Abstract
Evidence for transcription factor involvement in the initiation of DNA replication at certain replication origins in Saccharomyces cerevisiae mainly comes from an indirect assay which measures the mitotic stability of plasmids containing an autonomously replicating sequence (ARS), a selectable marker gene, and a centromere. In order to eliminate the effect of transcription factor binding to the selectable marker gene or centromere in such assays, we have adapted the DpnI assay to directly measure ARS replication activity in vivo by using ARS plasmids devoid of extraneous transcription elements. Using this assay, we found that the B3 element of ARS1, which serves as a binding site for the transcription factor Abf1p, does not stimulate ARS activity on plasmids lacking a centromere and a selectable marker gene. We also found with such plasmids that exogenous expression of the strong transcriptional activators Gal4 and Gal4-VP16 inhibited the replication activity of ARS1 when B3 was replaced by the Gal4 binding site, although these activators had previously been shown to stimulate replication activity in the stability assay. Moreover, a chromosomally inactive ARS, ARS301, which was active by itself on a plasmid, was inactivated by placing an Abf1p binding site in its vicinity. These results indicate that the sequences surrounding the ARS as well as properties of the ARS element itself determine its response to transcription factors.
Saccharomyces cerevisiae provides an excellent system for the analysis of eukaryotic replication origins for several reasons. First of all, the replication origin can be identified on a functional basis by analyzing it for DNA sequences that replicate in an autonomous fashion as plasmid DNA in yeast cells (autonomous replicating sequence [ARS]). When a DNA fragment with origin activity is ligated to a selectable marker gene, the resulting plasmid can transform yeast cells with high frequency (19, 43). Secondly, relative replication activity is easily estimated by measuring the mitotic stability of each ARS plasmid (24). This method has revealed that replication origins of budding yeast are compact (100 to 200 bp), with a modular structure consisting of an essential “core” sequence and auxiliary sequences which modulate replication activity. For example, ARS1, one of the best-characterized origins, contains two elements, A and B (10, 30). The A element contains a small sequence that is essential for origin activity and conserved among all ARS sequences (ARS consensus sequence [ACS]) (32). The A element functions as a recognition site for the origin recognition complex (ORC) involved in the initiation of DNA replication (3, 4). The B element consists of three subelements, B1, B2, and B3, each of which contributes to efficient replication although none is essential (30). One function of the B1 element is to enhance the binding of the ORC to the A element (34–36). The function of B2 remains unclear. The B3 element is a binding site for the transcription factor Abf1p (30). Other ARS elements so far analyzed are also characterized by the presence of an A element containing the ACS and auxiliary B elements (22, 23, 34, 48). Some of the B elements are interchangeable between different ARSs, although the sequences themselves are not well conserved (23, 34, 48). B3 can be replaced by the binding site for other transcription factors, like Rap1 or Gal4 (30). Moreover, the acidic activation domains of a number of transcription factors have been shown to activate replication when they were tethered to ARS1 (25).
For the analyses mentioned above, the mitotic stability of the ARS plasmid was used to measure the replication activity of each ARS element. The stability of the ARS plasmid depends not only on the replication activity of the ARS but also on the efficiency of segregation of the plasmid into daughter cells after each cell division (24). Thus, the test plasmid is composed of a centromere sequence to ensure plasmid segregation and a selectable marker gene. However, these requirements pose the problem that such artificially placed elements might influence the transcription factor dependency of ARS activity. As selectable genes have their own promoters, the transcription factors which bind to these promoters might affect ARS activity. The same could be true for the transcription factor Cbf1, which enhances centromere activity by binding to the centromere sequence (9). These considerations are especially relevant to transcription factors that can stimulate ARS activity even when located far away from the ARS. Indeed, Abf1 binding sites can stimulate ARS121 function at a considerable distance from the ARS (50). In addition, the presence of a centromere on an ARS plasmid was found to dramatically decrease the copy number of the ARS plasmid, suggesting that the centromere itself affects the replication efficiency of ARS (49).
The development of the two-dimensional gel method to detect replicating intermediates has enabled us to analyze origin activity on chromosomes (6). Interestingly, only some ARS elements act as active origins in their native chromosome positions, whereas all chromosomal origins so far analyzed show ARS activity (13, 14, 18, 33). Therefore, the chromosome location of certain ARS elements appears to repress their replication by an unknown mechanism. However, an alternative explanation would be that inactive origins on the chromosome are activated on plasmids by transcription factors that bind outside of the origin.
As a first step towards resolving these issues, we adapted the DpnI assay to directly measure the replication activity of ARS without the need for a selectable gene or centromere on the plasmid. Using the DpnI assay, we showed that transcription factor modulation of ARS activity was significantly affected by neighboring sequences. We also found that an inactive origin on the chromosome, ARS301, is active on the plasmid in the absence of other elements. Thus, its activity is apparently inhibited on the chromosome. As introduction of an Abf1p site into the ARS301 plasmid resulted in inhibition of replication activity from this origin in the DpnI assay, transcription factor binding near ARS sequences may provide one likely mechanism for the suppression of origin activity on the chromosome. These results strongly suggest that transcription factors modulate replication origin activity in a context-dependent manner.
MATERIALS AND METHODS
Strains and media.
SFY526 (MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 URA3::GAL1-lacZ′) was used for the DpnI assay and the β-galactosidase assay. YPD medium for yeast culture contained 2% glucose, 2% polypeptone, and 1% Bacto-Yeast Extract (Difco). YPAD was YPD supplemented with 20 mg of Ade per liter. Synthetic complete medium (SCM) for yeast culture consisted of glucose (2% [wt/vol]), yeast nitrogen base without amino acids (6.7 g/liter), isoleucine, lysine, tyrosine (30 mg/liter), arginine, histidine, methionine, tryptophan, uracil (20 mg/liter), valine (150 mg/liter), leucine (100 mg/liter), phenylalanine (50 mg/liter), threonine (200 mg/liter), and adenine (20 mg/ml).
Plasmids.
For the DpnI assay, only plasmids purified by two centrifugations on CsCl density gradients were used. The control plasmid for the DpnI assay, pHSG398− (46), was prepared from an Escherichia coli dam3 mutant strain whose DNA adenine methylase is defective. Plasmids pARS1/WTA, pARS1/Gal4#7, pARS1C/756,758, pARS1/LexA, pARS1/LexA,798-805, and pARS1C/798-805 were described previously (30). The plasmids pHK801 (WTA), -802 (mB3), -803 (B3/Gal4), -804 (B3/LexA), -805 (B3/LexA, mB2), and -806 (mB2) were constructed by inserting the EcoRI-HindIII fragments containing ARS1 from pARS1/WTA, pARS1C/756,758, pARS1/Gal4#7, pARS1/LexA, and pARS1/LexA,798-805, and pARS1C/798-805 between the EcoRI and HindIII sites of pBluescript SK(+) (pSK+) (Stratagene), respectively. Plasmid pHK801−A (ΔA) was constructed by removing the BglII-EcoRI fragment containing the A element from pHK801. The plasmids pYT528 and pYT530 were constructed by inserting the fragments containing ARS and CEN4 from pARS1/WTA and pARS1/Gal4#7, respectively, into pYC11 (45). The plasmids pARS1/Gal4X3G and pARS1/Gal4X5G were constructed by inserting in tandem two and four copies of the oligonucleotides containing high-affinity Gal4 binding sites (5′-TCGAGCGGAAGACTCTCCTCCGG-3′ and 5′-TCGACCGGAGGAGAGTCTTCCGC-3′) (17a, 28, 30) into the SalI site of pARS1/Gal4#7. Then the fragments containing ARS1 and CEN4 were transferred into pYC11 to give pYT530X3G and pYT530X5G, respectively. The pHK006 plasmid expressing the Gal4-VP16 fusion protein was constructed by inserting the XhoI-BamHI fragment of pSGGAL4-VP16 (17) containing the VP16 portion into pGBT9 (2). The pHK008 plasmid expressing the wild-type Gal4 protein was constructed by inserting the XhoI-BamHI fragment of pCL1 (15) containing part of Gal4 into pGBT9. The LexA-VP16 expression plasmid pHK035 was constructed by inserting the fragments containing the activation domain of VP16 from pSGGAL4-VP16 into pBTM116 (2) in a region located downstream of the coding sequence of the LexA DNA binding domain. The full-length coding sequence of rat c-Jun was obtained by PCR amplification with pRJ101 (37) as a substrate, and the resulting fragment was cloned into pGBT9 to give pHK009 expressing the Gal4–c-Jun fusion protein.
A 207-bp fragment containing ARS301 was obtained from a HindIII-BamHI digestion of pCS1 (39) (ARS301a) and cloned into pSK+ (pHK901). To make a plasmid containing ARS301 in the opposite orientation (pKH902), the same ARS301 sequence with BamHI and HindIII sites at each end was amplified by PCR with pCS1 as a substrate and cloned into pSK+. A 110-bp fragment (position 101 to 210 [39]) containing ARS301 was obtained by PCR and cloned into pSK+ to give pHK903 and -904.
DpnI assay.
Competent yeast cells were prepared as follows. Two days before transformation, a single colony of the strain SFY526 was inoculated into 10 ml of YPAD and grown for 1 day at 30°C. Ten milliliters of the overnight culture was inoculated into 40 ml of YPAD (total, 50 ml) and grown overnight at 30°C. The resulting overnight culture was inoculated into 200 ml of YPAD and grown for 3 h at 30°C. The culture was harvested and washed four times with sterile H2O and then resuspended in 25 ml of 0.1 M lithium acetate in TE buffer (pH 7.5), followed by gentle shaking for 45 min at 30°C. Then, 0.64 ml of 1 M dithiothreitol was added, and the suspension was shaken gently for 15 min at 30°C. The yeast cells were washed with 50 ml of ice-cold sterile H2O four times and with 20 ml of ice-cold 1 M sorbitol once. Finally, the cells were suspended in 0.7 ml of 1 M sorbitol and used for transformation.
The competent yeast cells were mixed with 1 μg each of the control plasmid DNA (pHSG398−), reporter plasmid DNA, and effector plasmid, transferred into an ice-cold disposable electroporation cuvette (0.2-cm gap) and pulsed at 1.5 kV, 25 μF, 400 Ω with a GENE Pulser (Bio-Rad). The transformed yeast cells were recovered by adding 1 ml of ice-cold 1 M sorbitol to the cuvette, 0.5 ml was inoculated into 5 ml of YPAD, and the cells were grown at 30°C for 12 h (fraction B). Then 200 μl of the overnight cultures was inoculated into 20 ml of SCM minus Trp and grown at 30°C for 36 h (fraction A). The cells harboring the effector plasmid grew about 10 generations during the 12- and 48-h periods after transformation as determined by the increase in the optical density of the culture at 600 nm.
From both fractions A and B, plasmids were isolated as follows. The cells were washed with sterile H2O four times and with 20 ml of a solution of 50 mM sodium phosphate (pH 5.6), 1.2 M sorbitol, 40 mM EDTA once. Then the cells were suspended in 1 ml of a solution containing 50 mM sodium phosphate (pH 5.6), 1.2 M sorbitol, and 10 mg of Zymolase 20T (Seikagaku Corporation)/ml and incubated at 37°C for 1 h. The cells were collected by centrifugation and suspended in 1 ml of 10 mM Tris-Cl (pH 8.0), 10 mM EDTA. For cell lysis, 200 μl of 10% sodium dodecyl sulfate (SDS) and 200 μl of 5 M potassium acetate were added and the suspension was allowed to stand on ice for 30 min. The supernatant was recovered by centrifugation at 13,000 × g for 15 min at 4°C. An equal volume of isopropanol was added to the supernatant and kept on ice for 30 min. The pellet was recovered by centrifugation and suspended with 0.4 ml of 10 mM Tris-Cl (pH 8.0), 10 mM EDTA. Then 20 μl of a 10-mg/ml concentration of RNase A was added and the solution was incubated for 1 h at 37°C. After the addition of 10 μl of 10% SDS and 10 μl of 20 mg of proteinase K/ml, the mixture was incubated for 1 h at 50°C. The DNA was precipitated by ethanol precipitation after extraction with phenol, phenol-chloroform, and chloroform and resuspended in 200 μl of TE (10 mM Tris-Cl [pH 8.0], 1 mM EDTA).
The DNA (10 μl) purified from the yeast was digested with 4 U of DpnI and 80 U of HindIII in 50 mM potassium acetate, 20 mM Tris acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol at 37°C overnight. The restriction endonuclease DpnI cleaves only methylated input DNA and leaves both newly replicated DNA and unmethylated control plasmid DNA intact. HindIII, which cuts the reporter and the control plasmid once, generates linearized fragments of the progeny DNA and the control plasmid DNA. Since the reporter plasmids are about 3.2 kb long and contain at least 16 DpnI cleavage sites, the DpnI-resistant molecules are easily separated from the DpnI-digested molecules during agarose gel electrophoresis. The reaction was terminated by the addition of loading buffer (2% Ficoll 400, 0.01 M Na2-EDTA, 0.1% SDS, 0.025% bromophenol blue, 0.025% xylene cyanol, pH 8.0), and the DNA was subjected to 0.8% agarose gel electrophoresis in TAE buffer (0.04 M Tris acetate, 0.02 M EDTA), transferred to a nylon membrane (Hybond N+ [Amersham]) by alkali blotting, and analyzed by hybridization. The probe DNA for hybridization was a mixture of two fragments: the NarI-NcoI fragment of pPyOICAT (31) containing parts of the chloramphenicol acetyltransferase gene for detection of the control plasmid and an SspI-BglI fragment of pSK+ containing the f1 ori for the detection of the reporter plasmids. The labeled DNAs were made by using rediprimer (Amersham).
The results of the DpnI assay were quantified either by densitometric scanning of X-ray film with a Quantity One scanner (PDI, Inc) or by BAS2000 image analysis (Fuji Photo Film Co. Ltd). After quantification, the value of the replicated band was normalized with that of the control band, and the relative replication activity was determined. At least two independent assays were performed for each experiment, and the average result was used to make the figures.
Plasmid stability assay.
The stability of the ARS was measured as described by Marahrens and Stillman (30). The loss rate of the each ARS plasmid was determined as described before (12).
RESULTS
DpnI assay.
To measure the replication activity of an ARS element without the influence of neighboring transcription factor binding sequences, we applied the DpnI assay, which has mostly been used to measure the replication activity of mammalian DNA viruses, such as simian virus 40 and polyomavirus. The DpnI assay relies on the observation that the DpnI restriction enzyme will only cut at its recognition sequence, GATC, when both DNA strands are fully methylated at position N6 of adenine by the Dam methylase of E. coli. Recognition sites that are methylated on only one strand (hemimethylated) or unmethylated on both strands (unmethylated) are refractive to cutting by DpnI. As yeast cells lack a Dam methylase, plasmids isolated from E. coli will become hemimethylated after one round of replication in yeast and, thus, will be resistant to DpnI restriction. In the DpnI assay, we measured the amount of replicated ARS plasmid in the cells 48 h after transfection (for details, see Materials and Methods). Since we needed neither a selectable marker gene nor a centromere sequence in this assay, we used plasmids containing only ARS1 or its derivatives (Fig. 1). Since the test plasmids contained multiple DpnI sites, only plasmids that had replicated all their DpnI sites were scored as replicated molecules.
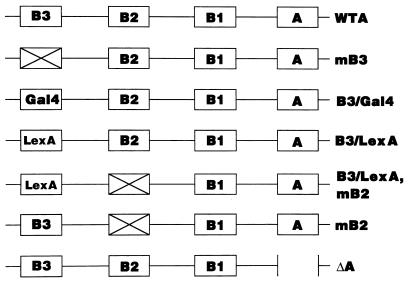
FIG. 1.
Schematic diagram of the ARS plasmids used in the DpnI assay. The A and B elements are shown by boxes. A cross represents the linker substitution of each element. In the B3/Gal4 and B3/LexA plasmids, the B3 element was replaced by the Gal4 and LexA binding sequence, respectively. In the mB3 plasmid, point mutations were introduced into the consensus sequence of the Abf1p binding site as described in Marahrens and Stillman (30). Each fragment containing an ARS was inserted into pSK+ as described in Materials and Methods.
When we transfected wild-type ARS1 (Fig. 1, WTA), we could detect a considerable number of DpnI-resistant molecules 48 h after transfection (Fig. 2A, lane 8), while no resistant molecules were observed 12 h after transfection (Fig. 2A, lane 4). This indicates that the DpnI-resistant molecules were those that had accumulated between 12 and 48 h after transfection. In other words, the DpnI assay mainly detected plasmids that had replicated during this period. However, even after 48 h, 40 to 60% of the ARS1 plasmid DNA was still DpnI sensitive (compare lanes 7 and 8), indicating that a significant portion had failed to replicate (see Discussion).
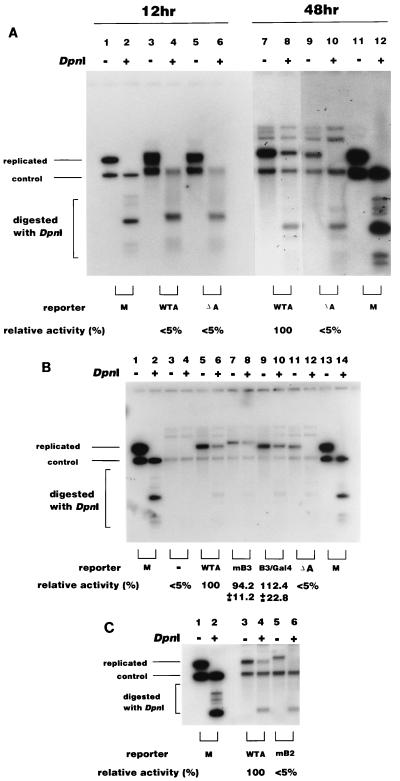
FIG. 2.
(A) DpnI assay for ARS plasmids in yeast cells. Plasmids containing wild-type ARS (WTA [pHK801], lanes 3, 4, 7, and 8) or deletions of the A element (ΔA [pKH801−A], lanes 5, 6, 9, and 10) were transfected into yeast cells together with the control plasmid. Twelve or 48 h after transfection, plasmid DNA was recovered, digested with HindIII or HindIII plus DpnI as indicated, and analyzed by Southern hybridization as described in Materials and Methods. The replication activities relative to the WTA plasmid were determined as described in Materials and Methods, and the average results of three independent experiments are indicated under each lane. <5%, replication was under the limit of detection. Mixtures of the WTA plasmid and control plasmids isolated from dam+ and dam3 mutant E. coli were also digested and used as markers (M; lanes 1, 2, 11, and 12). The bands visible above the replicated bands hybridized nonspecifically with the probe. (B) Effect of mutations in the B3 element. The indicated ARS plasmids (pHK801 [WTA], pHK802 [mB3], pHK803 [B3/Gal4], and pHK801-A [ΔA]) were transfected into yeast cells together with the control plasmid, and DNA was analyzed 48 h after transfection as described in Materials and Methods. The positions of DpnI-resistant replicated molecules and the control plasmid are shown. In lanes 3 and 4, no ARS plasmid was transfected. M, marker DNA as described for panel A. The relative replication activities were determined from three independent experiments as described for panel A. (C) Effect of a mutation in the B2 element. The replication activities of wild-type (WTA [pHK801], lanes 3 and 4) and the B2 mutant (mB2 [pHK806], lanes 5 and 6] were measured as described for panel A. The relative replication activities of three independent experiments are indicated under each lane. +, present; −, absent.
We tested the effect of deletion of the A element. The ARS plasmid harboring a deletion of the A element did not replicate at all (Fig. 2A, lane 10). This indicates that the DpnI-resistant plasmids were molecules that had undergone authentic DNA replication dependent on the A element.
Effect of mutations in the B elements on the DpnI assay.
The B3 element of ARS1 is the binding site for Abf1 and is required for efficient replication activity (30). Linker substitution or point mutations in B3 have been shown to cause a significant decrease in the stability of ARS plasmids (30). However, this result was obtained by the stability assay, which might be subject to interference from transcription factor binding to the centromere or selectable marker gene on the ARS plasmid. Since the DpnI assay enabled us to measure ARS activity without these elements, we tested the influence of the B3 mutations on plasmids containing only the ARS. We used two kinds of B3 mutation: point mutations (mB3 [Fig. 1]) and substitution mutations with a Gal4 binding site (B3/Gal4 [Fig. 1]). Surprisingly, both mutant ARS1 plasmids replicated as efficiently as the wild-type ARS1 plasmid in the DpnI assay (Fig. 2B, lanes 5 to 10). This result indicated that the B3 element was not functional in the absence of additional elements on the same plasmid.
We also analyzed the effect of mutations on the B2 element, which were previously reported to result in a significant loss of replication activity in the stability assay (30). We used an ARS1 plasmid harboring a linker substitution in the B2 element. This was the same mutation used by Marahrens and Stillman to show that ARS function is enhanced by the B2 element (30) except that our plasmid lacked a centromere and a selectable marker gene (mB2 [Fig. 1]). In contrast to B3 mutants, the mB2 plasmid barely replicated in the DpnI assay (Fig. 2C), showing that the B2 element is still required in the absence of other elements on the plasmid. In the stability assay, the plasmid having the B2 mutation still retained considerable replication activity, but the plasmid with B2 and B3 double mutations barely replicated (30). In our assay, the B2 mutation alone almost completely eliminated replication activity. Considering that B3 is not functional in our assay, the effect of the B2 mutation in our assay may be equivalent to that of the B2 and B3 double mutation in the stability assay. In summary, the DpnI assay showed that the B3 element does not contribute to the replication activity of an ARS plasmid lacking extraneous sequence elements. In contrast, the B2 element was found to be important for the activity of ARS under the same conditions.
Effect of exogenously expressed activators on the replication of ARS1 in the DpnI assay.
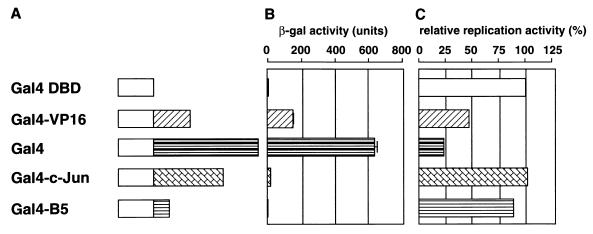
The B3 element can be replaced with the binding sites of other transcription factors, such as Rap1 or Gal4 (30). Recently, Li et al. showed that acidic transcription activation domains of a number of transcription factors fused to the DNA binding domain of Gal4 could stimulate the replication of ARS1 through a Gal4 binding site located in the B3 element (25). Since all the experiments were done with the stability assay, we analyzed the effect of exogenously expressed transcription factors on the replication activity of an ARS1 plasmid lacking other transcription factor recognition elements by the DpnI assay. We used a mutant ARS plasmid containing the Gal4 binding site instead of B3 (B3/Gal4 [Fig. 1]) and expressed the Gal4 fusion proteins exogenously. The fusion proteins used in our assay are shown in Fig. 3A, and their abilities to stimulate transcription are shown in Fig. 3B. All proteins contained the N-terminal 147 amino acids of Gal4, which comprises the sequence-specific DNA binding domain and the dimerization domain. Gal4-VP16 contains the acidic activation domain from VP16. The intact Gal4 protein also contains strong acidic activation domains for transcription activation. Both Gal4 and Gal4-VP16 show strong stimulation of transcription, with Gal4 being the stronger activator. Gal4–c-Jun carries the N-terminal 90-amino-acid region of c-Jun required for transcriptional activation in mammalian cells (44). This region only functions weakly in yeast cells (Fig. 3B). Gal4-B5 contains the DNA replication activation domain of transcription factor PEBP2αB1, which stimulates polyomavirus DNA replication but not transcription in mouse cells (11). Gal4-B5 did not stimulate transcription in yeast either (Fig. 3B).
FIG. 3.
Effect of expression of various Gal4 fusion activators on the replication of an ARS1 plasmid whose B3 element was replaced by a Gal4 binding site. (A) Schematic representation of the Gal4 fusion proteins. (B) Capacity of each fusion protein to stimulate transcription. The plasmids expressing the indicated Gal4 fusion proteins were transfected into the yeast SFY526 strain, which had the β-galactosidase gene linked to the GAL1 promoter, and the enzymatic activity of β-galactosidase in the transfected cells was assayed. (C) Capacity of each fusion protein to stimulate replication. The plasmids expressing the indicated Gal4 fusion proteins were cotransfected with the mutant ARS1 plasmid (B3/Gal4 [pHK803]) containing the Gal4 binding site in place of the B3 element. Replication activity was measured by the DpnI assay as described in Materials and Methods, and the results of at least two independent experiments and the relative amounts of the replicated molecules are indicated. Open bars and different patterns indicate the Gal4 DNA binding domain and the protein fused to the Gal4 DNA binding domain, respectively.
The effects of these transcriptional activators on the replication of ARS1 containing the Gal4 binding site are summarized in Fig. 3C. Expression of the Gal4 DNA binding domain did not affect the replication of the reporter plasmid, and the replication activity was almost the same as that of the wild-type ARS1 plasmid cotransfected with a plasmid expressing the Gal4 DNA binding domain (data not shown). The strong activators, Gal4 and Gal4-VP16, strongly inhibited replication, while the weak or nonfunctional activators, Gal4–c-Jun and Gal4-B5, did not have any notable effect on replication. The extent of inhibition roughly correlated with the strength of the transactivator in transcription. Gal4 and Gal4-VP16 inhibition of ARS1 activity occurred through the Gal4 binding site, since neither fusion protein could significantly inhibit the replication of a wild-type ARS1 plasmid lacking the Gal4 binding site (data not shown).
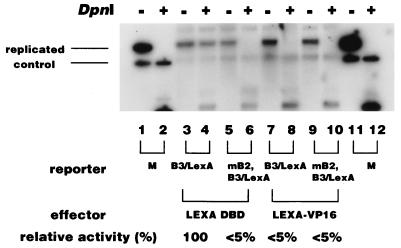
Marahrens and Stillman reported that a LexA-Gal4 fusion protein only slightly stimulated the replication of an ARS plasmid having a LexA binding site in B3, whereas significant stimulation was observed in the presence of a B2 mutation (30). Thus, we tested the effect of this activator on the replication of our reporter plasmid, whose B3 and B2 elements were replaced with the LexA binding site and mutated by a linker insertion, respectively (B3/LexA and mB2 [Fig. 1]). Consistent with the results shown in Fig. 2, the plasmid whose B3 site was replaced by the LexA binding site replicated well in the presence of the LexA DNA binding domain (Fig. 4, lane 4) whereas the additional presence of the B2 mutation almost abolished this replication activity (Fig. 4, lane 6). Expression of the LexA-VP16 fusion protein caused strong inhibition of the replication of the B3/LexA plasmid, and the level of inhibition was similar to that of the B3/Gal4 plasmid in the presence of Gal4-VP16 (Fig. 3C and 4, lanes 4 and 6). Moreover, LexA-VP16 did not stimulate the replication of the plasmid harboring the B2 mutation (Fig. 4, lane 10).
FIG. 4.
Effect of the LexA-VP16 fusion protein on the replication of the ARS1 plasmid containing the B2 mutation. The indicated ARS1 plasmids containing the LexA binding site in place of the B3 element (B3/LexA [pHK804] and B3/LexA, mB2 [pHK805]) were transfected together with plasmids expressing either the LexA DNA binding domain or the LexA-VP16 fusion protein, and replication activity was analyzed as described in Materials and Methods. The relative replication activities of two independent experiments are indicated under each lane. +, present; −, absent.
We also analyzed the effect of the Gal4 fusion protein in the stability assay. We used the ARS plasmid having CEN4 as a centromere, the LEU2 gene as a selectable marker, and ARS1 with the B3 region replaced by a single Gal4 binding site as an origin of replication (pYT530). The reporter plasmid with a single Gal4 binding site showed slightly decreased stability compared to the plasmid with the wild-type ARS1 sequence (pYT528) (Table 1). All of the fusion proteins were found to slightly increase the stability of ARS1 plasmids with a single Gal4 site, as reported previously (30, 39), whereas the wild-type ARS1 plasmid was not affected by Gal4-VP16 (Table 1). Thus, within the context of the stability assay, neither strong activators nor weak activators were able to inhibit replication.
TABLE 1.
Effect of GAL4 fusion proteins on the stability of ARS1 plasmids containing GAL4 binding sites
| Expt no. | Activator | Reporter | Mitotic stability loss per generation (%) (± SD) |
|---|---|---|---|
| 1 | pYT528 (WTA) | 5.2 ± 1.6 | |
| 2 | pYT530 (B3/Gal4) | 6.1 ± 0.9 | |
| 3 | pYT530X3G (B3/Gal4X3) | 6.1 ± 1.0 | |
| 4 | pYT530X5G (B3/Gal4X5) | 6.2 ± 1.0 | |
| 5 | Gal4-VP16 | pYT528 | 5.1 ± 1.1 |
| 6 | Gal4-DBD | pYT530 | 6.1 ± 2.2 |
| 7 | Gal4-VP16 | pYT530 | 5.1 ± 1.1 |
| 8 | Gal4 | pYT530 | 4.2 ± 1.5 |
| 9 | Gal4–c-Jun | pYT530 | 5.1 ± 1.5 |
| 10 | Gal4-RAD | pYT530 | 5.3 ± 1.6 |
| 11 | Gal4-DBD | pYT530XG3 | 5.4 ± 1.1 |
| 12 | Gal4-VP16 | pYT530XG3 | 7.2 ± 1.5 |
| 13 | Gal4 | pYT530XG3 | 6.3 ± 0.7 |
| 14 | Gal4–c-Jun | pYT530XG3 | 6.7 ± 1.3 |
| 15 | Gal4-RAD | pYT530XG3 | 7.4 ± 1.0 |
| 16 | Gal4-DBD | pYT530XG5 | 5.3 ± 2.1 |
| 17 | Gal4-VP16 | pYT530XG5 | 8.1 ± 1.7 |
| 18 | Gal4 | pYT530XG5 | 10.6 ± 1.4 |
| 19 | Gal4–c-Jun | pYT530XG5 | 7.4 ± 0.5 |
| 20 | Gal4-RAD | pYT530XG5 | 8.0 ± 1.2 |
These results indicated that an activator protein bound to the B3 element requires other elements outside of ARS to stimulate replication. Furthermore, binding of strong activators inhibits replication in the absence of other elements outside of ARS.
We also tested the influence of multiple Gal4 sites, since transcription factors synergistically activate transcription when their binding sites are multimerized. In the DpnI assay, we did not see a significant effect when the Gal4 site was multimerized. Even under these conditions, the same level of inhibition by Gal4 fusion proteins was observed (data not shown). Surprisingly, in the stability assay, multimerization of the Gal4 binding site caused an increase in the plasmid loss rate in the presence of Gal4 fusion proteins (Table 1, lines 7 to 10, 12 to 15, and 17 to 20). On the other hand, Gal4-DBD did not affect the loss rate (lines 6, 11, and 16), showing that the inhibitory effect depends on the nature of the protein domain fused to Gal4. Among the Gal4 fusion proteins tested, Gal4, which was the strongest transcriptional activator among those tested, showed the most severe effect. The reporter plasmid containing five Gal4 binding sites showed about a 2.5-fold increase in plasmid loss rate compared to the plasmid with a single Gal4 site (Table 1).
Activity of chromosomally inactive origins in the DpnI assay.
Certain ARS elements are functional as replication origins on plasmids but not on the chromosome. One possibility is that these ARS sequences are artificially activated by a neighboring element(s) positioned outside of the ARS sequence on the plasmid. The DpnI assay enabled us to test this possibility. We chose ARS301, which replicates as efficiently as ARS1 in the stability assay but poorly in its native chromosome position (14).
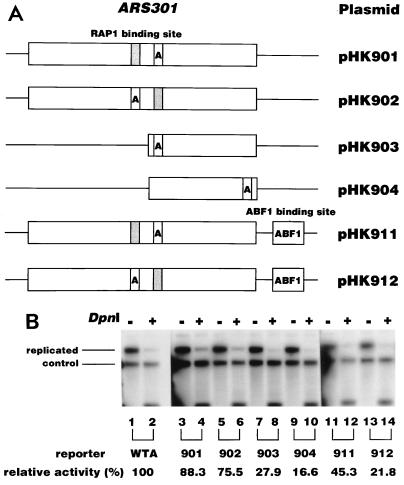
The 207-bp fragment of ARS301 was cloned into pSK+ in both orientations, and its replication activity was measured by the DpnI assay (Fig. 5A). This plasmid replicated as efficiently as ARS1 (Fig. 5B, lanes 2 and 4), showing that ARS301 possesses intrinsic replication origin activity. ARS301 contains a Rap1 binding site close to the essential ARS core sequence (Fig. 5A). Deletion of this portion caused a significant loss of replication activity (Fig. 5B, lanes 12 and 14), indicating that the deleted region including the Rap1 binding site contributes to replication activity. To test the effect of transcription factors on replication activity, the Abf1 binding site was inserted close to ARS301 (Fig. 5A). The introduction of an Abf1 binding site on either side of ARS301 caused a severe loss of replication activity, indicating that Abf1 has a negative effect on ARS301 replication. We also found that another chromosomally inactive origin, ARS608 (41), was also active without the need for any other element on the plasmid. However, the replication activity of ARS608 was not affected by introduction of the Abf1 binding site (data not shown).
FIG. 5.
Replication activity of chromosomally inactive ARS301 in the DpnI assay. (A) Schematic representation of the ARS301 plasmids used in the assay. The box indicates the DNA sequence derived from the yeast chromosome. The position of the ACS is indicated by the box containing the letter A. The shaded box and the box containing ABF1 indicate the binding site for Rap1p and Abf1p, respectively. (B) Replication activity of the ARS301 plasmid. The replication activities of the wild-type ARS1 plasmid and the ARS301 plasmid indicated in panel A were analyzed by the DpnI assay as described in Materials and Methods. The results obtained 48 h after transfection are indicated. The replication activities relative to that of the WTA plasmid were determined from two independent experiments and are indicated under each lane. +, present; −, absent.
Thus, ARS301 and ARS608 are intrinsically active, suggesting that they are inactivated by a still-unknown mechanism in their native chromosome positions. The inhibitory effect of the Abf1p binding site on ARS301 may indicate an involvement of this transcription factor in the origin inactivation process on the chromosome.
DISCUSSION
The involvement of transcription factors in the regulation of the ARS activity in budding yeast has been well documented (25, 30, 50). However, most of these results were obtained with the stability assay, which requires the use of ARS plasmids containing transcription elements. The influence of these elements on DNA replication as measured by the stability assay has never been fully taken into consideration. In this paper, we have been able to examine the replication activity of ARS plasmids containing only the ARS element by using the DpnI assay, which directly measures the number of replicated molecules during a given period. Using the DpnI assay, we found that the effects of transcription factors on the replication activity of ARS are potentiated by elements outside of the ARS, as well as by the nature of the ARS itself.
The binding site for the transcription factor Abf1, which is found in a subset of ARS elements, was shown by the stability assay to stimulate the ARS activity of ARS1 and ARS121 (30, 51). However, we found that the B3 element of ARS1, which is the binding site for Abf1, was not functional for the stimulation of DNA replication in the absence of other elements outside of the origin, suggesting that Abf1 cooperates with other transcription factors bound to the promoter region of the selectable gene or centromere region to activate ARS1 replication. This was in contrast to the B2 region, which was functional in the absence of elements outside of the ARS, indicating that B2 plays a distinct role independent of B3 in the replication of ARS1. We also found that strong transcription activators, such as Gal4 and Gal4-VP16, inhibited the replication activity via the Gal4 binding site positioned within the B3 element. On the other hand, neither Gal4 nor Gal4-VP16 inhibited the replication activity in the stability assay. Rather, replication activity was stimulated (Table 1) (25), suggesting that sequences adjacent to ARS converted potential inhibitors of replication into activators in the stability assay. These results clearly indicated that elements outside of ARS1 influence the function of transcription factors bound to the B3 element of ARS1.
We also found that introduction of an Abf1 binding site on either side of ARS301 caused a severe reduction in replication activity (Fig. 5). Thus, Abf1 by itself acts as an inhibitor of replication of ARS301. On the other hand, the Abf1p binding site barely affected the replication activity of ARS1 and ARS608 in the DpnI assay. Recently, Lin and Kowalski showed that introduction of the B3 element of ARS1 into ARS305 also caused inhibition of replication in the stability assay (26). In addition, the stimulation of DNA replication by a transcription factor in the stability assay appears to be emphasized by mutation of the B2 element (25, 30). These results stress not only the importance of the surrounding elements for activation of DNA replication by transcription factors but also the importance of sequences even inside the ARS. It is interesting to note that Abf1 works both positively and negatively in transcription, depending on the nature of the promoter and the other transcription factors in the transcription assay (7, 8).
We observed that multimerization of the Gal4 binding site lowered ARS plasmid stability (Table 1). This result, taken together with the results obtained by Lin and Kowalski mentioned above (26), indicates that transcription factors can inhibit ARS activity to some extent as measured by the stability assay. Therefore, the inhibitory effect observed in the DpnI assay was not due to the type of assay but rather to differences in origin context.
How do (strong) activators inhibit the initiation of replication at ARS? In the case of the Gal4 fusion proteins shown in Fig. 3, their capacities to inhibit DNA replication correlated well with their strengths as transcription activators, suggesting that inhibition involves the machinery of transcription. Since some ARS elements, including ARS1, contain a TATA-like sequence, and as the TATA binding protein (TBP) was shown to bind to these sequences in vitro (27), the recruitment of the transcriptional machinery, including TBP, to the ARS may interfere with the binding of replication factors to the DNA. Alternatively, strong activators could induce abortive transcription from neighboring cryptic promoters, resulting in inhibition of replication. Indeed, transcription entering ARS1 inhibits DNA replication (42, 47). The presence of a selectable marker gene with strong promoter activity close to the ARS might counteract the recruitment of transcription machinery to the ARS itself, thereby relieving the negative effect of certain transcription factors on DNA replication. Recently, Hu et al. suggested that the chromatin remodeling induced by transcription factors could stimulate DNA replication (20). Therefore, it is also possible that transcription factors induce changes in chromatin structure that inhibit the initiation of DNA replication, depending on the context of the replication origin.
On the chromosome, origins are located either between or within transcription units (40). For example, ARS1 is located downstream of the TRP1 gene. Thus, the transcription factors bound to the regulatory region of nearby genes might affect the replication activity of each origin. In the case of ARS1 on the chromosome, mutation of B3 caused a decrease in origin activity (29). In addition, Li and his colleagues showed that the strong acidic transcriptional activators stimulated the origin activity of ARS1 on the chromosome (20, 25). In our assay B3 required elements extraneous to the ARS to stimulate ARS1-dependent DNA replication, and strong activators inhibited the replication of ARS1, indicating that elements around ARS1 are crucial for the positive role played by B3 in the modulation of origin activity on the chromosome. It should be noted that Li and his colleagues mutated the B1 and B2 elements to reduce basal origin activity to a level that allowed clear stimulation by transcription factors (20, 25). Considering our results, it would be interesting to test the effect of strong transcription factors on wild-type ARS1 activity.
Some ARS elements are not used as origins on the chromosome but function quite well on plasmids. For this reason, some ARS elements are believed to be inactivated on the chromosome (13, 14, 18, 33). However, it may also be that inactive ARS elements are artificially activated on plasmids by neighboring sequences. Our results with the ARS elements ARS301 and ARS608, which are inactive on the chromosome, indicated that both of these elements are active when present alone on the plasmid and are indeed inactivated on the chromosome by an unknown mechanism. Inhibition of replication activity by transcription factors in this study suggests that transcription factors could play a role in the suppression of origin activity on the chromosome.
The replication origins on the budding yeast chromosome replicate sequentially during S phase in a defined pattern, i.e., some origins initiate replication early, while others are activated late in the cell cycle (16, 52). For example, ARS1 initiates relatively early in S phase while ARS301 initiates late when located on a plasmid (5). ARS608 on the chromosome also initiates late, with low efficiency (41, 52). Interestingly, treatment of cells with hydroxyurea, which blocks the progression of replication forks from early replicating origins, or with the DNA-damaging agent methyl methane sulfonate resulted in the inhibition of late-replicating origins. A mutation of the check point gene, RAD53, which encodes a protein kinase, releases the initiation block induced by hydroxyurea and methyl methane sulfonate. In addition, the rad53 mutation was found to enhance the frequency of initiation at late and/or weak origins (38, 40). These results indicate that the block to late and/or inefficient origins is an active process and may be involved in the cell’s surveillance of S-phase progression. Rad53 is required for all transcriptional responses to DNA replication blocks or DNA damage (1), suggesting that certain transcription factors are under the control of RAD53. Indeed, the transcription factor Crt1 was recently shown to be under the control of RAD53 protein kinases (21). These data, in conjunction with our results, raise the possibility that transcription factors under the control of RAD53 kinase are involved in the suppression of initiation from late and/or inefficient origins on the chromosome.
We observed that only a small portion of the transformed plasmids replicated (Fig. 2A). At 48 h after transformation, 40 to 50% of the plasmids were still DpnI sensitive. Plasmid-containing cells grew about 10 generations during the 12- to 48-h period after transformation (see Materials and Methods). If the plasmid DNA replicated once per generation, the amount of DNA should have increased about 1,000-fold during this period. This means that less than 0.1% of the plasmid actually replicated during the 12- to 48-h period after transformation. In other words, more than 99.9% of the transformed plasmids were not competent for replication and remained in the unreplicated state. This might reflect the chromatin structure of the ARS plasmid immediately after transformation. If the transformed ARS formed a nucleosome complex before ORC binding, the ORC complex might not have been able to bind to the ARS. Since the number of ORC complexes is limiting (about 600 molecules of ORC2p per cell [36]) compared to histone protein content, only a small portion of the transformed ARS plasmids may have been bound by ORC. It is also possible that the transformed DNA replicated only a few times during the 10 generations of growth. Alternatively, only a portion of the plasmid DNA molecules may have gained entry to the nuclei. In any case, a more detailed analysis will be required to clarify this issue.
The DpnI assay has some weaknesses compared to the stability assay. First, high transformation efficiency is required. Second, the DpnI assay requires more manipulation than the stability assay. Third, it is difficult to detect the replication of large plasmids (>5 kb), probably because of their low transformation efficiency. In spite of these weaknesses, the DpnI assay provides a direct and complementary tool to measure the replication activity of the ARS itself. Using this assay, we have shown that transcription factors have the potential to modulate the replication activity of the ARS elements both positively and negatively, depending on the nature of the transcription factors that bind in and around the ARS. Further study with the DpnI assay should tell us more about the role of transcription factors in the regulation of DNA replication in yeast cells.
ACKNOWLEDGMENTS
We thank Bruce Stillman and J. F. X. Diffley for providing plasmids and Hisao Masukata for furnishing us with the protocol for the DpnI assay in fission yeast before publication.
This study was supported in part by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science and Culture of Japan (to Y.M.).
REFERENCES
- 1.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple check-points and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 2.Bartel P, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 3.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Bousset K, Diffley J F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 7.Buchman A R, Kornberg R D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990;10:887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchman A R, Lue N F, Kornberg D. Connections between transcriptional activators, silencers, and telomers as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai M, Davis R W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 10.Celinker S E, Sweder K, Bailey J E, Campbell J L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L-F, Ito K, Murakami Y, Ito Y. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol Cell Biol. 1998;18:4165–4167. doi: 10.1128/mcb.18.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dani M G, Zakian V A. Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci USA. 1983;80:3406–3410. doi: 10.1073/pnas.80.11.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dershowitz A, Newlon C S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993;13:391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J, Broach J R, Newlon C S, Huberman J A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal origins of replication. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields S, Song O-K. A novel genetic system to detect protein-protein interaction. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 16.Friedman K L, Brewer B J, Fangman W L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 17.Fujii M, Tsuchiya H, Seiki M. HTLV-1 Tax has distinct but overlapping domains for transcriptional activation and enhancer specificity. Oncogene. 1991;6:2349–2352. [PubMed] [Google Scholar]
- 17a.Giniger E, Varnum S M, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 18.Greenfeder S A, Newlon C S. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 1992;3:999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao C-L, Carbon J. High-frequency transformation of yeast plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci USA. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y-F, Hao Z L, Li R. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 1999;13:637–642. doi: 10.1101/gad.13.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M, Zhou Z, Elledge S J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 22.Huang R Y, Kowalski D. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 1993;12:4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R Y, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 25.Li R, Yu D S, Tanaka M, Zheng L, Berger S L, Stillman B. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol Cell Biol. 1998;18:1296–1302. doi: 10.1128/mcb.18.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lue N F, Kornberg R D. A possible role for the yeast TATA-element-binding protein in DNA replication. Proc Natl Acad Sci USA. 1993;90:8018–8022. doi: 10.1073/pnas.90.17.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 29.Marahrens Y, Stillman B. Replicator dominance in a eukaryotic chromosome. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 31.Murakami Y, Asano M, Satake M, Ito Y. A tumor promoting phorbor ester, TPA, enhances polyomavirus replication by activating the function of the viral enhancer. Oncogene. 1990;5:5–13. [PubMed] [Google Scholar]
- 32.Newlon C S. Yeast chromosome replication and segregation. Microbiol Rev. 1988;52:568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newlon C S, Lipchitz L R, Collins I, Deshpande A, Devenish R J, Green R P, Klein H L, Palzkill T G, Ren R B, Synn S, et al. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. . (Erratum, 130:235, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowley A, Cocker J H, Harwood J, Diffley J F X. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai M, Okuda A, Hatayama K, Sato S, Nishi S, Muramatsu M. Structure and expression of the rat c-jun messenger RNA: tissue distribution and increase during chemical hepatocarcinogenesis. Cancer Res. 1989;49:5633–5637. [PubMed] [Google Scholar]
- 38.Santocanale C, Diffley F X. A Mec-1 and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 39.Santocanale C, Diffley J F X. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;23:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 40.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 41.Shirahige K, Iwasaki T, Rashid M B, Ogasawara N, Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder M, Sapolsky R J, Davis R W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stinchcomb D T, Struhl K, Davis R W. Isolation and characterization of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 44.Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988;332:649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeshita S M, Sato M, Toda M, Masahashi W, Hashimoto-Goto T. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka S, Halter D, Livingstone-Zatchej M, Reszel B, Thoma F. Transcription through the yeast origin of replication ARS1 ends at the ABFI binding site and affects extrachromosomal maintenance of minichromosomes. Nucleic Acids Res. 1994;22:3904–3910. doi: 10.1093/nar/22.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theis J F, Newlon C S. Domain B of ARS307 is modular and contributes to chromosomal replication origin function. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tschumper G, Carbon J. Copy number control by a yeast centromere. Gene. 1983;23:221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- 50.Walker S S, Francesconi S C, Eisenberg S. A DNA replication enhancer in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiltshire S, Raychaudhuri S, Eisenberg S. An Abf1p C-terminal region lacking transcriptional activation potential stimulates a yeast origin of replication. Nucleic Acids Res. 1997;25:4250–4256. doi: 10.1093/nar/25.21.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]