Highlights
-
•
Overexpression of the miR-200b/200a/429 cluster prevents mammary tumor initiation.
-
•
miR-200s may prevent mammary tumor initiation by suppressing Spp1, Saa1 and Saa2.
-
•
Overexpression of miR-200s does not impair normal mammary ductal development.
Keywords: miR-200, Saa1, Saa2, Spp1, Mammary tumor initiation, Transgenic mice
Abstract
The miR-200 family consists of five members expressed as two clusters: miR-200c/141 cluster and miR-200b/200a/429 cluster. In the mammary gland, miR-200s maintain epithelial identity by decreasing the expression of mesenchymal markers leading to high expression of epithelial markers. While the loss of miR-200s is associated with breast cancer growth and metastasis the impact of miR-200 expression on mammary tumor initiation has not been investigated. Using mammary specific expression of the miR-200b/200a/429 cluster in transgenic mice, we found that elevated expression miR-200s could almost completely prevent mammary tumor development. Only 1 of 16 MTB-IGFIRba429 transgenic mice (expressing both the IGF-IR and miR-200b/200a/429 transgenes) developed a mammary tumor while 100% of MTB-IGFIR transgenic mice (expressing only the IGF-IR transgene) developed mammary tumors. RNA sequencing, qRT-PCR, and immunohistochemistry of mammary tissue from 55-day old mice found Spp1, Saa1, and Saa2 to be elevated in mammary tumors and inhibited by miR-200b/200a/429 overexpression. This study suggests that miR-200s could be used as a preventative strategy to protect women from developing breast cancer. One concern with this approach is the potential negative impact miR-200 overexpression may have on mammary function. However, transgenic overexpression of miR-200s, on their own, did not significantly impact mammary ductal development indicating the miR-200 overexpression should not significantly impact mammary function. Thus, this study provides the initial foundation for using miR-200s for breast cancer prevention and additional studies should be performed to identify strategies for increasing mammary miR-200 expression and determine whether miR-200s can prevent mammary tumor initiation by other genetic alterations.
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that regulate mRNA translation by binding to mRNAs and in turn, prevent mRNA translation [1,2]. miRNAs are originally transcribed as long primary transcripts and these primary transcripts are processed in two cleavage steps mediated by Drosha and Dicer that produce a mature miRNA of 19–25 nucleotides [3], [4], [5], [6], [7]. Mature miRNAs are incorporated into a complex known as the RNA-induced silencing complex (RISC) [4]. Most miRNAs direct RISC to target mRNAs where they induce mRNA degradation or repress translation [3,4,8,9]. miRNAs incorporated into RISC bind primarily to 3′-UTRs of target mRNAs using the miRNA seed sequence found between nucleotides 2 and 8 of the miRNA [3,4,[8], [9], [10]]. However, several reports indicate that miRNAs can also bind mRNAs independent of the seed sequence [11], [12], [13]. miRNAs have been implicated in regulating a number of cancers including breast cancer. One family of miRNAs implicated in breast cancer is the miR-200 family. This family consists of 5 members organized into two clusters, the miR-200b/200a/429 cluster and the miR-200c/141 cluster [14], [15], [16]. These five miRNAs have highly similar seed sequences and the miR-200b, miR-200c, and miR-429 share a common seed sequence (AAUACUG) while miR-200a and miR-141 share the same seed sequence (AACACUG) [17]. The most completely characterized function of the miR-200 family is their role in maintaining epithelial identity. miR-200 family members reduce the expression of mesenchymal transcription factors such as Zeb1/2, Twist1/2, Snai1/2 [16,[18], [19], [20]] and increase the expression of epithelial genes such as E-cadherin [21,22].
The most studied function of the miR-200 family in breast cancer is the suppression of epithelial to mesenchymal transition (EMT) and thus tumor migration and metastasis. Work from our lab and others showed that increased expression of miR-200s in murine and human mammary tumor cells inhibited migration and invasion in vitro and metastasis in vivo [23], [24], [25], [26], [27], [28], [29]. miR-200s have also been reported to influence proliferation, stem/progenitor cell number and the expression of immune regulatory molecules [23,25,27,[30], [31], [32], [33], [34], [35], [36], [37]].
While the role of miR-200s in reducing breast cancer progression is well established, less is known about the role of miR-200s in inhibiting mammary tumor initiation. In vitro culture systems with normal mammary epithelial cells can be utilized to study tumor initiation however, the artificial environment including the lack of appropriate cell-cell and cell-ECM contacts as well as the loss of natural fluctuations in hormones, growth factors, nutrients and oxygen may influence cellular transformation. Thus, tumor initiation studies are typically performed in animal models with well-established tumor onset characteristics. Only 8 studies have examined miR-200 alterations in vivo using transgenic ([13,38]) or knockout ([39], [40], [41], [42], [43], [44]) models and none of these models altered miR-200 expression in the mammary gland. Thus, the impact of miR-200s on mammary tumor initiation have not been investigated in a relevant, in vivo environment.
In this study we have shown that overexpression of the miR-200b/200a/429 cluster in mammary epithelial cells almost completely suppressed mammary tumor development induced by the type I insulin-like growth factor receptor (IGF-IR). Saa1, Saa2, and Spp1 were identified as genes that may contribute to mammary tumor initiation that were suppressed by miR-200b/200a/429 overexpression. miR-200 overexpression in the mammary gland did not impair mammary ductal development suggesting that therapeutic use of miR-200s would not significantly impact normal mammary gland function.
Methods
Animals and ethics
Animals were housed and cared for following guidelines established by the Central Animal Facility at the University of Guelph and the guidelines established by the Canadian Council of Animal Care. This study was approved by the Animal Care Committee at the University of Guelph (AUP #3994).
TRE-200ba429 transgenic mice were generously donated by Dr. Ri Yiu (Northwestern University Feinberg School of Medicine, Chicago, IL, USA) and have been previously characterized [13]. As the background of the TRE-200ba429 mice was not pure, they were backcrossed into an FVB background. All tumor mice reported in this study were from backcross 6–8 which represents 98.45–99.6% FVB background. All mice used for the mammary gland development study were from backcross 8 or later.
MTB-IGFIR mice that overexpress the IGF-IR in mammary epithelial cells in a doxycycline inducible manner have been previously characterized by our lab [45,46]. MTB-IGFIRba429 mice that overexpress both IGF-IR and the miR-200b/200a/429 cluster in mammary epithelial cells in a doxycycline inducible manner were created by mating MTB-IGFIR mice with TRE-200ba429 mice. MTB-IGFIR and MTB-IGFIRba429 mice were placed on food supplemented with 2 g/kg of doxycycline when the mice were weaned at 21 days of age. Mice were monitored 2 times per week by palpating the mammary glands. Once a palpable mammary tumor was identified tumor size was measured using digital calipers. Mammary tumors were collected once they reached ∼10% of the mouse's body weight and mammary glands were collected from mice that did not develop mammary tumors 300 days after IGF-IR induction.
RNA extraction and RNA sequencing
RNA extraction was completed as previously described [23], and RNA sequencing and analysis was performed by Arraystar (Arraystar Inc, Rockville, MD). Fastq files were analyzed using Genialis software (Genialis Inc, Houston, TX) following the standard RNA-seq pipeline as previously described [30]. Hierarchical clustering was performed using Genialis software (Genialis Inc, Houston, TX) with all genes and Pearson distance measure and average linkage clustering. Pathway analysis was performed using Enrichr [47,48]. The data has been uploaded to GEO as GSE180264.
Real-time PCR
Quantitative real-time PCR was performed as described in Jones et al. [23] using primers for Saa1 (qMmuCID0007991), Saa2 (qMmuCED0026710), Spp1 (qMmuCED0040763), and Hprt (qMmuCED0045738). The expression of Saa1, Saa2, and Spp1 was presented relative to Hprt using CFX Maestro software version 2.2 (Bio-Rad Laboratories, Mississauga, ON). Primer efficiency for Saa1 was 109.1%, for Saa2, 109.7%, for Spp1, 104.5%, and for Hprt, 105.0%.
Histology and immunohistochemistry
Mammary glands were collected, fixed in 10% formalin overnight and embedded in paraffin. Sections were cut and stained with hematoxylin and eosin for histologic analysis. Immunohistochemistry was performed as previously described [49] using a 1:200 dilution of the anti-Spp1 antibody (cat# ab218237, Abcam, Toronto, ON) or a 1:200 Dilution of the anti-SAA1+SAA2 antibody (cat# ab199030). Slides were scanned using a Motic Easyscan digital slide scanner (Motic Richmond, BC).
Wholemount analysis
Wholemount analysis was performed on mammary glands from 55-day old and 75-day old mice as described in Moorehead et al. [50] except images were captured with a Canon EOS 6D camera (Canon Canada, Mississauga, ON) equipped with a 100 mm Canon marco lens (Canon Canada, Mississauga, ON). Duct length was determined by importing the images into Aperio ImageScope (Leica Biosystems, Concord, ON) and averaging the distance measured from the edge of the lymph node (closest to the nipple) to the tips of the three longest ducts.
miR-200b and miR-200c in situ hybridization
In situ hybridization was performed using a miR-200b (SR-mmu-miR-200b-3p-S1) or miR-200c (SR-mmu-miR-200c-3p-S1) probe from Advanced Cell Diagnostics (Newark, CA). Positive (SR-Mm-Snord85-S1) and negative control (SC-Scramble-S1) probes were also used but images using these probes were not included. In situ hybridization was performed as described in the miRNAscope HD (RED) Assay user manual using a 15 min incubation at 99 °C in 1x target retrieval reagent and a 30 min incubation with protease IV at 40 °C. Slides were scanned using a Motic Easyscan digital slide scanner (Motic Richmond, BC).
Statistics
For analysis comparing the means of two different groups, a Student's t-test was performed. For analyses comparing the means of three or more groups, an ANOVA followed by a Tukey's test was performed using GraphPad Prism 8.4.3 software (San Diego, CA). To compare survival curves a Log-rank (Mantel-Cox) test was performed using GraphPad Prism 9.1.2 software (San Diego, CA). Means were considered statistically different when p < 0.05.
Results
miR-200b, miR-200a and miR-429 are overexpressed in mammary epithelial cells of MTB-200ba429 transgenic mice
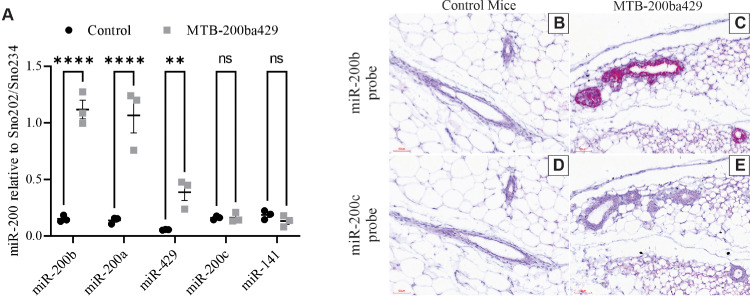
MTB-200ba429 mice are transgenic mice that overexpress the miR-200b/200a/429 cluster in a doxycycline inducible manner in mammary epithelial cells. When MTB-200ba429 mice are fed chow containing 2 g/kg of doxycycline beginning at 21 days of age, the levels of miR-200b, miR-200a and miR-429 are approximately 7-8-fold higher than the levels of these miRNAs in normal mammary glands of 55-day old mice (Fig. 1A). The re-expression of the miR-200b/200a/429 cluster did not significantly impact the re-expression of the other two miR-200 family members, miR-200c and miR-141 (Fig. 1A).
Fig. 1.
miR-200 expression in 55-day old control and MTB-200ba429 transgenic mice. (A) Expression of miR-200b, miR-200a, miR-429, miR-200c or miR-141 in virgin mammary glands from MTB-200ba429+ transgenic mice or control mice as determined by qRT-PCR, n = 3. (B) in situ hybridization for (B,C) miR-200b or (D,E) miR-200c in (B,D) control mammary glands or (C,E) mammary glands from MTB-200ba429+ mice. Positive staining for miR-200b or miR-200c appears red while the sections were counterstained with Gill's hematoxylin. Scale bars are 60 µm. ** indicates p < 0.01 and **** indicates p < 0.0001(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The increased expression of miR-200b (used as a surrogate for the miR-200b/200a/429 cluster; Fig. 1B,C) but not miR-200c (used as a surrogate for the miR-200c/141 cluster; Fig. 1D,E) was confirmed in mammary epithelial cells of MTB-200ba429 transgenic mice (Fig. 1C,E) compared to control mice (Fig. 1B,D) using in situ hybridization.
Transgenic expression of the miR-200b/200a/429 cluster prevents tumor initiation
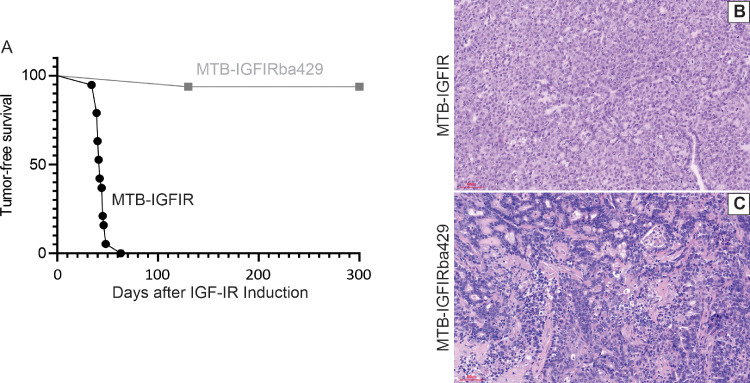
To determine whether miR-200s inhibit mammary tumor initiation and progression, TRE-200ba429 transgenic mice were mated with MTB-IGFIR transgenic mice. MTB-IGFIR transgenic mice overexpress the type I insulin-like growth factor receptor (IGF-IR) in mammary epithelial cells in a doxycycline inducible manner and IGF-IR overexpression induces rapid mammary tumor development in 100% of mice Fig. 2A and [45,46]. Crossing MTB-IGFIR and TRE-200ba429 transgenic mice produced mice (MTB-IGFIRba429 mice) that overexpressed both the IGF-IR transgene and the miR-200b/200a/429 transgene in the same mammary epithelial cells since both transgenes are driven by the reverse tetracycline transactivator and doxycycline. While MTB-IGFIR transgenic mice developed mammary tumors with 100% frequency and an average latency of ∼43 days post IGF-IR induction, overexpression of miR-200b/200a/429 in mammary epithelial cells overexpressing IGF-IR almost completely suppressed tumor incidence (Fig. 2A). Only 1 of 16 MTB-IGFIRba429 mice (6.25%) developed a mammary tumor and this tumor was first detected at 130 days post IGF-IR induction. The other 15 MTB-IGFIRba429 mice failed to develop mammary tumors by the study endpoint (300 days post IGF-IR induction). In addition, there were no hyperplastic lesions visible upon histologic analysis of mammary glands from MTB-IGFIRba429 300 days post IGF-IR induction. Mammary tumors that arose in MTB-IGFIR mice were typically solid sheets of epithelial cells with varying amount of necrosis (Fig. 2B) while the one tumor that arose in a MTB-IGFIRba429 mouse had tumor cells that appeared less densely packed (Fig. 2C). However, with only one tumor in the MTB-IGFIRba429 mice it is difficult to compare mammary tumor histology between MTB-IGFIR and MTB-IGFIRba429 mice.
Fig. 2.
Overexpression of the miR-200b/200a/429 prevents mammary tumor development induced by the IGF-IR transgene. (A) Tumor free survival curve for MTB-IGFIR (n = 19) and MTB-IGFIRba429+ (n = 16) transgenic mice. (B,C) Hematoxylin and eosin stained sections from (B) MTB-IGFIR tumor and (C) the only MTB-IGFIRba429 tumor. Scale bars are 100 µm.
Overexpression of miR-200b/200a/429 prevents IGF-IR induced hyperplasia but does not restore terminal end bud structure
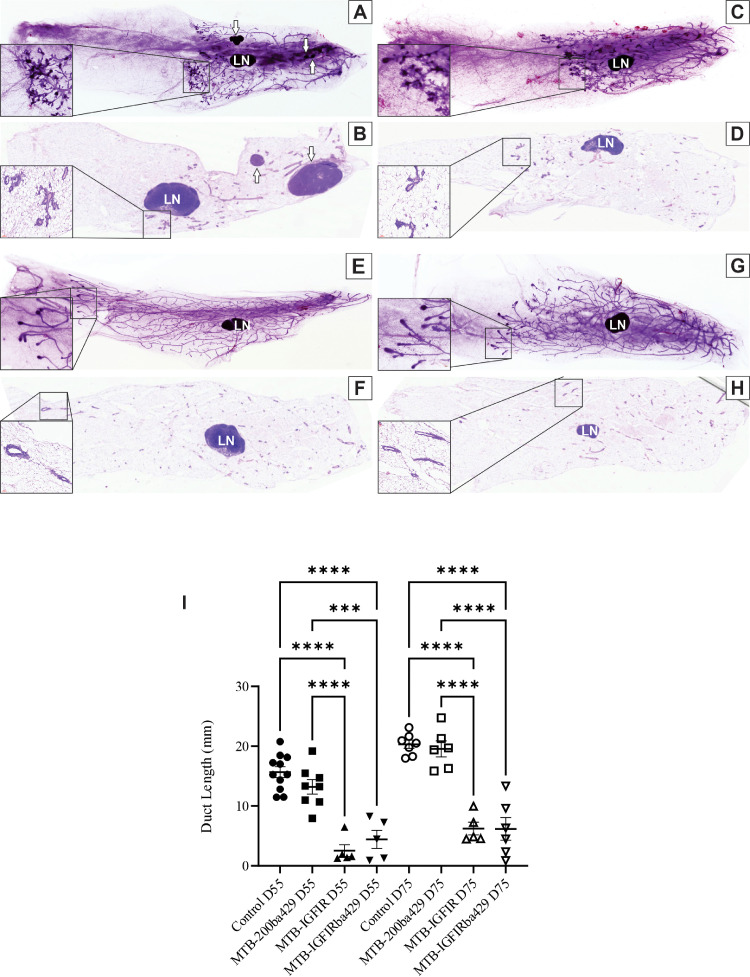
To better understand how overexpression of miR-200s inhibit mammary tumor initiation, mammary glands of 55-day old and 75-day old control, MTB-200ba429, MTB-IGFIR, and MTB-IGFIRba429 mice were evaluated using wholemount analysis and histology. By 55 days of age, or 34 days after induction of the IGF-IR transgene, MTB-IGFIR mice have hyperplastic lesions that are visible in mammary wholemounts and histologic sections (Fig. 3A,B, arrows indicate hyperplasia while LN indicates the lymph node found in the fourth mammary gland). In contrast, mammary glands from 55-day old MTB-IGFIRba429 mice did not display hyperplasia (Fig. 3C,D). The absence of hyperplastic lesions in the MTB-IGFIRba429 mice is consistent with the lack of tumor development observed in almost all MTB-IGFIRba429 mice.
Fig. 3.
Mammary ductal development in control, MTB-IGFIR, MTB-200ba429 and MTB-IGFIRba429 mice. (A,C,D,G) Wholemount analysis and (B,D,F,H) histologic analysis of 55-day old mammary glands from (A,B) MTB-IGFIR, (C,D) MTB-IGFIRba429, (E,F) control, and (G,H) MTB-200ba429 mice. Insets show a magnified version of the terminal end buds. White arrows indicate hyperplastic lesions while LN indicates the lymph node found in the 4th mammary gland. (I) Quantification of the ductal length in 55-day old (D55) and 75-day old (D75) mice. *** indicates p < 0.001 and **** indicates p < 0.0001.
Mammary ductal morphogenesis was significantly inhibited in both MTB-IGFIR (Fig. 3A,I) and MTB-IGFIRba429 mice (Fig. 3C,I) compared to control (Fig. 3E,I) and MTB-200ba429 mice (Fig. 3G,I) at both 55 and 75 days of age. Mammary wholemounts of both the MTB-IGFIR (Fig. 3A) and MTB-IGFIRba429 (Fig. 3B) mice revealed terminal end buds (TEBs) with irregular shapes compared to the smooth, bulbous shape of the TEBs in control (Fig. 3E) and MTB-200ba429 (Fig. 3G) mice. Therefore, miR-200b/200a/429 overexpression in IGF-IR transgenic mice can prevent mammary epithelial hyperplasia but cannot restore TEB structure or ductal elongation. However, overexpression of miR-200b/200a/429 in mammary epithelial cells on its own, does not significantly impair TEB structure or ductal morphogenesis.
RNA sequencing reveals genes potentially regulated by miR-200b/200a/429 that inhibit mammary tumor development
To investigate genes potentially regulated by miR-200b/200a/429 overexpression that prevented IGF-IR induced mammary tumor development, RNA sequencing was performed. Gene expression analysis was performed on day 55 mammary glands from control, MTB-200ba429, MTB-IGFIR and MTB-IGFIRba429 mice. Four mammary glands were collected from mice of each genotype. One of the MTB-IGFIRba429 mammary glands (mouse KW1084) was removed from the analysis as it consistently expressed lower levels of most transcripts compared to the remaining three mammary glands or had 0 reads for transcripts abundantly expressed in the mammary glands from the other three MTB-IGFIRba429 mice. For example, the counts per millions (CPM) for Krt8, a transcript expressed in mammary epithelial cells, was 0.39 CPM in mammary tissue from mouse KW1084 but exceeded 135 CPM in the other three mammary gland samples.
Hierarchical clustering revealed that mammary transcript expression did not segregate the mammary glands based on the genotype of the mouse from which they were derived (Supplemental File 1). Pairwise comparison of differentially expressed transcripts (log FC ≥ 1, FDR < 0.01) from mammary tissue from the various genotypes revealed a relatively small number of differentially expressed transcripts. For example, only 7 transcripts (miR200b, Cidea, Adtrp, Egfl6, Ces1f, Luzp2, Cox8b) were significantly upregulated and two transcripts (Sfrp2, Islr2) significantly downregulated in the MTB-200ba429 mammary glands compared to control mammary glands (Supplemental File 2) suggesting that overexpression of miR-200b/200a/429 only impacts a small number of genes during ductal morphogenesis. The fact that miR-200b was the most significant differentially expressed transcript (Supplemental File 2) further confirmed that miR-200b had been overexpressed in MTB-200ba429 mammary tissue.
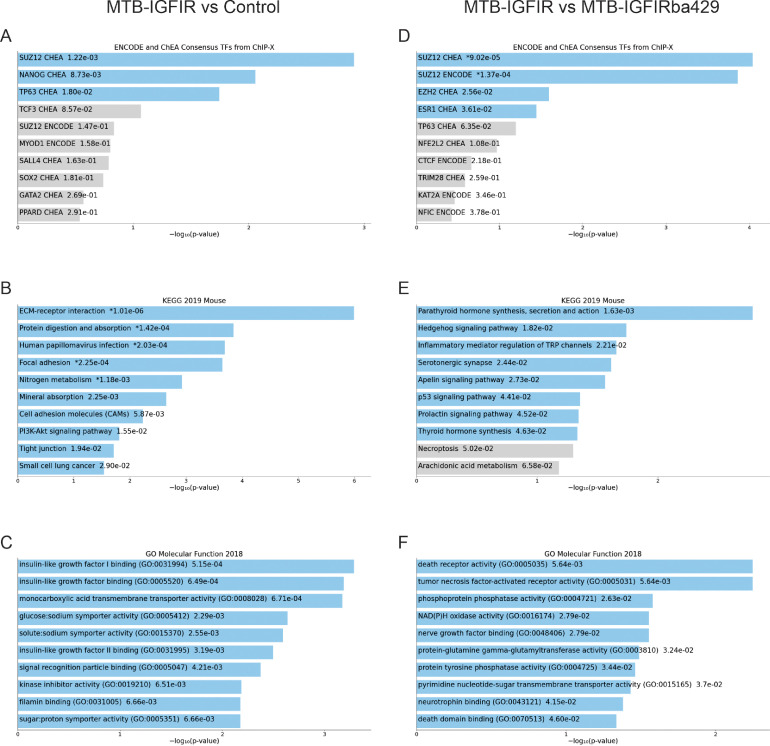
The greatest number of differentially expressed transcripts (log FC ≥ 1, FDR < 0.01) was observed when MTB-IGFIR mammary glands were compared to control mammary glands with 252 transcripts upregulated and 38 transcripts downregulated in MTB-IGFIR mammary glands compared to control mammary glands (Supplemental File 2). The fourth ranked differentially expressed transcript, based on FDR, in MTB-IGFIR mammary glands compared to control mammary glands was Igf1r (logFC 3.2, FDR 1.3 × 10−15) confirming that the IGF-IR transgene was overexpressed in MTB-IGFIR mice. Pathway analysis using only the 252 transcripts elevated in MTB-IGFIR mammary glands showed that the top Encode and ChEA pathway was SUZ12 while the top KEGG pathways was ECM-receptor interaction (Fig. 4A–C). The top Molecular Function was insulin-like growth factor I binding (Fig. 4C) which is consistent with hyperactivation of the IGF-IR due to transgenic overexpression.
Fig. 4.
Pathway analysis of RNA sequencing data using Enrichr. Top (A,D) ENCODE and ChEA Consensus TFs from ChiP-X, (B,E) KEGG 2019 Mouse, and (C,F) GO Molecular Function 2018 in (A-C) MTB-IGFIR 55-day old mammary glands compared to (D-F) control 55-day old mammary glands.
Transcripts differentially expressed in MTB-IGFIR mice compared to MTB-IGFIRba429 mice were also examined. The mammary glands from MTB-IGFIR mice contained 47 upregulated and 47 downregulated transcripts compared to MTB-IGFIRba429 mammary glands (Supplemental File 2). Pathway analysis of these 94 transcripts revealed the top Encode and ChEA pathway was SUZ12 while the top KEGG pathway and Molecular Function was Parathyroid hormone synthesis, secretion and action and death receptor activity, respectively (Fig. 4D–F).
The differentially expressed genes in the MTB-IGFIR vs control mammary glands were then compared to differentially expressed genes in the MTB-IGFIR vs MTB-IGFIRba429 mammary glands to identify genes consistently altered in mammary glands of mice that developed mammary tumors (MTB-IGFIR) compared to mammary glands of mice that rarely developed mammary tumors (control, MTB-IGFIRba429). Eighteen transcripts were identified, all of which were expressed at significantly higher levels in the MTB-IGFIR mammary glands compared to mammary glands from control or MTB-IGFIRba429 mice (Table 1). Of these 18 transcripts, 15 encoded genes, two encoded unclassified genes (GM42793, GM47585) and one coded for a long non-coding RNA (lncRNA, GM10384).
Table 1.
Predicted miR-200b, miR-200a, or miR-429 sites in transcripts differentially expressed in mammary glands from MTB-IGFIR vs Control and MTB-IGFIR vs MTB-IGFIRba429 mice.
| MTB-IGFIR vs Control |
MTB-IGFIR vs MTB-IGFIRba429 |
Predicted target of miR-200b/200a/429 |
|||||
|---|---|---|---|---|---|---|---|
| Transcript | LogFC | FDR | LogFC | FDR | TargetScan | miRDB | miRWalk |
| Saa2 | 3.9 | 9.2E-13 | 3.2 | 1.2E-21 | No | No | miR-200b, miR-200a |
| Saa1 | 3.6 | 1.4E-11 | 2.7 | 4.6E-10 | No | No | miR-200b |
| Slc26a9 | 3.1 | 9.4E-09 | 2.0 | 9.3E-03 | No | No | miR-200b, miR-200a, miR-429 |
| Gm42793 | 4.6 | 2.1E-07 | 4.2 | 1.9E-03 | – | – | – |
| Tmprss4 | 2.2 | 2.4E-07 | 1.7 | 6.2E-04 | miR-200a | No | miR-200b, miR-200a, miR-429 |
| Kctd14 | 1.6 | 1.2E-04 | 1.6 | 8.9E-03 | No | No | miR-200b, miR-200a, miR-429 |
| Spp1 | 2.8 | 3.3E-04 | 2.4 | 3.3E-05 | miR-200a | No | miR-200b |
| Car6 | 1.8 | 4.8E-04 | 3.2 | 6.3E-06 | No | No | miR-200b, miR-429 |
| Muc4 | 2.2 | 6.9E-04 | 1.8 | 7.8E-06 | No | No | miR-200b, miR-429 |
| Slc30a2 | 2.4 | 7.4E-04 | 2.1 | 8.9E-03 | miR-429 | No | miR-200b, miR-200a, miR-429 |
| Gm47585 | 3.5 | 1.4E-03 | 4.5 | 8.5E-03 | – | – | – |
| Proser2 | 1.3 | 1.6E-03 | 1.4 | 9.2E-04 | miR-200a | miR-200a | miR-200a |
| Rem2 | 1.6 | 1.7E-03 | 2.5 | 6.3E-06 | No | No | miR-200b, miR-200a |
| 9130230L23Rik | 1.8 | 2.1E-03 | 2.0 | 6.7E-03 | – | – | – |
| Slc35d3 | 2.2 | 2.2E-03 | 3.8 | 2.7E-05 | No | No | miR-200b, miR-429 |
| Duox1 | 2.0 | 3.2E-03 | 2.5 | 1.7E-03 | No | No | miR-200b, miR-200a |
| F2rl1 | 1.1 | 3.6E-03 | 1.3 | 2.8E-03 | No | No | miR-200b, miR-200a |
| Gm10384 | 1.8 | 3.8E-03 | 2.6 | 3.4E-08 | – | – | – |
Three databases (TargetScan, miRDB and miRWalk) were investigated for potential miR-200b, miR-200a or miR-429 binding sites. TargetScan uses the seed region of each miRNA to predict mRNA targets [51], [52], [53], [54], [55] and miRDB uses the target prediction tool MirTarget [56,57] while miRWalk searches for potential binding sites in 3′-UTR, 5′-UTR and coding regions using TarPmiR [58,59]. Only Proser2 was identified as a miR-200a target in all three databases while Tmprss4, Spp1, and Slc30a2 were identified as potential miR-200a or miR-429 targets in two of the databases (Table 1). The remaining transcripts were only identified as potential miR-200b, miR-200a or miR429 targets by miRWalk. Transcript 9130230L23Rik, the two unclassified genes and the lncRNA were not recognized by TargetScan, miRDB or miRWalk and thus information regarding their regulation by miR-200s was unavailable (Table 1).
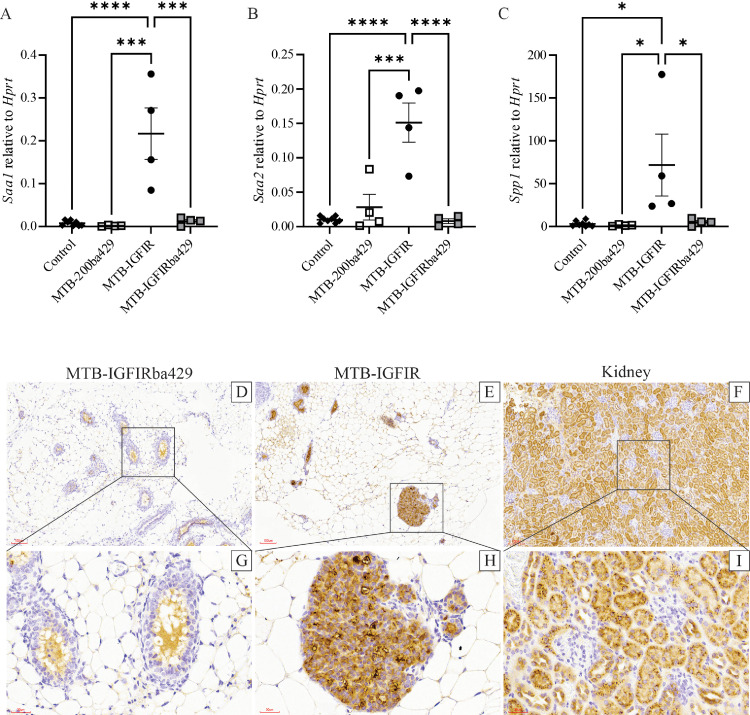
We then focused on Saa1, Saa2 and Spp1 since Saa1 and Saa2 were the two transcripts with the lowest FDR in Table 1 and we have previously shown that Spp1 was the most significant, differentially expressed gene in a DNA microarray analysis comparing MTB-IGFIR mammary tumors to mammary glands of control mice [60,61]. Quantitative real-time PCR confirmed the elevated levels of Saa1, Saa2, and Spp1 in mammary glands from 55-day old MTB-IGFIR mice compared to control, MTB-200ba429, and MTB-IGFIRba429 mammary glands (Fig. 5AC). Immunohistochemistry with a Spp1 antibody showed high levels of Spp1 staining in MTB-IGFIR hyperplastic lesions and very low levels in mammary epithelial cells of MTB-IGFIRba429 mice (Fig. 5A,B,D,E). Mouse kidney tissue served as a positive control for the Spp1 antibody (Fig. 5C,F). The antibody that detects both Saa1 and Saa2 did not detect Saa1/Saa2 protein in mammary ducts of MTB-IGFIRba429 mice or hyperplastic lesions of MTB-IGFIR mice. While the Saa1/Saa2 antibody did stain liver tissue, this antibody failed to detect Saa1/Saa2 in MTB-IGFIR hyperplastic lesions or control mammary epithelial cells (Supplemental File 3). The level of Saa1 and Saa2 transcripts were much lower (Fig. 5A,B) than the levels of Spp1 transcript (Fig. 5C) and thus the amount of Saa1 and Saa2 protein may be below the detection limit of this antibody in formalin-fixed, paraffin embedded mammary tissue.
Fig. 5.
Expression of Saa1, Saa2 and Spp1 in 55-day old mammary glands. Expression of (A) Saa1, (B) Saa2, and (C) Spp1 in mammary glands from 55-day old control, MTB-200ba429, MTB-IGFIR and MTB-IGFIRba429 mice as determined by quantitative real-time PCR. * indicates p < 0.05, *** indicates p < 0.001 and **** indicates p < 0.0001. Immunohistochemistry for Spp1 in mammary glands from 55-day old (D,G) MTB-IGFIRba429 or (E,H) MTB-IGFIR mice. (F,I) Kidney tissue from wild type mice served as a positive control. Scale bars for (D–F) are 100 µm and for (G-I) are 30 µm.
Discussion
The function of miR-200s in mammary tumor growth and metastasis have been extensively studied using human and murine mammary tumor cell lines. These studies from our lab [23], [24], [25] and others [62], [63], [64] generally show that miR-200s can inhibit the growth of primary mammary tumors and suppress metastatic spread. However, the ability of miR-200s to prevent mammary tumor initiation, has not been explored.
In this manuscript we have shown for the first time that overexpression of the miR-200b/200a/429 cluster in mammary epithelial cells significantly suppressed mammary tumor development induced by transgenic overexpression of IGF-IR. This finding is significant as although improving breast cancer therapy is an important clinical achievement, the ultimate goal in cancer biology is to prevent cancer development. Remarkably, an approximate 7-fold increase in the miR-200b/200a/429 cluster was sufficient to prevent mammary tumor development induced by a potent oncogene (IGF-IR) in ∼94% of the mice. Importantly, overexpression of the miR-200b/200a/429 cluster did not significantly impair mammary ductal development. The reason this observation is important is that although most women develop breast cancer after ductal development has been completed, the optimal window to administer preventative strategies remain unclear. Our lab and others have shown that mammary epithelial cells are particularly susceptible to transformation during puberty [65], [66], [67], [68], [69] and thus preventative strategies may need to be initiated during, or prior to the onset of, puberty for maximal efficacy. Two MTB-200ba429 and two control female mice were administered doxycycline supplemented food throughout their lifetime including during mating and lactation. The MTB-200ba429 females had the same litter size as the control mice and successfully nursed their offspring (unpublished observations). Thus, our data suggests that overexpressing miR-200s should not negatively affect mammary gland development or function.
Exactly how miR-200s prevents mammary tumor initiation remains incompletely defined however, our data suggest and important roles for Saa1, Saa2 and Spp1. These three genes were significantly elevated 34 days after the induction of the IGF-IR transgene when small, multifocal hyperplastic lesions were forming and co-expression of the miR-200b/200a/429 cluster with IGF-IR prevented the increase in Saa1, Saa2 and Spp1 expression. Saa1 and Saa2 genes encode serum amyloid A proteins Saa1 and Saa2, both of which are acute phase proteins [70]. Saa1 and Saa2 have been associated with pancreatic, renal, lung, colorectal, ovarian, oral, gastric, and breast cancer as well glioblastoma [71], [72], [73], [74], [75], [76], [77], [78]. Although the increase in Saa1 and Saa2 proteins are typically attributed to tumor-associated inflammation, studies have shown that Saa1 can regulate Akt signaling in tumor cells and siRNA knockdown of Saa1 in pancreatic cells inhibits migration/invasion and epithelial to mesenchymal transition [71,79]. In breast cancer, Saa1 was found to promote invasion [80,81]. However, it is also possible that Saa1 and Saa2 are elevated as part of the acute phase protein response [82,83] and thus may serve as biomarkers for mammary tumorigenesis but play no direct role in mammary tumor initiation.
Spp1 is secreted phosphoprotein 1 and is also known as osteopontin. This protein is an important regulator of bone but has also been implicated in breast cancer. Spp1 can be expressed by tumor cells as well as immune cells and fibroblasts where it can regulate process like EMT, angiogenesis and metastasis as well as modulate immune cell function [61,[84], [85], [86], [87]]. Spp1 contains an Arg-Gly-Asp (RGD) sequence allowing it to bind to several integrin receptors [88]. Another receptor for Spp1 is CD44 [89]. Following binding to either of these receptors, Spp1 can initiate signaling via the PI3K/Akt and Raf/MEK/ERK signaling pathways [90], [91], [92]. We have previously shown the importance of Spp1 in mammary tumorigenesis in MTB-IGFIR mice and in murine mammary tumor cell lines [60,61].
There is some indirect evidence that Spp1 regulates breast cancer initiation in women. Conditions that increase a women's chance of developing breast cancer such as high breast density, obesity and BRCA1 mutations have been associated with elevated Spp1 expression. Women with dense breasts have a 4-fold increase of developing breast cancer [93,94] and it has been shown that Spp1 is significantly elevated in dense breast tissue compared to normal breast tissue [95]. Similarly, obesity has been associated with an increased risk of developing breast cancer. One of the proteins elevated in the plasma of obese individuals is Spp1 [96]. Breast cancer risk is also elevated in women with BRCA1 mutations. Since BRCA1 can suppress Spp1 expression [97], women with BRCA1 mutations frequently express elevated levels of Spp1 and this increase in Spp1 expression may contribute to the elevated breast cancer risk associated with BRCA1 mutations.
There are multiple ways miR-200s could inhibit Saa1, Saa2 and Spp1 expression including, direct binding to Saa1, Saa2 and Spp1 mRNA and regulation of DNA or histone methylation in the promoters of these genes. Saa1, Saa2 and Spp1 have predicted miR-200b, miR-200a or miR-429 binding sites and thus miR-200s are predicted to directly regulate mRNA levels of these transcripts. With respect to methylation, our pathway analysis implicates SUZ12. SUZ12 is a component of the polycomb repressor complex 2 (PRC2) that mono-, di-, and trimethylates histone H3 on lysine 27 (H3K27) leading to chromatin compaction and suppression of transcription [98], [99], [100]. Saa1 [77] and Spp1 [101] have been shown to be regulated by H3K27 methylation and our previous study found that increased expression of miR-200s in murine and human breast cancer cells resulted in an elevation of H3K27me3 [24]. Therefore, miR-200s may directly target genes critical for tumor initiation and/or influence histone methylation to regulate gene expression.
Clinical application of increasing miR-200 expression to prevent breast cancer may be challenging, however, our work with transgenic mice overexpressing the miR-200b/200a/429 cluster did not find any significant impact on mammary ductal development or lactation suggesting that increasing miR-200s in the mammary gland will not impair mammary gland function. The biggest hurdle comes from identifying ways to increase miR-200 expression as our understanding of miR-200 regulation is poor. The expression of miR-200 family members do increase in mammary epithelial cells during pregnancy and lactation [102] and thus, lactogenic hormones potentially increase miR-200 expression. This is especially intriguing considering a full-term pregnancy and lactation reduced breast cancer risk [103], [104], [105]. An alternative way to clinically manipulate miR-200 expression or the expression of miR-200 regulated genes specifically in mammary epithelial cells would be through the injection of viral vectors or target inhibitors into the mammary ducts via the nipple. Mammary epithelial cells line the mammary ducts and administration of substances into the mammary duct will interact with mammary epithelial cells. This approach would also induce a localized increase in miR-200 or gene expression, eliminating potential side effects of systemically increasing miR-200 levels or miR-200 target genes.
In summary, our data shows that miR-200s can inhibit mammary tumor development potentially through the regulation Saa1, Saa2 and Spp1. Future studies will need to determine the exact mechanism through which miR-200s regulate the expression of these genes and whether miR-200s can suppress mammary tumor development induced by other oncogenes. Before miR-200s can be considered as a preventative strategy, approaches that can induce miR-200 expression in the mammary gland need to be identified. Fortunately, our data suggests that miR-200 overexpression in mammary epithelial cells should not significantly impair mammary development or function.
Ethics approval and consent to participate
Animals were housed and cared for following guidelines established by the Central Animal Facility at the University of Guelph and the guidelines established by the Canadian Council of Animal Care. This study was approved by the Animal Care Committee at the University of Guelph (AUP #3994).
Consent for publication
Not applicable.
Availability of data and materials
RNA sequencing has been uploaded to GEO under accession number GSE180264.
CRediT authorship contribution statement
Katrina L Watson: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Rui Yi: Resources, Writing – review & editing. Roger A Moorehead: Writing – original draft, Project administration, Formal analysis, Funding acquisition, Methodology, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding
This work was funded by a CHIR project grant (PJT-162218) awarded to RAM and a National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award numbers AR059697 and AR066703 to RY.
Acknowledgment
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101228.
Appendix. Supplementary materials
Supplemental File 1. Hierarchical clustering of RNA sequencing data of mammary glands from 55-day old control, MTB-200ba429, MTB-IGFIR and MTB-IGFIRba429 mice.
Supplemental File 2. Transcripts differentially expressed in mammary glands from 55-day old MTB-200ba429 mice vs control mice, MTB-IGFIR mice vs control mice and MTB-IGFIR mice vs MTB-IGFIRba429 mice. Log fold-change (LogFC) >1 and false discovery rate (FDR) < 0.01 were used as cut-offs.
Supplemental File 3. Immunohistochemistry using an antibody that detects both Saa1 and Saa2 in (A,D) mammary gland from a 55-day old MTB-IGFIRba429 mouse and (B,E) a hyperplastic lesion in the mammary gland of a 55-day old MTB-IGFIR mouse. (C,D) Liver tissue stained with the antibody served as a positive control. Scale bars for (D-F) are 100 µm and for (G-I) are 30 µm.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Mei J., Hao L., Wang H., Xu R., Liu Y., Zhu Y., Liu C. Systematic characterization of non-coding RNAs in triple-negative breast cancer. Cell Prolif. 2020;53:e12801. doi: 10.1111/cpr.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Di L.G., Calin G.A., Croce C.M. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res. C Embryo Today. 2006;78:180–189. doi: 10.1002/bdrc.20073. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Czech B., Hannon G.J. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hock J., Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoefert J.E., Bjerke G.A., Wang D., Yi R. The microRNA-200 family coordinately regulates cell adhesion and proliferation in hair morphogenesis. J. Cell Biol. 2018;217:2185–2204. doi: 10.1083/jcb.201708173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saini H.K., Enright A.J., Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genom. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer. 2012;132 doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 17.Trumbach D., Prakash N. The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis. Cell Tissue Res. 2015;359:161–177. doi: 10.1007/s00441-014-1911-z. [DOI] [PubMed] [Google Scholar]
- 18.Howe E.N., Cochrane D.R., Richer J.K. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J. Mammary Gland Biol. Neoplasia. 2012;17:65–77. doi: 10.1007/s10911-012-9244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radisky D.C. miR-200c at the nexus of epithelial-mesenchymal transition, resistance to apoptosis, and the breast cancer stem cell phenotype. Breast Cancer Res. 2011;13:110. doi: 10.1186/bcr2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., Goodall G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 21.Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Jones R., Watson K., Bruce A., Nersesian S., Kitz J., Moorehead R. Re-expression of miR-200c suppresses proliferation, colony formation and in vivo tumor growth of murine claudin-low mammary tumor cells. Oncotarget. 2017;8:23727–23749. doi: 10.18632/oncotarget.15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson K., Conquer-van Heumen G., Watson K.L., Roth M., Martin C.J., Moorehead R.A. Re-expression of miR-200s in claudin-low mammary tumor cells alters cell shape and reduces proliferation and invasion potentially through modulating other miRNAs and SUZ12 regulated genes. Cancer Cell Int. 2021;21:89. doi: 10.1186/s12935-021-01784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson K.L., Jones R.A., Bruce A., Moorehead R.A. The miR-200b/200a/429 cluster prevents metastasis and induces dormancy in a murine claudin-low mammary tumor cell line. Exp. Cell Res. 2018;369:17–26. doi: 10.1016/j.yexcr.2018.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Chen H., Li Z., Zhang L., Zhang L., Zhang Y., Wang Y., Xu M., Zhong Q. MicroRNA-200c inhibits the metastasis of triple-negative breast cancer by targeting ZEB2, an epithelial-mesenchymal transition regulator. Ann. Clin. Lab. Sci. 2020;50:519–527. [PubMed] [Google Scholar]
- 27.Knezevic J., Pfefferle A.D., Petrovic I., Greene S.B., Perou C.M., Rosen J.M. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene. 2015;34:5997–6006. doi: 10.1038/onc.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H.K., Park J.D., Choi S.H., Shin D.J., Hwang S., Jung H.Y., Park K.S. Functional link between miR-200a and ELK3 regulates the metastatic nature of breast cancer. Cancers (Basel) 2020;12:1225. doi: 10.3390/cancers12051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Y., Ma Y., Liu Y., Wang X.F. Effect of miR-200c on migration and proliferation of breast cancer MDA-MB-231 cells and BT-549 cells and the possible mechanism. Eur. Rev. Med. Pharmacol. Sci. 2020;24:735–739. doi: 10.26355/eurrev_202001_20053. [DOI] [PubMed] [Google Scholar]
- 30.Simpson K., Conquer-Van Heumen G., Watson K.L., Roth M., Martin C.J., Moorehead R.A. Re-expression of miR-200s in claudin-low mammary tumor cells alters cell shape and reduces proliferation and invasion potentially through modulating other miRNAs and SUZ12 regulated genes. Cancer Cell Int. 2021;21:89. doi: 10.1186/s12935-021-01784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphries B., Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S.P., Chiao E., Dirbas F.M., Somlo G., Pera R.A., Lao K., Clarke M.F. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V.G., Morton J., Sansom O., Schuler J., Stemmler M.P., Herzberger C., Hopt U., Keck T., Brabletz S., Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 34.Gill J.G., Langer E.M., Lindsley R.C., Cai M., Murphy T.L., Kyba M., Murphy K.M. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells. 2011;29:764–776. doi: 10.1002/stem.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G., Guo X., Hong W., Liu Q., Wei T., Lu C., Gao L., Ye D., Zhou Y., Chen J., Wang J., Wu M., Liu H., Kang J. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2858–2863. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Zhang Z., Xia S., Xing C., Ci X., Li X., Zhao R., Tian S., Ma G., Zhu Z., Fu L., Dong J.T. KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial-mesenchymal transition in epithelial cells. Mol. Cell Biol. 2013;33:4919–4935. doi: 10.1128/MCB.00787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grenda A., Krawczyk P. New dancing couple: PD-L1 and MicroRNA. Scand. J. Immunol. 2017;86:130–134. doi: 10.1111/sji.12577. [DOI] [PubMed] [Google Scholar]
- 38.Feng B., Cao Y., Chen S., Chu X., Chu Y., Chakrabarti S. miR-200b mediates endothelial-to-mesenchymal transition in diabetic cardiomyopathy. Diabetes. 2016;65:768–779. doi: 10.2337/db15-1033. [DOI] [PubMed] [Google Scholar]
- 39.Xu C., Shen W.B., Reece E.A., Hasuwa H., Harman C., Kaushal S., Yang P. Maternal diabetes induces senescence and neural tube defects sensitive to the senomorphic rapamycin. Sci. Adv. 2021;7:eabf5089. doi: 10.1126/sciadv.abf5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjerke G.A., Yi R. Integrated analysis of directly captured microRNA targets reveals the impact of microRNAs on mammalian transcriptome. RNA. 2020;26:306–323. doi: 10.1261/rna.073635.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran M., Lee S.M., Shin D.J., Wang L. Loss of miR-141/200c ameliorates hepatic steatosis and inflammation by reprogramming multiple signaling pathways in NASH. JCI Insight. 2017;2:e96094. doi: 10.1172/jci.insight.96094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren H., Tao C., Li K., Bi Y., Zheng X. Generation of a floxed allele of the mouse microRNA-200 clusters. Appl. Biochem. Biotechnol. 2017;182:1218–1228. doi: 10.1007/s12010-016-2394-z. [DOI] [PubMed] [Google Scholar]
- 43.Tao C., Ren H., Xu P., Cheng J., Huang S., Zhou R., Mu Y., Yang S., Qi D., Wang Y., Li K. Adipocyte miR-200b/a/429 ablation in mice leads to high-fat-diet-induced obesity. Oncotarget. 2016;7:67796–67807. doi: 10.18632/oncotarget.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasuwa H., Ueda J., Ikawa M., Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–73. doi: 10.1126/science.1237999. [DOI] [PubMed] [Google Scholar]
- 45.Jones R.A., Moorehead R.A. The impact of transgenic IGF-IR overexpression on mammary development and tumorigenesis. J. Mammary Gland Biol. Neoplasia. 2008;13:407–413. doi: 10.1007/s10911-008-9097-1. [DOI] [PubMed] [Google Scholar]
- 46.Jones R.A., Campbell C.I., Gunther E.J., Chodosh L.A., Petrik J.J., Khokha R., Moorehead R.A. Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene. 2007;26:1636–1644. doi: 10.1038/sj.onc.1209955. [DOI] [PubMed] [Google Scholar]
- 47.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linnerth N.M., Sirbovan K., Moorehead R.A. Use of a transgenic mouse model to identify markers of human lung tumors. Int. J. Cancer. 2005;114:977–982. doi: 10.1002/ijc.20814. [DOI] [PubMed] [Google Scholar]
- 50.Moorehead R.A., Fata J.E., Johnson M.B., Khokha R. Inhibition of mammary epithelial apoptosis and sustained phosphorylation of Akt/PKB in MMTV-IGF-II transgenic mice. Cell Death Differ. 2001;8:16–29. doi: 10.1038/sj.cdd.4400762. [DOI] [PubMed] [Google Scholar]
- 51.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W., Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding J., Li X., Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics. 2016;32:2768–2775. doi: 10.1093/bioinformatics/btw318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franks S.E., Campbell C.I., Barnett E.F., Siwicky M.D., Livingstone J., Cory S., Moorehead R.A. Transgenic IGF-IR overexpression induces mammary tumors with basal-like characteristics while IGF-IR independent mammary tumors express a claudin-low gene signature. Oncogene. 2012;31:3298–3309. doi: 10.1038/onc.2011.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh S., Thompson D.E., McConkey J., Murray P., Moorehead R.A. Osteopontin regulates proliferation, apoptosis, and migration of murine claudin-low mammary tumor cells. BMC Cancer. 2016;16:359. doi: 10.1186/s12885-016-2396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mekala J.R., Naushad S.M., Ponnusamy L., Arivazhagan G., Sakthiprasad V., Pal-Bhadra M. Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene. 2018;641:248–258. doi: 10.1016/j.gene.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 63.Piasecka D., Braun M., Kordek R., Sadej R., Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J. Cancer Res. Clin. Oncol. 2018;144:1401–1411. doi: 10.1007/s00432-018-2689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimono Y., Mukohyama J., Nakamura S., Minami H. MicroRNA regulation of human breast cancer stem cells. J. Clin. Med. 2015;5 doi: 10.3390/jcm5010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones R.A., Watson K.L., Campbell C.I., Moorehead R.A. IGF-IR mediated mammary tumorigenesis is enhanced during pubertal development. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0108781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Natarajan R., Aljaber D., Au D., Thai C., Sanchez A., Nunez A., Resto C., Chavez T., Jankowska M.M., Benmarhnia T., Yang J.A., Jones V., Tomsic J., McCune J.S., Sistrunk C., Doan S., Serrano M., Cardiff R.D., Dietze E.C., Seewaldt V.L. Environmental exposures during puberty: window of breast cancer risk and epigenetic damage. Int. J. Environ. Res. Public Health. 2020;17:96. doi: 10.3390/ijerph17020493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terry M.B., Michels K.B., Brody J.G., Byrne C., Chen S., Jerry D.J., Malecki K.M.C., Martin M.B., Miller R.L., Neuhausen S.L., Silk K., Trentham-Dietz A., Breast C., P. the Environment Research Environmental exposures during windows of susceptibility for breast cancer: a framework for prevention research. Breast Cancer Res. 2019;21:96. doi: 10.1186/s13058-019-1168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russo J., Tay L.K., Russo I.H. Differentiation of the mammary gland and susceptibility to carcinogenesis. Breast Cancer Res. Treat. 1982;2:5–73. doi: 10.1007/BF01805718. [DOI] [PubMed] [Google Scholar]
- 69.Russo J., Tay L.K., Ciocca D.R., Russo I.H. Molecular and cellular basis of the mammary gland susceptibility to carcinogenesis. Environ. Health Perspect. 1983;49:185–199. doi: 10.1289/ehp.8349185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sack G.H. In: Hoeger Ulrich, Harris Robin., editors. Vol. 94. Springer, Cham; 2020. Serum amyloid A (SAA) proteins; pp. 421–436. (Subcellular Biochemistry). [Google Scholar]
- 71.Zhang H., Xu Y., Deng G., Yuan F., Tan Y., Gao L., Sun Q., Qi Y., Yang K., Geng R., Jiang H., Liu B., Chen Q. SAA1 knockdown promotes the apoptosis of glioblastoma cells via downregulation of AKT signaling. J. Cancer. 2021;12:2756–2767. doi: 10.7150/jca.48419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gardner I.H., Siddharthan R., Watson K., Dewey E., Ruhl R., Khou S., Guan X., Xia Z., Tsikitis V.L., Anand S. A distinct innate immune signature of early onset colorectal cancer. Immunohorizons. 2021;5:489–499. doi: 10.4049/immunohorizons.2000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shang S., Li X., Gao Y., Guo S., Sun D., Zhou H., Sun Y., Wang P., Zhi H., Bai J., Ning S., Li X. MeImmS: predict clinical benefit of anti-PD-1/PD-L1 treatments based on DNA methylation in non-small cell lung cancer. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.676449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S., Cheng Y., Cheng G., Xu T., Ye Y., Miu Q., Cao Q., Yang X., Ruan H., Zhang X. High SAA1 expression predicts advanced tumors in renal cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.649761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren H., He G., Lu Z., He Q., Li S., Huang Z., Chen Z., Cao C., Wang A. Arecoline induces epithelial-mesenchymal transformation and promotes metastasis of oral cancer by SAA1 expression. Cancer Sci. 2021;112:2173–2184. doi: 10.1111/cas.14866. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Ding G., Sun J., Jiang L., Gao P., Zhou Q., Wang J., Tong S. Key pathways in prostate cancer with SPOP mutation identified by bioinformatic analysis. Open Med. (Wars) 2020;15:1039–1047. doi: 10.1515/med-2020-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yasukawa Y., Hattori N., Iida N., Takeshima H., Maeda M., Kiyono T., Sekine S., Seto Y., Ushijima T. SAA1 is upregulated in gastric cancer-associated fibroblasts possibly by its enhancer activation. Carcinogenesis. 2021;42:180–189. doi: 10.1093/carcin/bgaa131. [DOI] [PubMed] [Google Scholar]
- 78.Yi S., Zhou W. Tumorigenesis-related key genes in adolescents and young adults with HR(+)/HER2(-) breast cancer. Int. J. Clin. Exp. Pathol. 2020;13:2701–2709. [PMC free article] [PubMed] [Google Scholar]
- 79.Takehara M., Sato Y., Kimura T., Noda K., Miyamoto H., Fujino Y., Miyoshi J., Nakamura F., Wada H., Bando Y., Ikemoto T., Shimada M., Muguruma N., Takayama T. Cancer-associated adipocytes promote pancreatic cancer progression through SAA1 expression. Cancer Sci. 2020;111:2883–2894. doi: 10.1111/cas.14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J.Y., Lai Y.S., Chu P.Y., Chan S.H., Wang L.H., Hung W.C. Cancer-derived VEGF-C increases chemokine production in lymphatic endothelial cells to promote CXCR2-dependent cancer invasion and MDSC recruitment. Cancers (Basel) 2019;11:1120. doi: 10.3390/cancers11081120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hansen M.T., Forst B., Cremers N., Quagliata L., Ambartsumian N., Grum-Schwensen B., Klingelhofer J., Abdul-Al A., Herrmann P., Osterland M., Stein U., Nielsen G.H., Scherer P.E., Lukanidin E., Sleeman J.P., Grigorian M. A link between inflammation and metastasis: serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene. 2015;34:424–435. doi: 10.1038/onc.2013.568. [DOI] [PubMed] [Google Scholar]
- 82.Yang M., Liu F., Higuchi K., Sawashita J., Fu X., Zhang L., Zhang L., Fu L., Tong Z., Higuchi K. Serum amyloid A expression in the breast cancer tissue is associated with poor prognosis. Oncotarget. 2016;7:35843–35852. doi: 10.18632/oncotarget.8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fourie C., Shridas P., Davis T., de Villiers W.J.S., Engelbrecht A.M. Serum amyloid A and inflammasome activation: a link to breast cancer progression? Cytokine Growth Factor Rev. 2021;59:62–70. doi: 10.1016/j.cytogfr.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 84.Butti R., Nimma R., Kundu G., Bulbule A., Kumar T.V.S., Gunasekaran V.P., Tomar D., Kumar D., Mane A., Gill S.S., Patil T., Weber G.F., Kundu G.C. Tumor-derived osteopontin drives the resident fibroblast to myofibroblast differentiation through Twist1 to promote breast cancer progression. Oncogene. 2021;40:2002–2017. doi: 10.1038/s41388-021-01663-2. [DOI] [PubMed] [Google Scholar]
- 85.Kovacheva M., Zepp M., Schraad M., Berger S., Berger M.R. Conditional knockdown of osteopontin inhibits breast cancer skeletal metastasis. Int. J. Mol. Sci. 2019;20:4918. doi: 10.3390/ijms20194918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pang X., Gong K., Zhang X., Wu S., Cui Y., Qian B.Z. Osteopontin as a multifaceted driver of bone metastasis and drug resistance. Pharmacol. Res. 2019;144:235–244. doi: 10.1016/j.phrs.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 87.Zhao H., Chen Q., Alam A., Cui J., Suen K.C., Soo A.P., Eguchi S., Gu J., Ma D. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018;9:356. doi: 10.1038/s41419-018-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yokosaki Y., Tanaka K., Higashikawa F., Yamashita K., Eboshida A. Distinct structural requirements for binding of the integrins alphavbeta6, alphavbeta3, alphavbeta5, alpha5beta1 and alpha9beta1 to osteopontin. Matrix Biol. 2005;24:418–427. doi: 10.1016/j.matbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 89.Castello L.M., Raineri D., Salmi L., Clemente N., Vaschetto R., Quaglia M., Garzaro M., Gentilli S., Navalesi P., Cantaluppi V., Dianzani U., Aspesi A., Chiocchetti A. Osteopontin at the crossroads of inflammation and tumor progression. Mediat. Inflamm. 2017;2017 doi: 10.1155/2017/4049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robertson B.W., Bonsal L., Chellaiah M.A. Regulation of Erk1/2 activation by osteopontin in PC3 human prostate cancer cells. Mol. Cancer. 2010;9:260. doi: 10.1186/1476-4598-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurisetty V.V., Johnston P.G., Johnston N., Erwin P., Crowe P., Fernig D.G., Campbell F.C., Anderson I.P., Rudland P.S., El-Tanani M.K. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:7139–7149. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 92.Yuen H.F., Chan K.K., Grills C., Murray J.T., Platt-Higgins A., Eldin O.S., O'Byrne K., Janne P., Fennell D.A., Johnston P.G., Rudland P.S., El-Tanani M. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:380–391. doi: 10.1158/1078-0432.CCR-11-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boyd N.F., Martin L.J., Bronskill M., Yaffe M.J., Duric N., Minkin S. Breast tissue composition and susceptibility to breast cancer. J. Natl. Cancer Inst. 2010;102:1224–1237. doi: 10.1093/jnci/djq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pettersson A., Hankinson S.E., Willett W.C., Lagiou P., Trichopoulos D., Tamimi R.M. Nondense mammographic area and risk of breast cancer. Breast Cancer Res. 2011;13:R100. doi: 10.1186/bcr3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindahl G., Rzepecka A., Dabrosin C. Increased extracellular osteopontin levels in normal human breast tissue at high risk of developing cancer and its association with inflammatory biomarkers in situ. Front. Oncol. 2019;9:746. doi: 10.3389/fonc.2019.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cabia B., Andrade S., Carreira M.C., Casanueva F.F., Crujeiras A.B. A role for novel adipose tissue-secreted factors in obesity-related carcinogenesis. Obes. Rev. 2016;17:361–376. doi: 10.1111/obr.12377. [DOI] [PubMed] [Google Scholar]
- 97.El-Tanani M.K., Yuen H.F., Shi Z., Platt-Higgins A., Buckley N.E., Mullan P.B., Harkin D.P., Johnston P.G., Rudland P.S. Osteopontin can act as an effector for a germline mutation of BRCA1 in malignant transformation of breast cancer-related cells. Cancer Sci. 2010;101:1354–1360. doi: 10.1111/j.1349-7006.2010.01561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin C., Moorehead R.A. Polycomb repressor complex 2 function in breast cancer. Int. J. Oncol. 2020;57:1085–1094. doi: 10.3892/ijo.2020.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gautam N., Kaur M., Kaur S. Structural assembly of Polycomb group protein and Insight of EZH2 in cancer progression: a review. J. Cancer Res. Ther. 2021;17:311–326. doi: 10.4103/jcrt.JCRT_1090_19. [DOI] [PubMed] [Google Scholar]
- 100.Yoo K.H., Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int. J. Biol. Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang H., Jia P. MiR-153-3p inhibits osteogenic differentiation of periodontal ligament stem cells through KDM6A-induced demethylation of H3K27me3. J. Periodont. Res. 2021;56:379–387. doi: 10.1111/jre.12830. [DOI] [PubMed] [Google Scholar]
- 102.Roth M.J., Moorehead R.A. The miR-200 family in normal mammary gland development. BMC Dev. Biol. 2021;21:12. doi: 10.1186/s12861-021-00243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.MacMahon B., Cole P., Lin T.M., Lowe C.R., Mirra A.P., Ravnihar B., Salber E.J., Valaoras V.G., Yuasa S. Age at first birth and breast cancer risk. Bull. World Health Organ. 1970;43:209–221. [PMC free article] [PubMed] [Google Scholar]
- 104.Ewertz M., Duffy S.W., Adami H.O., Kvale G., Lund E., Meirik O., Mellemgaard A., Soini I., Tulinius H. Age at first birth, parity and risk of breast cancer: a meta-analysis of 8 studies from the Nordic countries. Int. J. Cancer. 1990;46:597–603. doi: 10.1002/ijc.2910460408. [DOI] [PubMed] [Google Scholar]
- 105.Husby A., Wohlfahrt J., Oyen N., Melbye M. Pregnancy duration and breast cancer risk. Nat. Commun. 2018;9:4255. doi: 10.1038/s41467-018-06748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1. Hierarchical clustering of RNA sequencing data of mammary glands from 55-day old control, MTB-200ba429, MTB-IGFIR and MTB-IGFIRba429 mice.
Supplemental File 2. Transcripts differentially expressed in mammary glands from 55-day old MTB-200ba429 mice vs control mice, MTB-IGFIR mice vs control mice and MTB-IGFIR mice vs MTB-IGFIRba429 mice. Log fold-change (LogFC) >1 and false discovery rate (FDR) < 0.01 were used as cut-offs.
Supplemental File 3. Immunohistochemistry using an antibody that detects both Saa1 and Saa2 in (A,D) mammary gland from a 55-day old MTB-IGFIRba429 mouse and (B,E) a hyperplastic lesion in the mammary gland of a 55-day old MTB-IGFIR mouse. (C,D) Liver tissue stained with the antibody served as a positive control. Scale bars for (D-F) are 100 µm and for (G-I) are 30 µm.
Data Availability Statement
RNA sequencing has been uploaded to GEO under accession number GSE180264.