) and 89.66% for subtype (κ = 0.85). Sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV) of the qualitative PCR were 82.29%, 100%, 44.44% and 100% respectively with an accuracy of 86.84%. RHA is a less time consuming and cheaper method for determination of HCV genotype and subtype yet results must be interpreted with caution and quality control monitoring should be strictly followed to ensure validity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-021-00729-9.
Keyword: Hepatitis C virus; Genotype; Subtype; Reverse hybridisation assay; Conventional sequencing
Hepatitis C virus (HCV) is a positive sense RNA virus of the family Flaviviridae. HCV infection leads to an asymptomatic course of disease and causes chronic infection. Earlier regimens for treatment of HCV infection were based on Interferon therapy where determination of HCV genotype and subtype was crucial. With the introduction of pan-genotypic drugs for management of HCV, determination of HCV genotype and subtype is no longer considered essential. But, given the emergence of resistance to directly acting antivirals (DAA) even in treatment naïve patients and the role of genotype and subtype in predicting the course of the disease and management of complications, accurate diagnosis is extremely crucial. Genotype and subtype of Hepatitis C virus (HCV) is commonly determined by reverse hybridisation assays (RHA) which are preferred over sequencing based techniques due to time constraints [1]. However, RHA may lead to incorrect determination of mixed infections, subtypes or even genotypes in up to 10% cases [2]. Thus, this study was undertaken as a pilot project to compare commercially available Reverse Hybridisation Assay with conventional sequencing for HCV genotype and subtype determination.
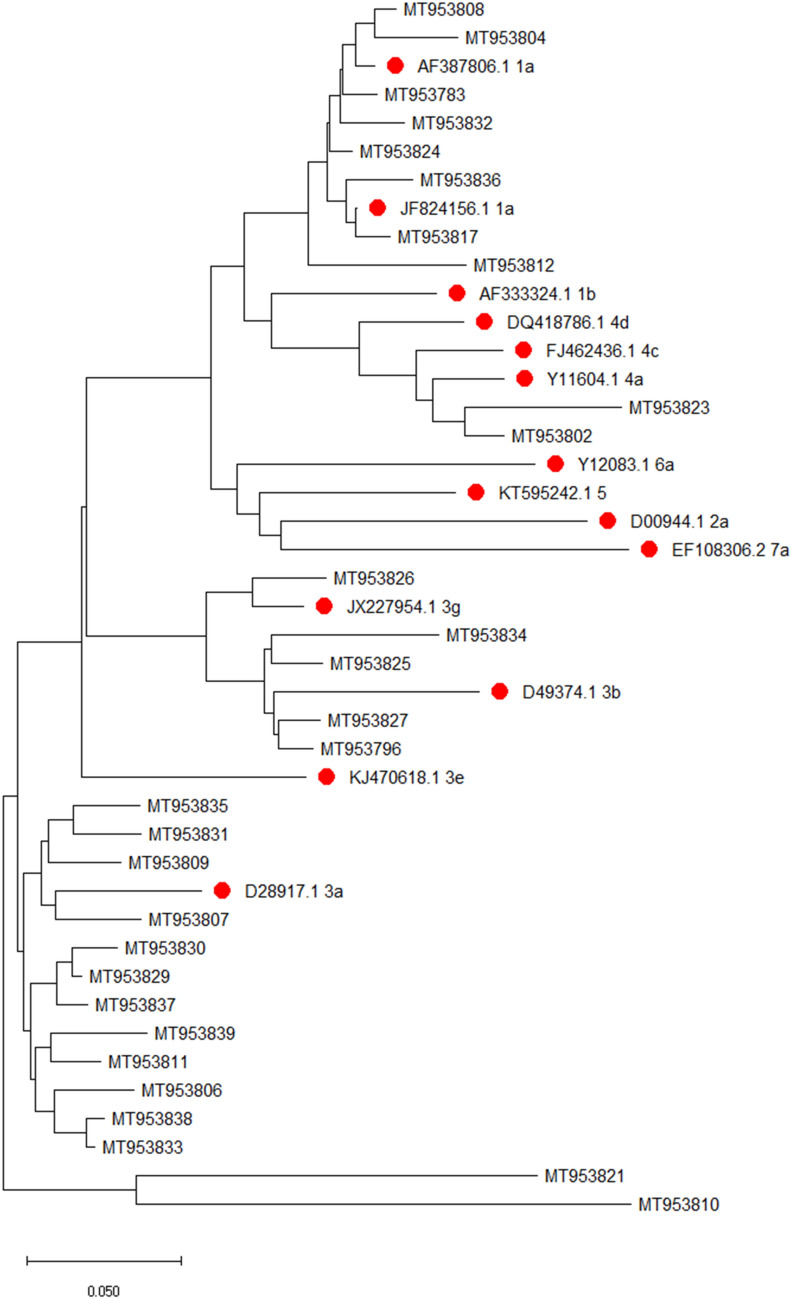
This study was done at a tertiary care centre in Uttarakhand protocol for which was approved by Institutional ethics committee (AIIMS/IEC/111/18). Samples were screened for anti-HCV antibody during the study period (2 months; June 2019 to July 2019). Anti-HCV antibody reactive cases were selected and informed written consent with relevant history collected. Serum was collected through standard aseptic laboratory procedures. All anti-HCV antibody testing was confirmed by 3rd generation ELISA (QUALISA™ HCV ELISA, Qualpro Diagnostics, Verna, Goa, India). First, viral RNA was extracted with QiAmp® Viral RNA Mini kit (Qiagen) strictly adhering to manufacturer’s instruction. All samples were processed for quantitative estimation of HCV RNA with Realstar® HCV RT-PCR 2.0 (Altona Diagnostics). BioRad CFX 96 Manager C1000 Touch™ real time PCR system (Version 3.0) was used for the quantitative detection of HCV RNA. Second, all samples were tested for qualitative estimation of HCV RNA and reverse hybridisation assay (HCV RNA Real Time Qualitative 2.0 with GEN-C 2.0, Nuclear Laser Medicine S.r.l) was done as explained in kit insert. Finally, c-DNA was synthesized (GoScript™ Reverse Transcription system, Promega, Madison, Wisconsin, USA) followed by nested PCR with primers using an optimised protocol as described by Verma et al. [3] targeting a 405 bp fragment of the 5’-NCR-Core region of HCV (GoTaq® Colourless Master Mix, Promega). Amplification was confirmed by gel electrophoresis. Samples with adequate amplification were used for sequencing reactions. Multiple sequence alignment was generated using MEGA-X software (version 10.0.5) (Fig. 1). The evolutionary history was inferred using the Neighbour-Joining method [4]. Reference sequences were described by Verma et al. [3], Lole et al. [5] and Prakash et al. [6] and global reference strains described by International Committee for Taxonomy of Viruses [7] were used. Confidence values were calculated by bootstrap analysis and consensus tree was produced by using MEGA-X [8]. Sequences were submitted to GenBank with accession numbers MT953783, MT953796, MT953802, MT953804, MT953806—MT953812, MT953817, MT953821, MT953823—MT953827, MT953829—MT953839.
Fig.1.
Phylogenetic tree of 29 strains under study alongside reference strains—global and Indian (pre-fixed with red dot) generated using MEGA-X (version 10.0.5)
Out of 38 anti-HCV antibody reactive patients, HCV RNA was quantitatively detected in 34 patients and qualitatively in 29 samples. Among 5 RNA negative samples (qualitatively), HCV RNA load was < 103 IU/mL. Sensitivity, specificity, NPV and PPV of the qualitative PCR were 82.29%, 100%, 44.44% and 100% respectively with an accuracy of 86.84% (Table 1).
Table 1.
Evaluation of qualitative PCR for detection of HCV RNA
| Quantiative (RealStar® HCV RT-PCR 2.0) | |||
|---|---|---|---|
| Qualitative (HCV RNA real time qualitative 2.0, nuclear laser medicine S.r.l) | Detected | Not detected | Total |
| Detected | 29 | 0 | 29 |
| Not detected | 5 | 4 | 9 |
| Total | 34 | 4 | 38 |
| 95% Confidence Interval | |||
| Sensitivity | 85.3% | 68.9% to 95.1% | |
| Specificity | 100% | 39.8% to 100% | |
| Positive predictive value (PPV) | 100% | – | |
| Negative predictive value (NPV) | 44.4% | 26.3% to 64.3% | |
| Accuracy | 86.8% | 71.9% to 95.6% | |
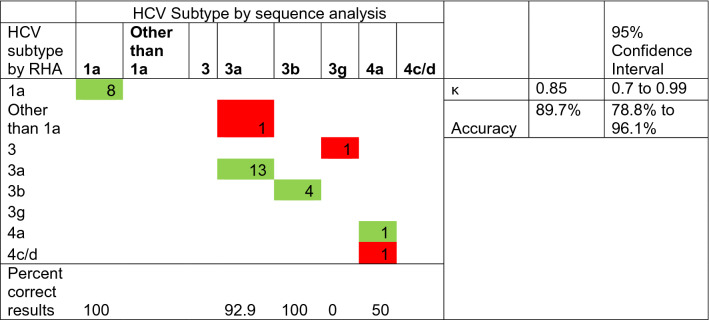
Further, out of the 29 samples processed for determination of HCV genotypes and subtypes, when compared to conventional sequencing, the accuracy of RHA results was 96.55% for determination of genotype (κ = 0.93) and 89.7% for subtype (κ = 0.85) (Tables 2 and 3). In our study the major subtype identified was 3a (48.3%) followed by 1a (27.6%). Other subtypes identified included 3b (13.8%), 3 g (3.5%) and 4a (6.9%).
Table 2.
Comparison of RHA and conventional sequencing approaches in determining HCV genotypes
Interpretation of κ-value: < 0.20 Poor, 0.21—0.40 Fair, 0.41—0.60 Moderate, 0.61—0.80 Good, 0.81—1.00 Very good agreement
*Cells in green denoting agreement
Table 3.
Comparison of RHA and conventional sequencing approaches in determining HCV subtypes
Interpretation of κ-value: < 0.20 Poor, 0.21—0.40 Fair, 0.41—0.60 Moderate, 0.61—0.80 Good, 0.81—1.00 Very good agreement
*Cells in green denoting agreement. Cells in red denoting disagreement
In this study, we evaluated a commercially available RHA for determination of HCV genotypes and subtypes. In this test, 5’ NCR and Core regions were targeted for generation of biotinylated amplicons using RT-PCR. These were hybridised to specific probes bound on a nitrocellulose strip by a poly T-tail, which formed purple-coloured precipitate upon reacting with the streptavidin–alkaline phosphatase complex. HCV is a positive-sense RNA virus with a single large Open Reading Frame (ORF) [9]. It is a highly mutagenic virus with rapid generation of different strains owing to its defective RNA-dependent RNA polymerase [10]. Yet, identical regions of co-linear genes are present across genotypes [11]. Based on this feature, partial sequences from sub-genomic regions were utilised to identify different HCV genotypes and subtypes [12]. While the 5’ NCR is most preferred for diagnosis of the HCV genotypes as it is highly conserved (92–98%), it cannot be utilised for accurate determination of subtypes of HCV due to insufficient sequence variation [13]. On the other hand, when used in conjunction with the Core region, the sequence information is sufficient to accurately determine not only the genotypes but also the subtypes of HCV [5, 14]. The other regions utilised for this purpose are the E1 and E2/NS1 as well as the NS5B regions. However, these regions are highly variable. Cia et al. observed that the amplification rate of sub-genomic regions is influenced by the viral load as well as the sequence conservations. The 5’NCR-Core regions are more conserved which gives better amplification and hence higher accuracy to determine the HCV genotypes and subtypes [15]. Another reason may be the lack of conservation in the primer-binding sites of the other regions that cause inadequate amplification when regions other than the 5’NCR-Core regions are targeted. Thus, the current research studied the 5’NCR-Core region of the HCV for conventional sequencing. As shown in Table 1, the qualitative RT-PCR had a sensitivity and specificity of 85.3% and 100% respectively, with an accuracy of 86.8%. The lower sensitivity of qualitative RT-PCR compared with quantitative RNA analysis can be explained by viral load of < 103 IU/mL in these samples. For four samples in which HCV RNA was not detected qualitatively or quantitatively, a history of completing course of directly acting antivirals was present.
As highlighted in Table 2, the accuracy of RHA for predicting the genotypes correctly was 96.6% with a κ score of 0.93. The results were 100% concordant for correctly determining the genotypes 1 and 4, while it was 94.7% in case of genotype 3. This is in contrast to a study where greater inaccuracy was observed in correctly determining Genotype 1 and 6 as compared to other genotypes [1].
For the prediction of subtypes, a concordance of 100% was observed both for subtypes 1a and 3b. But it was 92.9% and 50% in case of subtypes 3a and 4a respectively as shown in Table 3. While subtype 3a was misidentified as genotype 1 in one sample, one of the case of subtype 4a was misidentified as 4c/4d by RHA. The subtype 3 g though correctly identified up to genotype level could not be delineated into correct subtype by RHA. Thus, for subtype determination the overall accuracy was 89.7% with a κ value = 0.85. This may be explained by the fact that minor nucleotide variations among isolates in different geographical regions exist throughout the world, which may lead to misidentification when using a nucleotide pre-fixed technique such as RHA.
Thus it warrants caution when interpreting the result of RHA for determination of HCV genotypes and subtypes especially in the backdrop of low viral loads. This also highlights the importance of conventional sequencing techniques particularly for subtype determination. Zeuzem et al. in their study on 61 isolates found that RHA correctly identified each hepatitis C virus genotype (1, 2, and 3) but was insufficient for subtypes determination as HCV-1a isolates were incorrectly identified as HCV-1b and vice versa and HCV-2c isolates were misinterpreted as HCV-2a [16]. Similarly, Stelzl et al. in a similar study reported greater accuracy of sequencing based methods for determining HCV subtypes [17]. Knowledge about prevalent genotype and subtype in a particular geographical location is also important in order to choose the RHA which is able to clearly distinguish between them. In our study, isolates with subtype 3 g and 4a were also observed which could not be accurately identified by RHA. This may lead to inaccurate treatment initiation. This is echoed in a study on 134 subjects analysing the consequences of incorrect identification of HCV genotypes and subtypes where the results were found to be discordant in 15.7% cases which would have led to prescription of potentially suboptimal treatment regimens [18].
India being a resource poor country with a large population the benefits of any diagnostic test need to be weighed against the risks before their implantation and protocols to prevent inappropriate treatment regimens must be optimised accordingly. Thus, constant and vigilant quality monitoring is extremely essential when using such diagnostic techniques.
With the recent introduction of pan-genotypic drugs for treatment of HCV infection, the determination of genotype and subtype is mostly left out due to monetary as well as logistical issues. However, it is extremely crucial to determine the correct genotype and subtype. This will help us in predicting the course of disease associated with a particular genotype or subtype and evaluating the presence of recombinant strains, mixed infections and relapse and re-infections especially against the backdrop of emerging drug resistance even in treatment naïve subjects. The circulation of uncommon genotype 4 and subtypes such as 3 g also warrants caution while preparing management protocols.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all the staff of departments of Microbiology and Gastroenterology at the All India Institute of Medical Sciences, Rishikesh. Above all we would like to thank all the patients who have kindly consented to be a part of study and made this work possible.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
This pilot study was done at a tertiary care centre in Uttarakhand protocol for which was approved by Institutional ethics committee (AIIMS/IEC/111/18).
Consent to participate
Informed written consent was obtained from all participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goletti S, Zuyten S, Goeminne L, Verhofstede C, Rodriguez-Villalobos H, Bodeus M, et al. Comparison of Sanger sequencing for hepatitis C virus genotyping with a commercial line probe assay in a tertiary hospital. BMC Infect Dis. 2019;19(1):738. doi: 10.1186/s12879-019-4386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceccherini Silberstein F, Di Maio VC, Aragri M, Ciotti M, Cento V, Perno CF. Hepatitis C virus gene sequencing as a tool for precise genotyping in the era of new direct antiviral agents. Hepatology. 2016;63(3):1058–1059. doi: 10.1002/hep.27895. [DOI] [PubMed] [Google Scholar]
- 3.Verma V, Chakravarti A. Comparison of 5’ Noncoding-Core with 5’ Noncoding Regions of HCV by RT-PCR : Importance and Clinical Implications. Curr Microbiol. 2008;57:206–211. doi: 10.1007/s00284-008-9175-z. [DOI] [PubMed] [Google Scholar]
- 4.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 5.Lole KS, Jha JA, Shrotri SP, Tandon BN, Prasad VGM, Arankalle VA. Comparison of Hepatitis C virus genotyping by 5′ noncoding region- and core-based reverse transcriptase PCR assay with sequencing and use of the assay for determining subtype distribution in India. J Clin Microbiol. 2003;41(11):5240–5244. doi: 10.1128/JCM.41.11.5240-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash S, Shukla S, Ramakrishna V, Jain A. Distribution of hepatitis C genotypes in Uttar Pradesh, India; rare genotype 4 detected. J Med Virol. 2018;90(12):1875–1881. doi: 10.1002/jmv.25277. [DOI] [PubMed] [Google Scholar]
- 7.International Committee on Taxonomy of Viruses (ICTV). Confirmed HCV genotypes/subtypes (2019) [Internet]. 2019 [cited 2019 Sep 15]. Available from: https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/634/table-1---confirmed-hcv-genotypes-subtypes-may-2019
- 8.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naggie S, Wyles DL. Hepatitis C. In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas and Bennett’s Principles and Practices of Infectious Diseases. 9. Philadelphia: Elsevier; 2020. pp. 2040–2071. [Google Scholar]
- 10.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81(7):1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 11.Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, et al. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: Proposals for standardization. Arch Virol. 1998;143(12):2493–2503. doi: 10.1007/s007050050479. [DOI] [PubMed] [Google Scholar]
- 12.Simmonds P, Smith DB, McOmish F, Yap PL, Kolberg J, Urdea MS, et al. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75(5):1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Weck KE. Hepatitis C virus genotyping: Interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J Clin Microbiol. 2002;40(9):3127–3134. doi: 10.1128/JCM.40.9.3127-3134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: Quasispecies and genotypes. Semin Liver Dis. 1995;15(1):41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 15.Cai Q, Zhao Z, Liu Y, Shao X, Gao Z. Comparison of three different HCV genotyping methods: Core, NS5B sequence analysis and line probe assay. Int J Mol Med. 2013;31(2):347–352. doi: 10.3892/ijmm.2012.1209. [DOI] [PubMed] [Google Scholar]
- 16.Zeuzem S, Rüster B, Lee JH, Stripf T, Roth WK. Evaluation of a reverse hybridization assay for genotyping of hepatitis C virus. J Hepatol. 1995;23(6):654–661. doi: 10.1016/0168-8278(95)80030-1. [DOI] [PubMed] [Google Scholar]
- 17.Stelzl E, Van Der MC, Gouw R, Beld M, Grahovac M, Marth E, et al. Determination of the hepatitis C virus subtype: comparison of sequencing and reverse hybridization assays. Clin Chem Lab Med. 2007;45(2):167–170. doi: 10.1515/CCLM.2007.043. [DOI] [PubMed] [Google Scholar]
- 18.Polilli E, Cento V, Restelli U, Ceccherini-Silberstein F, Aragri M, Di Maio VC, et al. Consequences of inaccurate hepatitis C virus genotyping on the costs of prescription of direct antiviral agents in an Italian district. Clin Outcomes Res. 2016;8:467–473. doi: 10.2147/CEOR.S106238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.