Abstract
Background
Diagnosis of pancreatic cancer (Pca) is challenging. This study investigated the value of plasma-derived exosome miR-19b (Exo-miR-19b) in diagnosing patients with Pca.
Methods
Plasma was collected from 62 patients with Pca, 30 patients with other pancreatic tumor (OPT), 23 patients with chronic pancreatitis (CP), and 53 healthy volunteers. MiR-19b levels in plasma-derived exosomes were detected.
Results
Plasma-derived Exo-miR-19b levels normalized using miR-1228 were significantly lower in Pca patients than in patients with OPT, CP patients, and healthy volunteers. The diagnostic values of Exo-miR-19b normalized using miR-1228 were superior to those of serum cancer antigen 19-9 (CA19-9) in differentiating Pca patients from healthy volunteers (area under the curve (AUC): 0.942 vs. 0.813, p = 0.0054), potentially better than those of CA19-9 in differentiating Pca patients from CP patients (AUC: 0.898 vs. 0.792, p = 0.0720), and equivalent to those of CA19-9 in differentiating Pca patients from patients with OPT (AUC: 0.810 vs. 0.793, p = 0.8206). When normalized using Caenorhabditis elegans miR-39 (cel-miR-39), Exo-miR-19b levels in Pca patients were significantly higher than those in patients with OPT, CP patients, and healthy volunteers. The diagnostic values of Exo-miR-19b normalized using cel-miR-39 were equivalent to those of CA19-9 in differentiating Pca patients from healthy volunteers (AUC: 0.781 vs. 0.813, p = 0.6118) and CP patients (AUC: 0.672 vs. 0.792, p = 0.1235), while they were inferior to those of CA19-9 in differentiating Pca patients from patients with OPT (AUC: 0.631 vs. 0.793, p = 0.0353).
Conclusion
Plasma-derived Exo-miR-19b is a promising diagnostic marker for Pca. The diagnostic value of plasma-derived Exo-miR-19b normalized using miR-1228 is superior to that of serum CA19-9 in differentiating patients with Pca from healthy volunteers.
Keywords: pancreatic cancer, liquid biopsy, exosome, miRNA, biomarker, macrophages
Introduction
Pancreatic cancer (Pca) is a lethal disease with a 5-year survival rate of 10% and ranks as the fourth leading cause of cancer-related deaths in the United States (1). The difficulty in diagnosis of early stage diseases partly accounts for the poor prognosis of Pca. Several biomarkers for diagnosing Pca have been reported; however, most of these biomarkers have remained in the preclinical stage. Currently, only serum cancer antigen 19-9 (CA19-9) is proposed for the routine management of Pca. However, elevated CA19-9 is also observed in biliary infection or obstruction as well as other digestive cancers and inflammatory diseases and presents a moderate diagnostic value with a sensitivity and a specificity of 79% and 82%, respectively (2). Additionally, CA19-9 is not applicable for patients with negative expression of Lewis antigen, which is critical for CA19-9 biosynthesis, and the National Comprehensive Cancer Network guideline indicates that CA 19-9 is undetectable in Lewis (−) individuals (3). Notably, Luo et al. (4) reported that 8.4% of Pca patients (N = 1482) were Lewis (−). These results indicate that diagnosis of Pca based on CA19-9 will lead to missed diagnosis, and therefore, a more accurate circulating biomarker for Pca is urgently needed.
The application of liquid biopsy of circulating free DNA, tumor cells, or exosomes for cancer diagnosis has shown promise (5, 6), and among these markers, exosomes have been the subject of investigation. Exosomes are small (30–200 nm) vesicular structures that can carry pathogenic miRNAs, lncRNAs, mRNAs, DNA fragments, and proteins (7, 8). Several blood-derived exosome markers have been developed and show potential diagnostic value in Pca (5, 9). Circulating miRNAs serve as diagnostic biomarkers in multiple types of cancers, including biliary tract cancer (10), colorectal cancer (CRC) (11), Pca (5, 12), glioblastoma (13), prostate cancer (14), lung cancer (15), hepatocellular carcinoma (16), and other tumors (17). Unlike the multiple reports on circulating miRNAs, only a few studies have reported the diagnostic values of plasma- or serum-derived exosome miRNAs in Pca (18, 19).
Our previous study indicated that several plasma miRNAs were deregulated in Pca patients and presented diagnostic value12. Among the identified miRNAs, plasma miR-19b was significantly upregulated in patients with Pca and presented moderate diagnostic values in discriminating patients with Pca from those with chronic pancreatitis (CP) and pancreatic neuroendocrine tumor. Additionally, circulating exosomal miR-19b exhibited oncogenic functions in gastric cancer, lung adenocarcinoma, and esophageal squamous cell carcinoma (ESCC) (20–22).
In this study, we aimed to evaluate the potential diagnostic values of plasma-derived exosome miR-19b (Exo-miR-19b) in Pca. We investigated the expression levels and diagnostic values of Exo-miR-19b in patients with Pca, patients with CP, patients with other pancreatic tumor (OPT), and healthy volunteers.
Materials and Methods
Ethics Statement
This study was approved by the Medical Ethics Committee of Qilu Hospital of Shandong University. Written informed consent was obtained from all subjects.
Diagnostic Criteria for Pancreatic Diseases
Pca was cytologically or pathologically diagnosed depending on the cytological or histological examinations. OPTs were pathologically diagnosed depending on histological examinations of the resected specimen, including pancreatic neuroendocrine tumor, solid pseudopapillary tumor, serous or mucinous cystadenomas, intraductal papillary mucinous neoplasms, and epithelial cysts. CP was diagnosed on the basis of clinical diagnostic criteria or histological examinations.
Sample Collection and Exosome RNA Isolation
The serum CA19-9 levels of all included subjects could be obtained from the medical records; if not, the subjects were excluded. Pca patients undergoing neoadjuvant therapy were excluded. Peripheral venous blood (5 ml) was collected in sterile ethylene diamine tetraacetic acid-treated anticoagulant tubes before clinical intervention or surgery. The blood samples were centrifuged at 3,000 revolutions per minute (rpm) for 10 min; then plasmas were collected and stored at −80°C for further isolation of exosome. Exosomes were isolated from plasma using exoRNeasy Serum/Plasma Midi Kit (the kit can directly purify total exosomes RNA from plasma without the intermediate isolation of exosomes, Exiqon QIAGEN, #77044) according to the manufacturer’s instructions as reported by the previous study (23); then the RNA was harvested.
Quantitative Real-Time PCR for Detecting Plasma-Derived Exo-miR-19b
Synthetic Caenorhabditis elegans miR-39 (cel-miR-39, RiboBio, Guangzhou, China) at 30 nM was added to each exosome RNA sample for normalization before qRT-PCR. MiRNA was converted to cDNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The reverse transcription reactions were carried out at 16°C for 30 min, at 42°C for 30 min, and at 85°C for 5 min and held at 4°C. cDNA was stored at −20°C until use. A total of 20 μl of amplification system containing 1.33 μl of cDNA, 10 μl of TaqMan 2× Universal PCR Master Mix with no AmpErase UNG (Applied Biosystems), 1 μl of miRNA-specific probe, and 7.67 μl of nuclease-free water was used for analyzing the expression of miRNA. qRT-PCR ran on a Stepone Plus real-time PCR system (Applied Biosystems) and the reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. The cycle threshold (CT) values were calculated with SDS software (Applied Biosystems). All reactions were performed in triplicate. The expression levels of miRNA were normalized using the endogenous control (miR-1228) (24) or the exogenous control (cel-miR-39). ΔCT was calculated by subtracting CT values of the miRNA from CT values of the control. The relative expression levels of miRNA were calculated with the equation 2−ΔCT.
Statistical Analysis
All the statistical analyzes were performed by SPSS v.23.0 (IBM Corp., Armonk, NY). A two-sided p < 0.05 was considered as statistical significance. Continuous data were presented as the mean ± SD and analyzed using Student’s t-tests. Receiver operating characteristic (ROC) curves were created; and the area under the curve (AUC), sensitivity, and specificity were calculated to evaluate the diagnostic values of plasma-derived Exo-miR-19b using MedCalc Statistical Software version 19.0.4 (MedCalc Software bvba; http://www.medcalc.org). AUCs of plasma-derived Exo-miR-19b (AUC1) and CA19-9 (AUC2) were compared using Z tests.
Results
The Expression Levels of Plasma-Derived Exo-miR-19b in Patients With Pancreatic Cancer and Control Groups Normalized Using MiR-1228
Plasma samples were collected from 168 individuals, including 62 Pca patients, 30 patients with OPT, 23 CP patients, and 53 healthy volunteers. Exosomes RNA was extracted from the plasma samples, and Exo-miR-19b levels were determined by qRT-PCR.
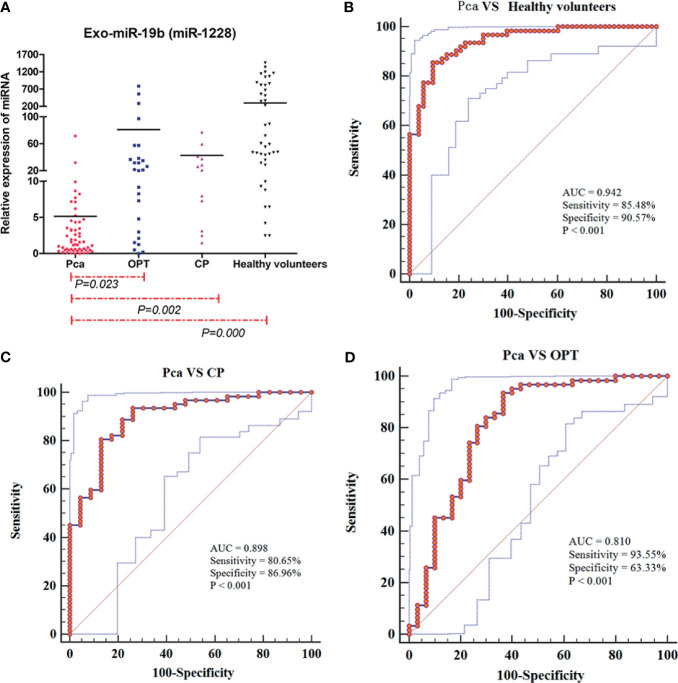
The levels of Exo-miR-19b in patients with Pca normalized using miR-1228 were significantly lower than the levels in patients with OPT, patients with CP, and healthy volunteers (p < 0.05, Figure 1A, Supplementary Material 1).
Figure 1.
Expression levels and diagnostic values of plasma-derived Exo-miR-19b normalized using miR-1228. (A) Exo-miR-19b levels were detected by qRT-PCR. (B) ROC for differentiating Pca patients from healthy volunteers. (C) ROC for differentiating Pca patients from CP patients. (D) ROC for differentiating Pca patients from patients with OPT. AUC, area under the curve; CP, chronic pancreatitis; OPT, other pancreatic tumor; Pca, pancreatic cancer; ROC, receiver operating characteristic.
The Diagnostic Value of Plasma-Derived Exo-miR-19b Normalized Using MiR-1228
Levels of plasma-derived Exo-miR-19b normalized using miR-1228 displayed diagnostic value in differentiating patients with Pca from patients with OPT (AUC = 0.810), patients with CP (AUC = 0.898), and healthy volunteers (AUC = 0.942) (Figures 1B–D, Supplementary Material 2).
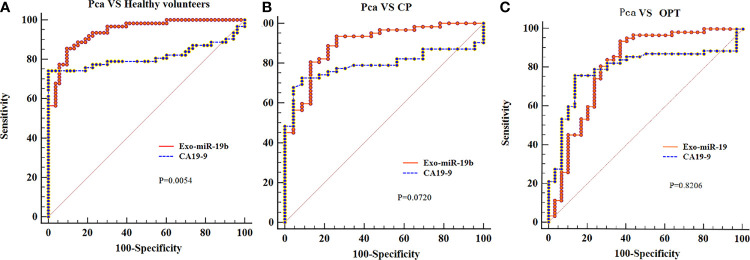
Exo-miR-19b was superior to serum CA19-9 in differentiating patients with Pca from healthy volunteers (AUC: 0.942 vs. 0.813, p = 0.0054), potentially better than CA19-9 in differentiating patients with Pca from CP (AUC: 0.898 vs. 0.792, p = 0.0720), and equivalent to CA19-9 in differentiating patients with Pca from patients with OPT (AUC: 0.810 vs. 0.793, p = 0.8206) (Figure 2, Supplementary Material 3).
Figure 2.
Comparison of the areas under the curves of CA19-9 with plasma-derived Exo-miR-19b levels normalized using miR-1228. (A) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from healthy volunteers. (B) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from CP. (C) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from OPT. AUC, area under the curve; CP, chronic pancreatitis; OPT, other pancreatic tumor; Pca, pancreatic cancer.
The Diagnostic Value of Plasma-Derived Exo-miR-19b Normalized Using cel-miR-39
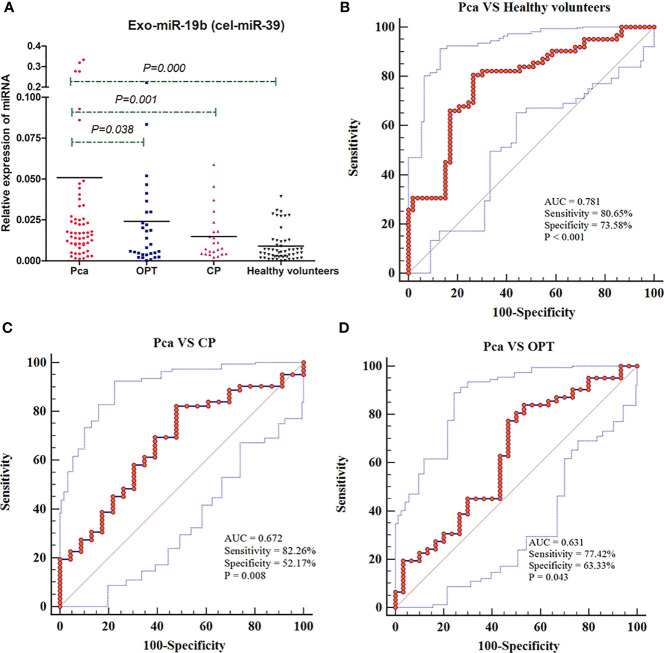
The levels of Exo-miR-19b normalized using cel-miR-39 were significantly higher in patients with Pca than in patients with OPT, patients with CP, and healthy volunteers (p < 0.05, Figure 3A, Supplementary Material 1).
Figure 3.
Expression levels and diagnostic values of plasma-derived Exo-miR-19b normalized using cel-miR-39. (A) Exo-miR-19b levels were detected by qRT-PCR. (B) ROC for differentiating Pca patients from healthy volunteers. (C) ROC for differentiating Pca patients from CP patients. (D) ROC for differentiating Pca patients from patients with OPT. AUC, area under the curve; CP, chronic pancreatitis; OPT, other pancreatic tumor; Pca, pancreatic cancer; ROC, Receiver operating characteristic.
Levels of plasma-derived Exo-miR-19b normalized using cel-miR-39 displayed diagnostic value in differentiating patients with Pca from patients with OPT (AUC = 0.631), patients with CP (AUC = 0.672), and healthy volunteers (AUC = 0.781) (Figures 3B–D, Supplementary Material 2).
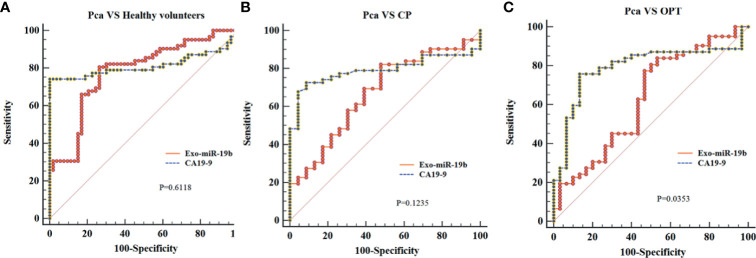
Exo-miR-19b was equivalent to CA19-9 in differentiating patients with Pca from healthy volunteers (AUC: 0.781 vs. 0.813, p = 0.6118) and patients with CP (AUC: 0.672 vs. 0.792, p = 0.1235), while it was inferior to CA19-9 in differentiating patients with Pca from patients with OPT (AUC: 0.631 vs. 0.793, p = 0.0353) (Figure 4, Supplementary Material 3).
Figure 4.
Comparison of the areas under the curves of CA19-9 with plasma-derived Exo-miR-19b levels normalized using cel-miR-39. (A) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from healthy volunteers. (B) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from CP. (C) AUCs of Exo-miR-19b and CA19-9 in discriminating patients with Pca from OPT. AUC, area under the curve; CP, chronic pancreatitis; OPT, other pancreatic tumor; Pca, pancreatic cancer.
Discussion
Although multiple studies are trying to find effective diagnostic markers for Pca, early diagnosis of Pca is still difficult (25). Several circulating exosomal miRNA biomarkers have been reported for the diagnosis of Pca19, including miR-1226 (26), miR-196a/1246 (27), miR-191/21/451a (28), and miR-10b/21/30c/161a/let-7a (29). However, few studies have investigated the diagnostic value of Exo-miR-19b (20), and no reports have been performed in Pca. In this study, we showed that plasma-derived Exo-miR-19b level normalized using miR-1228 was superior to that using serum CA19-9 in differentiating patients with Pca from healthy volunteers, potentially better than that using CA19-9 in differentiating patients with Pca from patients with CP, and equivalent to that using CA19-9 in differentiating patients with Pca from patients with OPT.
The selection of an appropriate endogenous control for normalization of circulating miRNA expression is crucial for obtaining reliable data. Several miRNAs are commonly used as endogenous controls for quantifying circulating miRNAs, such as miR-16, miR-223, let-7a, and RNU6B (24, 30). However, none of the endogenous miRNAs have been widely accepted. Our study used endogenous miR-1228 as a control for the quantification of plasma-derived Exo-miR-19b. MiR-1228 is widely involved in metabolism-related signalling pathways and organ morphology and not influenced by hemolysis (30, 31), indicating the suitability of miR-1228 as a housekeeping miRNA. Hu et al. (24) examined a large cohort of 544 subjects to identify a stable endogenous control for the quantification of circulating miRNAs in cancer patients. The authors found that miR-1228 functioned as a housekeeping gene and was stable in plasma samples from different kinds of tumors, including hepatocellular cancer, CRC, lung cancer, ESCC, gastric cancer, renal cancer, prostate cancer, and breast cancer. Duran-Sanchon et al. (31) verified the housekeeping role of miR-1228 in CRC by a large sample study; the authors showed that miR-1228 was an adequate endogenous control for circulating miRNA analysis in CRC and demonstrated a variability and stability superior to that of miR-16. Danese et al. (30) reported the expression levels and stability of miR-1228; the study found that miR-1228 displayed median PCR-derived cycle threshold values and was sufficiently homogenous and stable in exosomes, plasma, and tissues from CRC patients and healthy controls.
Previous studies on miR-19b functions indicated its oncogenic role (32, 33), suggesting that plasma-derived Exo-miR-19b would be upregulated in Pca patients compared with controls. However, our study found conflicting results. While the plasma-derived Exo-miR-19b level in Pca patients was significantly lower than that in the control subjects when normalized using miR-1228, the plasma-derived Exo-miR-19b in Pca patients was significantly higher than that in controls when normalized using cel-miR-39, which was consistent with the literature (20, 21). While the mechanism and reason underlying the differences in these results are not yet clear, some potential explanations are possible. First, some studies reported miR-1228 as a functional miRNA (34, 35), which might influence its value as an internal control. The role of miR-1228 in Pca has not been investigated, and the housekeeping role and the value of normalization for plasma-derived exosome miRNA need further experimental verification. Second, the use of an exogenous control has limitations. Exogenous controls have shown utility for quality control for RNA extraction and PCR, but useless in quality control for exosome extraction. However, quality control during extraction is important for exosome studies. Because of the differences between the expression profiles of miRNAs in plasma, serum, and blood cells (36), quality control of exosome extraction is necessary to eliminate the influence of blood components. Finally, the source of plasma-derived Exo-miR-19b is unclear. Besides Pca cells, blood cells, bone marrow mesenchymal stem cells, endothelial cells, and other cells secrete Exo-miR-19b [4-6]. The function of miR-19b or Exo-miR-19b in Pca cells might be not equal to that of plasma-derived Exo-miR-19b in Pca patients. Further studies are necessary to investigate the biological functions of plasma-derived Exo-miR-19b in Pca patients.
This study has some limitations. The control diseases are not comprehensive; acute pancreatitis, obstructive jaundice caused by benign diseases, and other digestive cancers are no included. CP is diagnosed according to either the clinical criteria or histological examinations. However, CP has a potential of malignant transformation; diagnosis with clinical criteria might miss cases with focal cancerization. The sample size is moderate; there is no stratified analysis of Pca cases; influences of jaundice, and locations and stages of the tumors on the levels of plasma-derived Exo-miR-19b are unknown. A larger sample muticenter study is helpful to disclose the diagnostic value of plasma-derived Exo-miR-19b.
In conclusion, we reported the diagnostic value of plasma-derived Exo-miR-19b in Pca. Our results showed that plasma-derived Exo-miR-19b level normalized using miR-1228 was superior to serum CA19-9 in differentiating patients with Pca from healthy volunteers, potentially better than CA19-9 in differentiating patients with Pca from patients with CP, and equivalent to CA19-9 in differentiating patients with Pca from patients with OPT.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Qilu Hospital of Shandong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JX proposed and designed the study. LW and JW wrote the draft. LW, JW, NY, FL, HZ, and SC collected and analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. JX and FL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81502051), the Shandong Provincial Natural Science Foundation, China (ZR2020MH256), Medical Health Science and Technology Project of Shandong Provincial Health Commission (2019WS386), Key Technology Research and Development Program of Shandong (2019GSF108065), and the China Postdoctoral Science Foundation (2018M632681).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Gabrielle White Wolf, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.739111/full#supplementary-material
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistic. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Goonetilleke KS, Siriwardena AK. Systematic Review of Carbohydrate Antigen (CA 19-9) as a Biochemical Marker in the Diagnosis of Pancreatic Cancer. Eur J Surg Oncol (2007) 33(3):266–70. doi: 10.1016/j.ejso.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 3.NCCN . NCCN Clinical Practice Guidelines in Oncology: Pancreatic Adenocarcinoma (Version 1.2020). Available at: http://www.nccn.org.
- 4.Luo G, Fan Z, Cheng H, Jin K, Guo M, Lu Y, et al. New Observations on the Utility of CA19-9 as a Biomarker in Lewis Negative Patients With Pancreatic Cancer. Pancreatology (2018) 18(8):971–6. doi: 10.1016/j.pan.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 5.Al-Shaheri FN, Alhamdani MSS, Bauer AS, Giese N, Büchler MW, Hackert T, et al. Blood Biomarkers for Differential Diagnosis and Early Detection of Pancreatic Cancer. Cancer Treat Rev (2021) 96:102193. doi: 10.1016/j.ctrv.2021.102193 [DOI] [PubMed] [Google Scholar]
- 6.Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov (2021) 11(4):858–73. doi: 10.1158/2159-8290.cd-20-1311 [DOI] [PubMed] [Google Scholar]
- 7.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902 [DOI] [PubMed] [Google Scholar]
- 8.Gurung S, Perocheau D, Touramanidou L, Baruteau J. The Exosome Journey: From Biogenesis to Uptake and Intracellular Signalling. Cell Commun Signal (2021) 19(1):47. doi: 10.1186/s12964-021-00730-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, et al. Early Detection of Pancreatic Cancer: Where Are We Now and Where are We Going? Int J Cancer (2017. a) 141(2):231–41. doi: 10.1002/ijc.30670 [DOI] [PubMed] [Google Scholar]
- 10.Ofoeyeno N, Ekpenyong E, Braconi C. Pathogenetic Role and Clinical Implications of Regulatory RNAs in Biliary Tract Cancer. Cancers (Basel) (2020) 13(1):12. doi: 10.3390/cancers13010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alves Dos Santos K, Clemente Dos Santos IC, Santos Silva C, Gomes Ribeiro H, de Farias Domingos I, Nogueira Silbiger V. Circulating Exosomal miRNAs as Biomarkers for the Diagnosis and Prognosis of Colorectal Cancer. Int J Mol Sci (2020) 22(1). doi: 10.3390/ijms22010346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Cao Z, Liu W, You L, Zhou L, Wang C, et al. Plasma miRNAs Effectively Distinguish Patients With Pancreatic Cancer From Controls: A Multicenter Study. Ann Surg (2016) 263(6):1173–9. doi: 10.1097/sla.0000000000001345 [DOI] [PubMed] [Google Scholar]
- 13.Ahmed SP, Castresana JS, Shahi MH. Glioblastoma and MiRNAs. Cancers (Basel) (2021) 13(7):1581. doi: 10.3390/cancers13071581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrighetti N, Beretta GL. miRNAs as Therapeutic Tools and Biomarkers for Prostate Cancer. Pharmaceutics (2021) 13(3):380. doi: 10.3390/pharmaceutics13030380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gayosso-Gómez LV, Ortiz-Quintero B. Circulating MicroRNAs in Blood and Other Body Fluids as Biomarkers for Diagnosis, Prognosis, and Therapy Response in Lung Cancer. Diagnostics (Basel) (2021) 11(3):421. doi: 10.3390/diagnostics11030421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Masaki T. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma: A Review. Cancers (Basel) (2021) 13(3):514. doi: 10.3390/cancers13030514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvão-Lima LJ, Morais AHF, Valentim RAM, Barreto E. miRNAs as Biomarkers for Early Cancer Detection and Their Application in the Development of New Diagnostic Tools. BioMed Eng Online (2021) 20(1):21. doi: 10.1186/s12938-021-00857-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Z, Jiang W, Zhang S, Li P. The State of the Art on Blood MicroRNAs in Pancreatic Ductal Adenocarcinoma. Anal Cell Pathol (Amst) (2019) 2019:9419072. doi: 10.1155/2019/9419072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ariston Gabriel AN, Wang F, Jiao Q, Yvette U, Yang X, Al-Ameri SA, et al. The Involvement of Exosomes in the Diagnosis and Treatment of Pancreatic Cancer. Mol Cancer (2020) 19(1):132. doi: 10.1186/s12943-020-01245-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A Serum Exosomal microRNA Panel as a Potential Biomarker Test for Gastric Cancer. Biochem Biophys Res Commun (2017) 493(3):1322–8. doi: 10.1016/j.bbrc.2017.10.003 [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, et al. A six-microRNA Panel in Plasma Was Identified as a Potential Biomarker for Lung Adenocarcinoma Diagnosis. Oncotarget (2017. b) 8(4):6513–25. doi: 10.18632/oncotarget.14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Q, Zhu Z, Song L, He Z. Transferred by Exosomes-Derived MiR-19b-3p Targets PTEN to Regulate Esophageal Cancer Cell Apoptosis, Migration and Invasion. Biosci Rep (2020) 40(11):BSR20201858. doi: 10.1042/bsr20201858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Zhou Y, Wang ZY, Yan BX, Zhou WF, Wang TT, et al. Exosomal microRNA Profiles From Serum and Cerebrospinal Fluid in Neurosyphilis. Sex Transm Infect (2019) 95(4):246–50. doi: 10.1136/sextrans-2018-053813 [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, et al. Human miR-1228 as a Stable Endogenous Control for the Quantification of Circulating microRNAs in Cancer Patients. Int J Cancer (2014) 135(5):1187–94. doi: 10.1002/ijc.28757 [DOI] [PubMed] [Google Scholar]
- 25.Li C, Li S, Zhang F, Wu M, Liang H, Song J, et al. Endothelial Microparticles-Mediated Transfer of microRNA-19b Promotes Atherosclerosis via Activating Perivascular Adipose Tissue Inflammation in Apoe(-/-) Mice. Biochem Biophys Res Commun (2018) 495(2):1922–9. doi: 10.1016/j.bbrc.2017.11.195 [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Wang J, Cui W, Liu Y, Zhou H, Wang Y, et al. Serum Exosomal miRNA-1226 as Potential Biomarker of Pancreatic Ductal Adenocarcinoma. Onco Targets Ther (2021) 14:1441–51. doi: 10.2147/ott.s296816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu YF, Hannafon BN, Zhao YD, Postier RG, Ding WQ. Plasma Exosome miR-196a and miR-1246 Are Potential Indicators of Localized Pancreatic Cancer. Oncotarget (2017) 8(44):77028–40. doi: 10.18632/oncotarget.20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, et al. An Elevated Expression of Serum Exosomal microRNA-191, - 21, -451a of Pancreatic Neoplasm Is Considered to be Efficient Diagnostic Marker. BMC Cancer (2018) 18(1):116. doi: 10.1186/s12885-018-4006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA Signature in Circulating Exosomes Is Superior to Exosomal Glypican-1 Levels for Diagnosing Pancreatic Cancer. Cancer Lett (2017) 393:86–93. doi: 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danese E, Minicozzi AM, Benati M, Paviati E, Lima-Oliveira G, Gusella M, et al. Reference miRNAs for Colorectal Cancer: Analysis and Verification of Current Data. Sci Rep (2017) 7(1):8413. doi: 10.1038/s41598-017-08784-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duran-Sanchon S, Vila-Navarro E, Marcuello M, Lozano JJ, Muñoz J, Cubiella J, et al. Validation of miR-1228-3p as Housekeeping for MicroRNA Analysis in Liquid Biopsies From Colorectal Cancer Patients. Biomolecules (2019) 10(1):16. doi: 10.3390/biom10010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuziwara CS, Kimura ET. Insights Into Regulation of the miR-17-92 Cluster of miRNAs in Cancer. Front Med (Lausanne) (2015) 2:64. doi: 10.3389/fmed.2015.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang LL, Wang XH, Sun BF, Zhang XD, Zhu XH, Yu ZJ, et al. Expression, Regulation and Mechanism of Action of the miR-17-92 Cluster in Tumor Cells (Review). Int J Mol Med (2017) 40(6):1624–30. doi: 10.3892/ijmm.2017.3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Dai J, Deng H, Wan H, Liu M, Wang J, et al. miR-1228 Promotes the Proliferation and Metastasis of Hepatoma Cells Through a P53 Forward Feedback Loop. Br J Cancer (2015) 112(2):365–74. doi: 10.1038/bjc.2014.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang L, Gao H, Wang L, Wang N, Zhang S, Zhou X, et al. Exosomes Derived From miR-1228 Overexpressing Bone Marrow-Mesenchymal Stem Cells Promote Growth of Gastric Cancer Cells. Aging (Albany NY) (2021) 13. doi: 10.18632/aging.202878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA Spectrum Between Serum and Plasma. PloS One (2012) 7(7):e41561. doi: 10.1371/journal.pone.0041561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.