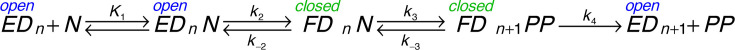Abstract
Nucleotide analogs are the cornerstone of direct acting antivirals used to control infection by RNA viruses. Here we review what is known about existing nucleotide/nucleoside analogs and the kinetics and mechanisms of RNA and DNA replication, with emphasis on the SARS-CoV-2 RNA dependent RNA polymerase (RdRp) in comparison to HIV reverse transcriptase and Hepatitis C RdRp. We demonstrate how accurate kinetic analysis reveals surprising results to explain the effectiveness of antiviral nucleoside analogs providing guidelines for the design of new inhibitors.
Keywords: Remdesivir, RNA-dependent RNA polymerase, Polymerase kinetics, Single-turnover kinetics, Nucleoside analogs, Enzyme specificity, Proofreading exonuclease, SARS CoV-2, Coronavirus
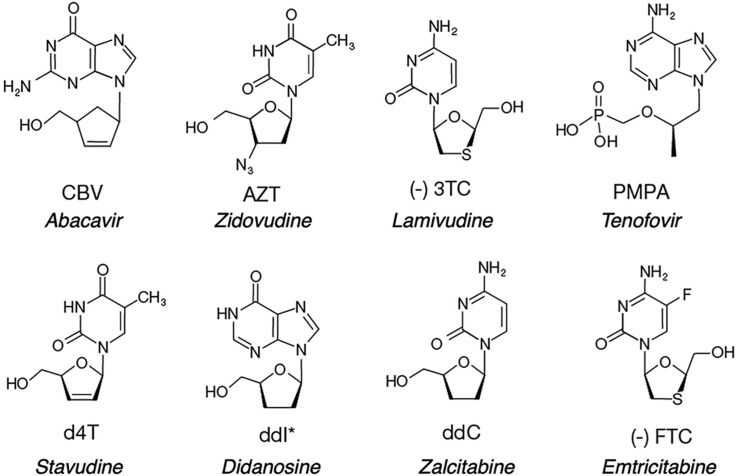
Eight nucleoside/nucleotide analogs have been approved by the FDA for treatment of HIV infections (Fig. 1 ). Although all analogs except for tenofovir are administered as nucleosides, they are all metabolized to nucleoside triphosphates to be active in vivo. Therefore, we will use the term nucleotide generically to refer to the analogs used to treat viral infections. Each of these analogs lacks a 3′OH and thereby acts as a chain terminator after being incorporated by HIV reverse transcriptase (HIVRT).
Fig. 1.
Structures of nucleoside/nucleotide analogs approved to treat HIV infections. Analogs are shown in their state before phosphorylation by cellular enzymes. Drug names are given in italics. Note that Tenofovir is a nucleotide analog with a phosphonate linkage to overcome the fact that cellular kinases are inefficient at adding the alpha phosphate to the structure lacking an intact ribose ring.
Although their chemical mechanisms of inhibition are identical and appear simple, the kinetic and thermodynamic basis for efficient incorporation of each analog is complex, as are the varied sets of mutations in HIVRT leading to drug resistance. The wealth of information gained from decades of work on HIVRT provides guidelines for the development of drugs to treat emerging viral infections, but certain features of nucleotide analogs directed against HIVRT do not apply in all cases. In particular, the long duration of treatment of HIV infections places exceptional demands for low toxicity, but the absence of a proofreading exonuclease in HIVRT simplifies the design requirements. In this review, we will highlight common features and point out unique aspects of the Hepatitis C virus (HCV) and the SARS CoV-2 RNA dependent RNA polymerases (RdRp) compared to HIVRT. We will highlight how nucleotide analogs used to inhibit the SARS CoV-2 RdRp must overcome unique features of the viral RdRp, including the presence of a proofreading exonuclease that would efficiently remove chain terminators after they are incorporated into the RNA strand.
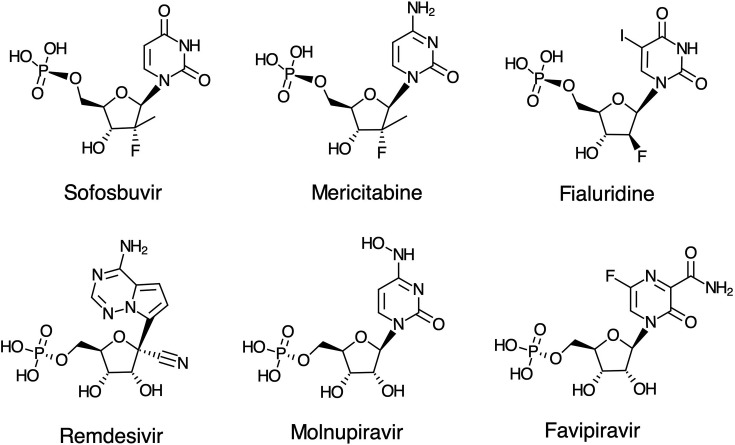
Nucleotide analogs directed against viral RdRps of HCV and SARS CoV-2 are shown in Fig. 2 . Sofosbuvir is used in combination therapy to effectively treat Hepatitis C viral infections, while Mericitabine failed because it is removed by an efficient ATP-dependent excision reaction [1]. Remdesivir is the only FDA approved, direct acting antiviral drug used to treat COVID-19 and acts as a delayed chain terminator [2], [3], [4]. Molnupiravir is in a Phase 3 clinical trial for COVID-19 and is thought to act by lethal mutagenesis, as suggested for a structurally similar analog, Favipiravir [5]. For comparison we also show Fialuridine, which led to the death of 5 out of 15 patients in a hepatitis B clinical trial [6]. It was later shown to be stably incorporated by the human mitochondrial DNA polymerase leading to lethal mutagenesis of the mitochondrial genome [7], [8]. Although Fialuridine was designed to be a “defacto chain terminator”, after incorporation by the human mitochondrial DNA polymerase, 60% of the time it was extended leading to lethal mutagenesis rather than being excised by the proofreading exonuclease. This study raises caution about the safety of nucleoside analogs that rely on lethal mutagenesis as a strategy to combat viral infections, and the importance of studies to accurately measure the kinetics of excision by the proofreading exonuclease of both viral (in the case of SARS CoV-2) and host polymerases.
Fig. 2.
Nucleoside analogs directed against RNA dependent RNA polymerases. Although each is administered as a prodrug, analogs are shown in their monophosphate form. Fialuridine is included here for comparison because it failed due to toxicity in a clinical trial for hepatitis B, which is a DNA virus.
1. Mechanistic basis for effective inhibition
Inhibition of HIVRT by nucleoside analogs lacking a 3′OH, thereby leading to chain termination, is a function of their efficiency in serving as substrates for incorporation by HIVRT. Therefore, we can estimate their effectiveness by the discrimination ratio (D) of k cat/K m values in comparing normal nucleotides (N) with nucleoside analogs (NA) in their triphosphate form.
| (1) |
Table 1 shows a summary of k cat, K m, and k cat/K m values for most of the nucleotide analogs along with the index by which HIVRT discriminates against the analogs relative to the normal nucleotides. The higher the value for D the less effective the analog as the enzyme exhibits a greater discrimination against the analog. Note the two of the analogs (AZT and PMPA) are actually slightly better substrates than the corresponding cognate nucleotide.
Table 1.
Kinetic parameters for incorporation of nucleotides and analogs.
| Nucleotide | kcat (s− 1) | Km (μM) | kcat/Km (μM− 1 s− 1) | D | Source |
|---|---|---|---|---|---|
| dTTP | 34 | 4.7 | 7.2 | Kellinger and Johnson [9] | |
| AZT | 29 | 3.5 | 8.3 | 0.9 | Kellinger and Johnson [9] |
| d4T | 1.4 | 1.2 | 1.2 | 6 | Vaccaro et al. [9a] |
| dCTP | 15 | 1.5 | 10 | Kellinger and Johnson [10] | |
| 3TC | 0.028 | 0.027 | 1 | 9.6 | Kellinger and Johnson [10] |
| dCTP | 23 | 30 | 0.8 | Feng et al. [10a] | |
| FTC | 0.08 | 1.4 | 0.06 | 13 | Feng et al. [10a] |
| dGTP | 24 | 14 | 1.7 | Ray et al. [11] | |
| CBV | 1 | 21 | 0.05 | 36 | Ray et al. [11] |
| dATP | 33 | 8.1 | 4.1 | Suo and Johnson [11a] | |
| ddATP | 55 | 54 | 1 | 4 | Suo and Johnson [11a] |
| PMPA | 49 | 58 | 0.84 | 0.8 | Suo and Johnson [11a] |
The discrimination against each analog is defined by Eq. (1) with reference to the corresponding canonical base. Didanosine (ddI) is metabolized to ddATP in the cell. We repeat values for dCTP to normalize the results from different labs.
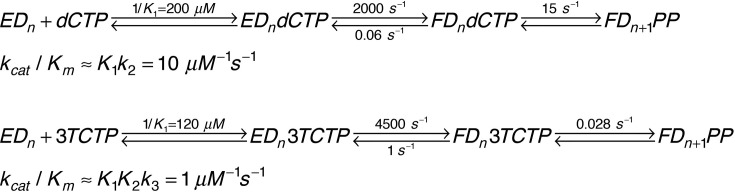
In earlier literature we developed a standard practice of referring to the kinetic parameters derived in single turnover kinetic measurements by the terms k pol (maximal rate) and K d (apparent K d for nucleotide concentration at the half-maximal rate) because it was believed that nucleotide binding occurred in a single step and single turnover experiments would provide a true K d for nucleotide binding, not a more complex K m term [12], [13], [14]. Moreover, we wanted to distinguish these results from the flawed steady state approaches used to examine DNA polymerase kinetics. However, we now know that nucleotide binding occurs in two steps: an initial weak nucleotide binding step followed by a fast conformational change step and then slower chemistry [10], [15], [16]. In either case, the single turnover kinetic analysis provides estimates of k cat and K m for incorporation representing parameters governing each cycle of nucleotide addition during processive synthesis [17]. However, in most cases the K m is not equal to the K d for nucleotide binding. This is an important distinction, especially when we attempt to understand selectivity against nucleotide analogs as explained below.
Three analogs (d4T, 3TC and FTC) exhibit K m values lower than for the corresponding canonical base pair. One would expect that an altered analog would bind less tightly than the normal nucleotide. This is most extreme in the case of 3TC (Fig. 1), which has a sulfur and the opposite stereochemistry in the ribose ring mimic. However, the K m for 3TC (27 nM) is much lower than that for dCTP (1.5 μM) for HIVRT. We can understand this result based on the two-step nucleotide binding reaction shown in Fig. 3 [10].
Fig. 3.
Pathway for nucleotide binding and incorporation. We show the pathway for nucleotide (N) binding to an enzyme-DNA complex with a primer n nucleotides in length (EDn). After binding, there is a change in enzyme structure from an open to a closed (FDn) state, leading to chemistry and release of pyrophosphate (PP).
When HIVRT copies DNA, pyrophosphate release (k 4) is faster than chemistry (k 3) and is largely irreversible in single turnover experiments. Therefore, the equation for the specificity constant for this model is reduced to that for a three-step reaction.
For a normal nucleotide, the fast conformational change step (k 2) is followed by the chemical reaction (k 3), but the rate of the reverse of the conformational change to allow release of bound nucleotide (k − 2) is slower than chemistry, so the two-step binding reaction does not come to equilibrium prior to chemistry [10]. Accordingly, when k − 2 < < k 3, the kinetic parameters for incorporation are approximated by:
In contrast, with a mismatched nucleotide or an analog, the rate of chemistry is much slower than the rate of nucleotide release [10]. When k − 2 > > k 3, the kinetic parameters for incorporation are approximated by:
Whereas the K m value for the correct nucleotide is a function of the relative rates of nucleotide binding and incorporation (K m = k cat/(k cat/K m)), with a nucleotide analog, the two-step nucleotide binding comes to equilibrium so K m = K d = 1/(K 1(1+K 2)). This makes nucleotide analogs more effective because the conformational change step leads to a lower K m to compensate for the slower chemistry step.
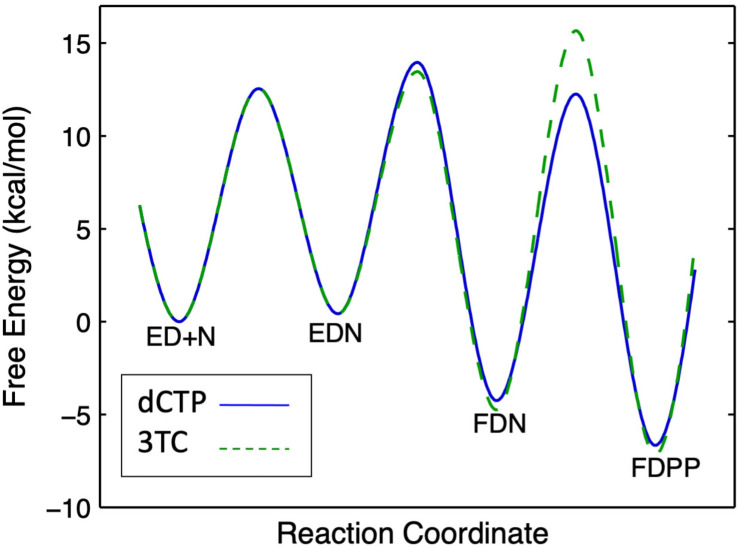
This can be illustrated by the free energy profile shown in Fig. 4 . Specificity is a function of all steps up to and including the highest barrier relative to the starting state (ED + N). For dCTP, the highest barrier is the conformational change step (EDN → FDN). In contrast, for 3TC-triphosphate (3TCTP) the highest barrier is the chemistry step (FDN → FDPP).
Fig. 4.
Free energy profiles. We compare dCTP (solid blue line) with 3TC (dashed green line). Note the change in specificity-determining step (highest barrier) in comparing dCTP with 3TC. This free energy profile was derived from the rate constants shown in Fig. 5.
As shown in Fig. 5 , the specificity constant for 3TCTP is only tenfold lower than that for dCTP even though its rate of incorporation is 650-fold slower. The slower rate of chemistry allows the conformational change to come to equilibrium, affording tighter nucleotide binding prior to the chemical step. This compensates for the slower chemistry in calculation of the specificity constant. Thus, 3TC is a surprisingly effective inhibitor even though it is a poor substrate. Although no other analog shows a K m that is lower than that for the cognate nucleotide, it is likely that the effect of the conformational change step to lower the K m will also apply to any analog with a slow rate of incorporation.
Fig. 5.
Rate constants comparing dCTP and 3TCTP. We show the rate constants governing incorporation of dCTP in comparison with 3TCTP along with the corresponding kcat/Km values. Note that even though 3TCTP is incorporated at a rate 650-fold slower than dCTP, the kcat/Km value is only tenfold lower [10].
The effect of the conformational change step also explains the small attenuation in k cat/K m values observed for AZT-resistant mutants of HIVRT [9]. As the chemistry step is made slower by the mutations, the conformational change reaches equilibrium leading to a lower K m which compensates for the slower rate of incorporation. Thus, the overall resistance to AZT is reduced to only a factor of two in comparing dTTP with AZT for wild-type versus mutant forms of HIVRT [10], [11].
2. Incorporation versus excision of nucleotide analogs
Chain terminators are effective in treating HIV infections because HIVRT lacks a proofreading function to remove the nucleotides after their incorporation. However, evolution of resistance to thymidine analogs is based in part on an ATP-dependent excision reaction in which the gamma phosphate of ATP reacts with the 3′-terminal nucleotide to form a dinucleotide tetraphosphate to remove the chain terminator [18]. After incorporation by an AZT-resistant HIVRT mutant, the half-life of AZT is approximately 2 min (A. Li and K. A. Johnson, unpublished).
ATP-dependent excision of the 3′-terminal nucleotide also occurs with the HCV RdRp, but the reaction catalyzed by the wild-type enzyme is 50-fold more efficient than that catalyzed by the thymidine analog resistant forms of HIVRT [1]. Accordingly, one might expect that no chain terminators could be used to treat HCV infections. As anticipated, Mericitabine (Fig. 2, a 2′F-2′-methyl cytosine analog) is efficiently removed with a half-life of less than a minute. Surprisingly, and for reasons we do not understand, the corresponding uracil analog (Sofosbuvir) is not removed by the ATP-dependent excision reaction after an hour of incubation. Further studies are needed to understand why Sofosbuvir is resistant to excision, while Mericitabine is not. ATP-dependent excision requires that the nucleotide remain in the polymerization site and to not translocate, so a more rapid or thermodynamically favorable translocation step after incorporation of Sofosbuvir could be responsible for the observed resistance to excision.
The role of the proofreading exonuclease in contributing to the effectiveness of nucleotide analogs used to treat COVID-19 remains to be explored, as described below. As a counterpoint to effectiveness, the toxicity of nucleoside analogs is a function of the selectivity of the proofreading exonuclease after their incorporation by the human mitochondrial DNA polymerase. For example, the toxicities of ddC and Fialuridine are attributable to their slow rates of excision after incorporation into mitochondrial DNA [7], [8].
3. Toxicity
HIV infections currently require continuous antiviral treatment for many years. Consequently, toxic side effects of the drugs and evolution of resistance become major limiting factors. For all but AZT, toxicity appears to be due to incorporation by the human mitochondrial DNA polymerase, so the major dose-limiting side effect is peripheral neuropathy, correlated with a reduction in the mitochondrial DNA content of peripheral nerves (reviewed in Ref. [8]). Interestingly, ddC is a better substrate for the mitochondrial polymerase than it is for HIVRT and is known to be so toxic that it is never used [8]. AZT was the first drug to be approved by the FDA and when it was the only drug available to treat AIDS, increased dosages attempting to overcome resistance led to lethal toxic side effects. AZT is a poor substrate for thymidine kinase, so the toxic side effects of AZT are due to competitive inhibition of thymidine kinase, leading to altered nucleotide pools [19], [20].
Toxicity of most deoxynucleotide analogs is due to their incorporation by the human mitochondrial DNA polymerase, Pol gamma [7], [8]. The biochemical basis for toxicity can be quantified and understood based on the ratio of values for discrimination for HIVRT versus Pol gamma. Because nucleotide analogs can be removed by the Pol gamma proofreading exonuclease, an additional correction factor needs be included according to the lifetime of a chain terminating nucleotide after incorporation by Pol gamma. Including both the relative values for discrimination and proofreading efficiency allows calculation of a therapeutic index, which is correlated with the toxicity of nucleoside analogs seen clinically [8], except for AZT as described above. Also note that cordycepin (3′ deoxyadenosine, not show) is sufficiently toxic that it is under consideration for anticancer therapy [21], but there may be multiple sites of action.
Although Remdesivir is effective in treating COVID-19 [22], [23], it also appears to be relatively toxic and is only administered for 5–10 days [24]. Longer treatments lead to generalized liver and kidney toxicity [25]. This generalized toxicity could be caused by multiple targets, which may not be surprising for an ATP analog. Nonetheless, because the time required for effective treatment of SARS CoV-2 infections is relatively short, toxicity is a less significant issue than it is for nucleoside analogs targeting HIVRT. We can expect less significant evolution of resistance because of the short treatment times. However, Remdesivir appears to be effective only if administered early during viral infection. Presumably, early treatment delays the rapid spread of the virus until the immune response takes effect. However, if given later Remdesivir appears to be less effective presumably because the virus has already caused significant damage to lungs and other tissues. Since 70% of those infected with SARS CoV-2 show little to no symptoms, decisions about when to administer Remdesivir must be based on projecting which patients are likely to develop more serious symptoms.
4. SARS CoV-2 RdRp kinetics and mechanism
When beginning work on any new enzyme it is always important to evaluate enzyme activity based on reasonable expectations for physiologically relevant rates. Previous preparations of the SARS CoV-2 RdRp showed poor activity that failed to meet this standard [4], [26]. For example, enzymes needing incubations of 15–30 min to extend a primer by 10 nt would require 1–2 months to replicate the 30 kb viral genome. Biochemical, structural, and kinetic studies are of questionable value when using an enzyme with such low activity. Care must be taken to develop and optimize expression and purification steps that yield active enzyme.
The SARS CoV-2 RdRp complex consists of nonstructural proteins (NSP) 12, 7 and 8. Because of insolubility of the SARS CoV RdRp NSP 12 when expressed in bacteria, most investigators rely on baculovirus-induced expression in insect cells [4], [26], [27], [28], [29], [30]. In our first attempts at bacterial expression of NSP12, we also noted that the protein was insoluble, confirming prior reports. However, we found that co-expression of NSP12, 7 and 8 with Tf, GroEL, and GroES chaperones yielded abundant soluble protein with high activity [3]. Furthermore, the enzyme was purified without the use of any tags, which can alter enzyme activity. Rather than rely upon a tagged enzyme, we found that optimization of expression represented an effective first purification step because the RdRp was expressed to where it represented large fraction of the soluble protein. Further purification through several column chromatography steps resulted in highly active enzyme, as show below. A parallel enzyme prepared with an 8xHis tag and an NSP7/8 fusion Shannon et al. [5] showed considerably lower activity. We have not repeated the attempted purification of the tagged protein, so our data do not definitively demonstrate a lower activity caused by the 8xHis tag (and/or the NSP7/8 fusion). Note that in our prior work with HIVRT, we also found it necessary to use untagged enzyme to achieve optimal activity [10]. Although there has been no systematic study of the effects on the His tag on polymerase activity, it is reasonable to suspect that a positively charged tag may have long range electrostatic interactions with negatively charged oligonucleotides that could interfere with activity.
Experiments to test for activity of the SARS CoV-2 RdRp complex were performed using the primer/template shown below, chosen for initial exploratory research because of some success with this sequence in earlier studies on the first SARS coronavirus [31], [32] and the duplex region was thought to be sufficiently long to fully occupy the polymerase RNA binding site.
[6-FAM]5′GUCAUUCUCCUAAGAAGCUA
3′CAGUAAGAGGAUUCUUCGAUAAUUUUAGUGUACCCCUAUC-5′
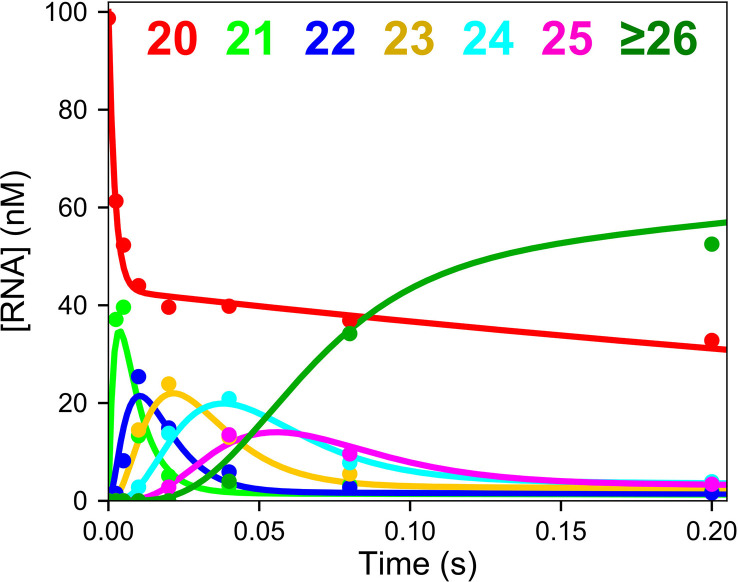
Fig. 6 shows the processive polymerization catalyzed by purified SARS CoV-2 RdRp complex [3]. The enzyme was preincubated with the primer/template, then the reaction was initiated by adding UTP, ATP and CTP. The RdRp catalyzed the incorporation of 12 nucleotides in less than 0.2 s. The observed average rate of polymerization was 300 nt/s, which is sufficient to replicate the 30 kb viral genome in less than 2 min. Thus, this preparation satisfies the expectations for a physiologically relevant rate of polymerization. In addition, global data fitting to account for the amplitudes as well as the rates of the reaction demonstrated that the enzyme was at least 60% active.
Fig. 6.
Kinetics of processive RNA polymerization. A solution of 2 μM NSP12/7/8 complex, 5 μM NSP8, 100 nM FAM-20/40 RNA, and 5 mM Mg2 + was mixed with 250 μM each of ATP, CTP, and UTP to start the reaction. Reaction products were resolved and quantified by capillary electrophoresis [33]. Smooth lines show the global fit of the data to define the rates of each sequential incorporation reaction [3].
Steady-state DNA polymerase measurements performed with DNA in excess over enzyme and following the fraction of DNA extended over the timescale of minutes provide estimates of k cat that are limited by the rate of DNA release [12], [14], [17]. Errors in k cat are carried over into correspondingly low K m estimates. Processivity can be defined as the ratio of the rate of polymerization divided by the DNA dissociation rate. Therefore, steady-state rate measurements typically underestimate k cat by a factor equal to the processivity. The higher the processivity of the enzyme, the greater the error introduced by steady-state methods applied to RNA and DNA polymerases. Particularly troublesome are “rate” measurements performed on the SARS CoV-2 RdRp based on the fraction of primer which was radio-labeled in the first step of the reaction and is subsequently extended [4]. The fraction of labeled primer that is extended is not a rate measurement, but rather is a measure of processivity, which is a function of the kinetic partitioning between extension versus dissociation of the RNA duplex from the polymerase. In the published studies, the inactive enzyme preparation and short oligonucleotide combined to yield low processivity [4]. At high nucleotide concentrations only 80% of the labeled primer was extended, which translates into a processivity of 4. In contrast, the processivity of a highly active SARS-CoV-2 RdRp was estimated to be 27,000 as described below [3].
To overcome the errors introduced by steady-state polymerase measurements, we perform experiments with an excess of enzyme over DNA and measure the rates of DNA extension on a ms timescale to capture the kinetics of polymerization occurring at the enzyme active site [3], [17]. Measurements of the nucleotide concentration dependence define k cat and k cat/K m for a single nucleotide incorporation providing parameters that govern each incorporation reaction during processive synthesis. The Michaelis constant is then defined by the ratio of k cat divided by k cat/K m. While k cat/K m provides a quantitative assessment of enzyme specificity, K m cannot be interpreted without additional information.
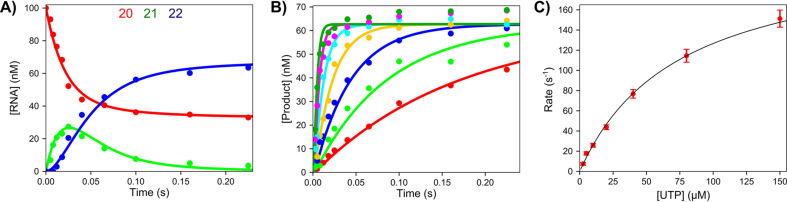
We applied these methods to investigate the kinetics of RNA polymerization by the SARS CoV-2 RdRp complex using the primer/template shown above [3]. Here we denote the active form of Remdesivir as Remdesivir triphosphate (RTP). Note that the template encodes for the incorporation of two UTPs followed by four ATPs (or RTPs). The kinetics of sequential nucleotide incorporations were resolved by fitting data based on computer simulation using numerical integration of the rate equations [17], [34], [35]. For example, the kinetics of incorporation of two UTP molecules are shown in Fig. 7 [3]. The time dependence of incorporation of the first and second UTP molecules are well resolved (Fig. 7A). Analysis of the concentration dependence of the only the first UTP incorporation, as illustrated in Fig. 7B and C, defines k cat and k cat/K m for the first UTP. More accurate global data fitting resolved the kinetics for each UTP.
Fig. 7.
Kinetics of sequential UTP incorporation. A mixture containing 2 μM NSP12/7/8 complex, 6 μM NSP8, 100 nM FAM-20/40 RNA, and 5 mM Mg2 + was mixed with varying concentrations of UTP (2.5–150 μM) to start the reaction. (A) Time dependence of conversion of 20 nt primer to 21 and then 22 nt by the incorporation of two UTP molecules at 20 μM UTP. (B) Concentration dependence of the first UTP incorporation. (C) UTP concentration dependence of the observed rate in (B) [3].
Using similar analysis, we examined the kinetics of incorporation of ATP (or RTP) after the incorporation of two UTPs. The fast initial incorporation of two UTPs led to a short, well-defined lag in the kinetics observed for the incorporation of ATP (or RTP). In fitting data by simulation, the known kinetics of UTP incorporation were included, so kinetic parameters for ATP (or RTP) were well resolved. In Table 2 , we summarized the kinetic parameters derived for the first UTP, ATP, or RTP [3].
Table 2.
Kinetic parameters for nucleotide incorporation.
| Nucleotide | kcat/Km (μM− 1 s− 1) | kcat (s− 1) | Km (μM) |
|---|---|---|---|
| UTP | 2.3 ± 0.2 | 308 ± 17 | 130 ± 9 |
| ATP | 0.74 ± 0.16 | 240 ± 30 | 320 ± 50 |
| RTP | 1.29 ± 0.06 | 68 ± 2 | 53 ± 2 |
Rate constants for incorporation are from T.L. Dangerfield, N.Z. Huang, K.A. Johnson, Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience (2020) 101849. Epub 2020/12/08. doi: 10.1016/j.isci.2020.101849. PubMed PMID: 33283177, PMCID: PMC7695572.
There are several important conclusions derived from these data. First, and most surprisingly, the specificity constant of RTP is nearly twice that for ATP, meaning that RTP is a better substrate for the RdRp than ATP. This accounts for how RTP can be an effective inhibitor in face of the 3 mM concentration of the competing ATP in vivo. For comparison, none of the eight nucleoside analogs approved for treatment of HIV infections show k cat/K m values greater than the cognate nucleotide.
Prior experiments designed to estimate competition between ATP and RTP revealed that RTP was favored over ATP by a factor of three [4] in spite of the meaningless rate measurements employed in that study. It is possible that at low nucleotide concentrations, an experiment looking for competition between the two substrates still provided what appears to be the correct answer despite the seriously flawed kinetic methods and largely inactive enzyme. These flaws invalidated the conclusions when they were published, but in retrospect, it is possible that competition between ATP and RTP still revealed the correct ratio of rate constants governing nucleotide extension even though the individual rate measurements of fraction extended per minute were uninterpretable and the enzyme was extremely inactive.
5. Structural basis for delayed inhibition by Remdesivir
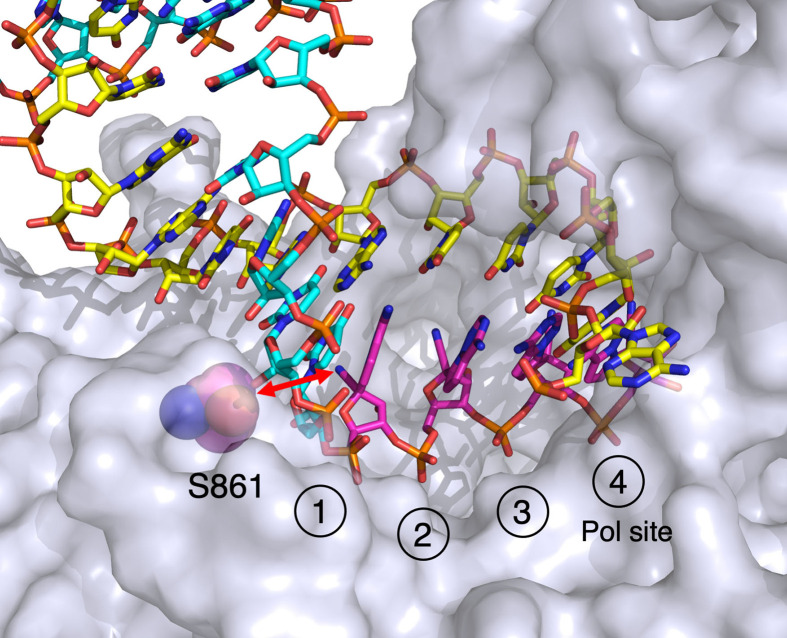
Our kinetic data showed that the SARS CoV-2 RdRp stalls after four molecules of remdesivir are incorporated, reaching completion in 20 s [3]. Based on this information we prepared samples for analysis of the Remdesivir-stalled structure by cryoEM. Enzyme (3.3 μM) preincubated with 200 nM RNA duplex was allowed to react with 8 μM UTP and 14 μM RTP for 20 s and then samples were applied to EM grids and rapidly frozen. Solution of the structure by cryoEM methods revealed the structural basis for the observed kinetic stalling of the polymerase (Fig. 8 ).
Fig. 8.
Structure of the stalled SARS CoV-2 RdRp complex with four molecules of Remdesivir. We show the structure of the stalled complex formed after incorporation of four molecules of RMP (magenta), labeled from 1 to 4 to give the order of incorporation. The RNA is in the pre-translocated state with the fourth RMP remaining in the active polymerase site (Pol site). The red arrow shows the likely steric clash between the cyano group of RMP with the beta carbon of S861 upon translocation. Drawn using Pymol from PDBID: 7L1F[2].
Fig. 8 gives a closeup of the active site of the enzyme showing four molecules of RMP (Remdesivir monophosphate) incorporated into the primer strand. The fourth and last RMP to be incorporated remains in the polymerase active site, in the pre-translocation state. This is unusual in that other structures of duplex RNA bound to the RdRp show the translocated state is preferred [26], [27], [29], [36] as is typical for DNA polymerases including HIVRT [37], [38]. Moreover, our kinetic analysis of DNA replication show that translocation is fast and thermodynamically favored [15]. In contrast, our structure shows that the polymerase stalled because the duplex RNA has failed to translocate to open the active site which is necessary to allow binding of the next nucleotide. Inspection of the structure reveals that translocation would be blocked by the steric interaction of the cyano group of RMP with the beta carbon of S861 (red arrow). Earlier studies had predicted this based on homology modeling and from structures of the enzyme with short oligonucleotides bound [28], [39]. Keeping with this conclusion, mutation S861G reduces the inhibitory effect of Remdesivir [40].
Note that our system showing the incorporation of four sequential molecules of RMP does not directly represent the likely stalled complex in vivo. Rather, we expect that a single RMP will be incorporated, followed by three normal nucleotides leading to the stalled complex. Further studies are needed to explore the kinetics of stalling after incorporation of a single RMP. Interestingly, by showing the incorporation of four RMPs, our structure affords the unique opportunity to show that the cyano group does not appear to contact the walls of the active site at any of the four sites. Steric blocking is predicted in attempts to translocate the RMP containing RNA beyond the observed state.
At longer times and with saturating concentrations of all four nucleotides, the Remdesivir-stalled complex is extended to full length product at a rate of 0.08 s− 1 [2], consistent with single molecule experiments showing that the Remdesivir-stalled RNA can be extended [41]. Thus, the steric block by RMP is not absolute. However, based on the analysis of the kinetics of proofreading in other systems [42], the significant slowing of the rate of extension would provide sufficient time for the exonuclease to efficiently remove the 3′-terminal nucleotide. It is likely that the polymerase idles in a futile cycle by incorporating then removing the 3′-terminal nucleotide in the stalled complex. Therefore, we expect that the exonuclease will make a major contribution toward effectiveness of nucleotide analogs, pointing to the importance of subsequent analysis of the kinetics of proofreading.
6. Role of the proofreading exonuclease in effectiveness of nucleotide analogs
SARS CoV-2 encodes a 3′–5′ proofreading exonuclease complex, consisting of catalytic subunit NSP14 and accessory subunit NSP10. Kinetic analysis of proofreading has shown that exonuclease specificity is determined by kinetic partitioning of primer/template in the polymerase site [42]. If the next round of polymerization is fast, polymerization continues. However, if the polymerase stalls, as it does after incorporating a mismatch, this gives time for the primer strand to flip from the polymerase site to the exonuclease site where the 3′-terminal base is rapidly hydrolyzed. The primer strand then returns to the polymerase active site where processive synthesis can continue. Thus, stalling of the polymerase provides the signal for the exonuclease to remove a nucleotide [14], [42]. Stalling caused by a chain terminator gives time for the primer strand to flip into the exonuclease site to effect removal of the nucleotide analog. Although the exonuclease reaction can be inhibited by the binding of the next correct nucleotide which stabilizes the binding of the primer at the polymerase site, this appears to vary depending on the nucleotide analog and the sequence context.
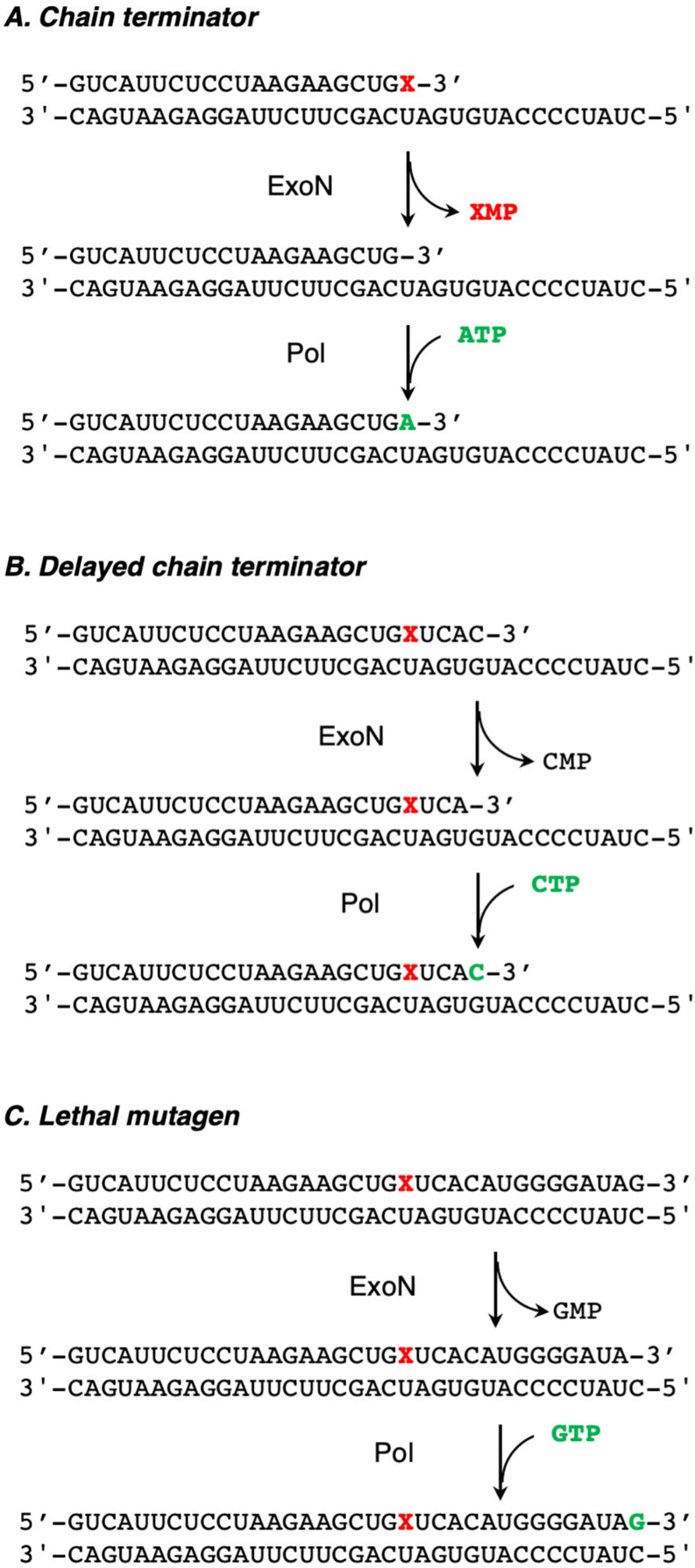
Various possible fates of a nucleotide analog are outlined in Fig. 9 . A simple chain terminator will be removed by the exonuclease. However, a delayed chain terminator will escape the exonuclease if it is extended more rapidly than it is excised. Remdesivir may not be intrinsically resistant to excision if it is rapidly extended by subsequent incorporation of a correct nucleotide. Once the polymerase stalls three residues after of the site of RMP incorporation, the 3′-terminal base would be removed by the exonuclease but then immediately replaced (Fig. 9B). As shown in Fig. 9C, if a nucleotide analog is readily extended and does not lead to significant stalling of the polymerase, it will fully escape exonuclease excision and may then have antiviral activity by lethal mutagenesis if it codes for mismatches in the next round of replication. The difference between the three classes of inhibitors could be quantified by measurements of the relative rates of extension versus excision using an RdRp reconstituted with the proofreading exonuclease (NSP10/14).
Fig. 9.
Role of the proofreading exonuclease. We illustrate three scenarios for the fate of a nucleotide analog after incorporation. (A) An obligate chain terminator is excised by the exonuclease, then replaced by a normal nucleotide. (B) A delayed chain terminator can be protected from the proofreading exonuclease by being buried by several normal nucleotides. (C) A lethal mutagen is stably incorporated and evades the proofreading function by fast extension, but in the next round of polymerization, copying the analog introduces mutations.
In the absence of the proofreading exonuclease complex, the delayed inhibition of replication by RTP can be overcome by the addition of higher concentrations of NTPs. Accordingly, it has been concluded that RTP may become stably incorporated into the viral genome and could cause some mutagenesis in the next round of RNA replication. However, studies to understand the biochemical fate of RTP after incorporation must be performed in the presence of the proofreading exonuclease. For example, it is known that mismatches that would be effectively removed by the proofreading exonuclease complex can be extended at higher concentrations of the next correct nucleotides, although at a slower rate than observed in the absence of the mismatch. Therefore, it is important to define the kinetics of extension versus excision after Remdesivir is incorporated in the presence of the proofreading exonuclease.
Remdesivir is fivefold more effective at inhibiting propagation of exonuclease deficient SARS coronavirus (compared to wild-type) in cell culture [43]. This observation suggests that after Remdesivir-induced stalling of replication, either the exonuclease can remove a significant fraction of the incorporated Remdesivir or that Remdesivir gains additional toxicity by becoming stably incorporated in the absence of the exonuclease. Each mechanism of inhibition categorized in Fig. 9 represents a theoretical extreme but, the potency of any nucleotide analog will depend on the kinetics on incorporation versus the relative rates of extension (when possible) and excision. For example, ddC is no longer used clinically to treat HIV because of toxic side effects. We noted that ddC is a better substrate for incorporation by the human mitochondrial DNA polymerase than it is for HIVRT and once it is incorporated into mitochondrial DNA it is not removed by the proofreading exonuclease [7]. This surprising result serves as one example of how important it will be to determine the kinetics of excision for any nucleotide analog to assess its antiviral potential and its mode of action against the SARS CoV-2 RdRp.
7. Summary and future directions
The wealth of information on the kinetic and structural basis for the effectiveness versus toxicity of nucleotide analogs provides guidelines for the development of new drugs to treat viral infections. The polymerase responsible for replication of the viral genome is the primary target for the development of new drugs, in part, because inhibitors directed against the polymerase active site are generally less susceptible to evolution of resistance. Nucleotide/nucleoside analogs and nonnucleoside inhibitors are the cornerstone of combination therapy for HIV and HCV infections [44], [45]. Similarly, the coronavirus RNA-dependent RNA polymerase (RdRp) is a prime target for drugs to combat the current COVID-19 pandemic [46]. Moreover, drugs developed now for COVID-19 are likely to be effective against future coronavirus outbreaks. The SARS-CoV-2 RdRp shows 96% identity and 98% similarity to the RdRp of the original SARS coronavirus, with mutations mostly on the surface of the proteins and none near the active sites [39]. However, development of new analogs still represents a significant challenge, especially given the potential for the proofreading exonuclease to remove conventional chain terminators. Although more data are required and there may be some exceptions among RdRps [47], it appears that RdRp enzymes from HCV and SARS CoV-2 prefer analogs containing a normal 3’OH. Clever chemical designs of Sofosbuvir and Remdesivir overcame this limitation using different means that were only fully understood after careful kinetic analysis. With Sofosbuvir, modifications at the 2′ position sufficiently alter the properties of the ribose ring to hinder the reaction of the 3’OH for subsequent polymerization, thus leading to chain termination. Remdesivir allows—and in fact may require—fast extension by the next correct base pair to escape excision by the proofreading exonuclease. The structure of Remdesivir is indeed unusual in that the addition of a cyano group at the 1′ position leads to delayed inhibition of translocation, effectively protecting RMP from excision by the proofreading exonuclease. However, Remdesivir is rather toxic which limits its more widespread use and restricts the allowable time for treatment. A new analog, Molnupiravir is thought to act by allowing fast extension and stable incorporation, but the modification of the uracil leads to lethal mutagenesis in the next round of incorporation, as suggested for Favipiravir [5]. Accurate kinetic studies are needed to establish the mechanistic basis for efficient inhibition of the SARS CoV-2. Moreover, the potential for host toxicity of a lethal mutagen is significant as learned from studies on the Fialuridine.
The kinetic basis for selectivity of the proofreading exonuclease is the largest unknown in understanding the effectiveness of nucleoside analogs for treating COVID-19. More studies are needed to establish the kinetic and mechanistic basis for selective removal or retention of nucleoside analogs by the exonuclease. The first necessary step is the assembly of a fully active RdRp complex with the NSP10/14 exonuclease subunits that meets standards for expected activity. The fact that no one has yet to publish a cryoEM structure of the RdRp in complex with the exonuclease suggests that reconstitution of an active polymerase/exonuclease complex will not be easy.
References
- 1.Villalba B., Li J., Johnson K.A. Resistance to excision determines efficiency of hepatitis C virus RNA-dependent RNA polymerase inhibition by nucleotide analogs. J. Biol. Chem. 2020;295:10112–10124. doi: 10.1074/jbc.RA120.013422. 32457046 Epub 2020/05/28. PMC7383390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552 e1544. doi: 10.1016/j.molcel.2021.01.035. 33631104 Epub 2021/02/26. PMC7843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23:101849. doi: 10.1016/j.isci.2020.101849. 33283177 Epub 2020/12/08. PMC7695572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. 32284326 Epub 2020/04/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shannon A., Selisko B., Le N.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. 32943628 Epub 2020/09/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colacino J.M. Mechanisms for the anti-hepatitis B virus activity and mitochondrial toxicity of fialuridine (FIAU) Antiviral Res. 1996;29:125–139. doi: 10.1016/0166-3542(95)00836-5. 79 [DOI] [PubMed] [Google Scholar]
- 7.Johnson A.A., Ray A.S., Hanes J., Suo Z.C., Colacino J.M., Anderson K.S., Johnson K.A. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:40847–40857. doi: 10.1074/jbc.M106743200. 9 [DOI] [PubMed] [Google Scholar]
- 8.Lee H., Hanes J., Johnson K.A. Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochemistry. 2003;42:14711–14719. doi: 10.1021/bi035596s. 14674745 Epub 2003/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellinger M.W., Johnson K.A. Role of induced fit in limiting discrimination against AZT by HIV reverse transcriptase. Biochemistry. 2011;50:5008–5015. doi: 10.1021/bi200204m. 21548586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Vaccaro J.A., Parnell K.M., Terezakis S.A., Anderson K.S. Mechanism of inhibition of the human immunodeficiency virus type 1 reverse transcriptase by d4TTP: an equivalent incorporation efficiency relative to the natural substrate dTTP. Antimicrob. Agents Chemother. 2000;44:217–221. doi: 10.1128/aac.44.1.217-221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellinger M.W., Johnson K.A. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7734–7739. doi: 10.1073/pnas.0913946107. 20385846 Epub 2010/04/14. doi: 0913946107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Feng J.Y., Shi J., Schinazi R.F., Anderson K.S. Mechanistic studies show that (-)-FTC-TP is a better inhibitor of HIV-1 reverse transcriptase than 3TC-TP. FASEB J. 1999;13:1511–1517. doi: 10.1096/fasebj.13.12.1511. [DOI] [PubMed] [Google Scholar]
- 11.Ray A.S., Murakami E., Basavapathruni A., Vaccaro J.A., Ulrich D., Chu C.K., Schinazi R.F., Anderson K.S. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry. 2003;42:8831–8841. doi: 10.1021/bi034435l. 1929 [DOI] [PubMed] [Google Scholar]
- 11a.Suo Z., Johnson K.A. Selective inhibition of HIV-1 reverse transcriptase by an antiviral inhibitor, (R)-9-(2-Phosphonylmethoxypropyl)adenine. J. Biol. Chem. 1998;273:27250–27258. doi: 10.1074/jbc.273.42.27250. [DOI] [PubMed] [Google Scholar]
- 12.Johnson K.A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. 47 [DOI] [PubMed] [Google Scholar]
- 13.Kati W.M., Johnson K.A., Jerva L.F., Anderson K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. 91 [PubMed] [Google Scholar]
- 14.Patel S.S., Wong I., Johnson K.A. Pre-steady-state kinetic-analysis of processive DNA-replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. 42 [DOI] [PubMed] [Google Scholar]
- 15.Dangerfield T.L., Johnson K.A. Conformational dynamics during high fidelity DNA replication and translocation defined using a DNA polymerase with a fluorescent artificial amino acid. J. Biol. Chem. 2021;296:100143. doi: 10.1074/jbc.RA120.016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai Y.-C., Johnson K.A. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson K.A. KinTek Corporation; Austin: 2019. Kinetic Analysis for the New Enzymology: Using Computer Simulation to Learn Kinetics and Solve Mechanisms. [Google Scholar]
- 18.Meyer P.R., Matsuura S.E., Mian A.M., So A.G., Scott W.A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. 2143 [DOI] [PubMed] [Google Scholar]
- 19.Lavie A., Schlichting I., Vetter I.R., Konrad M., Reinstein J., Goody R.S. The bottleneck in AZT activation. Nat. Med. 1997;3:922–924. doi: 10.1038/nm0897-922. 9256287 Epub 1997/08/01. [DOI] [PubMed] [Google Scholar]
- 20.Lavie A., Vetter I.R., Konrad M., Goody R.S., Reinstein J., Schlichting I. Structure of thymidylate kinase reveals the cause behind the limiting step in AZT activation. Nat. Struct. Biol. 1997;4:601–604. doi: 10.1038/nsb0897-601. 9253404 [DOI] [PubMed] [Google Scholar]
- 21.Khan M.A., Tania M. Cordycepin in anticancer research: molecular mechanism of therapeutic effects. Curr. Med. Chem. 2020;27:983–996. doi: 10.2174/0929867325666181001105749. 30277143 Epub 2018/10/03. [DOI] [PubMed] [Google Scholar]
- 22.Lamb Y.N. Remdesivir: first approval. Drugs. 2020;80:1355–1363. doi: 10.1007/s40265-020-01378-w. 32870481 Epub 2020/09/02. PMC7459246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malin J.J., Suarez I., Priesner V., Fatkenheuer G., Rybniker J. Remdesivir against COVID-19 and other viral diseases. Clin. Microbiol. Rev. 2020;34(1) doi: 10.1128/CMR.00162-20. 33055231 Epub 2020/10/16. PMC7566896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., et al. Remdesivir for 5 or 10 days in patients with severe Covid-19. N. Engl. J. Med. 2020;383:1827–1837. doi: 10.1056/NEJMoa2015301. 32459919 Epub 2020/05/28. PMC7377062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antinori S., Cossu M.V., Ridolfo A.L., Rech R., Bonazzetti C., Pagani G., Gubertini G., Coen M., Magni C., Castelli A., et al. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: clinical outcome and differences in post-treatment hospitalisation status. Pharmacol. Res. 2020;158:104899. doi: 10.1016/j.phrs.2020.104899. 32407959 Epub 2020/05/15. PMC7212963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. 32358203 Epub 2020/05/03. PMC7199908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. 32438371 Epub 2020/05/22. [DOI] [PubMed] [Google Scholar]
- 28.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428 e413. doi: 10.1016/j.cell.2020.05.034. 32526208 Epub 2020/06/12. PMC7242921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2020;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. 33232691 Epub 2020/11/25. PMC7666536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. 29279395 Epub 2017/12/28. PMC5777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. 2015;112:9436. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dangerfield T.L., Huang N.Z., Johnson K.A. High throughput quantification of short nucleic acid samples by capillary electrophoresis with automated data processing. Anal. Biochem. 2021;629:114239. doi: 10.1016/j.ab.2021.114239. 33979658 Epub 2021/05/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson K.A., Simpson Z.B., Blom T. FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. 19168024 Epub 2009/01/27. doi: S0003-2697(08)00848-8 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Johnson K.A., Simpson Z.B., Blom T. Global Kinetic Explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. 19154726 Epub 2009/01/22. doi: S0003-2697(08)00849-X [pii] [DOI] [PubMed] [Google Scholar]
- 36.Zhang W., Stephen P., Theriault J.F., Wang R., Lin S.X. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J. Phys. Chem. Lett. 2020;11:4430–4435. doi: 10.1021/acs.jpclett.0c00571. 32392072 Epub 2020/05/12. [DOI] [PubMed] [Google Scholar]
- 37.Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391(6664):251–258. doi: 10.1038/34593. 18 [DOI] [PubMed] [Google Scholar]
- 38.Huang H.F., Chopra R., Verdine G.L., Harrison S.C. Structure of a covalently trapped catalytic complex of HIV-I reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. 17 [DOI] [PubMed] [Google Scholar]
- 39.Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. 32283108 Epub 2020/04/14. PMC7151495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. 32967965 Epub 2020/09/25. PMC7681019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seifert M., Bera S.C., van Nies P., Kirchdoefer R.N., Shannon A., Le T.-T.-N., Grove T.L., Papini F.S., Arnold J.J., Almo S.C., Canard B., Depken M., Cameron C.E., Dulin D. bioRxiv; 2020. Signatures and Mechanisms of Efficacious Therapeutic Ribonucleotides Against SARSCoV-2 Revealed by Analysis of Its Replicase Using Magnetic Tweezers. [DOI] [Google Scholar]
- 42.Donlin M.J., Patel S.S., Johnson K.A. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. 53 [DOI] [PubMed] [Google Scholar]
- 43.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. 29511076 Epub 2018/03/08. PMC5844999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deval J., Selmi B., Boretto J., Egloff M.P., Guerreiro C., Sarfati S., Canard B. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using alpha-boranophosphate nucleotide analogues. J. Biol. Chem. 2002;277:42097–42104. doi: 10.1074/jbc.M206725200. 12194983 Epub 2002/08/27. M206725200 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Hurwitz S.J., Schinazi R.F. Practical considerations for developing nucleoside reverse transcriptase inhibitors. Drug Discov. Today Technol. 2012;9:e183–e193. doi: 10.1016/j.ddtec.2012.09.003. 23554824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buonaguro L., Tagliamonte M., Tornesello M.L., Buonaguro F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Med. 2020;18:185. doi: 10.1186/s12967-020-02355-3. 32370758 Epub 2020/05/07. PMC7200052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gizzi A.S., Grove T.L., Arnold J.J., Jose J., Jangra R.K., Garforth S.J., Du Q., Cahill S.M., Dulyaninova N.G., Love J.D., et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature. 2018;558:610–614. doi: 10.1038/s41586-018-0238-4. 29925952 Epub 2018/06/22. PMC6026066. [DOI] [PMC free article] [PubMed] [Google Scholar]