Abstract
Epistasis—gene–gene interaction—is common for mutations with large phenotypic effects in humans and model organisms. Epistasis impacts quantitative genetic models of speciation, response to natural and artificial selection, genetic mapping, and personalized medicine. However, the existence and magnitude of epistasis between alleles with small quantitative phenotypic effects are controversial and difficult to assess. Here, we use the Drosophila melanogaster Genetic Reference Panel of sequenced inbred lines to evaluate the magnitude of naturally occurring epistasis modifying the effects of mutations in jing and inv, two transcription factors that have subtle quantitative effects on head morphology as homozygotes. We find significant epistasis for both mutations and performed single marker genome-wide association analyses to map candidate modifier variants and loci affecting head morphology. A subset of these loci was significantly enriched for a known genetic interaction network, and mutations of the candidate epistatic modifier loci also affect head morphology.
Keywords: Drosophila Genetic Reference Panel, genome-wide association analyses, modifier loci, genetic interaction network
Introduction
Epistasis—gene–gene interaction—is common for mutations with large phenotypic effects. In humans, the same mutation for a rare disease can have different phenotypic manifestations in different patients, thought to be attributable to modifier variants in the different patient genetic backgrounds (Davidson et al. 2018; Bis-Brewer et al. 2020; Rahit and Tarailo-Graovac 2020; Sun et al. 2020). In model organisms, such as yeast, Arabidopsis, and Drosophila, genetic screens for mutations that enhance or suppress the effects of a mutation in a focal gene have been instrumental in deriving canonical genetic interaction networks and signaling pathways. However, the magnitude and even existence of epistasis between alleles with small quantitative phenotypic effects have been controversial since the earliest formulations of quantitative genetics theory (Fisher 1918; Wright 1931). Epistasis impacts quantitative genetic models of speciation, response to natural and artificial selection, genetic mapping, and personalized medicine (Phillips 2008; Mackay 2014). Unfortunately, the contribution of epistasis to quantitative genetic variation cannot be inferred from the relative magnitude of additive, dominance, and epistatic variance components, because additive and dominance variances are defined such that they contain most variance generated by epistatic effects for most allele frequencies, with very little contribution from the epistatic variance component (Falconer and Mackay 1996; Lynch and Walsh 1998; Hill et al. 2008; Huang and Mackay 2016).
Much of the evidence for epistasis for quantitative traits also comes from model organisms. In Drosophila melanogaster, epistasis between transposon-tagged mutations with subtle, quantitative phenotypic effects in different genes that were generated in a common genetic background has been demonstrated for metabolic phenotypes (Clark and Wang 1997), olfactory behavior (Fedorowicz et al. 1998; Anholt et al. 2003; Sambandan et al. 2006; Rollmann et al. 2007), locomotor behavior (van Swinderen and Greenspan 2005; Yamamoto et al. 2008), life span (Magwire et al. 2010), and aggressive behavior (Edwards et al. 2009; Zwarts et al. 2011). Furthermore, identical transposon-tagged mutations have different quantitative phenotypic effects in different wild-type genetic backgrounds (Rollmann et al. 2006; Magwire et al. 2010). However, these studies do not scale to large numbers of mutations; and interactions between mutations may not reflect interactions between naturally occurring variants.
Further evidence for strong and pervasive epistatic interactions comes from chromosome substitution lines, whereby single chromosomes (or chromosome segments) of one inbred strain are introgressed into the genetic background of another. Epistasis is indicated when the sum of the effects of all chromosomes (or chromosome segments) on a quantitative trait is significantly different from the effect estimated from the difference between the two parental genotypes (Edwards and Mackay 2009; Nadeau et al. 2012; Spiezio et al. 2012). Linkage mapping of quantitative trait loci (QTLs) in segregating populations derived from two inbred lines often detects epistatically interacting QTLs because epistasis is maximal when allele frequencies are intermediate (Flint and Mackay 2009; Mackay 2014). While chromosome substitution/introgression and QTL mapping can confirm the existence of epistasis, identifying the interacting loci is more challenging as large numbers of segregating progeny and multiple generations of recombination are needed to break down the linkage disequilibrium (LD) generated by crossing the inbred parental lines (Falconer and Mackay 1996; Lynch and Walsh 1998).
Mapping pairwise epistatic interactions in outbred populations or inbred lines derived from outbred populations has the potential to more precisely map interacting loci due to reduced LD relative to recent crosses of inbred lines, but suffers from low statistical power because of the exponentially larger number of tests needed than for single marker analysis [i.e., for n variants there are n(n−1)/2 possible pairwise epistatic interactions]. Therefore, methods that reduce the search space can increase the power to detect interacting loci. One such method stems from the realization that the main effect of a focal locus will differ for different allele frequencies of an epistatically interacting locus (Greene et al. 2009; Huang et al. 2012). Thus, marker associations for a quantitative trait will not replicate between populations with different allele frequencies in the presence of epistasis, and this can be exploited to restrict the genome-wide search for epistatic interactions using only the loci whose effects did not replicate as focal loci (Huang et al. 2012; Shorter et al. 2015). A second strategy that can only be implemented in model organisms is to cross (or introgress) mutant and wild-type loci of a focal gene into a mapping population. If the difference between the quantitative effects of the mutant and wild type varies significantly, there is epistasis, and the modifying loci can be mapped with the same power as for a single marker analysis of the same population. Many such studies in D. melanogaster have revealed extensive cryptic natural variation that modifies effects of mutations with large effects (Rendel 1959; Gibson and van Helden 1997; Polaczyk et al. 1998; Rutherford and Lindquist 1998; Gibson et al. 1999; Dworkin et al. 2009; Chow et al. 2016; Lavoy et al. 2018; Palu et al. 2019; Talsness et al. 2020). However, there are relatively few studies that have quantified the extent to which naturally occurring variation epistatically modifies the effects of mutations with subtle quantitative effects (Yamamoto et al. 2009; Swarup et al. 2012; He et al. 2016).
Here, we use the Drosophila Genetic Reference Panel (DGRP) of sequenced inbred lines (Mackay et al. 2012; Huang et al. 2014) to evaluate the magnitude of naturally occurring epistasis modifying the effects of mutations in the transcription factors jing and inv that have subtle quantitative effects on head morphology as homozygotes (Carreira et al. 2009). We find significant epistasis for both mutations and performed single marker genome-wide association (GWA) analyses to map candidate modifier variants and loci affecting head morphology. A subset of these loci was significantly enriched for a known genetic interaction network, and mutations of the candidate epistatic modifier loci also affect head morphology.
Materials and methods
Drosophila stocks
We used 197 DGRP (Mackay et al. 2012; Huang et al. 2014) lines (Supplementary Table S1); invected (invBG00846) and jing (jingBG02314) P[GT1]-element insertion lines in the Canton S (B) (CSB) coisogenic background (Magwire et al. 2010); and CSB coisogenic control lines to evaluate interactions between DGRP lines and inv and jing. To assess the effects of candidate epistatic modifier genes on head morphology, we obtained homozygous mutant lines of Minos Mi{ET1} insertion lines in five candidate genes (Mi{ET1}form3MB02055, Mi{ET1}kirreMB09143, Mi{ET1}klgMB05977, Mi{ET1}shakBMB03735, Mi{ET1}tkvMB02285), and their coisogenic w1118 control from the Bloomington Drosophila Stock Center. All stocks were reared on cornmeal-agar-molasses medium at 25°C, 60–75% relative humidity, and a 12-h light-dark cycle.
Head morphology analyses
We measured face width (the smallest distance between the eyes) and head width (the distance between the right and left side of the head capsule including the eyes) separately for males and females of each genotype tested. We collected 5- to 6-day-old flies from each genotype and froze them at −80°C. Head size (µm) was measured for decapitated heads fixed on a glass slide using O.C.T. Compound (Sakura Finetek Europe B.V.) using a stereo microscope (Leica M205C) connected to a computer. We measured face width of homozygous invBG00846 (N = 24 females, 23 males) and jingBG02314 (N = 20 females, 19 males) mutations and CSB (N = 19 females, 11 males); and head width of homozygous invBG00846 (N = 24 females, 23 males) and jingBG02314 (N = 20 females, 20 males) mutations and CSB (N = 19 females, 10 males). We used t-tests to assess the significance of the difference in head morphology between the mutations and wild type, separately for males and females.
We crossed homozygous invBG00846, jingBG02314, and CSB males to females of 197 DGRP lines and measured face width and head width of five F1 males and five F1 females from each of four replicate vials for each genotype and line (for a total of 23,640 flies). We used mixed-model ANOVAs to partition variation between sexes (S, fixed), F1 genotypes (invBG00846 and CSB or jingBG02314 and CSB) (G, fixed), DGRP lines (L, random), replicate vial within DGRP line [V(L), random] and all two- and three-way interactions: Y = µ + S + G + L + S×G + S×L + G×L + S×G×L + V(L) + S × V(L) + ε (Y is the Phenotype, µ the overall mean, and ε the residual variance). We also performed reduced analyses by genotype and by sex. We computed variance components for all random terms and computed the broad-sense heritabilities () of face and head width for the F1 crosses of each genotype to the DGRP lines as . We estimated the cross-sex genetic correlation () for the F1 crosses of each genotype to the DGRP lines as. The genetic correlation across sex and genotype () is . Mixed-model ANOVAs and estimates f variance components were performed using JMP® Pro, Version 14 (SAS Institute Inc., Cary, NC, USA).
We measured face width and head width of 10 males and 10 females in each of four replicate vials for five Minos Mi{ET1} insertion lines and their coisogenic w1118 control. We used mixed-model ANOVAs to partition variance between sexes (S, fixed), mutant and wild-type genotypes (G, fixed), the interaction between sex and genotype (S×G, fixed), replicate vial within genotype [V(G), random], and the interaction of sex and replicate vial within genotype [S × V(L), random]. We also ran reduced analyses separately for males and females.
GWA analyses
We performed GWA analyses using the DGRP GWA pipeline (http://dgrp2.gnets.ncsu.edu/). This pipeline implements a mixed-model analysis to evaluate the strength of association of alternative DGRP alleles with quantitative trait phenotypes for each of the ∼2 million variants for which the minor allele was present in at least four DGRP lines, while accounting for any effects of Wolbachia infection, karyotype of common polymorphic inversions and polygenic relatedness (Huang et al. 2014). We performed GWA analyses for the difference between DGRP/CSB and DGRP/jing and DGRP/CSB and DGRP/inv genotypes. These analyses specifically test for variants with nonadditive effects because the variation in the difference between the control and mutant phenotypes is equivalent to the genotype-by-DGRP line interaction term. All analyses were performed using genotype means, separately for males and females and for the average and difference between the two sexes. The difference between the two sexes models the S×G×L interaction effects in the analyses of differences between F1 DGRP/mutant and F1 DGRP/CSB genotypes.
We performed Biological Process Gene Ontology enrichment analyses for all candidate modifier genes using the PANTHER statistical over-representation test (Mi et al. 2013).
We annotated candidate modifier genes identified in the GWA analyses using complete genetic interaction networks from FlyBase (release r5.57), which were curated based on the literature. The genes are nodes in the network and the interactions are edges between the nodes. We mapped significant candidate genes from the GWA analyses onto this graphical representation of genetic networks and extracted subnetworks involving the candidate genes, with no missing nodes. We tested whether the maximum subnetwork is significantly greater than expected by chance using a permutation procedure (Morozova et al. 2015, 2018; Carbone et al. 2016).
Results
Quantitative genetic variation of head morphology
Previously, Carreira et al. (2009) showed that homozygous P[GT1]-element insertional mutations in jing (jingBG02314) and inv (invBG00846) affected face and head width compared to their coisogenic wild-type control genotype, Canton S B (CSB). We first showed that these homozygous mutations significantly reduce face width and head width in both sexes under our laboratory rearing conditions (Supplementary Table S1A and Figure S1).
Next, we crossed jingBG02314, invBG00846, and CSB females to males from 197 DGRP lines and measured face and head width on female and male F1 offspring (Supplementary Table S1, B and C). We partitioned variation in face and head width into the main effects of DGRP line, sex, and the DGRP line by sex interaction for each of the genotypes. The broad-sense heritabilities () were significant for each trait and genotype ( 0.377, 0.337, and 0.402, respectively, for face width in the DGRP lines crossed to CSB, jingBG02314, and invBG00846; and 0.431, 0.416, and 0.479, respectively, for head width in the DGRP lines crossed to CSB, jingBG02314, and invBG00846; Supplementary Table S2 and Figures S2 and S3). The line by sex interaction was also significant for each genotype and trait, indicating that the cross-sex genetic correlations, although high, are significantly different from unity (Supplementary Table S2). Thus, there is significant genetic variation in the magnitude of sexual dimorphism for head morphology for DGRP/CSB and DGRP/mutant F1 hybrids (Supplementary Figures S2 and S3). The phenotypic correlation between face and head width is also high for each sex and genotype (Supplementary Figure S4).
Given that there is genetic variation for head morphology for F1 CSB/DGRP and P[GT1]/DGRP genotypes, we next assessed whether the difference in head morphology traits between the mutations and CSB was constant or genetically variable among the DGRP lines; i.e., whether or not there is genotype-by-DGRP line interaction. We observed significant genotype by line—and genotype by sex by line—variation for face and head width (Supplementary Table S2). The genetic correlation across genotypes, lines, and sexes () is 0.817 and 0.801 for face width for jing and inv, respectively; and 0.735 and 0.687 for head width for jing and inv, respectively (Supplementary Table S2). Therefore, the effect of the jing and inv mutant alleles not only varies across the DGRP lines, but the magnitude and/or direction of this variation depends on sex (Figures 1 and 2), although the overall effect of CSB/DGRP vs mutant/DGRP genotype is highly significant for jing and inv face width and inv head width (but not jing head width). The variation in the effects of the mutations in the different DGRP genetic backgrounds is due to nonadditive gene action, due to DGRP alleles in jing or inv causing variation in the degree of dominance at these loci (allelic failure to complement) or by unlinked modifiers of the focal loci (epistatic failure to complement). Mapping the genetic modifiers enables us to distinguish whether dominance or epistasis causes the nonadditive genetic variance in head morphology.
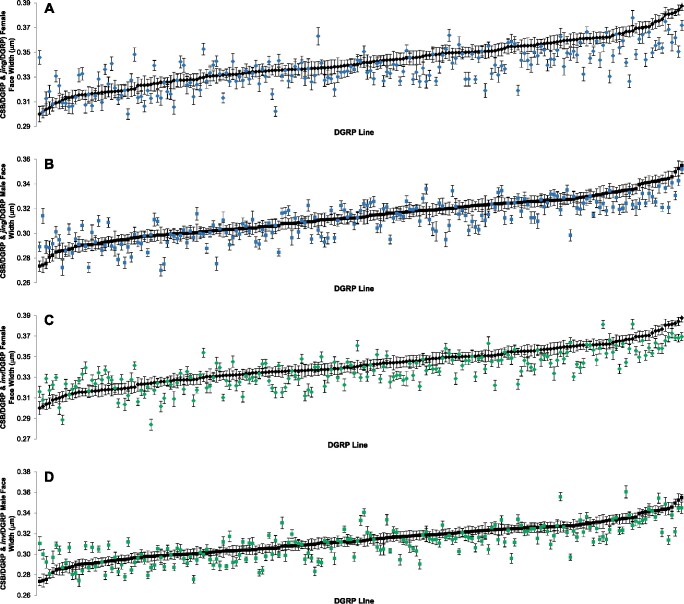
Figure 1.
Genotype by DGRP line interaction for face width. (A) Female CSB/DGRP and jing/DGRP. (B) Male CSB/DGRP and jing/DGRP. (C) Female CSB/DGRP and inv/DGRP. (D) Male CSB/DGRP and inv/DGRP. The DGRP lines are ordered from smallest to the largest CSB face width, and not the same for males and females. Error bars are SEM.
Figure 2.
Genotype by DGRP line interaction for head width. (A) Female CSB/DGRP and jing/DGRP. (B) Male CSB/DGRP and jing/DGRP. (C) Female CSB/DGRP and inv/DGRP. (D) Male CSB/DGRP and inv/DGRP. Male CSB/DGRP and inv/DGRP. The DGRP lines are ordered from smallest to the largest CSB face width, and not the same for males and females. Error bars are SEM.
GWA analyses of head morphology
We mapped modifiers of head morphology by performing GWA analyses for the difference in face width and head width line means between CSB/DGRP and mutant/DGRP genotypes. We performed these analyses for males, females, and the average and difference between the sexes, while accounting for any effects of Wolbachia infection, segregating polymorphic inversions, and polygenic relatedness (Huang et al. 2014). Variants associated with variation in the difference in line means between these genotypes are associated with the genotype-by-line interaction. Variants associated with the difference between males and females for the difference between CSB/DGRP and mutant/DGRP genotypes are associated with the genotype by line by sex interaction.
At a nominal P-value < 10−5, we identified 195 candidate modifier variants in or near (within 1 kb of the gene body) 147 genes for inv face width; 213 candidate modifier variants in or near 138 genes for inv head width; 660 candidate modifier variants in or near 365 genes for jing face width; and 72 candidate modifier variants in or near 47 genes for jing head width (Supplementary Table S3, A–D). Notably, the largest number of candidate modifier variants in each of these analyses was for the genotype by sex by line interaction variance, for which quantile–quantile plots show significant departure from random expectation below P < 10−5 (Supplementary Figure S5). None of the candidate modifier variants were located in or near jing or inv in the crosses of jing and inv, respectively, to the DGRP lines; therefore, all are candidate epistatic modifiers. However, jing is a candidate modifier gene for face width in the analyses of CSB/DGRP and inv/DGRP genotypes. As is typical for GWA analyses in the DGRP, the majority of associated variants are located in introns, upstream or downstream of candidate genes, or in intergenic regions, suggesting they might play a regulatory role. In total, we mapped 658 unique candidate genes, of which 36 overlapped between two or more analyses (Supplementary Table S3E). The candidate genes are highly enriched (Mi et al. 2013) for biological process Gene Ontology categories involved in development, differentiation, and morphogenesis (Supplementary Table S3F), which is intuitively reasonable for morphometric traits.
We asked whether the candidate modifier genes identified in the GWA analyses were known to interact in previously curated genetic interaction networks. Without computationally recruiting missing genes, we identified a significant (P = 0.001) network with 62 genes (Figure 3). Enrichment analysis of this network reveals enrichment of genes associated with development, differentiation, and morphogenesis; protein, signaling receptor, and transcription factor binding; and the Dpp-Scw, Dpp, TGFβ, Gbb, and Wnt signaling pathways (Supplementary Table S4).
Figure 3.
Genetic interaction network of candidate modifier loci from the GWA analyses. The network consists of 60 interacting genes (P < 0.001) with no missing genes.
Functional assessment
We cannot readily validate epistatic interactions between candidate DGRP modifier variants and genes. The former analysis would require that we engineer the major and minor DGRP candidate variants in the P[GT1] insertional mutation and the coisogenic CSB genotypes, while the latter would require that we generate P[GT1] mutations in the CSB background for the candidate modifier genes. However, we can assess whether homozygous mutations in the candidate modifier genes affect face and head width. This is weaker test because epistatic interactions can exist without a main effect, and different mutations in the same gene are likely to have different main and epistatic effects. We quantified face and head width for five Mi{ET1} mutations and their w1118 control genotype (Supplementary Tables S1D and S5). All mutations but kgl affected face width (Figure 4), and all significant mutations decreased face width relative to the control in at least one sex. In addition, all significant mutations had significant genotype by sex interactions. In contrast, only kirre was significant for head width (Supplementary Figure S6).
Figure 4.

Effects of homozygous mutations of candidate modifier genes on face width. The bars indicate the difference in face width of mutations in modifier genes from the control. Red denotes females and blue denotes males. Error bars are SEM. P-values from analyses of variance (Supplementary Table S5) for the effect of genotype pooled across sexes (G♀,♂), the genotype by sex interaction (G × S), and the effect of genotype for females (G♀) and males (G♂) are given in the table. ns: P > 0.05; *P, 0.05; **P < 0.01; ***P < 0.001.
Discussion
Epistasis impacts quantitative genetic models of speciation, response to natural and artificial selection, genetic mapping, and personalized medicine (Phillips 2008; Mackay 2014), but it is difficult to assess epistasis for quantitative traits and map the interacting variants/loci. QTL mapping in linkage populations has good power to detect QTL by QTL interactions, but is imprecise and hence challenging to identify interacting variants. Genome-wide pairwise epistasis screens in association mapping populations can potentially identify interacting loci/variants but suffer from a huge multiple testing penalty because of the large number of possible tests for epistasis. In D. melanogaster, a powerful strategy to convert a two-dimensional epistasis mapping screen to a one-dimensional screen, thus increasing the power to detect epistatic interactions, is to introgress or cross a candidate focal allele into the DGRP (Mackay et al. 2012; Huang et al. 2014) or another mapping population. However, most of these studies (Polaczyk et al. 1998; Dworkin et al. 2009; Chow et al. 2016; Lavoy et al. 2018; Palu et al. 2019; Talsness et al. 2020) utilized dominant-negative mutations with large effects or the GAL4-UAS system to over-express, mis-express or knockdown expression of the focal gene. Only a few studies utilized mutations with subtle, quantitative effects as focal genes (Yamamoto et al. 2009; Swarup et al. 2012; He et al. 2016). The advantage of the latter approach is that the epistatic interactions are more likely to mimic those between naturally occurring alleles; the disadvantage is these analyses require that the mapping population is crossed to both the focal mutant allele and a wild-type allele in the same coisogenic control background to account for naturally occurring quantitative genetic variation in the trait of interest. This design is an extension of the quantitative complementation test (Long et al. 1996; Pasyukova et al. 2000) to multiple wild-derived genotypes.
We performed a one-dimensional screen for epistasis for D. melanogaster face width and head width by crossing DGRP lines to mutations in jing and inv that were previously shown to have subtle, quantitative effects on these traits as homozygotes (Carreira et al. 2009), and their coisogenic control. Both jing and inv are transcription factors and are known to participate in epistatic interactions. jing is involved in axon guidance, central nervous system development, and other developmental processes and interacts with Egfr, hng1, hth, kay, lap, pbl, Pc, Ras85D, S, sim, slbo, sli, spi, tgo, trh, wg, and XNP (Larkin et al. 2021). inv is involved in neuroblast fate determination, neuron differentiation, and other developmental processes and interacts with en and ph-d (Larkin et al. 2021). Quantitative genetic analyses of face and head width for the F1 genotypes showed that both traits had significant genetic variation and significant DGRP line by mutant genotype interaction (i.e., epistasis), as well as significant genetic variation in sexual dimorphism and epistatic variation in sexual dimorphism. We performed GWA analyses and identified candidate modifier variants that were largely in presumed regulatory genomic regions. Candidate modifier genes within 1 kb of modifier variants were enriched for Gene Ontology categories involved in development, differentiation, and morphogenesis. A total of 62 of the candidate modifier genes mapped to a known genetic interaction network (P < 0.001) enriched for genes associated with development and morphogenesis, transcription factors, and canonical signaling pathways. Thus, variation in adult head morphology most likely results from subtle genetic variation affecting early development.
Three of the candidate modifier genes that epistatically interact with the jing mutation to affect head morphology, hth, pbl, and sli, are among those previously implicated to interact with jing (Larkin et al. 2021), but not via their effects on head morphology. In addition, 24 of the 36 candidate modifier genes (66.7%) present in more than one of the GWA analyses were found in crosses to both jing and inv; and sli was also identified in the crosses to inv. These observations suggest that, although inv and jing do not interact directly, they participate in the same genetic interaction network. They also suggest that genetic interaction networks derived from analyses of effects of null mutations on viability during development or mutations with clear qualitative adult phenotypes are at least in part the same as genetic interaction networks inferred from analyses of naturally occurring variation with small effects on quantitative traits in adults. Finally, common candidate modifier genes implicated by GWA analyses of F1 genotypes resulting from crosses of DGRP lines to genes that themselves affect the target phenotype suggests that extending these analyses to other genes affecting the same phenotype (including the candidate modifier genes that themselves affect head morphology) can further define trait-specific genetic interaction networks. For example, tkv is a candidate modifier gene that affects head morphology (Figure 4) and that is known to interact with arm, dally, dpp, nwk, and shn (Larkin et al. 2021); all of which are candidate modifier genes in the inferred jing/inv genetic interaction network for head morphology. This approach is applicable to any Drosophila quantitative trait.
Data availability
The authors confirm that all head morphology data necessary for confirming the conclusions of the article are present in Supplementary Table S1. Supplementary Table S2 gives the ANOVAs of head morphology traits from CSB/DGRP and mutant/DGRP F1 genotypes. Supplementary Table S3 presents the GWA analyses for modifier loci. Supplementary Table S4 shows the Gene Ontology enrichment analyses for candidate modifier genes in significant genetic interaction network. Supplementary Table S5 gives the ANOVAs of head morphology traits for Mi{ET1} mutations in candidate modifier genes and the w1118 control genotype. Supplementary Figure S1 shows the homozygous effects of homozygous jingBG02314 and invBG00846 mutations on head morphology. Supplementary Figure S2 shows the distribution of face width for F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S3 depicts the distribution of head width for F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S4 shows the phenotypic correlations (rP ± SE) of face width and head width in F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S5 shows quantile–quantile (Q-Q) plots of –log10P-values and plots of locations of nominally significant (P < 10−5) variants across the Drosophila genome. Supplementary Figure S6 depicts the effects of homozygous mutations of candidate modifier genes on head width.
The DGRP lines are available from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/Browse/DGRP.php). Raw sequence data are available at the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra). The SRA Accession numbers are: DGRP_21, SRX021040; DGRP_26, SRX021056; DGRP_28, SRX021783 and SRX021782; DGRP_31, SRX155996; DGRP_32, SRX155997; DGRP_38, SRX025317; DGRP_40, SRX021235; DGRP_41, SRX021791 and SRX021790; DGRP_42, SRX021255; DGRP_45, SRX021261; DGRP_48, SRX155989; DGRP_49, SRX021267; DGRP_57, SRX021296; DGRP_59, SRX021327; DGRP_69, SRX023449; DGRP_73, SRX023450; DGRP_75, SRX021384; DGRP_83, SRX023456; DGRP_85, SRX021490; DGRP_88, SRX021495; DGRP_91, SRX021503; DGRP_93, SRX021504; DGRP_100, SRX156026; DGRP_101, SRX020747; DGRP_105, SRX020745; DGRP_109, SRX020746 and SRX156025; DGRP_129, SRX020748; DGRP_136, SRX020753; DGRP_138, SRX021008; DGRP_142, SRX020759; DGRP_149, SRX020760; DGRP_153, SRX021514; DGRP_158, SRX021009; DGRP_161, SRX020761; DGRP_176, SRX020763 and SRX020762; DGRP_177, SRX021026; DGRP_181, SRX020912; DGRP_189, SRX155979; DGRP_195, SRX021039; DGRP_208, SRX005977; DGRP_208, SRX015853; DGRP_217, SRX021041; DGRP_223, SRX155994; DGRP_227, SRX021042; DGRP_228, SRX021043; DGRP_229, SRX021052; DGRP_233,SRX021061; DGRP_235, SRX021053; DGRP_237, SRX023423 and SRX023422; DGRP_239, SRX021054; DGRP_256, SRX021055; DGRP_280, SRX021058; DGRP_287, SRX021059; DGRP_301, SRX155995 and SRX005978; DGRP_301, SRX157787; DGRP_303, SRX155978 and SRX155977 and SRX005986 and SRX005985; DGRP_303, SRX157789 and SRX157788; DGRP_304SRX156009 and SRX156008 and SRX005988 and SRX005987; DGRP_304, SRX015854; DGRP_306SRX156007 and SRX006140 and SRX006139; DGRP_306, SRX157798 and SRX157797 and SRX015855 and SRX016258 and SRX016257; DGRP_307, SRX156012 and SRX156011 and SRX006188 and SRX006187 and SRX006186; DGRP_307, SRX015860; DGRP_309, SRX021060; DGRP_310, SRX021080; DGRP_313, SRX006277, SRX006276 and SRX006275; DGRP_313, SRX015856; DGRP_315, SRX156010 and SRX006143 and SRX006142 and SRX006141; DGRP_315, SRX015859; DGRP_317, SRX021081; DGRP_318, SRX021082; DGRP_319, SRX155981; DGRP_320, SRX021063; DGRP_321, SRX021094; DGRP_324, SRX006144 and SRX006145 and SRX155982; DGRP_324, SRX015974; DGRP_325, SRX021793; DGRP_332, SRX021095; DGRP_335, SRX021097; DGRP_335, SRX157913; DGRP_336, SRX021096; DGRP_338, SRX021097; DGRP_340, SRX156030; DGRP_348, SRX156029; DGRP_350, SRX021100; DGRP_352, SRX021101; DGRP_354, SRX156027; DGRP_355, SRX156028; DGRP_356, SRX023833; DGRP_357, SRX006146 and SRX006147; DGRP_357, SRX015861; DGRP_358, SRX006283 and SRX006282; DGRP_358, SRX015862; DGRP_359, SRX023424; DGRP_360, SRX155999 and SRX006309; DGRP_360, SRX016258 and SRX016257; DGRP_361, SRX155984; DGRP_362, SRX006288 and SRX006287; DGRP_362, SRX157914 and SRX157915; DGRP_365SRX006291, SRX006290 and SRX006289; DGRP_365, SRX015863; DGRP_367, SRX021103; DGRP_370, SRX021104; DGRP_371, SRX021257 and SRX156000; DGRP_373, SRX023425 and SRX155983; DGRP_374, SRX023427; DGRP_375, SRX006150, SRX006149 and SRX006148; DGRP_375, SRX015864; DGRP_377, SRX023834; DGRP_379, SRX006293 and SRX006292; DGRP_379, SRX015865; DGRP_380, SRX006303, SRX006302, SRX006301 and SRX006300; DGRP_380, SRX015866; DGRP_381, SRX021112; DGRP_382, SRX156013l DGRP_383, SRX021113; DGRP_385, SRX159098; DGRP_386, SRX021798 and SRX021797; DGRP_390, SRX156014; DGRP_391, SRX023452, SRX006152 and SRX006151; DGRP_391, SRX015867; DGRP_392, SRX021157; DGRP_395, SRX156015; DGRP_397, SRX156017; DGRP_399, SRX006154 and SRX006153; DGRP_399, SRX015868; DGRP_405, SRX021242; DGRP_406, SRX021254; DGRP_409, SRX021243 and SRX156016; DGRP_426, SRX021245; DGRP_427, SRX006155; DGRP_427, SRX016041; DGRP_437, SRX006156 and SRX156001; DGRP_437, SRX016042; DGRP_439, SRX021244; DGRP_440, SRX021246; DGRP_441, SRX023835; DGRP_443, SRX021260; DGRP_461, SRX021262; DGRP_486, SRX006157; DGRP_486, SRX016043; DGRP_491, SRX021268; DGRP_492, SRX021270; DGRP_502, SRX021271; DGRP_505, SRX156002; DGRP_508, SRX021272; DGRP_509, SRX021273; DGRP_513; SRX021282; DGRP_517, SRX024363 and SRX024362; DGRP_517, SRX016210 and SRX016209; DGRP_528, SRX155985; DGRP_530, SRX156031; DGRP_531, SRX021290; DGRP_535, SRX021293; DGRP_551, SRX156034; DGRP_555, SRX006159; DGRP_555, SRX016072; DGRP_559, SRX156032; DGRP_563, SRX023836; DGRP_566, SRX156033; DGRP_584, SRX155987 and SRX155986; DGRP_589, SRX023837; DGRP_595, SRX021328; DGRP_596, SRX156004; DGRP_627, SRX155988; DGRP_630, SRX156003; DGRP_634, SRX156018; DGRP_639, SRX006161 and SRX006160; DGRP_639, SRX016339; DGRP_642, SRX021331; DGRP_646, SRX021332; DGRP_703, SRX021508; DGRP_705, SRX006162; DGRP_705, SRX016134; DGRP_707, SRX006163; DGRP_707, SRX016135; DGRP_712, SRX006164; DGRP_712, SRX016136; DGRP_714, SRX006166 and SRX006165; DGRP_714, SRX016137; DGRP_716, SRX021380; DGRP_721, SRX021381; DGRP_727, SRX021382; DGRP_730, SRX006308; DGRP_730, SRX016138; DGRP_732, SRX006167; DGRP_732, SRX157997; DGRP_737, SRX023451; DGRP_738, SRX021383; DGRP_748, SRX156019 and SRX156020; DGRP_757, SRX021385; DGRP_761, SRX021386; DGRP_765, SRX006169 and SRX006168; DGRP_765, SRX016176; DGRP_774, SRX006170 and SRX156022; DGRP_774, SRX016175 and SRX158004 and SRX158005; DGRP_776, SRX021387; DGRP_783, SRX023455; DGRP_786, SRX006171; DGRP_786, SRX016177; DGRP_787, SRX021388; DGRP_790, SRX021389; DGRP_796, SRX021390; DGRP_799, SRX006172 and SRX006173; DGRP_799, SRX016178; DGRP_801, SRX021391 and SRX156021; DGRP_802, SRX025318 and SRX156005; DGRP_804, SRX021399; DGRP_805, SRX021400; DGRP_808, SRX021402 and SRX155992; DGRP_810, SRX021418; DGRP_812, SRX021419; DGRP_818, SRX021478; DGRP_819, SRX156006; DGRP_820, SRX006174 and SRX006175; DGRP_820, SRX016179; DGRP_821, SRX155990 and SRX155991; DGRP_822, SRX021476; DGRP_832, SRX021477; DGRP_837, SRX021479; DGRP_843, SRX156036; DGRP_849, SRX156035; DGRP_850, SRX155993; DGRP_852, SRX006304 and SRX006305; DGRP_852, SRX016300 and SRX016301, DGRP_853, SRX021491 and SRX155976; DGRP_855, SRX021563; DGRP_857, SRX021492; DGRP_859, SRX006176; DGRP_859, SRX016184; DGRP_861, SRX021493; DGRP_879, SRX021494; DGRP_882, SRX021496; DGRP_884, SRX021498; DGRP_887, SRX021527; DGRP_890, SRX021499; DGRP_892, SRX023838; DGRP_894, SRX021528; DGRP_897, SRX023457; DGRP_900, SRX156023; DGRP_907, SRX021500; DGRP_908, SRX021501; DGRP_911, SRX021502; and DGRP_913, SRX156024. The genotypes, quality scores, phenotypes, and web-based analysis tools are available from the DGRP website (http://dgrp2.gnets.ncsu.edu). All codes used to analyze these data are given in https://github.com/qgg-lab/DGRPHeadMorphologyEpistasis.
Funding
This research was supported by the National Institutes of Health grant R01 GM 45146 to T.F.C.M and R.R.H.A.
Conflicts of interest
The authors declare that there is no conflict of interest.
Supplementary Material
Literature cited
- Anholt RRH, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, et al. 2003. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 35:180–184. [DOI] [PubMed] [Google Scholar]
- Bis-Brewer DM, Fazal S, Züchner S.. 2020. Genetic modifiers and non-Mendelian aspects of CMT. Brain Res. 1726:146459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone MA, Yamamoto A, Huang W, Lyman RA, Meadors TB, et al. 2016. Genetic architecture of natural variation in visual senescence in Drosophila. Proc Natl Acad Sci U S A. 113:E6620–E6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira VP, Mensch J, Fanara JJ.. 2009. Body size in Drosophila: genetic architecture, allometries and sexual dimorphism. Heredity (Edinb). 102:246–256. [DOI] [PubMed] [Google Scholar]
- Chow CY, Kelsey KJP, Wolfner MF, Clark AG.. 2016. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum Mol Genet. 25:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Wang L.. 1997. Epistasis in measured genotypes: Drosophila P-element insertions. Genetics. 147:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BA, Hassan S, Garcia EJ, Tayebi N, Sidransky E.. 2018. Exploring genetic modifiers of Gaucher disease: the next horizon. Hum Mutat. 39:1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I, Kennerly E, Tack D, Hutchinson J, Brown J, et al. 2009. Genomic consequences of background effects on scalloped mutant expressivity in the wing of Drosophila melanogaster. Genetics. 181:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Mackay TFC.. 2009. Quantitative trait loci for aggressive behavior in Drosophila melanogaster. Genetics. 182:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Zwarts L, Yamamoto A, Callaerts P, Mackay TFC.. 2009. Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol. 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC.. 1996. Introduction to Quantitative Genetics. 4th ed.Longman, Harlow: Essex. [Google Scholar]
- Fedorowicz GM, Fry JD, Anholt RRH, Mackay TFC.. 1998. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics. 148:1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans Roy Soc Edinburgh. 52:399–433. [Google Scholar]
- Flint J, Mackay TFC.. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, van Helden S.. 1997. Is function of the Drosophila homeotic gene Ultrabithorax canalized? Genetics. 147:1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Wemple M, van Helden S.. 1999. Potential variance affecting homeotic Ultrabithorax and Antennapedia phenotypes in Drosophila melanogaster. Genetics. 151:1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH.. 2009. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 4:e5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhou S, St. Armour GE, Mackay TFC, Anholt RRH.. 2016. Epistatic partners of neurogenic genes modulate Drosophila olfactory behavior. Genes Brain Behav. 15:280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM.. 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4:e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Mackay TFC.. 2016. The genetic architecture of quantitative traits cannot be inferred from variance component analysis. PLoS Genet. 12:e1006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu D, Anholt RRH, et al. 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A. 109:15553–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, dos Santos G, et al. ; FlyBase Consortium. 2021. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49:D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoy S, Chittoor-Vinod VG, Chow CY, Martin I.. 2018. Genetic modifiers of neurodegeneration in a Drosophila model of Parkinson's disease. Genetics. 209:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Mullaney SL, Mackay TFC, Langley CH.. 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics. 144:1497–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B.. 1998. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer. [Google Scholar]
- Mackay TFC. 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 15:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature. 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Yamamoto A, Carbone MA, Roshina NV, Symonenko AV, et al. 2010. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 6:e1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, A, Muruganujan PD, Thomas PD.. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41:D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Huang W, Pray VA, Whitham T, Anholt RRH, et al. 2015. Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult Drosophila. BMC Genomics. 16:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Hussain Y, McCoy LJ, Zhirnov EV, Davis MR, et al. 2018. A Cyclin E centered genetic network contributes to alcohol-induced variation in Drosophila development. G3 (Bethesda). 8:2643–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Forejt J, Takada T, Shiroishi T.. 2012. Chromosome substitution strains: gene discovery, functional analysis, and systems studies. Mamm Genome. 23:693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palu RAS, Ong E, Stevens K, Chung S, Owings KG, et al. 2019. Natural genetic variation screen in Drosophila identifies Wnt signaling, mitochondrial metabolism, and redox homeostasis genes as modifiers of apoptosis. G3 (Bethesda). 9:3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova EG, Vieira C, Mackay TFC.. 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics. 156:1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 9:855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaczyk PJ, Gasperini R, Gibson G.. 1998. Naturally occurring genetic variation affects Drosophila photoreceptor determination. Dev Genes Evol. 207:462–470. [DOI] [PubMed] [Google Scholar]
- Rahit KMTH, Tarailo-Graovac M.. 2020. Genetic modifiers and rare Mendelian disease. Genes (Basel). 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendel JM. 1959. Canalization of the scute phenotype of Drosophila. Evolution. 13:425–439. [Google Scholar]
- Rollmann SM, Magwire MM, Morgan TJ, Ozsoy ED, Yamamoto A, et al. 2006. Pleiotropic fitness effects of the Tre1-Gr5a region in Drosophila melanogaster. Nat Genet. 38:824–829. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Yamamoto A, Goossens T, Zwarts L, Callaerts-Végh Z, et al. 2007. The early developmental gene Semaphorin 5c contributes to olfactory behavior in adult Drosophila. Genetics. 176:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S.. 1998. Hsp90 as a capacitor for morphological evolution. Nature. 396:336–342. [DOI] [PubMed] [Google Scholar]
- Sambandan D, Yamamoto A, Fanara JJ, Mackay TFC, Anholt RRH.. 2006. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics. 174:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Couch C, Huang W, Carbone MA, Peiffer J, et al. 2015. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc Natl Acad Sci U S A. 112:E3555–E3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiezio SH, Takada T, Shiroishi T, Nadeau JH.. 2012. Genetic divergence and the genetic architecture of complex traits in chromosome substitution strains of mice. BMC Genet. 13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Guo Y, Lan X, Jia J, Cai X, et al. 2020. PhenoModifier: a genetic modifier database for elucidating the genetic basis of human phenotypic variation. Nucleic Acids Res. 48:D977–D982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, Harbison ST, Hahn LE, Morozova TV, Yamamoto A, et al. 2012. Extensive epistasis for olfactory behaviour, sleep and waking activity in Drosophila melanogaster. Genet Res (Camb). 94:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsness DM, Owings KG, Coelho E, Mercenne G, Pleinis JM, et al. 2020. A Drosophila screen identifies NKCC1 as a modifier of NGLY1 deficiency. Elife. 9:e57831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swinderen B, Greenspan RJ.. 2005. Flexibility in a gene network affecting a simple behavior in Drosophila melanogaster. Genetics. 169:2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1931. Evolution in Mendelian populations. Genetics. 16:97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Anholt RRH, Mackay TFC.. 2009. Epistatic interactions attenuate mutations affecting startle behaviour in Drosophila melanogaster. Genet Res (Camb). 91:373–382. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Zwarts L, Callaerts P, Norga K, Mackay TFC, et al. 2008. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc Natl Acad Sci U S A. 105:12393–12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwarts L, Magwire MM, Carbone MA, Versteven M, Herteleer L, et al. 2011. Complex genetic architecture of Drosophila aggressive behavior. Proc Natl Acad Sci U S A. 108:17070–17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all head morphology data necessary for confirming the conclusions of the article are present in Supplementary Table S1. Supplementary Table S2 gives the ANOVAs of head morphology traits from CSB/DGRP and mutant/DGRP F1 genotypes. Supplementary Table S3 presents the GWA analyses for modifier loci. Supplementary Table S4 shows the Gene Ontology enrichment analyses for candidate modifier genes in significant genetic interaction network. Supplementary Table S5 gives the ANOVAs of head morphology traits for Mi{ET1} mutations in candidate modifier genes and the w1118 control genotype. Supplementary Figure S1 shows the homozygous effects of homozygous jingBG02314 and invBG00846 mutations on head morphology. Supplementary Figure S2 shows the distribution of face width for F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S3 depicts the distribution of head width for F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S4 shows the phenotypic correlations (rP ± SE) of face width and head width in F1 genotypes from crosses of 197 DGRP lines to CSB, jingBG02314, and invBG00846. Supplementary Figure S5 shows quantile–quantile (Q-Q) plots of –log10P-values and plots of locations of nominally significant (P < 10−5) variants across the Drosophila genome. Supplementary Figure S6 depicts the effects of homozygous mutations of candidate modifier genes on head width.
The DGRP lines are available from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/Browse/DGRP.php). Raw sequence data are available at the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra). The SRA Accession numbers are: DGRP_21, SRX021040; DGRP_26, SRX021056; DGRP_28, SRX021783 and SRX021782; DGRP_31, SRX155996; DGRP_32, SRX155997; DGRP_38, SRX025317; DGRP_40, SRX021235; DGRP_41, SRX021791 and SRX021790; DGRP_42, SRX021255; DGRP_45, SRX021261; DGRP_48, SRX155989; DGRP_49, SRX021267; DGRP_57, SRX021296; DGRP_59, SRX021327; DGRP_69, SRX023449; DGRP_73, SRX023450; DGRP_75, SRX021384; DGRP_83, SRX023456; DGRP_85, SRX021490; DGRP_88, SRX021495; DGRP_91, SRX021503; DGRP_93, SRX021504; DGRP_100, SRX156026; DGRP_101, SRX020747; DGRP_105, SRX020745; DGRP_109, SRX020746 and SRX156025; DGRP_129, SRX020748; DGRP_136, SRX020753; DGRP_138, SRX021008; DGRP_142, SRX020759; DGRP_149, SRX020760; DGRP_153, SRX021514; DGRP_158, SRX021009; DGRP_161, SRX020761; DGRP_176, SRX020763 and SRX020762; DGRP_177, SRX021026; DGRP_181, SRX020912; DGRP_189, SRX155979; DGRP_195, SRX021039; DGRP_208, SRX005977; DGRP_208, SRX015853; DGRP_217, SRX021041; DGRP_223, SRX155994; DGRP_227, SRX021042; DGRP_228, SRX021043; DGRP_229, SRX021052; DGRP_233,SRX021061; DGRP_235, SRX021053; DGRP_237, SRX023423 and SRX023422; DGRP_239, SRX021054; DGRP_256, SRX021055; DGRP_280, SRX021058; DGRP_287, SRX021059; DGRP_301, SRX155995 and SRX005978; DGRP_301, SRX157787; DGRP_303, SRX155978 and SRX155977 and SRX005986 and SRX005985; DGRP_303, SRX157789 and SRX157788; DGRP_304SRX156009 and SRX156008 and SRX005988 and SRX005987; DGRP_304, SRX015854; DGRP_306SRX156007 and SRX006140 and SRX006139; DGRP_306, SRX157798 and SRX157797 and SRX015855 and SRX016258 and SRX016257; DGRP_307, SRX156012 and SRX156011 and SRX006188 and SRX006187 and SRX006186; DGRP_307, SRX015860; DGRP_309, SRX021060; DGRP_310, SRX021080; DGRP_313, SRX006277, SRX006276 and SRX006275; DGRP_313, SRX015856; DGRP_315, SRX156010 and SRX006143 and SRX006142 and SRX006141; DGRP_315, SRX015859; DGRP_317, SRX021081; DGRP_318, SRX021082; DGRP_319, SRX155981; DGRP_320, SRX021063; DGRP_321, SRX021094; DGRP_324, SRX006144 and SRX006145 and SRX155982; DGRP_324, SRX015974; DGRP_325, SRX021793; DGRP_332, SRX021095; DGRP_335, SRX021097; DGRP_335, SRX157913; DGRP_336, SRX021096; DGRP_338, SRX021097; DGRP_340, SRX156030; DGRP_348, SRX156029; DGRP_350, SRX021100; DGRP_352, SRX021101; DGRP_354, SRX156027; DGRP_355, SRX156028; DGRP_356, SRX023833; DGRP_357, SRX006146 and SRX006147; DGRP_357, SRX015861; DGRP_358, SRX006283 and SRX006282; DGRP_358, SRX015862; DGRP_359, SRX023424; DGRP_360, SRX155999 and SRX006309; DGRP_360, SRX016258 and SRX016257; DGRP_361, SRX155984; DGRP_362, SRX006288 and SRX006287; DGRP_362, SRX157914 and SRX157915; DGRP_365SRX006291, SRX006290 and SRX006289; DGRP_365, SRX015863; DGRP_367, SRX021103; DGRP_370, SRX021104; DGRP_371, SRX021257 and SRX156000; DGRP_373, SRX023425 and SRX155983; DGRP_374, SRX023427; DGRP_375, SRX006150, SRX006149 and SRX006148; DGRP_375, SRX015864; DGRP_377, SRX023834; DGRP_379, SRX006293 and SRX006292; DGRP_379, SRX015865; DGRP_380, SRX006303, SRX006302, SRX006301 and SRX006300; DGRP_380, SRX015866; DGRP_381, SRX021112; DGRP_382, SRX156013l DGRP_383, SRX021113; DGRP_385, SRX159098; DGRP_386, SRX021798 and SRX021797; DGRP_390, SRX156014; DGRP_391, SRX023452, SRX006152 and SRX006151; DGRP_391, SRX015867; DGRP_392, SRX021157; DGRP_395, SRX156015; DGRP_397, SRX156017; DGRP_399, SRX006154 and SRX006153; DGRP_399, SRX015868; DGRP_405, SRX021242; DGRP_406, SRX021254; DGRP_409, SRX021243 and SRX156016; DGRP_426, SRX021245; DGRP_427, SRX006155; DGRP_427, SRX016041; DGRP_437, SRX006156 and SRX156001; DGRP_437, SRX016042; DGRP_439, SRX021244; DGRP_440, SRX021246; DGRP_441, SRX023835; DGRP_443, SRX021260; DGRP_461, SRX021262; DGRP_486, SRX006157; DGRP_486, SRX016043; DGRP_491, SRX021268; DGRP_492, SRX021270; DGRP_502, SRX021271; DGRP_505, SRX156002; DGRP_508, SRX021272; DGRP_509, SRX021273; DGRP_513; SRX021282; DGRP_517, SRX024363 and SRX024362; DGRP_517, SRX016210 and SRX016209; DGRP_528, SRX155985; DGRP_530, SRX156031; DGRP_531, SRX021290; DGRP_535, SRX021293; DGRP_551, SRX156034; DGRP_555, SRX006159; DGRP_555, SRX016072; DGRP_559, SRX156032; DGRP_563, SRX023836; DGRP_566, SRX156033; DGRP_584, SRX155987 and SRX155986; DGRP_589, SRX023837; DGRP_595, SRX021328; DGRP_596, SRX156004; DGRP_627, SRX155988; DGRP_630, SRX156003; DGRP_634, SRX156018; DGRP_639, SRX006161 and SRX006160; DGRP_639, SRX016339; DGRP_642, SRX021331; DGRP_646, SRX021332; DGRP_703, SRX021508; DGRP_705, SRX006162; DGRP_705, SRX016134; DGRP_707, SRX006163; DGRP_707, SRX016135; DGRP_712, SRX006164; DGRP_712, SRX016136; DGRP_714, SRX006166 and SRX006165; DGRP_714, SRX016137; DGRP_716, SRX021380; DGRP_721, SRX021381; DGRP_727, SRX021382; DGRP_730, SRX006308; DGRP_730, SRX016138; DGRP_732, SRX006167; DGRP_732, SRX157997; DGRP_737, SRX023451; DGRP_738, SRX021383; DGRP_748, SRX156019 and SRX156020; DGRP_757, SRX021385; DGRP_761, SRX021386; DGRP_765, SRX006169 and SRX006168; DGRP_765, SRX016176; DGRP_774, SRX006170 and SRX156022; DGRP_774, SRX016175 and SRX158004 and SRX158005; DGRP_776, SRX021387; DGRP_783, SRX023455; DGRP_786, SRX006171; DGRP_786, SRX016177; DGRP_787, SRX021388; DGRP_790, SRX021389; DGRP_796, SRX021390; DGRP_799, SRX006172 and SRX006173; DGRP_799, SRX016178; DGRP_801, SRX021391 and SRX156021; DGRP_802, SRX025318 and SRX156005; DGRP_804, SRX021399; DGRP_805, SRX021400; DGRP_808, SRX021402 and SRX155992; DGRP_810, SRX021418; DGRP_812, SRX021419; DGRP_818, SRX021478; DGRP_819, SRX156006; DGRP_820, SRX006174 and SRX006175; DGRP_820, SRX016179; DGRP_821, SRX155990 and SRX155991; DGRP_822, SRX021476; DGRP_832, SRX021477; DGRP_837, SRX021479; DGRP_843, SRX156036; DGRP_849, SRX156035; DGRP_850, SRX155993; DGRP_852, SRX006304 and SRX006305; DGRP_852, SRX016300 and SRX016301, DGRP_853, SRX021491 and SRX155976; DGRP_855, SRX021563; DGRP_857, SRX021492; DGRP_859, SRX006176; DGRP_859, SRX016184; DGRP_861, SRX021493; DGRP_879, SRX021494; DGRP_882, SRX021496; DGRP_884, SRX021498; DGRP_887, SRX021527; DGRP_890, SRX021499; DGRP_892, SRX023838; DGRP_894, SRX021528; DGRP_897, SRX023457; DGRP_900, SRX156023; DGRP_907, SRX021500; DGRP_908, SRX021501; DGRP_911, SRX021502; and DGRP_913, SRX156024. The genotypes, quality scores, phenotypes, and web-based analysis tools are available from the DGRP website (http://dgrp2.gnets.ncsu.edu). All codes used to analyze these data are given in https://github.com/qgg-lab/DGRPHeadMorphologyEpistasis.