Abstract
Human papilloma virus genotype 16 (HPV-16), a predominant etiological cause of cervical cancer (CC) vary in inflicting oncogenicity according to their geographical distribution and mutational changes. With no published data from central India, the present study aimed to genetically analyze HPV-16 E6/E7 variant obtained from CC women of Chhattisgarh. In twenty one CC patients, PCR amplified E6/E7 genes were decoded by DNA sequencing to study phylogenetic relatedness, mutational changes and their in-silico effect on protein structure. E6 analysis revealed nineteen sequences exhibited intratypic variation. L83V mutation was observed in 76.2% sequences followed by S71C seen in 28.6% sequences. Mutations of E41G, A46G, F47V, R77S, L99V and Q107K were observed in three sequences each. C140 Stop codon mutation has caused early truncation of E6 in three sequences to produce the conformational structural change. In contrast, E7 was relatively more conserved showing D4E (4.7%), G88R (23.8%), I93T (9.5%) and C94S (9.5%) mutations. Other than L83V and S71C, E6 and E7 mutations were reported for the first time from India. E6/E7 nonsynonmous mutations have a spectrum of biological effect in progression of CC. Phylogenetic analysis revealed ten sequence belonged to Asian while eleven to European sublineage to show CC cases in Chhattisgarh are a mix of Asian and European lineage. Asian sequences showing higher frequency of L83V mutations and exclusive presence of S71C and C140 Stop codon mutations may be linked with higher oncogenicity. Various E6/E7 mutational data may prove useful for development of better diagnostic and vaccine for the region of Chhattisgarh.
Keywords: Human papilloma virus genotype 16, Cervical cancer, E6/E7 oncoprotein, Nonsynomous mutations
Introduction
Cervical cancer is the fourth most common cancer among women causing 604,100 cases and 341,831 deaths worldwide in 2020 [1]. In India, every year, 96,922 women are diagnosed with cervical cancer and 60,078 die from the disease [2]. HPV-16 is the most predominant genotype among 14 high risk HPV primarily responsible for causing 60–75% CC consequent upon their persistent cervix infection among women worldwide [3, 4]. Persistent infection of HPV-16 further depends on intratypic variants which confer variable degree of oncogenicity and disease outcome according to their geographical distribution of lineage, sublineage and mutational changes in E6 and E7 oncoprotein [3, 5]. Intratypic HPV-16 variants differ in nucleotide sequence by no more than 2% in the coding region and 5% in the non-coding region of the viral genome with respect to the prototype [3]. Whole genome sequencing of HPV-16 has classified its variants into four main lineage (A/B/C/D) and 10 sublineages namely A [A1-A3 (European) and A4 (Asian)], B [B1 (African-1, Afr 1a) and B2 (African 1, Afr 1b)], C (African-2, Afr 2a) and D [(D1, North American NA1), D2 (Asian-American, AA2) and D3 (Asian-American(AA1)]. The empirically defined differences of 1.0–10.0% and 0.5–1.0% were used in classifying lineage and sublineages, respectively [6].
HPV-16 E6, a transcriptional transactivator oncoprotein facilitates ubiquitin mediated proteosomal degradation of the tumor suppressor protein, p53. E7 oncoprotein bind and inactivate pRb to trigger the cell cycle progression [7]. Any nucleotide mutational changes in E6 and E7 oncoprotein may lead to altered biological function and could ultimately affect oncogenic potential of the respective HPV-16 variant strains [8]. Analysis of E6 and E7 gene of HPV-16 intratypic variants are thus very vital to measure and understand HPV associated risk for the development of invasive CC [9].
Although intratypic HPV-16 variants of various lineage, sublineage and mutational changes have been reported worldwide, in India, few studies have been published so far focusing north, east and southern part of India leaving Central India unstudied for analyzing intratypic HPV-16 E6/E7 mutations [3, 10, 11]. Considering no data published from central India, this study has been undertaken to characterize E6 and E7 open reading frame (ORF) of HPV-16 variants and the in-silico effect of the mutational changes on their protein structure from CC cases among women of Chhattisgarh visiting tertiary care center of All India Institute of Medical Sciences (AIIMS), Raipur, Chhattisgarh.
Materials and methods
Sample collection
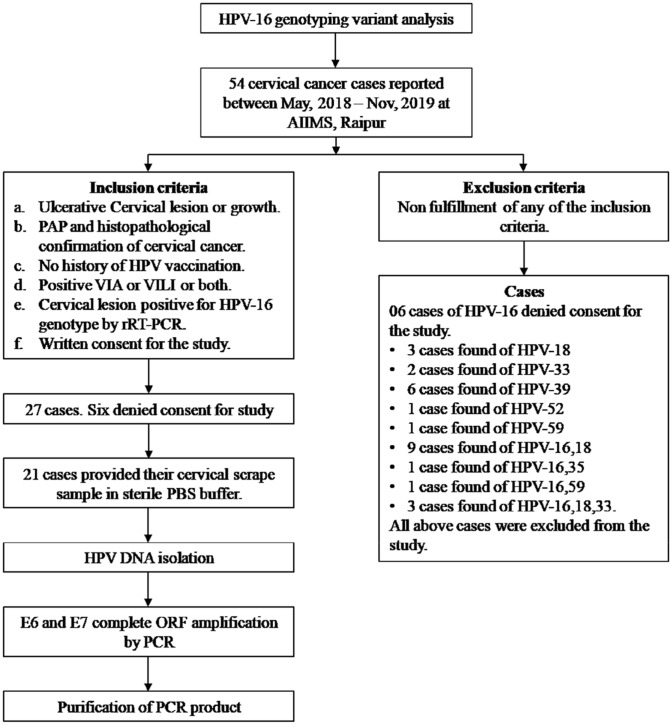
Fifty four CC cases reported between May, 2018 to Jun, 2019 in the department of Gynecology, AIIMS, Raipur were assessed for inclusion, exclusion and subsequent methodology (Fig. 1). Twenty one women fulfilling inclusion criteria and willingly signed patient consent were included in the study. Institutional ethical approval was obtained.
Fig. 1.
Schematic flow chart to explain inclusion, exclusion and total number of cases processed in the present study
Cervical scrape specimen were obtained on cervical brush in 1X phosphate buffered saline (PBS-pH7.2) from ectocervix and endocervix region containing exfoliated epithelial cells by Gynecologist from these twenty one women.
DNA isolation
HPV-16 DNA was isolated from cervical scrape specimens using QIAGEN DNA Mini Kit (catalogue number 51304), Germany as per the strict adherence to manufacturer instructions.
E6 and E7 ORF amplification
E6 gene (nucleotide [nt] 83 to 559) of 477 bp and E7 gene of 297 bp (nt 562 to 858) were amplified using primers, PCR condition and thermal cycling as described earlier [10].
HPV-16 E6/E7 variant ORF sequencing
All the PCR amplicons were purified and sequenced in ABI 3130 sequencer (ABI, USA) as described earlier [12] using Big Dye Terminator Cycle sequencing kit (Applied Biosystem, Thermo Scientific) having fluorescence labelled dideoxy nucleotide to determine DNA nucleotide sequence from both the directions.
NCBI BLAST analysis and gene bank submission
NCBI online BLAST 2.0 software server (http://www.ncbi.niti.gov/BLAST/) was used to confirm genotype and amplified region. The accession number provided for 21 E6 ORF sequences were MN987212-MN987228 and MN701613-MN701616. The accession number provided for 21 E7 ORF sequences were MN943490-MN943495, MN881074-881080, MN881087, MN881089-MN881091, MN881093, MW388714-MN388716.
Multiple sequence and phylogenetic analysis
This was performed by Clustal W and MEGA-X version 10.0.5 software Hi 9 using Neighbour-joining method and the Kimura 2- Parameter model with boot-strap resampling (1000replicates) [13] against reference sequence of HPV-16 NC_001526.4.
Inter Pro Scan and 3D Modeling of E6 and E7
HPV-16 E6/E7 protein sequences were retrieved from Uniprot (https://www.uniprot.org/uniprot/P03126.fasta). Motif and domain searches were made on EBI server (http://www.ebi.ac.uk/Tools/InterProScan/) using Inter Pro Scan [14]. 3D model of E6 protein (E6 wild type, E6 truncated and E6-958 variant) and E7 proteins (E7 wild type and E7-143 variant) were constructed using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER).
Results
E6 ORF sequence analysis
Nineteen sequences (19/21) showed at least one or more than one nucleotide changes in the ORF whereas two sequences matched the reference prototype. A total of twelve polymorphic sites consist of eleven (11/12) non-synonymous and one (1/12) synonymous mutation (silent) (Table 1, Fig. 2). Sixteen sequences (76.2%) contained L83V (TTG → GTG) AA change. Six sequences (28.6%) showed mutational changes of S71C. E41G, A46G, F47V, R77S, L99V and Q107K mutation were found respectively in at least three sequences. Interestingly, mutation of C140 Stop codon in three sequence has reduced truncated E6 gene by 12 AA (Table 1). The prominent non-synonymous AA changes of D4Y and E148K were observed in one sequence only. Silent mutation of V42V (GTA → GTG) was detected in one sequence.
Table 1.
Mutational changes in various amino acid (AA) positions of E6 gene in comparison to reference sequence NC_001526
| AA position | 4 | 41 | 42 | 46 | 47 | 71 | 77 | 83 | 99 | 107 | 140 | 148 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NC 001526 |
D GAT |
E GAG |
V GTA |
A GCT |
F TTT |
S TCT |
R AGA |
L TTG |
L TTG |
Q CAA |
C TGC |
E GAA |
| Nt position | 114 | 225 | 229 | 240 | 244 | 315 | 334 | 350 | 400 | 422 | 523 | 545 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV-801 | – | – | – | – | – | – | – |
V GTG |
– | – |
Stop TGA |
– |
| HPV-290 | – | – | – | – | – | – | – |
V GTG |
– | – |
Stop TGA |
– |
| HPV-142 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-254 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-270 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-659 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-01 | – |
G GGG |
– |
G GGT |
– | – | – | – | – | – | – | – |
| HPV-720 | – |
G GGG |
– |
G GGT |
– | – | – | – | – | – | – | – |
| HPV-740 | – | – | – | – | – | – | – |
V GTG |
V GTG |
K AAA |
– | – |
| HPV-713 | – | – | – | – | – | – | – |
V GTG |
V GTG |
K AAA |
– | – |
| HPV-02 | – | – | – | – |
V GTT |
– |
S AGT |
V GTG |
– | – | – | – |
| HPV-143 | – | – | – | – |
V GTT |
– |
S AGT |
V GTG |
– | – | – | – |
| HPV-958 | – |
G GGG |
V GTG |
G GGT |
V GTT |
– |
S AGT |
V GTG |
V GTG |
K AAA |
– | – |
| HPV-03 | – | – | – | – | – | – | – |
V GTG |
– | – | – | – |
| HPV-646 | – | – | – | – | – | – | – | – | – | – | – |
K AAA |
| HPV-487 |
Y TAC |
– | – | – | – | – | – |
V GTG |
– | – |
Stop TGA |
– |
| HPV-625 | – | – | – | – | – | – | – |
V GTG |
– | – | – | – |
| HPV-674 | – | – | – | – | – | – | – | – | – | – | – | – |
| HPV-725 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-726 | – | – | – | – | – |
C TGT |
– |
V GTG |
– | – | – | – |
| HPV-467 | – | – | – | – | – | – | – | – | – | – | – | – |
| U89348 | – | – | – | – | – | – | – |
V GTG |
– | – | – | – |
Fig. 2.
Mutational changes in E6 leading to change of various AA. Number shows the AA positions and all single capital letter indicate AA. A, Alanine; D, Aspartic acid; C, Cysteine; E, Glutamic acid; F, phenylalanine; G, Glycine; K, Lysine; L, Leucine; Q, Glutamine; R, Arginine; S, Serine; V, Valine
E7 ORF sequence analysis
E7 gene was found comparatively more conserved as only four non-synonymous and 2 synonymous mutations were observed (Table 2).The most frequent missense mutations observed was G88R in five strains (23.8%). Two missense mutation I93T and C94S in two sequences (9.5%) each and one sequence harbouring D4E mutation were also observed. Two sequences showed two silent mutations C91C and P92P respectively (Table 2).
Table 2.
Mutational changes in various AA position of E7 gene in comparison to reference sequence NC_001526
| AA position | 4 | 88 | 91 | 92 | 93 | 94 |
|---|---|---|---|---|---|---|
| NC 001526 |
D GAT |
G GGA |
C TGC |
P CCC |
I ATC |
C TGT |
| U89348 | – | – | – | – | – | – |
| Nt position change | 573 | 823 | 834 | 836 | 838 | 841 |
|---|---|---|---|---|---|---|
| HPV-646 | – |
R CGA |
– | – | – | – |
| HPV-713 | – | – | – | – | – | |
| HPV-958 | – |
R CGA |
– | – | – | – |
| HPV-01 | – | – |
C TGT |
P CCA |
T ACC |
S AGT |
| HPV-02 | – | – | – | – | – | – |
| HPV-03 | – | – | – | – | – | – |
| HPV-142 | – | – | – | – | – | – |
| HPV-254 | – | – | – | – | – | – |
| HPV-270 |
E GAG |
– | – | – | – | – |
| HPV-290 | – | – | – | – | – | – |
| HPV-487 | – | – | – | – | – | – |
| HPV-725 | – | – | – | – | – | – |
| HPV-467 | – | – | – | – | – | – |
| HPV-659 | – | – | – | – | – | – |
| HPV-143 | – | – |
C TGT |
P CCA |
T ACC |
S AGT |
| HPV-625 | – | – | – | – | – | – |
| HPV-674 | – | – | – | – | – | – |
| HPV-720 | – |
R CGA |
– | – | – | – |
| HPV-726 | – |
R CGA |
– | – | – | – |
| HPV-740 | – |
R CGA |
– | – | – | – |
| HPV-801 | – | – | – | – | – |
E6 and E7 ORF phylogenetic analysis
Phylogenetic analysis revealed all sequences belonged to A lineage (Figs. 3, 4). Ten sequences belonged to A4 (Asian) sublineage and 11 sequences to European sublineage of A1-A3. 81.8 and 70% L83V mutations were found in Asian and European lineage respectively. Second highest mutation of S71C and early truncation of E6 was observed only in Asian lineage while found absent in European lineage.
Fig. 3.
Phylogenetic analysis of 21 HPV E6 ORF sequences in comparison of various lineage and sublineage reference E6 gene sequences using the Neighbor-Joining algorithm in MEGA software (version 7.0). Bootstrap analysis of 1000 replicates were performed on each tree to determine the confidence. All twenty one sequences were marked in dark circle. Among, the 18 reference sequences NC 001526, U89348 and K02718 represented reference sequences of HPV-16. The rest of the sequences represents various lineage (A/B/C/D) and sublineage reference sequences namely MH921951 and LC368988 (A1), HQ537752 and AF536179 (A2), HQ537758 and HQ644236 (A3), AF534061 and LC456628(A4), AF536180(B1), HQ644298(B2), AF472509(C), HQ644257(D1), AY68579(D2), AF4022678(D3), AF472508 (Af-1)
Fig. 4.
Phylogenetic analysis of 21 HPV E7 ORF sequences and 18 reference representative E7 gene sequences of various lineage and sublineage conducted using the Neighbor-Joining algorithm in MEGA software (version 7.0). Bootstrap analysis of 1000 replicates was performed on each tree to determine the confidence. All twenty one sequences were marked in dark circle. The reference and various lineage and sublineage sequences are similar as described with their accession number in the legend of Fig. 3
Domain prediction and Modeling
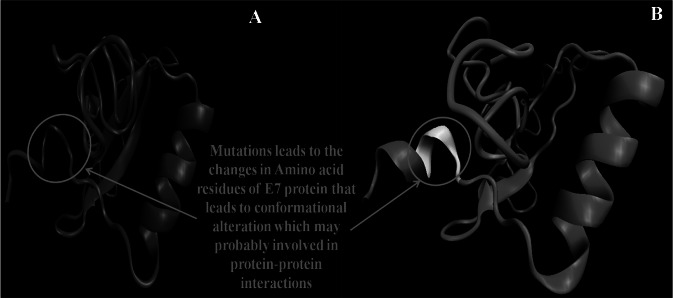
E6 protein showed two conserved domains; AA 6–86 and 87–115 (#IPR038575). These domains have two-conserved zinc-binding CXXC motifs (37 AA) ranged from AA 37–73 and 110–146. E7 protein showed the conserved domains: 1–98 AA (#IPR000148). 3D modeling of the E6 wild type, truncated, E6-958, E7 wild type and E7-143 proteins found C-score within the range of (− 5, 2), where a C-score of a higher value indicated a model with a higher confidence and vice-versa (Table 3). Reference and E6-958 showed that last 12 AA of second domain made a hook like structure to possibly interact to the AA residues (76, 79 and 80) of preceding domain (Fig. 5), which was found missing in the truncated E6 protein. 3D model of E6-958 has shown some conformational changes probably due to various point mutations (Table 1). 3D model of E7-143 has shown some AA variation at protein tail and may alter the conformation to involve in protein–protein interaction (Fig. 6).
Table 3.
Modeling parameters for HPV16 through I-TASSER
| S. no. | Selected protein model | Amino acids length | Estimated TM-Score | Estimated RMSD value (Å) | C-Score |
|---|---|---|---|---|---|
| 1 | E6 (Wild Type) | 1–158 | 0.90 ± 0.06 | 2.3 ± 1.8 | 1.33 |
| 2 | E6 (Truncated) | 1–146 | 0.89 ± 0.07 | 2.2 ± 1.7 | 1.29 |
| 3 | E6 (958 Variant) | 1–158 | 0.90 ± 0.06 | 2.3 ± 1.8 | 1.32 |
| 4 | E7 (Wild type) | 1–92 | 0.52 ± 0.15 | 7.1 ± 4.2 | − 1.55 |
| 5 | E7 (143 Variant) | 1–92 | 0.54 ± 0.15 | 6.8 ± 4.1 | − 1.41 |
Fig. 5.
3D Models of E6 proteins (Wild type, Truncated and Variant-958) of HPV-16. A E6 wild type protein, Yellow color indicates the chain of 12 AA, which was absent in the truncated protein. Last AA residue of wild type E6 may interact to the AA residues (76, 79 and 80). B E6 truncated protein in which last 12 AA were missing due to the early truncation. This truncation inhibiting the interaction with N-terminal domain. C E6-958 Variant protein in which AA changes due to point mutations could leads to conformational changes
Fig. 6.
3D mode A shows E7 wild type protein whereas B represent E7-143 variant showing some AA variation at the protein tail and may affect its conformations
Discussion
This study was done to understand the molecular epidemiology of E6/E7 HPV-16 variants from Chhattisgarh to unearth the mutational pattern, their effect on the protein structure and phylogenetic analysis to provide the explanation for the HPV-16 induced CC. The non-synonymous nucleotide changes observed in E6 and E7 were eleven and four respectively. These mutation were compared with previous reported studies.
The prominent E6 mutation of L83V was detected in 76.2% sequences. Earlier Indian studies has reported L83V frequency of 72.3% from North, East and South India, 100% from North and 70% from South and East India respectively [3, 10, 11]. High frequency of L83V was also reported from other countries like China, France, England and European countries [9, 15–18]. Finding of L83V as a sole mutation in one sequence while in others in co-existence with other co-mutations strongly indicate its role in invasive CC in women of Chhattisgarh while other co-mutations may act as precipitating factor in progression of the disease [15]. This correlation was in line of agreement with earlier studies on Swedish and British women [9, 16]. In contrast, Italian and South China women found L83V a low risk HPV-16 variant [8, 16]. Thus, our and other earlier reported studies adequately link L83V mutation with the progression of the CC. Our second most frequent E6 mutation of S71C (28.6%) was also reported by Pillai et al. This mutation occurs at site of T-cell epitope and may help virus in evasion of the immune response. Other than these two mutation, no other similarity were found between our and earlier Indian studies to substantiate the fact of existence of HPV-16 variant differ geographically and underscore the importance of such molecular epidemiological studies to unearth different variants of HPV-16. The other common mutation of D25E in E6 genomic region reported earlier from Japan and Thailand was not found in our sequences [19, 20]. The most important mutational finding of substitution of C → A at codon position 140 has changed the Cysteine (TGC) to stop codon (TGA). This stop codon at AA140 has resulted in the early truncation of the E6 protein by 12 AA.
Figure 5 illustrated the 3D model of E6 wild type, E6 truncated and E6-958 variant. Among E6 and E6-958, the presence of last 12 AA formed a hook like complete structure at C-terminal domain to form the interaction channel with N-terminal domain. This particular interaction is found missing in E6 truncated variant due to absence of these last 12AA. E6 protein zinc binding CXXC motif is of 37 AA encompassing AA37-73 and 110–146. These CXXC motifs interact to E7 carboxyl terminus to trigger a cellular defense mechanism known as “trophic sentinel response”[21]. This “trophic sentinel response” triggers has the characteristics of eliminating the deviant cell (having the aberrant cellular and / or viral DNA synthesis in differentiated keratinocytes that presumably lack environmental mitogen stimulation) through cell type specific abortive processes including cell death, differentiation and senescence [21].
So, truncation of 110–146 motif prevent CXXC motif interaction to E7 carboxyl terminus to inhibit triggering of “trophic sentinel response”. Hence, the absence of “trophic sentinel response” results in the uninterrupted aberrant cellular and/or viral DNA synthesis in differentiated keratinocytes, all leading to eventually help in the progression of CC(Table 4).
Table 4.
Amino acid changes and their biological significances
| Gene | AA changes | Biological significance | References |
|---|---|---|---|
| E6 | D4Y | Non synonymous mutation and occurred in the region that encode the alpha helix | [22] |
| E41G | Involved in zinc finger motif-1 | [23] | |
| V42 | Involved in zinc finger motif-1 | [23] | |
| A46G | Involved in zinc finger motif-1 | [23] | |
| F47V | Participate in the dimerization of the E6 N-terminal domain and important for the p53 degradation | [24] | |
| S71C | T-cell epitope | [3] | |
| R77S | Abrogation of growth arrest and lost the ability to maintain episomes | [23] | |
| L83V | E6 trancriptional transactivation, T-cell epitope, p53 degradation and important for the stability of E6 protein | [3, 25] | |
| L99V | Important for the stability of E6 protein | [25] | |
| Q107K | Involved in zinc finger motif-2 | [23] | |
| C140stop | E6 trancriptional transactivation, p53 binding and degradation, B and T cell epitope | [3] | |
| E148K | Significant to p53 binding and involved in PDZ domain interaction | [23] | |
| E7 | D4E | Increases electrophoretic mobility of E7 protein | [26] |
| G88R | pRB binding and degradation | [26] | |
| I93T | pRB binding and degradation | [26] | |
| C94S | Involved in zinc finger motif-1 | [26] |
This truncation could be an enhancement factor in disease progression by transcription transactivation, p53 binding and degradation. Other prominent missense mutation of R77S, L106V, Q107K, F47V, A46G, E41G and double mutation of D4Y have various biological effects of involvement in zinc finger motif-1, dimerization of E6 N-terminal domain, B and T cell epitope, transcriptional transactivation and p53 inactivation (Table 4). The specific mutation of L83V, F47V and C140Stop codon may help in progression of the cervical cancer by enhancing the p53degradation leading to inhibit activation of apoptosis promoting gene bax [3, 24]. This all ultimately led to inhibition of p53 induced activation of tumour suppressor gene p21. Thus, HPV-16 E6 variants effecting one or more of the biological function pose a high risk of persistence and progression of CC. We propose early screening of HPV-16 infected women for these mutations to initiate early treatment for preventing progression of cervical lesion to invasive CC.
In contrast, our finding of low polymorphism in E7 also reiterated the earlier findings that E7 is more conserved in comparison to E6 [3, 10, 11, 27]. The possible reason cited for E6 having higher mutational frequency than E7 may be more of a regional characteristic exhibited by the HPV-16 variants circulating in Chhattisgarh. Ours and other published studies from different part of the world have attributed geographical conditions and ethnic population as major factor responsible for showing variation in E6/E7 mutational pattern [3, 4, 10, 11, 27–30]. Interestingly, on the three sequences showing E6 truncation, no changes were seen in E7. Importantly, our reported four nonsynonymous mutation in E7 of D4E, G88R, I93T, C94S were also not reported earlier from India. These mutations have a biological role in electrophoretic mobility of E7, zinc finger motif-1, pRB binding and degradation in progression of CC (Table 4). The previously reported mutation of N29S from Asian countries was not seen in our sequences [28]. This observation was in line of finding of Pandey et al. and disagreement with Pillai et al. who have found this mutation albeit in one sequence. HPV-16 E6/E7 common mutation of D32E/N29S reported earlier from Beijing was also not found in our study [29]. In contrast to our finding of more conserved E7, some studies have reported a high frequency of E7 mutation [3, 30]. However, these studies E7 reported mutations were found restricted to specific nucleotide positions in contrast to E6 where in more number of nucleotides show simultaneous mutational changes. Nonetheless, majority of the published studies have the finding of higher frequency of E6 mutation than E7 [3, 4, 10, 27, 28]. Hence, all these observations strongly point out mutational characteristic of HPV-16 E6/E7 be more of a regional characteristic of HPV-16 variant prevalent specifically in the ethnic population inhabiting particular geographical conditions.
E7 protein modelling on the other hand revealed that few AA mutational variation in E7 variants in comparison to reference E7 sequence may altered the conformation of E7 protein to involve more protein–protein interaction and have a biological role in electrophoretic mobility of E7, zinc finger motif-1, pRB binding and degradation to ultimately help in progression to CC (Table 4).
In E6 gene, the nucleotide position (Nt) 116 to 223, 245 to 313 and 423 to 522 were appeared conserved whereas in E7, conserved region was observed from Nt position 575 to 821. These conserved region may be an ideal target for silencing using either short interfering RNA or ribozyme treatment [31] and a vaccine candidate by exploring their immunogenic therapeutic role.
The E6 and E7 phylogenetic sequence analysis showed that the HPV-16 variant circulated in central India is a mixed herediatary linkage of Asian and European strains. In contrast to earlier Indian study of Pandey et al. which has found complete absence of Asian lineage in North India (Delhi, Aligarh), we have found both Asian(47.6%) and European(52.4%) lineage in our study. The higher frequency of L83V in Asian lineage and complete absence of truncated sequence in European lineage showed that Asian lineage be more oncogenic in central part of India.
The limitation of the present study includes analysis of lower number of CC cases. However, considering the various reasons like majority of Chhattisgarh women population belongs to tribals, low literacy rate, unawareness of CC and its risk factor, unemployment, unwillingness to visit gynecologist for periodical gynecological examination due to various local myths and unwillingness for participation in present study eventually led to lower number of CC cases for analysis of HPV-16 variants. Nevertheless, low number of twenty CC cases has also been reported from the Republic of Congo [27]. Moreover, considering the aspect of no molecular epidemiology data from Central India, analysis of HPV-16 intratypic variants to unravel the mutational changes in entire E6 and E7 gene of HPV-16 variants and their potential association with CC progression in HPV infected host cell appeared to be useful in providing the insight of HPV-16 intratypic strains circulating in Chhattisgarh.
In conclusion, our study has found high prevalence of L83V mutation followed by S71C in E6 and other first time reported mutation from India to propose that L83 V mutation may prefer to cause oncogenic progression in co-existence of other mutations found in the study. The unreported mutations of D4Y, E41G, A46G, F47T, R77S, L99V, Q107K, E148K in E6 gene and D4E, I93T, C94S in E7 have the various degree of biological effect in development of invasive and metastatic cervical cancer. Phylogenetic analysis revealed mixed circulation of Asian and European lineage. Finding of higher L83V and exclusive mutation of S71C and truncation mutation further propose higher oncogenecity associated with Asian lineage. The mutational epidemiological and phylogeny finding may prove a baseline data for future studies pertaining to periodical monitoring of HPV-16 variants using more number of cases and development of diagnostic, vaccine and other therapeutic approaches.
Acknowledgements
We sincerely thank Ms Priyanka Singh and Ms Somya Sharma, both Research Assistant, Viral Research Diagnostic Laboratory, AIIMS, Raipur for their technical support in the study.
Authors contribution
Conceptualization: SSN, AB, PD, SA; Methodology: SSN, KS, PS, PA; Formal analysis and investigation: SSN, KS, PS, PA, NH; Writing - SSN, PS; Original draft preparation: SSN, PS; Writing - review and editing: SSN, AB, SA; Funding acquisition: SSN; Supervision: SSN.
Funding
This study has been financially supported by SERB-DST fund (EMR/2016/005428).
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Yes.
Consent to participate
Yes.
Consent for publication
Yes.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. https://gco.iarc.fr/today/data/factsheets/cancers/23-cervix-uteri-fact-sheet.pdf. Int. Agency Res. Cancer. Cervix uteri Globocan fact sheet, 2020. https://gco.iarc.fr/today/data/factsheets/cancers/23-cervix-uteri-fact-sheet.pdf.
- 2.ICO/IARC. India: Human Papillomavirus and related cancers, fact sheet 2019. ICO/IARC Information Centre on HPV and Cancer. https://hpvcentre.net/statistics/reports/IND_FS.pdf. 2020.
- 3.Pillai MR, Hariharan R, Babu JM, Lakshmi S, Chiplunkar SV, Patkar M, et al. Molecular variants of HPV-16 associated with cervical cancer in Indian population. Int J Cancer. 2009;125:91–103. doi: 10.1002/ijc.24322. [DOI] [PubMed] [Google Scholar]
- 4.Negi SS, Bhargava A, Singh P, Aggarwal S, Hussain N, Das P. Predominance of high-risk human papillomavirus genotype 16 and 39 in women with premalignant and malignant cervical pathology from Raipur, Chhattisgarh: Clinical evaluation of tagging oligonucleotide cleavage and extension mediated genotyping assay. Indian J Med Microbiol. 2019;37:255–262. doi: 10.4103/ijmm.IJMM_19_162. [DOI] [PubMed] [Google Scholar]
- 5.Rader JS, Tsaih S-W, Fullin D, Murray MW, Iden M, Zimmermann MT, et al. Genetic variations in human papillomavirus and cervical cancer outcomes. Int J Cancer. 2019;144:2206–2214. doi: 10.1002/ijc.32038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Pan Y, Gao W, Ke Y, Lu Z. Whole-genome analysis of human papillomavirus types 16, 18, and 58 isolated from cervical precancer and cancer samples in Chinese women. Sci Rep. 2017;7:263. doi: 10.1038/s41598-017-00364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan PKS, Ching WL, Tak HC, Li WWH, Lo KWK, Chan MYM, et al. Human papillomavirus type 16 intratypic variant infection and risk for cervical neoplasia in Southern China. J Infect Dis. 2002;186:696–700. doi: 10.1086/342048. [DOI] [PubMed] [Google Scholar]
- 9.Londesborough P, Linda HO, Terry G, Cuzick J, Wheeler C, Singer A. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int J Cancer. 1996;69:364–368. doi: 10.1002/(SICI)1097-0215(19961021)69:5<364::AID-IJC2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Pande S, Jain N, Prusty BK, Bhambhani S, Gupta S, Sharma R, et al. Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in North India. J Clin Microbiol. 2008;46:1060–1066. doi: 10.1128/JCM.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathish N, Abraham P, Peedicayil A, Sridharan G, Chandy G. HPV 16 E6 sequence variations in Indian patients with cervical neoplasia. Cancer Lett. 2005;229:93–99. doi: 10.1016/j.canlet.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 12.Negi SS, Singh P, Bhargava A, Chandrakar S, Gaikwad U, Das P, et al. Effective pragmatic approach of diagnosis of multidrug-resistant tuberculosis by high-resolution melt curve assay. Int J Mycobacteriol. 2018;7:228–235. doi: 10.4103/ijmy.ijmy_100_18. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma D, Bisht D. Secretory proteome analysis of streptomycin-resistant mycobacterium tuberculosis clinical isolates. SLAS Discov. 2017;22:1229–1238. doi: 10.1177/2472555217698428. [DOI] [PubMed] [Google Scholar]
- 15.Zhe X, Xin H, Pan Z, Jin F, Zheng W, Li H, et al. Genetic variations in E6, E7 and the long control region of human papillomavirus type 16 among patients with cervical lesions in Xinjiang, China. Cancer Cell Int. 2019;19:65. doi: 10.1186/s12935-019-0774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zehbe I, Wilander E, Delius H, Tommasino M. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 1998;58:829–833. [PubMed] [Google Scholar]
- 17.Cornet I, Gheit T, Iannacone MR, Vignat J, Sylla BS, Del Mistro A, et al. HPV16 genetic variation and the development of cervical cancer worldwide. Br J Cancer. 2013;108:240–244. doi: 10.1038/bjc.2012.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grodzki M, Besson G, Clavel C, Arslan A, Franceschi S, Birembaut P, et al. Increased risk for cervical disease progression of french women infected with the human papillomavirus type 16 E6-350G variant. Cancer Epidemiol Biomarkers Prev. 2006;15:820–822. doi: 10.1158/1055-9965.EPI-05-0864. [DOI] [PubMed] [Google Scholar]
- 19.Vaeteewoottacharn K, Jearanaikoon P, Ponglikitmongkol M. Co-mutation of HPV16 E6 and E7 genes in Thai squamous cervical carcinomas. Anticancer Res. 2003;23:1927–1931. [PubMed] [Google Scholar]
- 20.Matsumoto K, Yoshikawa H, Nakagawa S, Tang X, Yasugi T, Kawana K, et al. Enhanced oncogenicity of human papillomavirus type 16 (HPV16) variants in Japanese population. Cancer Lett. 2000;156:159–165. doi: 10.1016/S0304-3835(00)00457-2. [DOI] [PubMed] [Google Scholar]
- 21.Munger K, Baldwin A, Edward KM, Hayakawa H, Nguyen CL, Owen M, Grace M, Huh KW. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Li Q, Huang J, Li J, Yang F, Min X, et al. E6 and E7 gene polymorphisms in human papillomavirus Type-6 identified in Southwest China. Virol J. 2019;16:114. doi: 10.1186/s12985-019-1221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster SA, Demers GW, Etscheid BG, Galloway DA. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Cherry JJ, Dineen JV, Androphy EJ, Baleja JD. Determinants of stability for the E6 protein of papillomavirus type 16. J Mol Biol. 2009;386:1123–1137. doi: 10.1016/j.jmb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boumba LMA, Hilali L, Mouallif M, Moukassa D, Ennaji MM. Specific genotypes of human papillomavirus in 125 high-grade squamous lesions and invasive cervical cancer cases from Congolese women. BMC Public Health. 2014;14:1320. doi: 10.1186/1471-2458-14-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gheit T, Cornet I, Clifford GM, Iftner T, Munk C, Tommasino M, et al. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol Biomarkers Prev. 2011;20:1315–1321. doi: 10.1158/1055-9965.EPI-10-1187. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Zhao J, Liao Q. Association between human papillomavirus (HPV) type 16 infection and E6/E7 gene variant and the cervical lesions in Beijing. Chin J Exp Clin Virol. 2007;21:32–34. [PubMed] [Google Scholar]
- 30.de Boer MA, Peters LAW, Aziz MF, Siregar B, Cornain S, Vrede MA, et al. Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol Oncol. 2004;94:488–494. doi: 10.1016/j.ygyno.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.