Abstract
We present a Chemistry and Structure Screen Integrated Efficiently (CASSIE) approach (named for Greek prophet Cassandra) to design inhibitors for cancer biology and pathogenesis. CASSIE provides an effective path to target master keys to control the repair-replication interface for cancer cells and SARS CoV-2 pathogenesis as exemplified here by specific targeting of Poly(ADP-ribose) glycohydrolase (PARG) and ADP-ribose glycohydrolase ARH3 macrodomains plus SARS CoV-2 nonstructural protein 3 (Nsp3) Macrodomain 1 (Mac1) and Nsp15 nuclease. As opposed to the classical massive effort employing libraries with large numbers of compounds against single proteins, we make inhibitor design for multiple targets efficient. Our compact, chemically diverse, 5000 compound Goldilocks (GL) library has an intermediate number of compounds sized between fragments and drugs with predicted favorable ADME (absorption, distribution, metabolism, and excretion) and toxicological profiles. Amalgamating our core GL library with an approved drug (AD) library, we employ a combined GLAD library virtual screen, enabling an effective and efficient design cycle of ranked computer docking, top hit biophysical and cell validations, and defined bound structures using human proteins or their avatars. As new drug design is increasingly pathway directed as well as molecular and mechanism based, our CASSIE approach facilitates testing multiple related targets by efficiently turning a set of interacting drug discovery problems into a tractable medicinal chemistry engineering problem of optimizing affinity and ADME properties based upon early co-crystal structures. Optimization efforts are made efficient by a computationally-focused iterative chemistry and structure screen. Thus, we herein describe and apply CASSIE to define prototypic, specific inhibitors for PARG vs distinct inhibitors for the related macrodomains of ARH3 and SARS CoV-2 Nsp3 plus the SARS CoV-2 Nsp15 RNA nuclease.
Keywords: Chemical library, Drug discovery, DNA replication and repair, Sars-COV2, Cancer
1. Introduction
Because of the high failure rate and extremely high cost of clinical trials, many drugs going into clinical trials are often modifications of approved drugs. For example, we identified the antibiotic Novobiocin (NVB) in a high throughput screen, as a DNA polymerase theta (POLθ) synthetically lethal with Homologous Recombination (HR) deficiency and thus a candidate target for HR-deficient cancers (Zhou et al., 2021). However, it is difficult to alter the complex structure of approved drugs and natural products for a new target (Atanasov et al., 2021; Omanakuttan et al., 2012). On the other side of the size range, fragment screening where crystals are probed with small molecules was designed to identify novel chemical structures (Murray & Blundell, 2010; Wilson 3rd et al., 2021), but obtaining larger compounds with higher affinity and selectivity plus with the proper orientation as indicated by the fragment-bound structures has proved problematic for drug discovery. Conceptually, an ideal process would be to start drug discovery using a Goldilocks (GL) compound library that is pre-selected for ideal drug characteristics and that is sized bigger than fragments but smaller than approved drugs. This intermediate size enables chemical elaboration to improve molecule properties, either for binding or for drug characteristics. To exemplify the versatility of our approach, we present our GLAD library (our GL library combined with an Approved Drug or AD library) and our Chemistry and Structure Screen Integrated Efficiently (CASSIE) pipeline and then their application to define lead inhibitors for two distinct target families: ADP ribosylase macrodomains and SARS CoV-2 Nsp15 endoribonuclease.
Controlling repair-replication and innate immune responses, macrodomains are an important drug target family: they counteract ADP-ribosylation in DNA repair and in viral infection, and thus are a fundamental cellular response to stress. In the context of DNA damage, they regulate efficient targeting and kinetics for the DNA damage response and are a target for synthetic lethality in DNA repair deficient cancer cells. Trapped PARP1 and DNA adducts facilitate MRE11 and RAD51 recruitment (Moiani et al., 2018; Syed & Tainer, 2018) and stall replication forks. Their repair involves removal of poly-ADP-ribosylation (PARylation) by macrodomain-containing glycohydrolases such as poly(ADP-ribose) glycohydrolase (PARG) (Houl et al., 2019). In the host antiviral environment, viral pathogens such as SARS CoV-2 employ a related macrodomain, Nsp3 Mac1, to remove ADP-ribosylation to dysregulate host immune response (Brosey et al., 2021). These features make macrodomains a focus for structure-based drug discovery as promising therapeutic targets both for cancer and antiviral agents. A key issue for DNA repair and virus macrodomain targets concerns homology of viral and human macrodomains and its impact on inhibitor binding pockets.
As part of an effort to develop inhibitors for additional SARS CoV-2 non-structural proteins (Nsp), we also applied our CASSIE approach for the SARS CoV-2 endoribonuclease Nsp15. Nsp15 is implicated in viral pathogenicity and evading host innate immune response as defective Nsp15 led to reduced viral pathogenicity in CoV models (Ancar et al., 2020; Deng et al., 2019; Kindler et al., 2017; Yuen et al., 2020). Its nidoviral uridylate-specific endoribonuclease (NendoU) function may regulate viral RNA synthesis, limit recognition of viral RNA by cellular sensors, and block early, protective antiviral host cell response. In particular, Nsp15 may help CoV evade host detection by trimming long 5′-polyuridine (polyU) tracts, a pathogen-associated molecular pattern (PAMP) that cells use to detect viruses (Hackbart, Deng, & Baker, 2020). Nsp15 NendoU activity is necessary and sufficient for inhibiting eIF2α-dependent and -independent stress granules (SGs) that function as an antiviral hub and whose suppression is needed for CoV replication (Gao et al., 2021). Thus, an Nsp15i could be of value by critically combining a direct anti-viral effect by enabling the early, protective immune response to viruses that thereby averts later excessive dysregulated damaging immune activation (Channappanavar et al., 2016; Channappanavar & Perlman, 2017); consistently, an Nsp15 natural product inhibitor (Nsp15i) efficiently neutralized SARS-CoV-2 (Hong et al., 2021). Yet, Nsp15 is understudied and virus-specific with no viable clinical drug, and the few cell-active inhibitors identified lack specificity (Choi et al., 2021; Hong et al., 2021). Thus, we apply CASSIE to target Nsp15, as it is essential in CoV biology (Kim et al., 2020; Xu et al., 2021) and as it could be an independent or anti-CoV cocktail target.
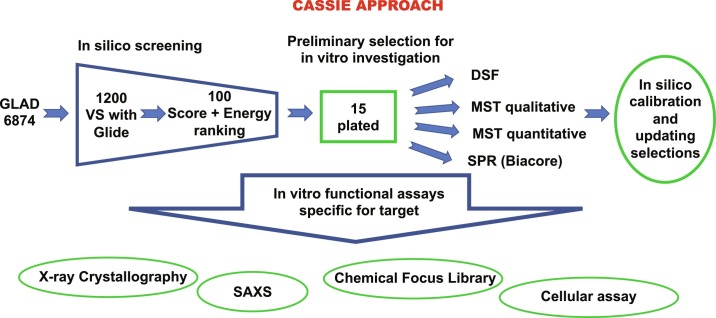
As an overview of our methodology, we begin by harnessing Evolutionary Trace (ET) methods to distinguish conserved and variable sequence areas of the active site pocket as shown for PARG and Nsp3 Mac1 (Brosey et al., 2021). Based upon an evolutionary perspective of surface sequence conservation, we focus on sites offering optimal specificity. Next, we use our CASSIE approach of in silico screening of our selective GLAD library against available protein structures and experimental validation by binding assays (Fig. 1 ). By employing a chemically-diverse, tractably-sized 5000 compound GL library of compounds larger than fragments but smaller than drugs, we collapse potentially immense chemical library space into a sparse set of cell-friendly chemotypes. Our tactical approach for efficient computational screens is to identify the top 15–100 candidates for binding measurements. Top binding candidates are employed for crystallization experiments to obtain X-ray crystal structures of inhibitor candidates with micromolar or better binding. When human structures are not available, useful crystallographic feedback is obtained by employing avatars that incorporate essential target site features (Moiani et al., 2018). We use structural results to provide a rational basis to build focused chemical libraries to improve binding affinity plus favorable ADME (absorption, distribution, metabolism, and excretion) properties and toxicological chemotypes. We show here how our GLAD library and CASSIE pipeline can provide inhibitor tools for cell biology and leads for preclinical drug discovery.
Fig. 1.
Schematic flow-chart representation of the CASSIE approach.
2. Targeting evolutionary conservation and identifying functional sites
The multiple interfaces, that DNA repair proteins have with their substrate and with other DNA repair and replication proteins and that viral proteins have for pathogen–host interactions, pose a challenge for in silico screening. It is key to define a target functional site and to identify residues relevant to the ligand docking volume. While structural and biological data may help define target sites, evolutionary sequence conservation complements the identification of novel regions of functional relevance and helps to prioritize sites. ET analysis combines phylogenetic relationships and sequence information to identify residues and regions of functional importance (Lichtarge, Bourne, & Cohen, 1996; Mihalek, Res, & Lichtarge, 2004). Importantly, ET scores protein residues according to the phylogenetic distances of the homologous sequences that vary at that residue and outputs these scores in a range of 0 (most important) to 100 (least important). These scores can be subsequently mapped to available structures or protein structure predictions (Lua & Lichtarge, 2010).
Clusters of high-ranking residues signify functionally important regions, such as active sites or critical allosteric sites (Lees-Miller et al., 2020), with confidence in these clusters made stronger by the independence of ET calculation from available structural information. Notably, these identified residues can be input into the in silico screening programs to define Virtual High-Throughput Screening (VHTS) receptor space. See Section 5 for VHTS input into the docking program, GLIDE. Here, we outline how to use the ET server (http://evolution.lichtargelab.org/) (Lichtarge et al., 1996; Mihalek et al., 2004).
2.1. Identify homologous sequences by BLAST searches
The ET server uses the Blastall 2.2.15 program (Altschul et al., 1997) and the NCBI non-redundant (nr) UniRef90, and Uniref100 databases (Maglott, Ostell, Pruitt, & Tatusova, 2007; Suzek et al., 2015). Typically, at least 50 homologous sequences in an alignment are used to calculate the ET scores. The exact number depends on the availability of homologous sequences and the sequence selection procedure. Technically, we can run ET with as many sequences as we like. The more sequences we use, the slower it is to align them and score (these relationships are exponential rather than linear). In the ET server, the user can define the maximum number of sequences (the default is 500 from the BLAST search). We typically start with 5000 sequences from BLAST.
2.2. Select sequences for the multiple sequences alignment (MSA)
Synthetic constructs, mutant sequences, and fragments of sequences should not be included in the MSA. Therefore, ET embodies filtering algorithms to remove such BLAST hits. Furthermore, redundant sequences may not be necessary or may bias the ET outcome to functions specific to the over-represented phylogenetic branches, so redundancy should be avoided. Scripts embedded to the ET server may select representative homologs at various phylogenetic distances that have the fewest alignment gaps compared to the query sequence. Typically, this process chooses approximately 160 sequences.
2.3. Align the sequences using MUSCLE alignment program
In the extremely rare cases that technical errors appear using MUSCLE alignment program (Edgar, 2004), the ET pipeline may turn to ClustalW alignment program (Thompson, Higgins, & Gibson, 1994). The steps 2.1–2.3 may be skipped if users prefer to input manually curated MSA.
2.4. Run ET analysis
We used the option “position-specific gap-reducing real-valued trace.” Although the ET algorithm may proceed with less homologous sequences, it is recommended to use MSA with at least five homologous sequences, although such small numbers of sequences may result in low resolution for ET scores (many ties in the residue ranks). Low resolution of ET ranks may also be obtained for larger number of homologous sequences, when they have high percentage identity compared to the query sequence. The ET analysis outputs scores for each residue in the range of 0 (most important) to 100 (least important).
2.5. Color available structures according to ET scores
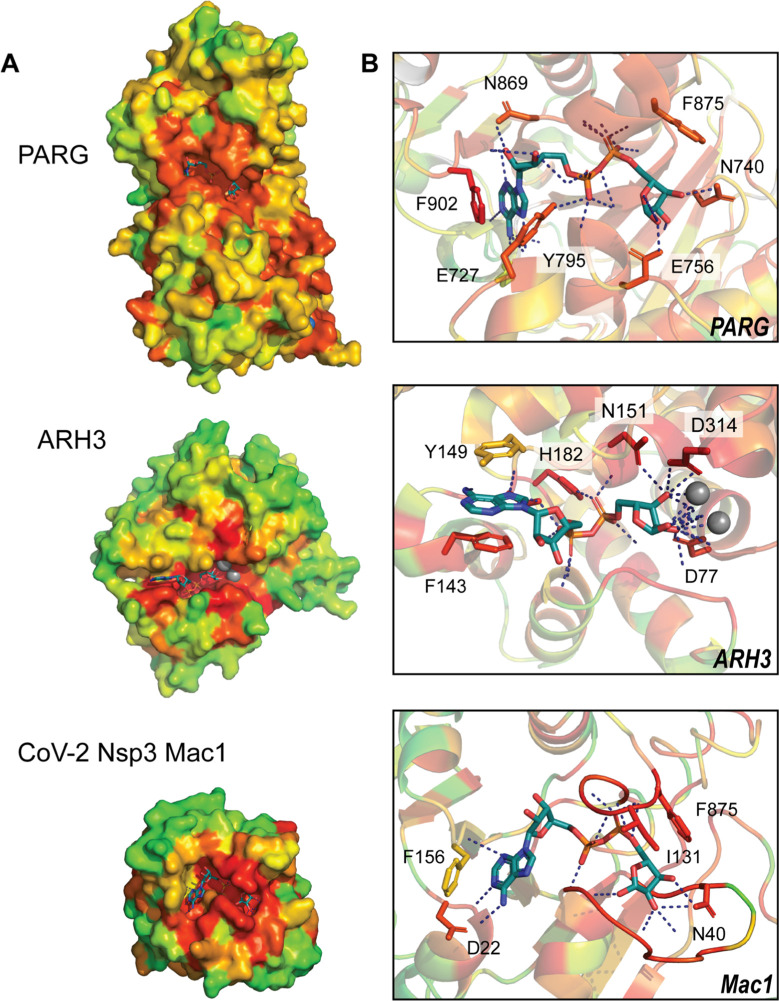
This can be done with the PyMOL program and the PyETV plugin (Lua & Lichtarge, 2010). Fig. 2 shows the PDB structures: 4B1G (human PARG), 6D36 (human ARH3), and 6W02 (CoV-2 Mac1), colored by prioritization scores (red notes the most important and green notes the least important residues). The PyETV plugin offers additional schemes to represent the important residues and additional functions, such as measuring the z-score for the non-random clustering of the top ET residues.
Fig. 2.
Evolutionary trace analysis identifies functional residues within PARG, ARH3, and CoV-2 Nsp3 Mac1 active sites. High-ranking ET values (red-orange) cluster at the active sites of each enzyme. Inspection of residues within the active sites (right panel) shows greatest conservation among residues coordinating the ADPr pyrophosphate linker and terminal ribose and increased variation within the adenine pocket.
2.6. Structural analysis of ET results on PARG, ARH3, and SARS CoV-2 Nsp3 Mac1
Applying this approach to PARG as a key repair-replication regulator and for comparative exemplary macrodomains ARH3 and SARS CoV-2 Nsp3 Mac1, highlights the active site as a shared primary region of functional significance in all three enzymes (Fig 2). Inspection of the active site reveals greater importance among residues coordinating the ADP ribose (ADPr) pyrophosphate linker and terminal ribose group relative to those stabilizing the adenine moiety. This is underscored by the unique binding conformations assumed by aromatic residues within the adenine pocket (c.f. PARG—phenylalanine 902, tyrosine 795; ARH3—phenylalanine 143, tyrosine 149; SARS CoV-2 Mac1—phenylalanine 156). These structural analyses by ET generally identify the active sites and other targets for screening. In this example, they suggest that selective targeting of the adenine pocket can be leveraged to identify ligands specific to each enzyme.
2.7. ET analysis of SARS CoV-2 proteins
We note that ET analyses for all SARS CoV-2 proteins, are available through an interactive GUI (http://cov.lichtargelab.org/) (Wang et al., 2021).
3. GLAD library
3.1. GLAD library description
The GLAD library has been assembled based upon experimental feedback over the last 5 years to consist of 6874 compounds and is divided into two major families as discussed in the introduction GL (Goldilocks) and AD (Approved Drugs).
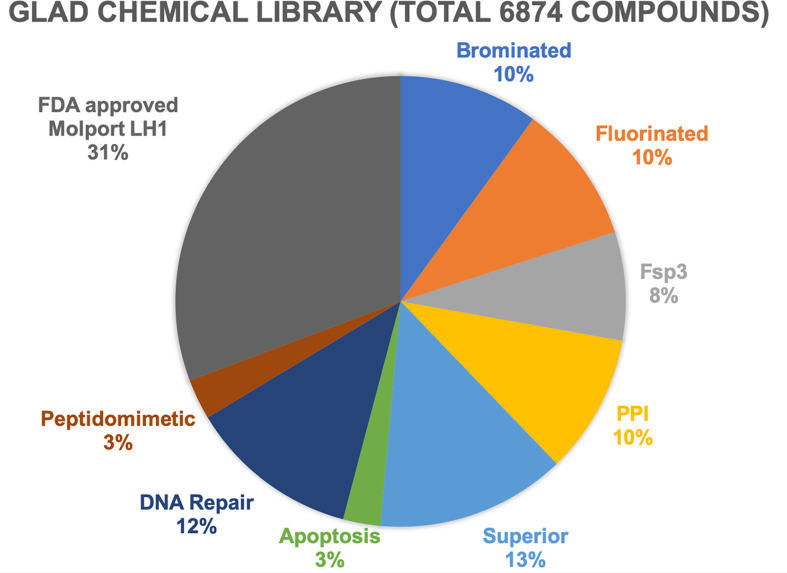
The GL library is subdivided into compound families (Fig. 3 ). The GL library was first assembled with 2500 compounds and then expanded based upon experimental results in two different steps to 4763. The initial GL compounds were selected to ensure chemical diversity using calculated two-dimensional fingerprints and the Tanimoto similarity index (Bajusz, Racz, & Heberger, 2015). Additionally, compounds were selected to minimize potential compound promiscuity (Bruns & Watson, 2012; Dahlin et al., 2015), assay interference (e.g., fluorescence), and with predicted favorable physicochemical properties (size, solubility, cellular permeability, etc.) (Lipinski, Lombardo, Dominy, & Feeney, 2001; Veber et al., 2002). The GL library includes compounds that are brominated, fluorinated, Protein–Protein Interaction disruptors (PPI), Fsp3 (containing sp3 hybridized carbon units), Superior (compounds optimized for solubility, low toxicity and cell permeability), peptidomimetic, and/or are targeting proteins involved in apoptosis and DNA Repair. The GL library was optimized for the percentage of each compound subfamily to its current composition (Fig. 3). The last expansion (2111 compounds), called AD (Approved Drugs), provides larger soluble compounds including Vitamins, lipids, natural compounds and additional human metabolites representing circa 31% of the full GLAD library. A complete virtual entity of the library has been assembled for in silico study. For experimental measurements, copies of the library are stored both in powder form and as DMSO solutions for in vitro and in vivo experiments.
Fig. 3.
Pie chart of GLAD library and its subfamilies distribution. Schematic representation of the distribution of 6874 GLAD compounds in its subfamilies with explicit percentage: AD (FDA approved library and others), Brominated, Fluorinated, Fsp3 carbon hybridization, PPI (Protein–Protein Interaction disruptors), Superior (optimized for Cell assay), compounds targeting Apoptosis and DNA Repair proteins (ligands involved or tested in our in-house DNA Repair system), Peptidomimetic.
3.2. Representation of the virtual GLAD library for in silico virtual screening
The material library is accompanied by a full collection of structure data format (.sdf) files representing all compounds with related information such as chemical identification, calculated properties, molecular weight, and SMILES. These are provided by our supplier (https://lifechemicals.com/).
3.3. Ligand files preparation
Upload the .sdf files for the full GLAD library to Maestro.
3.4. Optimize the ligands for in silico study
Run LigPrep program (Chen & Foloppe, 2010). In the program, we use the force field OPLS4 (Lu et al., 2021), which generates multiple conformers for each ligand to be incorporated in the GLAD in silico library for Virtual Screening. This collection of compounds is supplemented with additional compound controls that are known to bind to the target. Several human metabolites are already present in the AD subdivision.
4. Protein target and grid preparation
To reduce the computational complexity, we generate a ligand receptor grid onto the protein target site of interest. We use a rigid docking site with flexible ligands (Schnecke, Swanson, Getzoff, Tainer, & Kuhn, 1998). The PDB structure selected for in silico study needs to be prepared, optimized and minimized (Nguyen et al., 2021). If a protein target has an allosteric mechanism and the active site where the grid is centered has more than one preferential conformation, then multiple target structures need to be prepared in parallel to run multiple virtual screens (Moiani et al., 2018).
4.1. Prepare the protein structure
In the Protein Preparation Wizard (Sastry, Adzhigirey, Day, Annabhimoju, & Sherman, 2013), we strip waters and non-metal ligands from the structure. We keep metals and assign with proper charges. We change non-standard amino acids to standard amino acids, like selenomethionine to methionine. We either complete missing loops or cap termini resulting from missing loops with N-terminal acetyl and C-terminal amide capping groups and which gives the proper zwitterionic charge.
4.2. Generate a grid in the target area
This is typically an active site or protein-protein interaction surface. In the Receptor grid generation, we pick residues in the active site. The program will build a cube, typically with a range of dimensions of 10–15 Å on each side. If using a large grid (e.g., double the size), the calculations may take 1–2 days or longer, depending on computing power. Multiple grids can be generated and targeted at the same time. Results from multiple calculations and multiple grids requires a more accurate evaluation to generate the proper ranking of results.
4.3. Grids for macrodomains
For PARG, we used three structures (PDB ID 6OAK and PDB ID 4B1H with two conformations of phenylalanine 902) with the cubic dimensions (12 Å on each side) around phenylalanine 902, tyrosine 795, and phenylalanine 875 (based on the human structure). The Receptor grid generation program calculated the midpoint between the three c-alpha and centered the grid around the midpoint. We also generated grids for MAC1 (PDB ID 7KG3) and for ARH3 (PDB ID 6D36).
4.4. Grids for Nsp15 trimeric assembly
For Nsp15 (PDB ID 6WLC), we centered our grid on only Tyrosine 343 as it interacts with tipiracil and UMP (Kim et al., 2020, Kim et al., 2021). Notably, we generated the grid on a Nsp15 homotrimer, as the active site is near the subunit interface.
5. Virtual screen and ranking parameter selections
Next, we dock our GLAD library into the selected cubic grids of the protein using Glide program (Friesner et al., 2006). Glide allows the ligand to adopt multiple conformations but the protein is kept rigid. The virtual screening parameters are optimized using a workflow implemented in the Schrödinger package
5.1. Run a virtual screen calculation
In the Glide docking program we use “standard precision” and a parallelizing multiprocessor cluster and enable the MMGBSA scoring calculation in Prime. MM-GBSA energies (ΔG) are calculated from binding energies including covalent, van der Waals (VDW), coulombic, solvation, hydrogen bonding as well as from packing and energy differences related to strain in protein and ligand (Genheden & Ryde, 2015). As needed for speed, we use VHTS receptor space, which offers a lower precision but faster calculation.
5.2. Identify the top 10% compounds based on the docking score
Some ligands may occur multiple times because they bind in different conformations.
5.3. Rescore these top 10% compounds with the MMGBSA energy score
We have the most leads when we combine the docking score (weighted 70%) with MMGBSA energy (weighted 30%) (Moiani et al., 2009; Moiani, Cavallotti, Famulari, & Schmuck, 2008). We find both top binders but also more chemically diverse compounds in the list.
5.4. Remove compound redundancies
When the compound appears multiple times in the list because of different conformations, we typically remove all but the top scoring conformation.
5.5. Pick top scoring ligands with unique chemical features
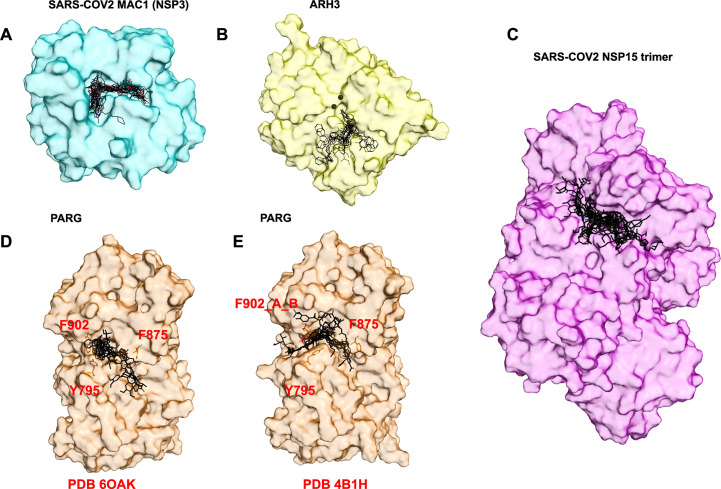
We analyze the subfamily type of the GLAD library in the full list of results, view the ligands docked on available structures, and select about 15 top in silico leads, which contain the highest number of unique chemical features, also known as chemical singletons. Fig. 4 (A–E) presents our selection for the macrodomain targets and Nsp15 for each of them a top 15 compounds were selected per ranking, with chemical singleton redundancies cleaned. The target is presented in surface representation to better display the population of top in silico leads.
Fig. 4.
Surface presentation of target proteins in complex with top 15 in silico leads. (A) MAC1 from PDB ID 7KG3 surface representation in complex with top 15 in silico results. (B) ARH3 from PDB ID 6D36 surface representation, magnesium ions black sphere, in complex with top 15 in silico results. (C) Nsp15 from PDB ID 6WLC) surface representation, in complex with top 15 in silico results. (D and E) PARG from three structures PDB ID 6OAK and PDB ID 4B1H with dual conformation a and b for amino acid F902 surface representation with key residue in stick in complex with top 15 results.
5.6. Final selection for in vitro experiments
The final selection of 15 is plated and moved to in vitro structural, binding and functional assays (see 6, 7).
5.7. PARG in silico leads
We merged the top compounds from in silico virtual screens performed on three different conformational structure of human PARG. Although we used different structures, some compounds were found in both docking analyses (Fig. 4D and E). After merging, the top ranked were 90% AD, and only two compounds were from the GL, a PPI and a DNA Repair uracil derivative. Interestingly, atracurium, in the top 15, has chemical similarity to a small unit of branched PAR.
5.8. ARH3 in silico leads
We merged the results obtained from targeting two grids simultaneously. To target the adenosyl clamp between phenylalanine 127 and tyrosine 133, the two grids were centered close to metal active site, specifically at residue asparagine 135. The results were grouped together and properly re-ranked as described in Section 5.3. The top 15 in silico leads’ distribution shows 50/50 ratio between GL and AD. Specifically the highest scoring AD are drugs approved for infection and for targeting the immune system, which is interesting in light of the immune evasion role of this viral enzyme. The GL top in silico leads are peptidomimetics, PPI, Superior, and molecules targeting DNA Repair proteins.
5.9. SARS CoV-2 MAC1 (Nsp3) in silico leads
After applying our ranking methods on the results from Glide, we analyzed the chemotype of top 15 in silico leads. The top two compounds are part of the DNA Repair subfamily of GL, and specifically are uracil derivatives. ADPr scored as third in the ranking, matching the high affinity for MAC1, proven by in vitro measurements (in Section 7) and shown in Fig. 5A. Top in silico leads include peptidomimetics and PPI disruptors. More than 50% of top results were AD, including an anticancer drug (Daunorubicin) and an antiviral drug (Valganiclovir). None of the compounds bound MAC1, based on Microscale Thermophoresis (MST) binding results except ADPr. The graphic model of top silico leads for Mac1 is shown in Fig. 4A.
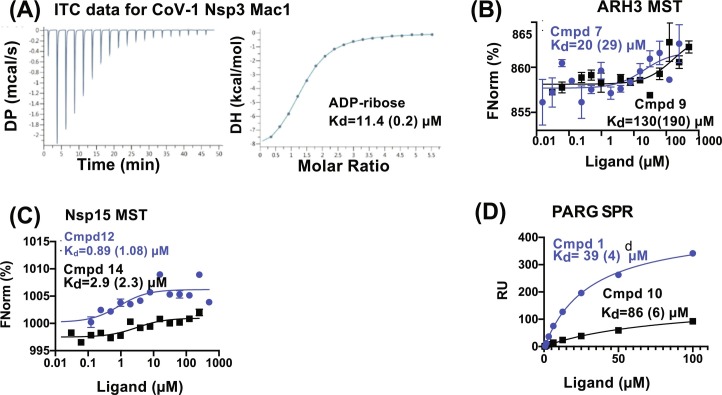
Fig. 5.
Representative binding curves for in vitro binding experiments for CoV-1 Nsp3 Mac1, ARH3, Nsp15 and PARG. (A) ITC data for CoV-1 Nsp3 Mac1 binding to ADP-ribose. (B) MST data for ARH3 binding to two of the top scored compounds. Binding was detected for 3 of the 15 compounds with binding constants ranging from 20 to 400 μM. (C) MST data for Nsp15 binding to two of the top scored compounds. Binding was detected for 12 of the 15 compounds with binding constants ranging from 180 nM to 400 μM. (D) SPR data for PARG binding to two of the top scored compounds. Binding was detected for 6 of the 16 compounds with binding constants ranging from 23 to 1000 μM.
Panel (A): Reproduced from Brosey, C. A., Houl, J. H., Katsonis, P., Balapiti-Modarage, L. P. F., Bommagani, S., Arvai, A., et al. (2021). Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Progress in Biophysics and Molecular Biology. 163, 171–186.
5.10. SARS CoV-2 Nsp15 in silico leads
The analysis of top in silico leads shows 70% are AD, while the rest include a PPI and three DNA Repair subfamily uracil derivatives. Among the AD results, we found certain peptides like Goserelin and Desmopressin or complex molecules like Anidulafungin (an antifungal drug). Interestingly, we found some high molecular weight compounds as top in silico leads with an extended binding surface as shown in Fig. 4C.
6. Fast qualitative and quantitative in vitro binding assays of top in silico results
As part of CASSIE (Fig. 1), we validate our in silico hits with binding assays. The top in silico selections are physically plated at suitable concentrations for qualitative binding. Compounds are prepared from the mother solution (directly plated by supplier) and made uniform at 10 mM concentration in DMSO. Typically, the in vitro techniques selected for binding studies are Microscale thermophoresis (MST) or Surface Plasmon Resonance (SPR, Biacore). Measuring ligand-protein affinity constants ranging from nM to mM, MST tracks thermophoresis, the movement of fluorescently labeled molecules in and out of a small temperature gradient. When a protein binds a ligand, there may be changes in protein conformation or even the protein hydration shell. These changes can alter the rate of movement (thermophoresis) of the protein–ligand complex as compared to the control, protein without added ligand. This is monitored by changes in fluorescence, with varying ligand concentrations to generate a binding curve from which affinity is calculated. The deflections in the binding curves can be either positive or negative as the deflection is dependent on the relative rates of the protein without ligand vs the protein ligand complex. However, if there is no change in the thermophoresis rates upon a binding event, this event will be silent or undetected using MST. SPR tracks ligand binding by measuring changes in polarized light striking the target protein or protein complex attached to a sensor surface. The protein is usually covalently attached to a sensor surface or captured using an affinity tag such as his6 or streptavidin. These two binding assays, one detecting changes in thermophoresis of the protein by binding ligand and the other detecting changes in target mass upon binding, provide complementary information and validation of the small molecule in silico lead. The selection of top compounds from in silico are run in parallel to a well-known binder for the enzyme and compared. Compounds identified with adequate affinity move to a quantitative test, such as isothermal calorimetry (ITC) which usually requires added material to ensure reproducibility of multiple measurements.
6.1. MST assay
We typically measure MST on a Monolith NT1.15. Proteins are labeled with Atto488 NHS-ester (ATTO-TEC, GBH) according to the manufacturer's protocol with labeling efficiency 1:1 protein-to-dye ratio. Ligands are serially diluted into solution of PBS, 0.5 mM fresh TCEP, 0.01% Tween-20 with the Atto488-labeled protein. Samples are loaded into standard capillaries and each set analyzed in triplicate. Data fitting with the MO Affinity Analysis software for a series of compounds is performed at the lowest fixed power and time which gives the largest signal changed based on the either known binders or the most consistent change for that series. For ARH3 analysis, ADPr was used as a control and the settings were at high power with thermophoresis at 20 s. For Nsp15, the most consistent change for the series was with medium power with thermophoresis at 10 s. Capillaries with measure intensities greater than 1% DMSO or with intensities (±) 5% of the overall average are excluded from the analysis. Binding was labeled as not detected (ND) if the data could not be fit to the Kd model, or the calculated response amplitude was less than 2.
For ARH3, binding was detected for 3 of the 15 compounds with binding constants ranging from 20 to 400 μM. For Nsp15 binding was detected for 12 of the 15 compounds with binding constants ranging from 180 nM to 400 μM. For PARG there was no detectable change in the thermophoresis based on the above criteria and as a result, SPR was used.
6.2. SPR assay
We typically measure SPR on a Biacore T200 with proteins attached to the sensor surface. For PARG, 1 mg/ml protein was covalently attached to a Biacore CM5 chip using amine coupling via an EDC/NHS (N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride/N-hydroxysuccinimide) reaction. A control channel is treated similarly, except using ethanolamine instead of protein. The final relative response unit (RU) was 899.7. Serially dilute ligand starting at 100 μM was passed over the surface at 30 μL/min with a contact time of 120 s. The data was fit to a steady state affinity model with the Biacore Evaluation 3.0 software using blank subtracted RU values at 4 s before injection stop for a single run. For PARG, binding was detected for 6 of the 16 compounds with binding constants ranging from 23 to 1000 μM.
6.3. Limitation of these two assays and data evaluation for top in silico leads
MST in qualitative mode is a relatively quick method and needs a low quantity of material (including protein) but the data are not extremely reliable, particularly if the binding data extrapolated fluctuation range is over half millimolar. MST in quantitative mode would be more appropriate and needs multiple measurements in the titration, which requires more time and material with more reproducibility but the final binding Kd is still not precise.
SPR offers better data if the system is properly calibrated. The protein needs to be anchored to the chip and a calibration test implemented. Following these steps, the system can efficiently produce multiple, reliable, and accurate affinity binding measurements. ITC is the best but requires time for calibration and optimization plus an abundant quantity of material.
If micromolar or better binders are not discovered within the top 15, then we recommend testing compounds in multiple binding assays and to test all of the top 100 compounds. In silico docking scores do not directly correlate to experimental affinities.
7. Structural analysis of top in silico compounds
After in vitro binding assay validations of the in silico data, we proceed to structure-based optimizations. Crystallography is an optimal structural validation for precision; however, co-crystal structures may be difficult to obtain with preliminary candidates. With the CASSIE approach, inhibition of function and low micromolar binding are more effectively evaluated with minimal effort and protein.
7.1. What to look for in initial co-crystal structures
In binding pockets, we use crystal structures strategically to identify two different kind of subsites: anchor points and induced plasticity. This approach is based upon our discovery of anchored plasticity to gain 20,000-fold between active site pockets formed by invariant sequences in nitric oxide synthase (NOS) and the ability to selectively inhibit MRE11 endonuclease and exonuclease activities (Garcin et al., 2008; Shibata et al., 2014). Tactically, the larger sized AD library components can identify inducible plasticity regions within binding sites. The intermediate sized GL library components are better at identifying potential anchor points in binding sites that can be used for focused libraries to optimize affinity and drug-like properties by growing compounds into surface subsites with plasticity. Thus, this combination of data enables focused compound libraries to employ a systematic strategy of retaining anchor interactions while growing compounds in directions defined by a vector from the anchor site to flexible subsites. Focused libraries are designed from lead compounds by adding new chemical moieties that can bind in neighboring regions. Target flexibility or plasticity, if identified by structural analysis, can also be harnessed to provide additional regions for growing compounds to increase specificity. By employing this anchored plasticity approach CASSIE provides a systematic approach to enlarging binding pock subsites for increased affinity. This can span from creating new pockets for inhibitors as we did for Map kinase (Perry, Harris, Moiani, Olson, & Tainer, 2009) to building upon substrate analogue structures (Daniels et al., 2000; Doi et al., 2006) and learning how inhibitors displace bound water molecules (Putnam, Arvai, Bourne, & Tainer, 2000). Although we currently use crystal structures and temperature factors, as employed for EXO5 nuclease (Hambarde et al., 2021) to identify target plasticity, we expect to be able to use atomic structures combined with SAXS to identify key plasticity associated with allosteric mechanisms (Brosey & Tainer, 2019; Hammel et al., 2021; Hammel & Tainer, 2021). Observed plasticity can even be used to apply inhibitor doorstoppers to mimic a protein inhibitor and block an enzyme's active site closing as needed for strong DNA binding, by uracil-DNA glycosylase (Nguyen et al., 2021; Putnam et al., 1999).
8. Summary and prospects for advances
Urgent demands for efficient drug design stem from the SARS CoV-2 pandemic (200 million cases and 4.4 million deaths as of August 24, 2021, Johns Hopkins Center for Systems Science and Engineering, https://coronavirus.jhu.edu/map.html) and cancer (19 million new cases and 10 million deaths) (Sung et al., 2021). These diseases underscore the merit of new approaches that reduce bottlenecks and improve efficiency for drug discovery. Yet, meeting this need is challenging as both cancer cells and pathogens have a mutator phenotype that can promote selection of resistance to both the host immune system and drug therapies. For cancer drugs, relevant new mechanistic insights have come from targeting poly (ADP-ribose) polymerase (PARP). PARP inhibitors (PARPi) are successful for ovarian cancer and breast cancer indications, but resistance often develops (Li et al., 2020), suggesting a need to also be able to inhibit additional pathway proteins or complementary pathways.
Interestingly, the clinical success of PARPi's can depend upon trapping PARP1 on the damaged DNA, and this allosteric capacity has been structurally characterized for different PARPi (Zandarashvili et al., 2020). Indeed, we also know from biology that catalytically inactive proteins that bind and hold damaged DNA can control damage outcomes, as seen for ATL that forms a complex with alkylated base damage and thereby alters repair pathway outcome from base repair to the nucleotide excision repair pathway (Tubbs et al., 2009). Analogously, XPG impacts multiple repair pathways by binding and sculpting DNA junctions and protein partners in addition to its nuclease activity (Tsutakawa et al., 2020), FEN1 sculpts 5’ flaps to avoid template switching at replication forks (Perry et al., 2006, Trego et al., 2011, Tsutakawa et al., 2017), GRB2 adaptor protein efficiently brings MRE11 nuclease to DNA breaks (Ye et al., 2021), XRCC1 links MRE11 and PolQ to promote alternative end joining of DNA breaks (Eckelmann et al., 2020), SLX4IP binds and maintains SLX4-XPF-ERCC1 complex for inter-strand crosslink repair (Zhang et al., 2019), and acetylation targets oxidative base repair initiation to open chromatin (Bacolla et al., 2021). In extreme examples, transient polyvalent binding by flexible proteins and RNA can create phase separated condensates of functional proteins for processes such as repair, as seen for DNA break repair protein KU and its complex with long non-coding RNA (Thapar et al., 2021). Furthermore, combining structural and computational methods is proving enabling to identify and target molecular mechanisms as well as to understand the mechanisms where point mutations can cause different human diseases (Yan et al., 2019).
All of the above noted observations support the value of molecular mechanistic knowledge for drug discovery. In fact, most successful new drugs result from molecular and mechanistic targeting that requires chemical, biophysical, biochemical and structural knowledge. It therefore becomes important to develop systematic strategies to attain this information efficiently. As a result of this reasoning, we developed the CASSIE pipeline and GLAD library to these potential overcome rate-limiting steps in developing inhibitor tools and preclinical drug candidates.
In Greek myth, Cassandra consistently made accurate predictions that were unfortunately not believed. CASSIE predictions, however, do not require faith as they can be tested and validated by practical levels of time and effort. In contrast, high throughput screens with overly large libraries can in practice be high input with challenges to effectively handle output. The CASSIE pipeline and GLAD library approach provide a manageable number of compounds, hits, and validation measurements within weeks. Getting compounds into cells and crystal structures in minimal time frames enables molecular targets and mechanisms to be efficiently tested directly and thereby provides a critical enabling path for preclinical drug discovery.
Acknowledgments
We thank Chi-Lin Tsai for comments. We acknowledge National Institute of Health (NIH) grants (P01 CA092584, R35CA220430), R01 GM141226 (to I.K.K.), a Robert A. Welch Chemistry Chair, and Cancer Prevention and Research Institute of Texas (CPRIT) RP180813. X-ray diffraction data was collected at SSRL beamline 12-2 and ALS beamlines 12.3.1 [supported by NIH project ALS-ENABLE (P30 GM124169) and the Integrated Diffraction Analysis Technologies (IDAT) program] and 8.3.1 (supported by NIH R01 GM124149 and P30 GM124169). Nsp15 research was supported by the U.S. Department of Energy (DOE) Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on response to coronavirus disease 2019, with funding provided by the Coronavirus CARES Act.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancar R., Li Y., Kindler E., Cooper D.A., Ransom M., Thiel V., et al. Physiologic RNA targets and refined sequence specificity of coronavirus EndoU. RNA. 2020;26:1976–1999. doi: 10.1261/rna.076604.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov A.G., Zotchev S.B., Dirsch V.M., The International Natural Product Sciences Taskforce, Supuran C.T. Natural products in drug discovery: Advances and opportunities. Nature Reviews. Drug Discovery. 2021;20:200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Sengupta S., Ye Z., Yang C., Mitra J., De-Paula R.B., et al. Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin. Nucleic Acids Research. 2021;49:221–243. doi: 10.1093/nar/gkaa1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz D., Racz A., Heberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? Journal of Cheminformatics. 2015;7:20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey C.A., Houl J.H., Katsonis P., Balapiti-Modarage L.P.F., Bommagani S., Arvai A., et al. Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Progress in Biophysics and Molecular Biology. 2021;163:171–186. doi: 10.1016/j.pbiomolbio.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey C.A., Tainer J.A. Evolving SAXS versatility: Solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Current Opinion in Structural Biology. 2019;58:197–213. doi: 10.1016/j.sbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns R.F., Watson I.A. Rules for identifying potentially reactive or promiscuous compounds. Journal of Medicinal Chemistry. 2012;55:9763–9772. doi: 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host & Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Seminars in Immunopathology. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.J., Foloppe N. Drug-like bioactive structures and conformational coverage with the LigPrep/ConfGen suite: Comparison to programs MOE and catalyst. Journal of Chemical Information and Modeling. 2010;50:822–839. doi: 10.1021/ci100026x. [DOI] [PubMed] [Google Scholar]
- Choi R., Zhou M., Shek R., Wilson J.W., Tillery L., Craig J.K., et al. High-throughput screening of the ReFRAME, Pandemic Box, and COVID Box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin J.L., Nissink J.W., Strasser J.M., Francis S., Higgins L., Zhou H., et al. PAINS in the assay: Chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. Journal of Medicinal Chemistry. 2015;58:2091–2113. doi: 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D.S., Mol C.D., Arvai A.S., Kanugula S., Pegg A.E., Tainer J.A. Active and alkylated human AGT structures: A novel zinc site, inhibitor and extrahelical base binding. The EMBO Journal. 2000;19:1719–1730. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., van Geelen A., Buckley A.C., O'Brien A., Pillatzki A., Lager K.M., et al. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. Journal of Virology. 2019;93:e02000–e02018. doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y., Katafuchi A., Fujiwara Y., Hitomi K., Tainer J.A., Ide H., et al. Synthesis and characterization of oligonucleotides containing 2'-fluorinated thymidine glycol as inhibitors of the endonuclease III reaction. Nucleic Acids Research. 2006;34:1540–1551. doi: 10.1093/nar/gkl061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelmann B.J., Bacolla A., Wang H., Ye Z., Guerrero E.N., Jiang W., et al. XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer. 2020;2 doi: 10.1093/narcan/zcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gao B., Gong X., Fang S., Weng W., Wang H., Chu H., et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathogens. 2021;17 doi: 10.1371/journal.ppat.1008690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin E.D., Arvai A.S., Rosenfeld R.J., Kroeger M.D., Crane B.R., Andersson G., et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nature Chemical Biology. 2008;4:700–707. doi: 10.1038/nchembio.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambarde S., Tsai C.L., Pandita R.K., Bacolla A., Maitra A., Charaka V., et al. EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart. Molecular Cell. 2021;81:2989–3006. doi: 10.1016/j.molcel.2021.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M., Rashid I., Sverzhinsky A., Pourfarjam Y., Tsai M.S., Ellenberger T., et al. An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIalpha within a flexible DNA repair complex. Nucleic Acids Research. 2021;49:306–321. doi: 10.1093/nar/gkaa1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M., Tainer J.A. X-ray scattering reveals disordered linkers and dynamic interfaces in complexes and mechanisms for DNA double-strand break repair impacting cell and cancer biology. Protein Science. 2021;30:1735–1756. doi: 10.1002/pro.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Seo S.H., Woo S.J., Kwon Y., Song M., Ha N.C. Epigallocatechin gallate inhibits the uridylate-specific endoribonuclease Nsp15 and efficiently neutralizes the SARS-CoV-2 strain. Journal of Agricultural and Food Chemistry. 2021;69:5948–5954. doi: 10.1021/acs.jafc.1c02050. [DOI] [PubMed] [Google Scholar]
- Houl J.H., Ye Z., Brosey C.A., Balapiti-Modarage L.P.F., Namjoshi S., Bacolla A., et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nature Communications. 2019;10:5654. doi: 10.1038/s41467-019-13508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Communications Biology. 2021;4:193. doi: 10.1038/s42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathogens. 2017;13 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller J.P., Cobban A., Katsonis P., Bacolla A., Tsutakawa S.E., Hammel M., et al. Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Progress in Biophysics and Molecular Biology. 2020;163:87–108. doi: 10.1016/j.pbiomolbio.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu Z.Y., Wu N., Chen Y.C., Cheng Q., Wang J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Molecular Cancer. 2020;19:107. doi: 10.1186/s12943-020-01227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. Journal of Molecular Biology. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lu C., Wu C., Ghoreishi D., Chen W., Wang L., Damm W., et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. Journal of Chemical Theory and Computation. 2021;17:4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- Lua R.C., Lichtarge O. PyETV: A PyMOL evolutionary trace viewer to analyze functional site predictions in protein complexes. Bioinformatics. 2010;26:2981–2982. doi: 10.1093/bioinformatics/btq566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez gene: Gene-centered information at NCBI. Nucleic Acids Research. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalek I., Res I., Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. Journal of Molecular Biology. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- Moiani D., Cavallotti C., Famulari A., Schmuck C. Oxoanion binding by guanidiniocarbonylpyrrole cations in water: A combined DFT and MD investigation. Chemistry. 2008;14:5207–5219. doi: 10.1002/chem.200701745. [DOI] [PubMed] [Google Scholar]
- Moiani D., Ronato D.A., Brosey C.A., Arvai A.S., Syed A., Masson J.Y., et al. Targeting allostery with avatars to design inhibitors assessed by cell activity: Dissecting MRE11 endo- and exonuclease activities. Methods in Enzymology. 2018;601:205–241. doi: 10.1016/bs.mie.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiani D., Salvalaglio M., Cavallotti C., Bujacz A., Redzynia I., Bujacz G., et al. Structural characterization of a Protein A mimetic peptide dendrimer bound to human IgG. The Journal of Physical Chemistry. 2009;113:16268–16275. doi: 10.1021/jp909405b. [DOI] [PubMed] [Google Scholar]
- Murray C.W., Blundell T.L. Structural biology in fragment-based drug design. Current Opinion in Structural Biology. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Nguyen M.T., Moiani D., Ahmed Z., Arvai A.S., Namjoshi S., Shin D.S., et al. An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation. Progress in Biophysics and Molecular Biology. 2021;163:143–159. doi: 10.1016/j.pbiomolbio.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omanakuttan A., Nambiar J., Harris R.M., Bose C., Pandurangan N., Varghese R.K., et al. Anacardic acid inhibits the catalytic activity of matrix metalloproteinase-2 and matrix metalloproteinase-9. Molecular Pharmacology. 2012;82:614–622. doi: 10.1124/mol.112.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.J., Harris R.M., Moiani D., Olson A.J., Tainer J.A. p38alpha MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses. Journal of Molecular Biology. 2009;391:1–11. doi: 10.1016/j.jmb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.J., et al. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nature Structure & Molecular Biology. 2006;13(5):414–422. doi: 10.1038/nsmb1088. 1088. [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Arvai A.S., Bourne Y., Tainer J.A. Active and inhibited human catalase structures: Ligand and NADPH binding and catalytic mechanism. Journal of Molecular Biology. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Shroyer M.J., Lundquist A.J., Mol C.D., Arvai A.S., Mosbaugh D.W., et al. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. Journal of Molecular Biology. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Schnecke V., Swanson C.A., Getzoff E.D., Tainer J.A., Kuhn L.A. Screening a peptidyl database for potential ligands to proteins with side-chain flexibility. Proteins. 1998;33:74–87. [PubMed] [Google Scholar]
- Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt C. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A., Tainer J.A. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in dna replication and repair. Annual Review of Biochemistry. 2018;87:263–294. doi: 10.1146/annurev-biochem-062917-012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R., Wang J.L., Hammel M., Ye R., Liang K., Sun C., et al. Mechanism of efficient double-strand break repair by a long non-coding RNA. Nucleic Acids Research. 2021;49:1199–1200. doi: 10.1093/nar/gkaa1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trego K.S., et al. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle. 2011;10(12):1998–2007. doi: 10.4161/cc.10.12.15878. 15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa S.E., Sarker A.H., Ng C., Arvai A.S., Shin D.S., Shih B., et al. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:14127–14138. doi: 10.1073/pnas.1921311117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa S.E., et al. Phosphate steering by Flap Endonuclease 1 promotes 5’-flap specificity and incision to prevent genome instability. Nature Communication. 2017 doi: 10.1038/ncomms15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs J.L., Latypov V., Kanugula S., Butt A., Melikishvili M., Kraehenbuehl R., et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Wang C., Konecki D.M., Marciano D.C., Govindarajan H., Williams A.M., Wastuwidyaningtyas B., et al. Identification of evolutionarily stable functional and immunogenic sites across the SARS-CoV-2 proteome and greater coronavirus family. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.M., 3rd, Deacon A.M., Duncton M.A.J., Pellicena P., Georgiadis M.M., Yeh A.P., et al. Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA damage response. Progress in Biophysics and Molecular Biology. 2021;163:130–142. doi: 10.1016/j.pbiomolbio.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Pei G., Liu H., Ju X., Wang J., Ding Q., et al. Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome. Cell Reports. 2021;36:109482. doi: 10.1016/j.celrep.2021.109482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Dodd T., He Y., Tainer J.A., Tsutakawa S.E., Ivanov I. Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural & Molecular Biology. 2019;26:397–406. doi: 10.1038/s41594-019-0220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Xu S., Shi Y., Bacolla A., Syed A., Moiani D., et al. GRB2 enforces homology-directed repair initiation by MRE11. Science Advances. 2021;7 doi: 10.1126/sciadv.abe9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerging Microbes & Infections. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandarashvili L., Langelier M.F., Velagapudi U.K., Hancock M.A., Steffen J.D., Billur R., et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science. 2020;368 doi: 10.1126/science.aax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chen Z., Ye Y., Ye Z., Cao D., Xiong Y., et al. SLX4IP acts with SLX4 and XPF-ERCC1 to promote interstrand crosslink repair. Nucleic Acids Research. 2019;47:10181–10201. doi: 10.1093/nar/gkz769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gelot C., Pantelidou C., Li A., Yucel H., Davis R.E., et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nature Cancer. 2021;2:598–610. doi: 10.1038/s43018-021-00203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]