Abstract
The time of dim light melatonin onset (DLMO) is the gold standard for circadian phase assessment in humans, but collection of samples for DLMO is time and resource intensive. Numerous studies have attempted to estimate circadian phase from actigraphy data, but most of these studies have involved individuals on controlled and stable sleep-wake schedules, with mean errors reported between 0.5 and 1 hours. We found that such algorithms are less successful in estimating DLMO in a population of college students with more irregular schedules: mean errors in estimating the time of DLMO are approximately 1.5–1.6 hours. We reframed the problem as a classification problem and estimated whether an individual’s current phase was before or after DLMO. Using a neural network, we found high classification accuracy of about 90%, which decreased the mean error in DLMO estimation - identifying the time at which the switch in classification occurs - to approximately 1.3 hours. To test whether this classification approach was valid when activity and circadian rhythms are decoupled, we applied the same neural network to data from inpatient forced desynchrony studies in which participants are scheduled to sleep and wake at all circadian phases (rather than their habitual schedules). In participants on forced desynchrony protocols, overall classification accuracy dropped to 55–65% with a range of 20–80% for a given day; this accuracy was highly dependent upon the phase angle (i.e., time) between DLMO and sleep onset, with highest accuracy at phase angles associated with nighttime sleep. Circadian patterns in activity, therefore, should be included when developing and testing actigraphy-based approaches to circadian phase estimation. Our novel algorithm may be a promising approach for estimating the onset of melatonin in some conditions and could be generalized to other hormones.
Keywords: melatonin, circadian rhythm, actigraphy, machine learning, classification, shiftwork, biological clocks
Introduction
Circadian rhythms are oscillations in behavior and physiology with a period of approximately 24 h. At the cellular level, these rhythms are driven by transcriptional-translational feedback loops1, resulting in oscillations both in transcription of approximately 50% of genes in at least one organ2 and in expression of many drug targets. This motivates the growing interest in chronotherapeutic approaches, in which drug intake is timed to maximize efficacy and minimize side effects3.
Misalignment between internal circadian rhythms and the environment, which occurs with jetlag, shiftwork, and circadian disorders, has been linked to adverse health outcomes, including cardiovascular disease, metabolic disease, and cancer4. Methods are needed, therefore, to shift the phase of the circadian clock to align with the environment. Both the magnitude and direction of the phase shift caused by both photic and non-photic (including pharmaceutical) interventions depends on the phase (i.e., time) at which the intervention is delivered5–8. Thus, we must be able to precisely assess an individual’s current circadian phase9 before applying interventions.
The gold standard for circadian phase assessment in humans is dim light melatonin onset (DLMO). This phase assessment technique requires the collection of multiple hours of blood or saliva samples under low light conditions, usually in an inpatient setting, for later melatonin assay. This method is time-consuming and resource-intensive. Furthermore, it does not allow for circadian phase to be assessed continuously or in real-time, limiting its usefulness for timing interventions. These limitations motivate the need for an alternative method of circadian phase estimation. Recent research has focused on two main approaches, using a genetic assay or less invasive actigraphy data10.
Several methods have been proposed to estimate circadian phase using panels of circadian-cycling genes and proteins (reviewed in Crnko et al.11). Machine learning algorithms12 have reduced the number of genes used for these panels from over 10013 to about a dozen genes in monocytes extracted from whole blood14. Most of these approaches have mean errors between 1 and 2 hours (Table S1). These methods still have the limitations that the collection of samples is invasive and results are not available in real-time.
Actigraphy data can be collected non-invasively in real-time. These data can include activity counts, light levels, skin temperature, and/or heart rate. There are three main model categories for estimating circadian phase using actigraphy data: limit cycle oscillator models, regression models, and neural network models15; their errors are between 0.4 and 1.1 h (Table S2). Limit cycle models use actigraphy data as input to differential equations-based models, most commonly the Kronauer model16 or its amended version which includes a non-photic component17, to predict how the series of light inputs shifts oscillator phase18. Some regression models have used derived markers of actigraphy rhythms, such as the time of peak activity or the time when wrist temperature begins to increase, to estimate DLMO19. Another approach has used an adaptive notch filter (ANF) to derive harmonic estimates to estimate phase shifts20. The limit cycle, linear regression, and ANF models all require several days of data for (i) the limit cycle result to be insensitive to initial conditions or (ii) to define average activity markers, limiting their utility for a phase assessment that can be performed quickly (with the goal of real-time assessment). Other regression models have used light and skin temperature recordings from multiple skin electrodes over a 24 h period to estimate circadian phase with a mean error of 0.7 h21. Recent neural networks have had success in prediction when the model’s exact functional form is unknown22. Using the same input data but increasing the flexibility of the model by training a neural network, this mean error was decreased to 0.4 h23. A similar approach in a different population, but with seven days of actigraphy recordings, found a mean error of 0.6 h in a population with habitual day active schedules24, with similar accuracies when using just a single wrist temperature sensor. These studies are limited due to small sample sizes (n<25) and the fact that participants follow regular, controlled schedules. Moreover, the same neural network model produced mean errors of over 2 h for participants on night shift schedules25, suggesting that the model does not generalize well to populations with less regular schedules or when the individual is awake at circadian phases usually associated with sleep. More recent work has again explored using differential equations-based models with some success in shift workers, although absolute mean errors remained higher26–28 than those reported in these models for participants with aligned schedules.
We therefore tested the generalizability of neural network-based models to a population of college students who had a high range in regularity in their sleep schedules29. We also developed a novel classification-based approach, which we tested in the college student population and in participants on inpatient forced desynchrony protocols in which sleep and wake occurred at all circadian phases to determine the effect of circadian misalignment on DLMO estimation.
Materials and Methods
The studies were approved by the Partners Human Research Committee and were performed according to the principles outlined by the Helsinki Declaration. Appropriate informed consent was obtained. Details of each study are in the original publications30–35.
Actigraphy Data from a College Student Population
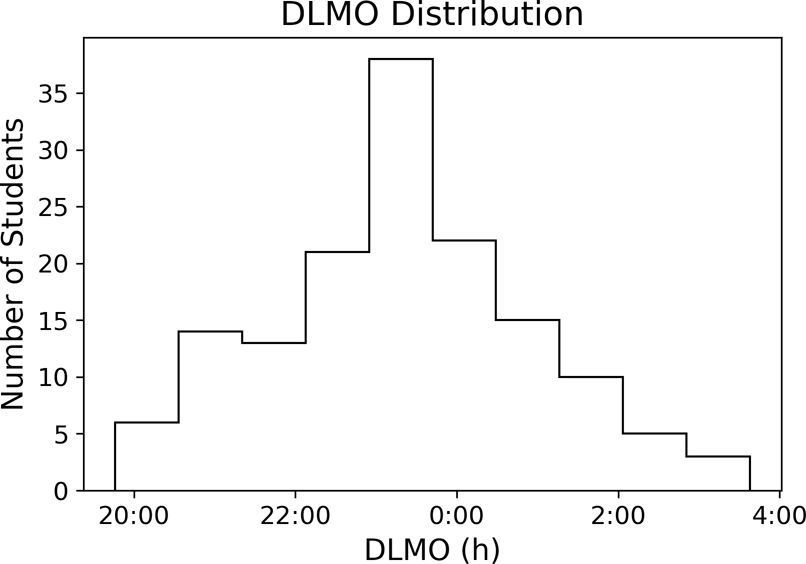
Data are from 174 participants from cohorts of ~30 undergraduate students each across six different semesters for ~30 days while students lived at school30,36. Participants completed demographic, psychological, and sleep habit questionnaires at the beginning of the study (“static data”). For all days, participants wore an actiwatch (Motionlogger, AMI, Ardsley, NY) that collected minute-by-minute light levels and activity counts (zero crossing mode) in addition to a Q-sensor (Affectiva, Boston, MA) that collected skin temperature readings at 8 Hz. At one time during these ~30 days, participants were admitted to an inpatient facility for DLMO assessment. This data set was chosen because there was a wide distribution of DLMO values (Figure 1) with mean DLMO value of 23.3±1.58 h and range of ~8 h, broader than the ~5 h range which is typical of the adult population37.
Figure 1. DLMO in the Study of College Students.
Histogram showing the distribution of the timing of DLMO in this population.
Actigraphy Data from Forced Desynchrony Protocols
Data are from four inpatient forced desynchrony (FD) protocols. During an FD, participants are placed on a non-24-h schedule that is outside of the range of entrainment of the circadian clock, allowing for the decoupling of rest-activity patterns and circadian rhythms. The four FD protocols are summarized in Table S3. Minute-by-minute light and activity data and hourly blood melatonin data from the FD portion of the study were used. For each day, we defined the phase angle of entrainment as the difference between DLMO and scheduled sleep start.
DLMO Determination
DLMO was calculated by linearly interpolating the time at which melatonin levels crossed the 5 pg/mL threshold in saliva or 10 pg/mL threshold in blood 29.
Data Cleaning and Standardization
We wanted our method to be capable of estimating DLMO using only a limited timeframe of data, so we restricted our input data to the 24 hours immediately before the day of DLMO assessment. For training and testing our model, we considered only participants who had complete light and activity recordings for this day. For skin temperature data, we computed the average value for each minute and linearly interpolated missing values. Data were removed from further analysis if more than a minute of skin temperature recordings were missing from the end of the 24 h period or if total activity, light, or temperature recordings for the 24 h period were less than 3 standard deviations below the population mean, suggesting device malfunction. Data from 27 participants were removed.
To reduce the number of model inputs, we computed the average value of light, activity, and skin temperature within a 30-minute bin. We then standardized these binned inputs by participant for each data type (see Supplementary Information). We also compared performance using the median instead of the average.
The data were randomly split into a training (n=118) and independent test set (n=29), where we performed 10-fold cross-validation within the training set (which was randomly split into 10 test sets for cross-validation) to fit the hyperparameters of the model.
Continuous Neural Networks
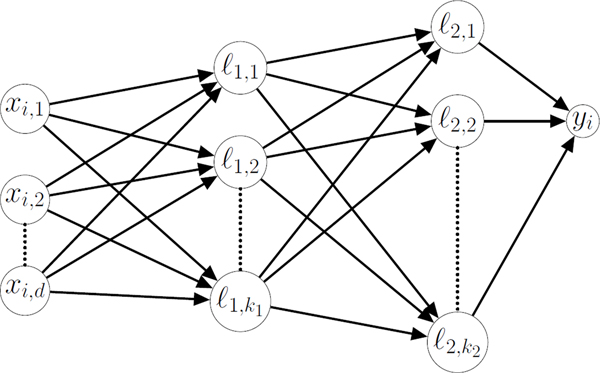
We first trained (using the college student data) a neural network to continuously estimate time of DLMO. We implemented a basic feedforward hidden layer neural network architecture with one and two hidden layers with hyperbolic tangent activation functions in the keras package in Python (Figure 2). To increase robustness, we also introduced dropout into these models, where models are randomly thinned during training to reduce overfitting to particular data features38. We varied the hyperparameters of the number of nodes in each layer (between 10 and 100 in multiples of 10 for the single hidden layer model, and all combinations of between 10 and 50 in multiples of 10 for the two hidden layer model) and dropout rate (between 0.1 and 0.5 in multiples of 0.1). For each of these models we trained the weights of the model using Adam optimization (learning rate = 0.005 for 500 iterations) to minimize the sum of the squared error between our estimated DLMO and the experimentally measured DLMO value on each of the cross-validation splits. We then chose the hyperparameter with the lowest mean cost function for our final model architecture, retrained this model on the full training set, and used the weights from training to evaluate the model on the test set.
Figure 2. Example Neural Network.
A schematic of a neural network, where x is a vector of input data for d input features where xi, v is the vth feature for the ith participant and yi is the network output for the ith participant. In the case of the continuous neural network, yi is a continuous variable representing the network’s prediction of the time of DLMO for the ith participant. In the case of the classification neural network, yi falls between 0 and 1 and if yi< 0.5, the timepoint is classified as falling before DLMO. Nodes lm,n represent hidden nodes in the network, where m is the layer of the node, and n is the index of the node within each layer. The value of each node is determined by for bn a fit constant. The number of nodes in each layer, km for bn a fit constant. The number of nodes in each layer, km, is a hyperparameter which was selected through cross validation.
We also tested the performance of long short-term memory (LSTM) architectures fitting a single layer of LSTM units (where we fit this hyperparameter between 10 and 50 in multiples of 10), which include recurrent loops with memory lags. LSTMs have had success when analyzing time series data39 such as our 24 h actigraphy recordings, and recurrent neural networks, such as LSTMs, have been previously applied to the college student dataset40
Classification Approach to DLMO Estimation
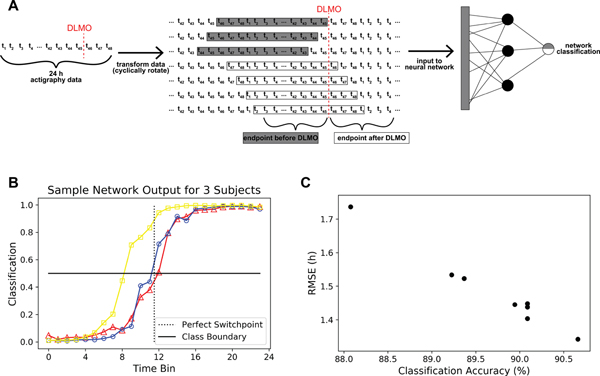
We also developed a novel approach to estimate the timing of DLMO, based on classification. Using this approach, we could effectively increase our sample size using the existing data. While our population was larger than those used to fit previous phase estimation models, it was still small for machine learning applications; the number of samples in our model was smaller than the number of parameters in the model because of the parameters for the nodes in the hidden layers. Rather than estimating the time of DLMO directly, we predicted whether the last timepoint in a 24 h series of data falls before or after DLMO; we shifted a 24 h interval of data (Figure 4A), where for each participant we have 6 h (12 datapoints in 30-minute bins) of samples where the last timepoint occurs before DLMO and 6 h of samples where the last timepoint occurs after DLMO. This approach created a balanced dataset and allowed us to increase the number of samples in our dataset 24-fold.
Figure 4. Outputs of the Classification Algorithm and Relationship to DLMO Estimation Accuracy A.
A schematic for the process of using actigraphy data in our classification algorithm. We use 24 h of actigraphy data, where data are binned into 30-minute time-bins for a total of 48 datapoints for each datatype, so ti represents the ith 30-minute bin of activity, light, and skin temperature data. We create a classification problem, where we predict whether the endpoint of an interval of 24 h of data falls before or after DLMO using a neural network. B. Classification neural network outputs for three test participants. For each participant, each line shows the 24 predictions of whether the time series falls before or after DLMO by rotating our 24 h series of data. Outputs less than 0.5 are classified as occurring before DLMO and outputs greater than 0.5 are classified as occurring after DLMO. A perfectly accurate classifier would have the switch in classification between time-bins 11 and 12. C. Comparison of classification accuracy versus the resulting root mean squared error in DLMO estimation based on where the switch in classification occurs for the different models in Table 3.
We compared the same neural network structures, but with rectified linear unit (ReLU, outputs the greater of the input and 0) activation between layers. To make the networks predict a classification we introduced a sigmoidal nonlinearity in the output, and instead of using the squared error loss as our cost function, we used cross entropy loss to train the model with stochastic gradient descent (learning rate=0.001 for 100 iterations).
Our trained model outputs class labels for each 24 h time series (rather than a continuous-time estimation). From these class labels, we could then estimate a value for DLMO by taking the midpoint of the times that the class labels switch from before to after DLMO.
Circadian Rhythmicity of Activity Levels
The circadian phase of each actigraphy data point was calculated using circadian period (i.e., slope) and starting phase (i.e., intercept) from the linear regression of DLMO collected during the FD portion of each experiment. Data were then assigned to 3-h circadian time bins (one-eighth of the participant’s circadian period) and 3-h wake duration time bins. Within each time bin, we used Zero Inflated Poisson (ZIP)-based statistics on activity levels for each participant. We then averaged bins across participants within each of the FD protocols. We tested for differences by circadian phase and time awake within each of the 4 FD protocols using repeated-measures ANOVA using SAS 9.4.
Results
Previous Methods Do Not Generalize to the College Student Population
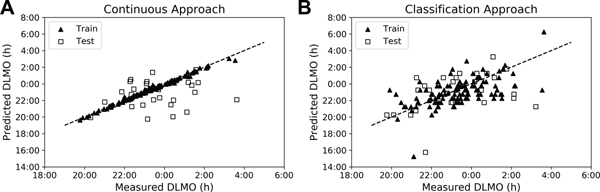
We first tested whether machine learning, actigraphy-based models like those used in more tightly-controlled, small-sample-size populations23,24 would extend to college students living at their school on self-selected, sleep-wake schedules. We found root mean squared errors (RMSE) of approximately 1.6 h for most models, with the best performing models (LSTMs) having errors of about 1.5 h for the average features (Table 1). When using the median (instead of average) features, (i) there was not much improvement except in the case of the full data set with all the demographic and actigraphy features, where errors decreased to about 1.4 h, but (ii) there were more consistent results across the models because the model predicted the mean DLMO of the dataset (Table S6, Figure S2). The Pearson’s R for the relationship between experimentally measured DLMO and the continuous model estimated phase in the training set was 0.99 (p<<<0.01), it was 0.17 (p=0.38) for the test set (Figure 3A). These errors are higher than those reported in the populations with more regular schedules and perform only minimally better than simply predicting the population mean, suggesting that these models do not generalize well to populations with irregular schedules.
Table 1. Performance of Continuous Neural Network Models for DLMO Estimation in College Students.
The root-mean-squared-error (RMSE) in cross-validation (mean ± standard deviation) and on the independent test set between the DLMO estimated by our models and the experimentally measured DLMO on different subsets of the data. Static data were collected only once per study; they include demographic data and baseline questionnaires.
| Cross-Validation Results | ||||||
| Model | Full Dataset | Actigraphy Data | Activity & Skin Temperature | Activity & Light | Light & Skin Temperature | Static Data |
| Single Layer | 1.68±0.44 (30 nodes) |
2.00±0.88 (30 nodes) |
2.62±1.69 (10 nodes) |
1.68±0.37 (10 nodes) |
2.32±1.22 (10 nodes) |
2.66±1.67 (10 nodes) |
| Single Layer with Dropout | 1.54±0.42 (p=0.3) |
1.77±0.74 (p=0.4) |
2.52±1.99 (p=0.1) |
1.57±0.35 (p=0.1) |
1.82±0.69 (p=0.1) |
1.80±0.38 (p=0.3) |
| Double Layer | 1.58±0.31 (30x10 nodes) |
1.62±0.38 (10x10 nodes) |
2.24±0.42 (10x30 nodes) |
1.58±0.31 (50x10 nodes) |
1.73±0.67 (30x10 nodes) |
2.34±1.50 (10x10 nodes) |
| Double Layer with Dropout | 1.59±0.30 (p=0.1) |
1.62±0.34 (p=0.1) |
1.82±0.46 (p=0.5) |
1.59±0.30 (p=0.1) |
1.81±0.69 (p=0.2) |
1.64±0.38 (p=0.1) |
| LSTM | 1.94±1.07 (10 units) |
1.57±0.30 (50 units) |
1.57±0.31 (20 units) |
1.57±0.31 (20 units) |
1.57±0.30 (40 units) |
- |
| LSTM with Dropout | 1.41±0.30 (p=0.5) |
1.51±0.31 (p=0.4) |
1.55±0.33 (p=0.5) |
1.57±0.30 (p=0.1) |
1.57±0.31 (p=0.1) |
- |
| Test Results | ||||||
| Model | Full Dataset | Actigraphy Data | Activity & Skin Temperature | Activity & Light | Light & Skin Temperature | Static Data |
| Single Layer | 2.15 | 2.17 | 8.29 | 1.59 | 2.58 | 1.88 |
| Single Layer with Dropout | 1.65 | 1.74 | 8.13 | 1.66 | 2.16 | 1.89 |
| Double Layer | 1.58 | 1.58 | 9.20 | 1.58 | 1.58 | 1.62 |
| Double Layer with Dropout | 1.64 | 1.65 | 5.56 | 1.65 | 1.73 | 1.62 |
| LSTM | 1.58 | 1.57 | 1.58 | 1.57 | 1.58 | - |
| LSTM with Dropout | 1.47 | 1.51 | 1.56 | 1.58 | 1.58 | - |
Figure 3. Neural Network Based Estimations of DLMO.
Scatter plot showing experimentally measured DLMO vs. model estimated phase for the training and test sets with the single layer model for continuous neural networks (A) and classification neural networks (B). The dashed line represents perfect estimation.
A Classification Approach Improves Phase Estimation Accuracy in the College Student Population
The classification approach could predict whether a timepoint was before or after DLMO based on 24 h of data with high accuracy (~90%) for both the training and test set (Table 2, Table S4) for both average and median features. Predictions were more consistent when using the median features (Tables S7–S8).
Table 2. Performance of Classifiers for DLMO Estimation in College Students.
The classification accuracy of whether a timepoint falls before or after experimentally observed DLMO in cross-validation (mean ± standard deviation) and on the independent test set on different subsets of the data.
| Cross-Validation Results | |||||
| Model | Full Dataset | Actigraphy Data | Activity & Skin Temperature | Activity & Light | Light & Skin Temperature |
| Single Layer | 89.1±3.7 (100 nodes) |
89.4±3.6 (90 nodes) |
88.8±3.4 (50 nodes) |
89.8±3.4 (90 nodes) |
87.0±3.0 (30 nodes) |
| Single Layer with Dropout | 89.3±3.4 (p=0.4) |
89.3±3.2 (p=0.3) |
89.1±2.9 (p=0.5) |
89.5±3.8 (p=0.5) |
87.1±3.4 (p=0.2) |
| Double Layer | 88.9±3.6 (50x50 nodes) |
89.3±3.6 (30x50 nodes) |
89.2±3.5 (30x40 nodes) |
90.0±3.9 (40x40 nodes) |
86.8±3.7 (30x40 nodes) |
| Double Layer with Dropout | 89.3±3.3 (p=0.4) |
89.5±3.4 (p=0.3) |
88.7±3.2 (p=0.1) |
89.6±3.8 (p=0.4) |
87.1±2.9 (p=0.2) |
| Test Results | |||||
| Model | Full Dataset | Actigraphy Data | Activity & Skin Temperature | Activity & Light | Light & Skin Temperature |
| Single Layer | 90.7 | 90.2 | 89.1 | 89.9 | 86.1 |
| Single Layer with Dropout | 89.9 | 90.2 | 89.8 | 89.8 | 85.8 |
| Double Layer | 90.1 | 90.9 | 89.4 | 90.5 | 86.4 |
| Double Layer with Dropout | 89.2 | 89.8 | 88.8 | 90.1 | 85.1 |
A sample output of the classification neural network is shown in Figure 4B; we have presented the data so that the switch in classification should occur between the eleventh and twelfth time-bin for perfect accuracy. We found that the changes in class prediction usually only crossed the decision boundary once; in cases where this was not true, we defined the switch as the first time that the decision boundary was crossed for simplicity. The RMSE for these estimations decreased in all models when using the average features (Table 3) with the best performing models producing an RMSE of 1.3–1.4 hours (R=0.59 (p<<<0.01) in the training set and R=0.40 (p=0.03) in the test set (Figure 3B)). Even seemingly small improvements in classification accuracy can lead to large improvements in RMSE (Figure 4C), which suggests that the errors in the classification occur near the decision boundary. In 16% of participants, the change in classification occurs within 15 minutes of measured DLMO, and in 35% of participants, the switch occurs within 30 minutes. However, the classification approach did not improve over the continuous approach when using the median features, suggesting that the increased variance provided by the average is important to the success of the classification approach (Table S9).
Table 3. Classification Performance vs. RMSE in Estimated DLMO.
A comparison of classification accuracy (predicting if a timepoint falls before or after DLMO) to the resulting RMSE between estimated DLMO, calculated based on the switch in classification, and the experimentally measured DLMO for different classification models.
| Model | Full Dataset | Actigraphy Dataset | ||
|---|---|---|---|---|
| Test Accuracy | Test RMSE | Test Accuracy | Test RMSE | |
| Single Layer | 90.7 | 1.34 | 90.2 | 1.36 |
| Single Layer with Dropout | 89.9 | 1.45 | 90.2 | 1.42 |
| Double Layer | 90.1 | 1.40 | 90.9 | 1.30 |
| Double Layer with Dropout | 89.2 | 1.53 | 89.8 | 1.82 |
| Logistic Regression | 88.1 | 1.74 | 90.4 | 1.33 |
| SVM (linear kernel) | 89.4 | 1.52 | 90.7 | 1.37 |
| SVM (polynomial kernel) | 90.1 | 1.44 | 89.9 | 1.45 |
| SVM (RBF kernel) | 90.1 | 1.45 | 90.5 | 1.39 |
Accuracy of the Classification Approach Depends on the Phase Angle Between DLMO and Sleep Onset
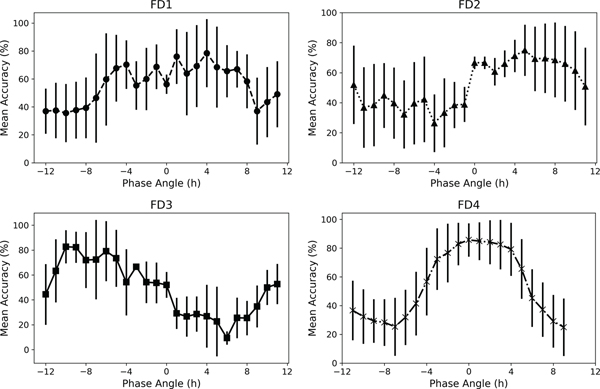
We fit the model to the training set of light and activity patterns in the college sleep students and tested the performance in the FD datasets. In the FD data, overall classification accuracy dropped to approximately 50–60% (Table 4), preventing a meaningful determination of DLMO based on switches between class labels. Within individual participants, we found that classification accuracy could vary between 20% and 80% depending upon the day in the protocol, probably due to the phase angles between sleep onset and DLMO; a FD schedule includes all possible phase angles, while the phase angle is typically 2.2 ± 1.0 h in aligned populations41. Mean classification accuracy was higher for days where the phase angles were those of alignment (approximately −4 to 4 h) (Figure 5).
Table 4. Performance of Classification Models for DLMO Estimation in Forced Desynchrony (FD) Protocols.
Classification accuracy performance on four different FD protocols to test for generalization of these models originally fit to activity and light level data from the college student population.
| Model | FD1 | FD2 | FD3 | FD4 |
|---|---|---|---|---|
| Single Layer | 54.1 | 52.3 | 51.8 | 57.2 |
| Single Layer with Dropout | 54.9 | 53.1 | 51.0 | 56.7 |
| Double Layer | 53.4 | 54.3 | 52.1 | 57.4 |
| Double Layer with Dropout | 54.8 | 53.6 | 51.6 | 57.0 |
Figure 5. Classification Accuracy in Forced Desynchrony (FD) Protocols is Dependent on Phase Angle Between Sleep Onset and DLMO.
The mean (standard deviation) accuracy of the neural networks classification for days relative to the phase angle (between sleep onset and DLMO) for each of the 4 different FD protocols.
To further explore these findings, we analyzed the activity data relative to circadian and wake duration bins. There were significant differences in activity levels by circadian phase (p<0.007) and wake duration (p<0.001) for all four FD studies (Figure S3). The circadian rhythm of activity was highest at circadian times 13.5–19.5 h and lowest at −1.5–0.5 h relative to DLMO.
Discussion
Our classification-based method showed improvement over the continuous approaches in college students on self-selected schedules, suggesting that our model may be more generalizable since the range of observed phases (Figure 1) is also broader in college students compared to healthy participants studied under inpatient conditions. The range of errors in estimating circadian phase in both our continuous models (which were based on previous approaches23,24) and classification models, however, are greater than previous studies19,21,23,24. While our and other methods were successful in estimating DLMO in populations on “normal” schedules, they were less successful when circadian phase and rest-activity rhythms were misaligned (as is common with rotating shiftwork and circadian disorders); in such conditions, phase shifting interventions may be the most valuable. Therefore, there is still a need for further improvement in circadian phase estimation in misaligned conditions.
Desired Accuracy for Clinical Utility
Repeated measures of DLMO in this college population42 and studies on the variance of DLMO assessment show typical fluctuations of less than an hour43. Therefore, a single phase estimate may be useful for clinical and other applications. The accuracy required for phase estimation methods to be clinically useful, however, may depend on the application9. Errors of approximately one hour may be acceptable to predict cognitive performance, but efficacy of phase shifting decreases from about 80% to 60% if errors in the timing of a light pulse increase from 1 to 2 hours15, resulting in possible errors in the shifted phase44.
Further Improvements to the Classification Approach
Although our population is larger than in previous studies, training the model on still larger populations could be beneficial for refining the model, as machine learning approaches have greatest success on large datasets because of the large number of parameters that are needed in these models. One possible reason the classification approach was more successful in estimating DLMO than the continuous models was because that approach increased our dataset size. Using this same expanded dataset but predicting time to DLMO from a specific time based on 24 h of data in a continuous framework, did not improve errors in DLMO estimation; this suggests the classification approach confers additional benefits beyond just more effectively using data.
Our classification accuracy may have performed poorly on the FD protocols because our training set of college students was not representative of these misaligned schedules, so using broader training data may lead to improvements. Some initial experiments in training on the FD datasets suggest some limited generalizability to the population of college students with approximately 70% accuracy. This is still lower than attained in our models trained in the college student population (as reported above), suggesting that other differences (i.e., controlled light and temperature environments) between the inpatient and outpatient populations may be important to the higher accuracy for models trained in the college population. To what extent classification accuracy depends on the training and testing demographic populations (e.g., circadian phase tends to be earlier in older populations45) should be explored.
Larger populations may also allow for other types of machine learning models to be implemented. Convolutional neural networks are highly effective in classifying images46, and may also be effective here if classification is dependent on the location of a specific feature of the actigraphy data waveform. Recent research has explored physics-informed neural networks, which, by introducing physical laws as constraints, can solve differential equations47. Based on the success of the differential equation-based approaches in estimating circadian phase18, adding known dynamics about the impact of light in creating phase shifts in combination with neural networks may be a promising direction to pursue.
As these networks are further developed, other features of the input data could be considered. In our analysis, we did not find improvements when using smaller time-bins for the data (Tables S5 and S10), but the length of the time-bin does put a lower bound on accuracy. For data to correspond to a circadian cycle, we chose to use a 24 h interval of data. Requiring 24 h of data limits the ability to immediately assess circadian phase so shorter time intervals are preferrable, but we found that accuracy decreased with decreasing time intervals of data (Figure S1). Incorporating data from when actigraphy and a single DLMO measurement were available in addition to current actigraphy data may also be valuable for future predictions, given the relative consistency of DLMO42.
Significance of Activity Rhythms and Challenges Under Circadian Misalignment
Our results demonstrate that activity levels may be important both for circadian phase estimation and other applications using activity as an input because of its circadian variation. Lowest accuracy in our classification approach was seen across models using average features when activity was not included as a model input, unlike previous studies that suggested skin temperature data was more important for prediction accuracy21,23.
Circadian variation may explain why the actigraphy-based approaches are highly limited in their predictive ability under circadian misalignment; FD protocols result in activity levels at circadian phases different than “normal”: higher levels during scheduled wake during the circadian “night” and lower levels during scheduled sleep during the circadian “day”. Circadian misalignment is common in individuals working night shifts and those with circadian rhythm sleep disorders: two populations that may need accurate phase assessments for interventions. Thus, a different data source or methodology may be required for phase assessment in these populations because of these changes in timing of behaviors and/or different light sensitivities in people with circadian disorders48,49.
We have presented a novel, machine learning approach in using actigraphy to estimate DLMO that is effective in outpatient, circadian-aligned populations. The classification-based approach introduced here is not limited to the prediction of the rise in melatonin but could also be used to estimate the timing of other hormones.
Supplementary Material
Acknowledgements
This work was supported by NIH T32-HL007901(LSB, AWM), R01GM105018, R00HL119618(AJKP), K24HL105664(EBK), K01HL146992 (AWM), P01AG009975 (CAC, EBK), and F32DK107146 (AWM), R21-NR018974 (MSH); Harvard Catalyst| The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102); MIT Media Lab Consortium, Samsung Electronics, and NEC Corporation (AS); NASA 80NSSC20K0576(MSH). LKB was supported in part by 2R01AG044416-06 and 1R01OH011773.
Conflicts of Interest
LSB, AWM, and FJD declare no conflicts of interest. MSH declares no conflicts of interest directly related to the study; she has provided paid limited consulting for The MathWorks, Inc. (Natick, MA, USA) and she was paid by the Fund for Scientific Research – FNRS (Belgium) for a grant review. AJKP is an investigator on projects supported by the CRC for Alertness, Safety, and Productivity, and he has received research funding from Versalux and Delos. LKB was on the scientific board of CurAegis and received funding from Puget Sound Pilots and Boston Children’s Hospital. AS has received travel reimbursement or honorarium payments from Gordon Research Conferences, Pola Chemical Industries, Leuven Mindgate, American Epilepsy Society, IEEE, and Association for the Advancement of Artificial Intelligence. She has received research support from Microsoft, Sony Corporation, NEC Corporation, and POLA chemicals, and consulting fees from Gideon Health and Suntory Global Innovation Center. EBK has received travel reimbursement from the Sleep Research Society, the National Sleep Foundation, the Santa Fe Institute, the World Conference of Chronobiology, the Gordon Research Conference, and the German Sleep Society (DGSM); she was paid by the Puerto Rico Trust for a grant review, and has consulted for the National Sleep Foundation and Sanofi-Genzyme. CAC reports grants to BWH from FAA, NHLBI, NIA, NIOSH, NASA, and DOD; is/was a paid consultant to Emory University, Inselspital Bern, Institute of Digital Media and Child Development, Klarman Family Foundation, Physician’s Seal, Sleep Research Society Foundation, Tencent Holdings Ltd, Teva Pharma Australia, and Vanda Pharmaceuticals Inc, in which Dr. Czeisler also holds an equity interest; received travel support from Bloomage International Investment Group, Inc., UK Biotechnology and Biological Sciences Research Council, Bouley Botanical, Dr. Stanley Ho Medical Development Foundation, European Biological Rhythms Society, German National Academy of Sciences (Leopoldina), National Safey Council, National Sleep Foundation, Stanford Medical School Alumni Association, Tencent Holdings Ltd, and Vanda Pharmaceuticals Inc; receives research/education support through BWH from Cephalon, Mary Ann & Stanley Snider via Combined Jewish Philanthropies, Harmony Biosciences LLC, Jazz Pharmaceuticals PLC Inc, Johnson & Johnson, NeuroCare, Inc., Philips Respironics Inc/Philips Homecare Solutions, Regeneron Pharmaceuticals, Regional Home Care, Teva Pharmaceuticals Industries Ltd, Sanofi SA, Optum, ResMed, San Francisco Bar Pilots, Sanofi, Schneider, Simmons, Sysco, Philips, Vanda Pharmaceuticals; is/was an expert witness in legal cases, including those involving Advanced Power Technologies, Aegis Chemical Solutions LLC, Amtrak; Casper Sleep Inc, C&J Energy Services, Dallas Police Association, Enterprise Rent-A-Car, Espinal Trucking/Eagle Transport Group LLC/Steel Warehouse Inc, FedEx, Greyhound Lines Inc/Motor Coach Industries/FirstGroup America, PAR Electrical Contractors Inc, Product & Logistics Services LLC/Schlumberger Technology Corp/Gelco Fleet Trust, Puckett Emergency Medical Services LLC, Union Pacific Railroad, and Vanda Pharmaceuticals; serves as the incumbent of an endowed professorship provided to Harvard University by Cephalon, Inc.; and receives royalties from McGraw Hill, and Philips Respironics for the Actiwatch-2 and Actiwatch Spectrum devices. CAC’s interests were reviewed and are managed by the Brigham and Women’s Hospital and Mass General Brigham in accordance with their conflict of interest policies.
Data Availability Statement
We will make the datasets available to other investigators upon request. Outside investigators should submit their request in writing to Dr. Klerman. The request must be in accordance with MGB Policies, Harvard Medical School (HMS), and Harvard University guidelines. Such datasets will not contain identifying information per the regulations outlined in HIPAA. Per standard MGB policies, we will require a data-sharing agreement from any investigator or entity requesting the data.
References
- 1.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 2.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballesta A, Innominato PF, Dallmann R, Rand DA, Lévi FA. Systems chronotherapeutics. Pharmacol Rev. 2017;69(2):161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron KG, Reid KJ. Circadian misalignment and health. Int Rev Psychiatry. 2014;26(2):139–154. doi: 10.3109/09540261.2014.911149.Circadian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boivin Diane B., Duffy Jeanne F., Kronauer Richard E., Czelsler Charles A.. Dose-response relationships for resetting of human circadian clock by light. Nature. 1996;379(8):540–542. https://www.nature.com/articles/379540a0.pdf. [DOI] [PubMed] [Google Scholar]
- 6.Duffy JF, Kronauer RE, Czeisler CA. Phase-shifting human circadian rhythms: Influence of sleep timing, social contact and light exposure. J Physiol. 1996;495(1):289–297. doi: 10.1113/jphysiol.1996.sp021593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronauer JRE, Czeisler CA, Megan E. Phase-amplitude resetting of the human circadian pacemaker via bright light : A further analysis. J Biol Rhythms. 1985;9(3):295–314. [DOI] [PubMed] [Google Scholar]
- 8.Breslow ER, Phillips AJK, Huang JM, St. Hilaire MA, Klerman EB. A mathematical model of the circadian phase-shifting effects of exogenous melatonin. J Biol Rhythms. 2013;28(1):79–89. doi: 10.1177/0748730412468081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klerman EB, Rahman SA, St. Hilaire MA What time is it? A tale of three clocks, with implications for personalized medicine. J Pineal Res. 2020;68(4):1–5. doi: 10.1111/jpi.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijk DJ, Duffy JF. Novel approaches for assessing circadian rhythmicity in humans: A review. J Biol Rhythms. 2020;35(5):421–438. doi: 10.1177/0748730420940483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crnko S, Schutte H, Doevendans PA, Sluijter JPG, van Laake LW. Minimally invasive ways of determining circadian rhythms in humans. Physiology. 2021;36:7–20. doi: 10.1152/physiol.00018.2020. [DOI] [PubMed] [Google Scholar]
- 12.Hughey JJ, Hastie T, Butte AJ. ZeitZeiger: Supervised learning for high-dimensional data from an oscillatory system. Nucleic Acids Res. 2016;44(8):e80. doi: 10.1093/nar/gkw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda HR, Chen W, Minami Y, et al. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci. 2004;101(31):11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittenbrink N, Kunz D, Kramer A, et al. High-accuracy determination of internal circadian time from a single blood sample. J Clin Invest. 2018;128(9):3826–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone JE, Postnova S, Sletten TL, Rajaratnam SMW, Phillips AJK. Computational approaches for individual circadian phase prediction in field settings. Curr Opin Syst Biol. 2020;22:39–51. doi: 10.1016/j.coisb.2020.07.011. [DOI] [Google Scholar]
- 16.Kronauer RE, Forger DB, Jewett ME. Quantifying human circadian pacemaker response to brief, extended, and repeated light stimuli over the phototopic range. J Biol Rhythms. 1999;14(6):500–515. doi: 10.1177/074873099129001073. [DOI] [PubMed] [Google Scholar]
- 17.St. Hilaire MA, Klerman EB, Khalsa SBS, Wright KP, Czeisler CA, Kronauer RE. Addition of a non-photic component to a light-based mathematical model of the human circadian pacemaker. J Theor Biol. 2007;247(4):583–599. doi: 10.1016/j.jtbi.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woelders T, Beersma DGM, Gordijn MCM, Hut RA, Wams EJ. Daily light exposure patterns reveal phase and period of the human circadian clock. J Biol Rhythms. 2017;32(3):274–286. doi: 10.1177/0748730417696787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonmati-Carrion MA, Middleton B, Revell V, Skene DJ, Rol MA, Madrid JA. Circadian phase asessment by ambulatory monitoring in humans: Correlation with dim light melatonin onset. Chronobiol Int. 2014;31(1):37–51. doi: 10.3109/07420528.2013.820740. [DOI] [PubMed] [Google Scholar]
- 20.Yin J, Julius A, Wen JT, Oishi MMK, Brown LK. Actigraphy-based parameter tuning process for adaptive notch filter and circadian phase shift estimation. Chronobiol Int. 2020;37(11):1552–1564. doi: 10.1080/07420528.2020.1805460. [DOI] [PubMed] [Google Scholar]
- 21.Kolodyazhniy V, Späti J, Frey S, et al. Estimation of human circadian phase via a multi-channel ambulatory monitoring system and a multiple regression model. J Biol Rhythms. 2011;26(1):55–67. doi: 10.1177/0748730410391619. [DOI] [PubMed] [Google Scholar]
- 22.Ramsundar B, Eastman P, Walters P, Pande V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More. O’Reilly Media, Inc.; 2019. [Google Scholar]
- 23.Kolodyazhniy V, Späti J, Frey S, et al. An improved method for estimating human circadian phase derived From multichannel ambulatory monitoring and artificial neural networks. Chronobiol Int. 2012;29(8):1078–1097. doi: 10.3109/07420528.2012.700669. [DOI] [PubMed] [Google Scholar]
- 24.Stone JE, Ftouni S, Lockley SW, et al. A neural network model to predict circadian phase in normal living conditions. Sleep Med. 2017;40(2017):e315. doi: 10.1016/j.sleep.2017.11.927. [DOI] [Google Scholar]
- 25.Stone JE, Phillips AJK, Ftouni S, et al. Generalizability of a neural network model for circadian phase prediction in real-world conditions. Sci Rep. 2019;9:11001. doi: 10.1038/s41598-019-47311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng P, Walch O, Huang Y, et al. Predicting circadian misalignment with wearable technology : validation of wrist-worn actigraphy and photometry in night shift workers. Sleep. 2020;44(2):1–8. doi: 10.1093/sleep/zsaa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone JE, Aubert XL, Maass H, et al. Application of a limit-cycle oscillator model for prediction of circadian phase in rotating night shift workers. Sci Rep. 2019;9:11032. doi: 10.1038/s41598-019-47290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St. Hilaire MA, Lammers-van der Holst HM, Chinoy ED, Isherwood CM, Duffy JF. Prediction of individual differences in circadian adaptation to night work among older adults: application of a mathematical model using individual sleep-wake and light exposure data. Chronobiol Int. 2020;37(9–10):1404–1411. doi: 10.1080/07420528.2020.1813153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips AJK, Clerx WM, O’Brien CS, et al. Irregular sleep/wake patterns are associated with poorer academic performance and delayed circadian and sleep/wake timing. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-017-03171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sano A, Taylor S, McHill AW, et al. Identifying objective physiological markers and modifiable behaviors for self-reported stress and mental health status using wearable sensors and mobile phones: Observational study. J Med Internet Res. 2018;20(6):e210. doi: 10.2196/jmir.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHill AW, Phillips AJK, Czeisler CA, et al. Later circadian timing of food intake is associated with increased body fat. Am J Clin Nutr. 2017;106(5):1213–1219. doi: 10.3945/ajcn.117.161588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen DA, Wang W, Wyatt JK, et al. Uncovering residual effects of chronic sleep loss on human performance. Sci Transl Med. 2010;2(14):14ra3. doi: 10.1126/scitranslmed.3000458.Uncovering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grady S, Aeschbach D, Wright KP, Czeisler CA. Effect of modafinil on impairments in neurobehavioral performance and learning associated with extended wakefulness and circadian misalignment. Neuropsychopharmacology. 2010;35(9):1910–1920. doi: 10.1038/npp.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gronfier C, Wright KP, Kronauer RE, Czeisler CA. Entrainment of the human circadian pacemaker to longer-than-24-h days. Proc Natl Acad Sci. 2007;104(21):9081–9086. doi: 10.1073/pnas.0702835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHill AW, Hull JT, Wang W, Czeisler CA, Klerman EB. Chronic sleep curtailment, even without extended (>16-h) wakefulness, degrades human vigilance performance. Proc Natl Acad Sci. 2018;115(23):6070–6075. doi: 10.1073/pnas.1706694115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHill AW, Phillips AJK, Czeisler CA, et al. Later circadian timing of food intake is associated with increased body fat. Am J Clin Nutr. 2017;106(5):1213–1219. doi: 10.3945/ajcn.117.161588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongrain V, Lavoie S, Selmaoui B, Paquet J, Dumont M. Phase relationships between sleep-wake cycle and underlying circadian rhythms in morningness-eveningness. J Biol Rhythms. 2004;19(3):248–257. doi: 10.1177/0748730404264365. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. doi: 10.1214/12-AOS1000. [DOI] [Google Scholar]
- 39.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 40.Wan C, McHill AW, Klerman EB, Sano A. Sensor-based estimation of dim light melatonin onset (DLMO) using features of two time scales. ACM Trans Comput Healthc. 2021:In press. [Google Scholar]
- 41.Wright KP, Gronfier C, Duffy JF, Czeisler CA. Intrinsic period and light intensity determine the phase relationship between melatonin and sleep in humans. J Biol Rhythms. 2005;20(2):168–177. doi: 10.1177/0748730404274265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHill AW, Sano A, Hilditch CJ, et al. Robust stability of melatonin circadian phase, sleep metrics, and chronotype across months in young adults living in real-world settings. J Pineal Res. 2021;70(3):e12720. doi: 10.1111/jpi.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klerman H, St Hilaire MA, Kronauer RE, et al. Analysis method and experimental conditions affect computed circadian phase from melatonin data. PLoS One. 2012;7(4):e33836. doi: 10.1371/journal.pone.0033836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown LS, Klerman EB, Doyle FJ III. Compensating for sensor error in the model predictive control of circadian clock phase. IEEE Control Syst Lett. 2019;3(4):853–858. doi: 10.1109/LCSYS.2019.2919438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duffy JF, Zitting K-M, Chinoy ED. Aging and circadian rhythms. Sleep Med Clin. 2015;10(4):423–434. doi: 10.1016/j.jsmc.2015.08.002.Aging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krizhevsky BA, Sutskever I, Hinton GE. ImageNet classification with deep convolutaionl neural networks. Commun ACM. 2017;60(6):84–90. [Google Scholar]
- 47.Raissi M, Perdikaris P, Karniadakis GE. Physics informed deep learning (Part I): Data-driven discovery of nonlinear partial differential equations. arXiv. 2017:1–22. [Google Scholar]
- 48.Watson LA, Phillips AJK, Hosken IT, et al. Increased sensitivity of the circadian system to light in delayed sleep–wake phase disorder. J Physiol. 2018;596(24):6249–6261. doi: 10.1113/JP275917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbott SM, Choi J, Wilson J, Zee PC. Melanopsin-dependent phototransduction is impaired in delayed sleep–wake phase disorder and sighted non–24-hour sleep–wake rhythm disorder. Sleep. 2021;44(2):1–9. doi: 10.1093/sleep/zsaa184. [DOI] [PubMed] [Google Scholar]
- 50.Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. 2017;114(20):5312–5317. doi: 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun R, Kath WL, Iwanaszko M, Kula-eversole E, Abbott SM. Universal method for robust detection of circadian state from gene expression. Proc Natl Acad Sci. 2018;(23):1–10. doi: 10.1073/pnas.1800314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We will make the datasets available to other investigators upon request. Outside investigators should submit their request in writing to Dr. Klerman. The request must be in accordance with MGB Policies, Harvard Medical School (HMS), and Harvard University guidelines. Such datasets will not contain identifying information per the regulations outlined in HIPAA. Per standard MGB policies, we will require a data-sharing agreement from any investigator or entity requesting the data.