Abstract
Objective:
Preoperative differentiation between parotid Warthin’s tumor (WT) and pleomorphic adenoma (PMA) is crucial for treatment decisions. The purpose of this study was to establish and validate an MRI-based radiomics nomogram for preoperative differentiation between WT and PMA.
Methods and materials:
A total of 127 patients with histological diagnosis of WT or PMA from two clinical centres were enrolled in training set (n = 75; WT = 34, PMA = 41) and external test set (n = 52; WT = 24, PMA = 28). Radiomics features were extracted from axial T1WI and fs-T2WI images. A radiomics signature was constructed, and a radiomics score (Rad-score) was calculated. A clinical factors model was built using demographics and MRI findings. A radiomics nomogram combining the independent clinical factors and Rad-score was constructed. The receiver operating characteristic analysis was used to assess the performance levels of the nomogram, radiomics signature and clinical model.
Results:
The radiomics nomogram incorporating the age and radiomics signature showed favourable predictive value for differentiating parotid WT from PMA, with AUCs of 0.953 and 0.918 for the training set and test set, respectively.
Conclusions:
The MRI-based radiomics nomogram had good performance in distinguishing parotid WT from PMA, which could optimize clinical decision-making.
Keywords: Parotid neoplasms, Radiomics, Magnetic resonance imaging
Introduction
Parotid gland pleomorphic adenoma (PMA) and Warthin’s tumor (WT) are the two most common benign tumours of the parotid gland.1The biological behaviours of the two tumours are different, with the result that the treatment and prognosis also differ.2 PMA is susceptible to malignant transformation and recurrence, and superficial or total parotidectomy is, therefore, recommended.2 However, malignant transformation and recurrence of WT seldom occurs, and tumorectomy is considered sufficient.2 Therefore, the preoperative discrimination of WT from PMA is crucial for choosing the most appropriate surgical treatment for patients.
As a novel non-invasive technique, radiomics aims to extract quantitative and higher dimensional data from digital biomedical images, facilitating a full exploration of intratumoural information and changes.3,4 Different imaging modalities, including CT, MRI, PET and ultrasound, can be used as the basis for extracting these quantitative data.4 Previous studies have suggested that there is a strong correlation between these high dimensional data and tissue heterogeneity at the cellular level.5,6 Radiomics has been widely used in oncology studies and applied successfully to differentiating between early and advanced stage diseases, differentiating benign and malignant tumours, predicting treatment outcome and assessing cancer genetics.7 Radiomics has also been successfully applied to the assessment of head and neck tumours,8,9 and previous studies have reported the value of texture analysis of CT or MR images for the discrimination of WT from PMA.10,11 Compared with texture analysis, radiomics analysis can provide a more comprehensive description of the tumour by extracting many more statistical features. To the best of our knowledge, no study has focused on the value of radiomics for discriminating between WT and PMA. As a simple graphical representation of a statistical predictive model, nomogram uses biological and clinical variables to determine a model that generates a probability of a clinical event.12,13 It has been widely used in radiomics studies.141516
In this current study, we aimed to establish an MRI radiomics-based prediction nomogram for discriminating between WT and PMA, and then to assess its validity.
Methods and materials
Patients
The institutional review board of two clinical centres approved this retrospective study and an institutional review board waiver was obtained. In the current study, all the enrolled patients with histological diagnosis of WT or PMA on surgically resected specimens were from two clinical centres. Consecutive patients presenting between January 2015 and June 2020 were identified from Centre 1, and those presenting between January 2013 and June 2020 were identified from Centre 2. These two patient groups were used to form a training set and external test set, respectively. The inclusion criteria were: 1) patients with histologically confirmed WT or PMA and complete clinical data; and 2) patients with an MRI examination including axial T1-weighted imaging (T1WI) and fat-saturated T2-weighted imaging (fs-T2WI) performed less than 7 days before surgical resection. The exclusion criteria included: (1) tumours with a maximum diameter <5 mm, and (2) the existence of imaging artifacts making the images unsatisfactory for radiomics analysis. The exclusion criteria for a maximum tumor diameter at 5 mm were set to minimize the influence of partial volume effects, which might distort the true tissue-specific image texture.10 A total of 127 patients met the above criteria, with 75 being enrolled in the training set (WT = 34, PMA = 41) and 52 in the external test set (WT = 24, PMA = 28). The clinical data of the patients included gender, age and smoking history.
MRI acquisition
MRI was performed using one of two 3.0-T MRI scanners with a matched eight-channel phased-array coil (GE 3.0-Tesla Signa scanner, GE Healthcare, Milwaukee, WI, USA; Siemens 3.0-Tesla Skyra scanner, Siemens, Erlangen, Germany). All patients underwent axial T1WI and fs-T2WI sequences. The T1WI was acquired using the following parameters: echo time 11 ms; repetition time 420 or 500 ms; number of excitations 2; slice thickness 4 mm; slice spacing 1 mm; acquisition matrix 320 × 256 or 320 × 203; and field of view 22 cm. The parameters for the fs-T2WI sequence included: echo time 102 or 103 ms; repetition time 3600 or 3000 ms; number of excitations 2; slice thickness 4 mm; slice spacing 1 mm; acquisition matrix 320 × 256 or 320 × 203; field of view 22 cm.
MRI feature evaluation and clinical model construction
Two oral and maxillofacial radiologists with 8 and 12 years of experience who were blinded to the pathology results analysed the MRI features in consensus. The MRI features analysed were: (1) the maximum diameter of the tumour; (2) well-demarcated or poorly demarcated tumour margin; (3) heterogeneous appearance (10% of the tumour having a different signal17 ); (4) growth pattern (focal/ multifocal); (5) presence of cystic or necrotic regions (an area with hypointensity on T1WI and hyperintensity on fs-T2WI); (6) lobulated appearance of the tumour; and (7) parotid tail involvement. In case of multiple nodules within a single patient, the nodule with biggest volume was included in the analysis.
The clinical data and MRI features of the training set were compared between WT and PMA using univariate analysis. The significant variables identified in the univariate analysis were input into a multiple logistic regression analysis to identify independent factors for building a clinical model.
Tumour segmentation and radiomics feature extraction
The segmentation of the tumours was performed by two radiologists who manually delineated regions of interest (ROIs) defining the tumour contour in a slice-by-slice manner on T1WI and fs-T2WI using 3D Slicer software (v.4.10.2, https://www.slicer.org; Figure 1). The segmentation data were then input into 3D Slicer to extract the radiomics features.
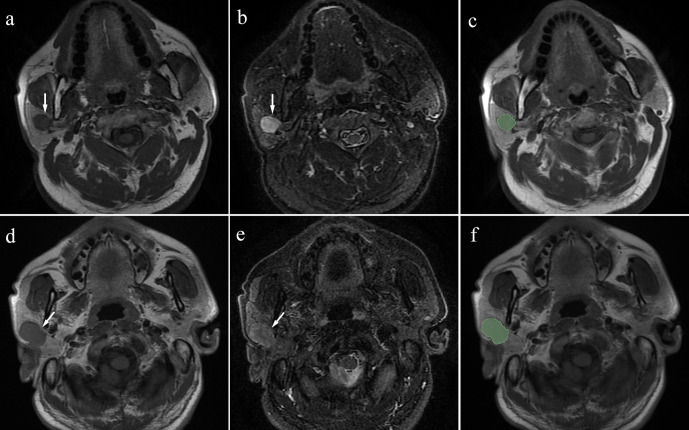
Figure 1.
a: T1-weighted image; b: fat-saturated T2 weighted image) Case 1: Pleomorphic adenoma in a 63-year-old male. A mass can be seen in the right parotid (arrow). (c) Manual segmentation of the mass. (d, e) (d: T1-weighted image; e: fat-saturated T2 weighted image) Case 2: Warthin’s tumour in a 51-year-old male. A mass can be seen in the right parotid (arrow). (f) Manual segmentation of the mass.
Eight feature groups (a total of 851 radiomics features), including shape (14 features), first-order statistics (18 features), gray level dependence matrix (GLDM, 14 features), gray level co-occurrence matrix (GLCM, 24 features), gray level run length matrix (GLRLM, 16features), gray level size zone matrix (GLSZM, 16 features), neighbouring gray tone difference matrix (NGTDM, five features) and wavelet features (744 features), were extracted from the ROIs for each MRI sequence using 3D Slicer. The definitions of feature groups were supplied in “Supplementary Material 1”. The details and formulas for these features are shown both in the supplementary materials of a previous radiomics study18 and at https://pyradiomics.readthedocs.io/en/latest/features.html.
A total of 1702 radiomics features were finally extracted from the T1WI and fs-T2WI sequences of each subject. Details of the feature extraction are provided in Supplementary Table 1. Considering different protocol parameters from different MRI scanners, a number of preprocessing procedures were applied. The “μ ± 3σ” method was used to correct for the effects of different MR scanners and acquisition protocols and normalize the image intensities.19 Meanwhile, “N4ITK” bias field correction was applied to correct the intensity non-uniformities caused by the inhomogeneity of the scanner’s magnetic field during image acquisition.20 Resampling (voxel size = 1×1×1 mm3) was performed to ensure the conservation of scales and directions when deriving the 3D features.21 Z-score normalization was also performed as preprocessing steps for data to guarantee the repeatability of the results.
The intraobserver and interobserver reliabilities of the radiomics features were estimated using 20 randomly chosen images. To evaluate interobserver reliability, the tumour segmentation was independently performed by two radiologists (Readers 1 and 2) over the same period. Reader one then repeated the same procedure at a 2-week interval to evaluate intraobserver reliability. Reader one then performed the remaining image segmentations. The agreement was evaluated using the intraclass correlation coefficient (ICC). Features with ICCs > 0.75 were considered to show good agreement and were selected for further analysis, whereas features with ICCs ≤ 0.75 were excluded.
Feature selection and development of the radiomics signature
Two-step feature selection methods were used to minimize overfitting22 and identify the features that were most effective for discriminating between WT and PMA. One-way analysis of variance (ANOVA) was first performed to select statistically significant features for the training set. The selected features were then input into a least absolute shrinkage and selection operator (LASSO) regression algorithm23 to identify the most effective features (with non-zero coefficients) for discriminating between WT and PMA. The tuning regularization parameter λ, which controls the strength of regularization (a method commonly used for alleviating overfitting in machine learning24,25), was chosen using 10-fold cross-validation via the one-standard error of the minimum criteria (the 1-SE criteria, a simpler model). A radiomics signature was constructed based on the finally selected features, and a radiomics score (Rad-score) was generated for each patient using a linear combination of the values of the selected features weighted by their corresponding non-zero coefficients.
Radiomics nomogram construction
A radiomics nomogram incorporating the Rad-score and independent clinical factors was constructed using multivariate logistic regression. A radiomics nomogram score (Nomo-score) was then generated for each patient using the Rad-score and independent clinical factors. To investigate the performance characteristics of the radiomics nomogram, calibration curves were graphically generated for both the training and test sets.
The discrimination performance of the different models
The area under the curve (AUC) of the receiver operating characteristics (ROC) curve was calculated for both the training set and test set, to assess the discrimination performance of the clinical model, radiomics signature,and radiomics nomogram. The sensitivity, specificity and accuracy of the three models were also calculated. Decision curve analysis (DCA) was conducted to estimate the clinical utility of the three models by quantifying their net benefits at different threshold probabilities when applied to the test set.
Statistical analysis
SPSS v.17.0 (SPSS Inc., Chicago, IL, USA) and R statistical software (v.3.3.3; https://www.r-project.org) were used for statistical analysis. A two-sided P-value of <0.05 was considered significant. The chi-square test and Fisher’s exact test were used for qualitative data, and the independent samples t-test was used for quantitative data. The R packages used were: (1) the “pROC” package for the ROC curves, (2) the “rms” package for the nomograms and calibration curves, (3) the “glmnet” package for LASSO logistic regression, and (4) the “rmda” package to perform the DCA. All the packages can be downloaded at https://cran.r-project.org/web/packages/.
Results
Clinical model construction
The characteristics of the patients in the training and test sets are detailed in Table 1. In the training set, gender, age, smoking history, growth pattern, lobulated appearance and parotid tail involvement showed significant differences between the WT and PMA groups. After multiple logistic regression analysis, only age (p < 0.001, odds ratio = 0.910, CI, 0.866 to 0.955) remained an independent predictor in the clinical model.
Table 1.
Clinical factors of the training and test sets
| Clinical factors | Training set (n = 75) | Test set (n = 52) | ||||
|---|---|---|---|---|---|---|
| WT (n = 34) |
PMA (n = 41) |
P1 | WT (n = 24) |
PMA (n = 28) |
P2 | |
| Gender (M/F) | 25/9 | 18/23 | 0.010 | 23/1 | 12/16 | <0.001 |
| Age, year | 59.21 ± 11.66 | 44.27 ± 13.80 | <0.001 | 53.04 ± 13.87 | 43.57 ± 10.54 | 0.007 |
| Smoking history (absent /present) | 16/18 | 29/12 | 0.037 | 5/19 | 24/4 | <0.001 |
| Maximum diameter, mm | 25.22 ± 6.05 | 26.43 ± 8.84 | 0.483 | 27.75 ± 11.86 | 26.79 ± 10.47 | 0.756 |
| Margin (well defined/ill defined) | 27/7 | 33/8 | 0.908 | 19/5 | 23/5 | 1.000 |
| Heterogeneous appearance (absent /present) | 14/20 | 24/17 | 0.134 | 15/9 | 15/13 | 0.516 |
| Growth pattern (focal/ multifocal) | 25/9 | 39/2 | 0.021 | 19/5 | 27/1 | 0.132 |
| Cystic or necrotic areas (absent /present) | 15/19 | 24/17 | 0.213 | 11/13 | 19/9 | 0.109 |
| Lobulated appearance (absent /present) | 20/14 | 14/27 | 0.033 | 18/6 | 13/15 | 0.036 |
| Parotid tail involvement (absent /present) | 13/21 | 26/15 | 0.030 | 4/20 | 22/6 | <0.001 |
F: female; M: male; PMA: pleomorphic adenoma; WT: Warthin’s tumour.
Numerical data are presented as mean± standard deviation, categorical data as numbers (n).
P1: the P-value of comparison between WT and PMA in training set; P2: the P-value of comparison between WT and PMA in test set.
Feature selection and development of the radiomics signature
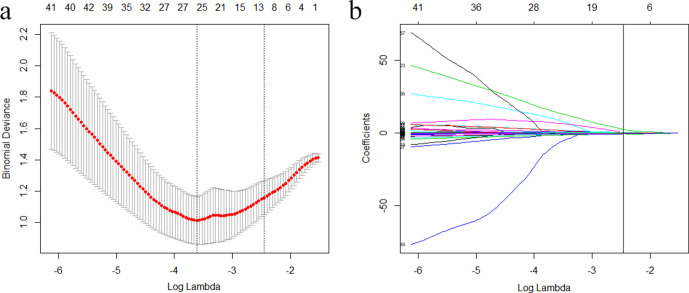
In total, 1521 features with satisfactory inter- and intraobserver agreements were included in further analysis (Supplementary Table 2). A total of 952 features showed significant differences between WT and PMA based on a one-way ANOVA applied to the training set. After application of the LASSO regression model, 12 features with non-zero coefficients were finally selected to build the radiomics signature with an optimal regulation weight λ of 0.086 under the 1-SE criterion (Figure 2, Table 2). The Rad-score was calculated using the following formula: Rad-score = −1.467 – (A × 0.459) + (B × 2.509) – (C × 0.179) + (D × 0.467) + (E × 0.144) – (F × 0.131) + (G × 0.029) – (H × 0.075) + (I × 0.067) – (J × 0.136) – (K × 0.568) – (L × 0.534). The variables A to L represent the selected radiomics features. There was a significant difference in the Rad-score between WT and PMA in the both training and test sets (Table 3).
Figure 2.
Selection of radiomics features via the least absolute shrinkage and selection operator (LASSO) regression algorithm. (a) Tuning parameter (λ) selection in LASSO model used tenfold cross-validation via 1-standard error criterion. The optimal values of the LASSO tuning parameter (λ) are indicated by the dotted vertical lines (the right one), and a value λ of 0.086 was chosen. (b) LASSO coefficient profiles of the 952 radiomics features. A coefficient profile plot was generated versus the selected log λ value using tenfold cross-validation. Twelve radiomics features with non-zero coefficients were finally selected.
Table 2.
Radiomics feature selection results
| Variables | Sequences | Radiomics feature name |
|---|---|---|
| A | T1WI | Correlation. GLCM. Wavelet HHL |
| B | fs-T2WI | Imc2. GLCM. Wavelet LLH |
| C | fs-T2WI | Kurtosis. First order. Original |
| D | fs-T2WI | Kurtosis. First order. Wavelet HLL |
| E | fs-T2WI | Correlation. GLCM. Wavelet LLH |
| F | fs-T2WI | Size Zone Non-Uniformity Normalized. GLSZM. Wavelet LLH |
| G | fs-T2WI | Cluster Shade. GLCM. Wavelet HLH |
| H | fs-T2WI | Cluster Shade. GLCM. Wavelet HHL |
| I | fs-T2WI | MCC. GLCM. Wavelet HHL |
| J | fs-T2WI | Cluster Shade. GLCM. Wavelet LLL |
| K | fs-T2WI | Gray Level Non-Uniformity Normalized. GLRLM. Wavelet LLL |
| L | fs-T2WI | Gray Level Non-Uniformity Normalized. GLSZM. Wavelet LLL |
GLCM: Gray level co-occurrence matrix; GLSZM: Gray level size zone matrix; GLRLM: gray level run length matrix; T1WI: T1-weighted images; fs-T2WI: fat-saturated T2 weighted images.
Table 3.
The results of Rad-score and Nomo-score in the training and test sets
| Training set (n = 75) | Test set (n = 52) | |||||
|---|---|---|---|---|---|---|
| WT (n = 34) |
PMA (n = 41) |
P1 | WT (n = 24) |
PMA (n = 28) |
P2 | |
| Rad-score | −0.457 ± 0.632 | 0.757 ± 0.530 | <0.001 | −0.395 ± 0.685 | 0.750 ± 0.605 | <0.001 |
| Nomo-score | −3.229 ± 2.927 | 2.866 ± 2.248 | <0.001 | −2.424 ± 2.627 | 2.900 ± 2.735 | <0.001 |
Nomo-score: nomogram score.; PMA: pleomorphic adenoma;Rad-score: radiomics score;WT: Warthin’s tumour.
Numerical data are presented as mean± standard deviation, categorical data as numbers (n).
P1: the P-value of comparison between WT and PMA in training set; P2: the P-value of comparison between WT and PMA in test set.
Radiomics nomogram construction
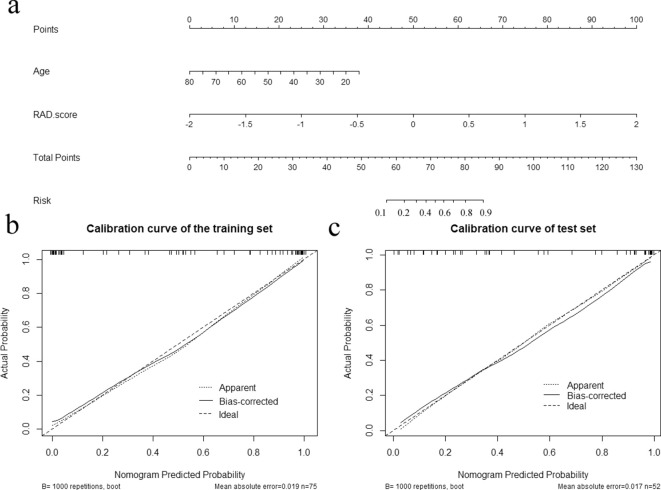
The radiomics nomogram was constructed incorporating age and Rad-score (Figure 3a). The Nomo-score for this nomogram was calculated using the following formula: Nomo-score = 3.942+(Rad-score ×3.898) – (Age × 0.091). The Nomo-score showed a significant difference between WT and PMA in both the training and test sets (Table 3), while the calibration curves showed that the radiomics nomogram had good calibration in both the training and test sets (Figure 3b and c).
Figure 3.
Developed radiomics nomogram and calibration curves for the radiomics nomogram. (a) Age and Rad-score were used for building the radiomics nomogram. Calibration curves for the radiomics nomogram in the training (b) and test (c) sets. Calibration curves indicate the goodness of fit of the nomogram. The 45° straight line represents the perfect match between the actual (Y-axis) and nomogram-predicted (X-axis) probabilities. A closer distance between two curves indicates higher accuracy
The discrimination performance of the different models
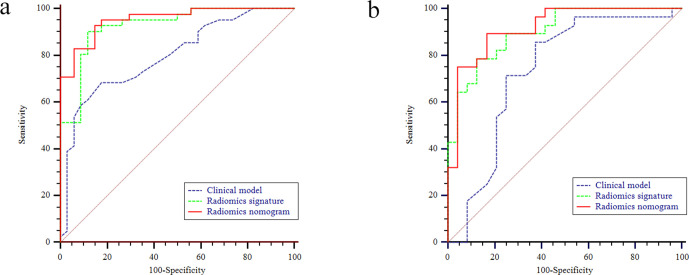
The discrimination performance of each model is shown in Table 4. The ROC curves of each model for both training and test sets are shown in Figure 4. For both training and test sets, the radiomics nomogram had better discrimination performance and a higher AUC than the clinical model (p < 0.001 in the training set; p = 0.009 in the test set).
Table 4.
Diagnostic performance of the clinical model, the radiomics signature and the radiomics nomogram
| Model | AUC (95% CI) | Sensitivity %* | Specificity %* | Accuracy %* |
|---|---|---|---|---|
| Training set (n = 75) | ||||
| Clinical model | 0.797 (0.689 to 0.881) | 68.29 (28/41) | 82.35 (28/34) | 74.67 (56/75) |
| Radiomics signature | 0.926 (0.842 to 0.974) | 90.24 (37/41) | 88.24 (30/34) | 89.33 (67/75) |
| Radiomics nomogram | 0.953 (0.878 to 0.989) | 92.68 (38/41) | 85.29 (29/34) | 89.33 (67/75) |
| Test set (n = 52) | ||||
| Clinical model | 0.734 (0.593 to 0.847) | 85.71 (24/28) | 62.50 (15/24) | 75.00 (39/52) |
| Radiomics signature | 0.902 (0.787 to 0.967) | 78.57 (22/28) | 87.50 (21/24) | 82.69 (43/52) |
| Radiomics nomogram | 0.918 (0.808 to 0.976) | 89.29 (25/28) | 83.33 (20/24) | 86.54 (45/52) |
CI: confidence interval;AUC: area under the curve
Numbers in parentheses were used to calculate percentages
Figure 4.
The receiver operating characteristic (ROC) curves of the clinical model, the radiomics signature and the radiomics nomogram in the training (a) and test (b) sets, respectively.
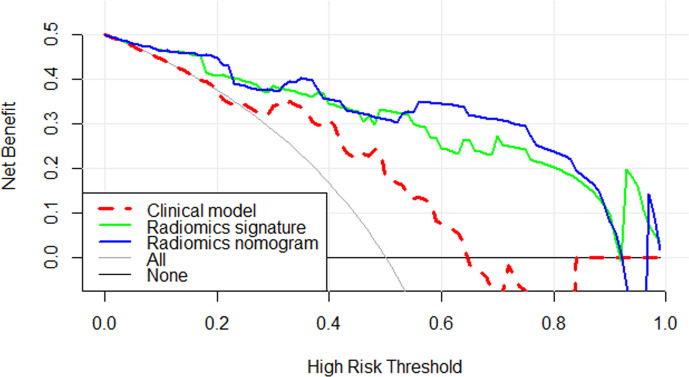
The DCA of the three models (Figure 5) indicates that for the differentiation of WT from PMA, the radiomics nomogram had a higher overall net benefit than the other models across the majority of the range of reasonable threshold probabilities. Therefore, we believe that the radiomics nomogram developed in our study could be a reliable clinical diagnostic tool for discriminating between WT and PMA.
Figure 5.
Decision curve analysis for three models. The y-axis indicates the net benefit; x-axis indicates threshold probability. The red-dotted line, green line and blue line represent net benefit of the clinical model, the radiomics signature and the radiomics nomogram, respectively. The radiomics nomogram had a higher overall net benefit in differentiating WT from PMA than the clinical model and simple diagnoses such as all PMA (gray line) or all WT patients (black line) across the full range of threshold probabilities at which a patient would be diagnosed as PMA. WT = Warthin’s tumour; PMA = Pleomorphic adenoma.
Discussion
The differentiation between WT and PMA is necessary because of their different biological behaviours and treatments. In the current study, we developed an MRI radiomics-based prediction nomogram for discriminating between WT and PMA. Our results show that this preoperative MRI-based radiomics nomogram had excellent performance for differentiating WT from PMA in both the training and test sets, with AUCs of 0.953 and 0.918, respectively. Furthermore, DCA showed that this nomogram was clinically useful.
It can be challenging to distinguish between WT and PMA on the basis of traditional CT and MR imaging, because the imaging characteristics of these two tumours are generally very similar.26 Our results showed that there were no significant differences between WT and PMA in regard to tumour size, margin, heterogeneous appearance and cystic or necrotic formation. Only a lobulated appearance was more common in the WT groups. Previous studies have reported that some demographic features are associated with WT, such as male predominance, higher incidence in the fifth and sixth decades of life and a smoking history.1,27,28 Furthermore, WT has a tendency to show multiplicity, bilaterality and parotid tail involvement.1,29 Our results are similar to those of previous studies, with the multiple logistic regression analysis showing age to be an independent predictor of WT. Our clinical model demonstrated that a parotid tumour in an old age patient was more likely to be WT than PMA.
Fine needle aspiration (FNA) biopsy has been increasingly used to obtain tissue for pathological analysis of parotid tumours.27 However, some disadvantages of FNA have been reported, such as facial nerve palsy and high rates of insufficient diagnostic aspirations.30–32 Recently, diffusion-weighted imaging (DWI) has been used to evaluate parotid gland tumours, with some studies demonstrating that DWI is of value for distinguishing PMA from WT.2,27,33,34 However, analyses in areas of tumour-related cyst formation, necrosis and haemorrhage were excluded from these studies. In the current study, the tumour segmentations included the entire tumour volume, without excluding any tumour areas.
Radiomics can non-invasively extract a high throughput of quantitative information from traditional medical images, making it possible to evaluate the internal textures of tumours that cannot be captured by visual assessment. Some previous studies incorporating radiomics or radiomics nomograms have reported good predictive values for the differential diagnosis of tumours.15,16,35–37 However, at the time of writing, only texture analysis has been applied to the diagnosis of parotid gland tumour. A previous study demonstrated that MRI texture analysis based on T1WI could be used to distinguish WT from PMA.10 Al Ajmi et al11 also found that multienergy CT texture analysis could quantitatively discriminate WT from PMA, with accuracy, sensitivity and specificity of 93%, 92% and 94%, respectively. In the current study, the radiomics nomogram model we created provided a comprehensive description of the tumours by extracting more statistical features than used in conventional texture analysis. Our results showed that this radiomics nomogram combining age and radiomics signature had good preoperative prediction performance. Additionally, we used an external validation method to validate the constructed nomogram in the current study, which is a stronger design for evaluating model performance.38
Some limitations of this study should be noted. First, the retrospective design of this study brings the potential for selection bias. A second limitation is the relatively small sample of external test data. Multicentre studies with larger samples are needed to further validate this nomogram. Third, the radiomics signature built in this study was only based on non-contrast-enhanced MRI. Further advanced MRI acquisitions such as contrast-enhanced T1WI or DWI are needed to achieve a high-level of evidence for clinical application.
Conclusion
In conclusion, we developed and validated a radiomics nomogram incorporating a radiomics signature and age. This nomogram is an effective tool to preoperatively and non-invasively distinguish WT from PMA and could optimize clinical decision-making.
Footnotes
Ethical Standards and Patient Consent: We declare that all human have been approved by the Institutional Review Board of the Affiliated Hospital of Qingdao University and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We declare that all patients gave informed consent prior to inclusion in this study.
The authors Ying-mei Zheng and Jiao Chen contributed equally to the work.
Contributor Information
Ying-mei Zheng, Email: luck1219@163.com.
Jiao Chen, Email: chenjiaocn@126.com.
Qi Xu, Email: xuqiniuniu@163.com.
Wen-hui Zhao, Email: 569360472@qq.com.
Xin-feng Wang, Email: Wangxinfeng666@163.com.
Ming-gang Yuan, Email: ymg13314613@126.com.
Zong-jing Liu, Email: lzj17853297156@126.com.
Zeng-jie Wu, Email: 15269263997@126.com.
Cheng Dong, Email: derc007@sina.com.
REFERENCES
- 1.Patel DK, Morton RP. Demographics of benign parotid tumours: Warthin’s tumour versus other benign salivary tumours. Acta Otolaryngol 2016; 136: 83–6. doi: 10.3109/00016489.2015.1081276 [DOI] [PubMed] [Google Scholar]
- 2.Liu Yi‐Jui, Lee Yi‐Hsiung, Chang Hing‐Chiu, Chung Hsiao‐Wen, Wang Chih‐Wei, Juan Cheng‐Hsuan, Liu YJ, Lee YH, Chang HC, Chung HW, Wang CW, Juan CH, et al. Imaging quality of propeller diffusion‐weighted MR imaging and its diagnostic performance in distinguishing pleomorphic adenomas from Warthin tumors of the parotid gland. NMR Biomed 2020; 33: e4282. doi: 10.1002/nbm.4282 [DOI] [PubMed] [Google Scholar]
- 3.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon SH, Kim J, Joung J-G, Cha H, Park W-Y, Ahn JS, et al. Correlations between metabolic texture features, genetic heterogeneity, and mutation burden in patients with lung cancer. Eur J Nucl Med Mol Imaging 2019; 46: 446–54. doi: 10.1007/s00259-018-4138-5 [DOI] [PubMed] [Google Scholar]
- 6.Choi E-R, Lee HY, Jeong JY, Choi Y-L, Kim J, Bae J, et al. Quantitative image variables reflect the intratumoral pathologic heterogeneity of lung adenocarcinoma. Oncotarget 2016; 7: 67302–13. doi: 10.18632/oncotarget.11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol 2016; 61: R150–66. doi: 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong AJ, Kanwar A, Mohamed AS, Fuller CD. Radiomics in head and neck cancer: from exploration to application. Transl Cancer Res 2016; 5: 371–82. doi: 10.21037/tcr.2016.07.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buch K, Fujita A, Li B, Kawashima Y, Qureshi MM, Sakai O. Using texture analysis to determine human papillomavirus status of oropharyngeal squamous cell carcinomas on CT. American Journal of Neuroradiology 2015; 36: 1343–8. doi: 10.3174/ajnr.A4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fruehwald-Pallamar J, Czerny C, Holzer-Fruehwald L, Nemec SF, Mueller-Mang C, Weber M, et al. Texture-based and diffusion-weighted discrimination of parotid gland lesions on Mr images at 3.0 Tesla. NMR Biomed 2013; 26: 1372–9. doi: 10.1002/nbm.2962 [DOI] [PubMed] [Google Scholar]
- 11.Al Ajmi E, Forghani B, Reinhold C, Bayat M, Forghani R. Spectral multi-energy CT texture analysis with machine learning for tissue classification: an investigation using classification of benign parotid tumours as a testing paradigm. Eur Radiol 2018; 28: 2604–11. doi: 10.1007/s00330-017-5214-0 [DOI] [PubMed] [Google Scholar]
- 12.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. JCO 2008; 26: 1364–70. doi: 10.1200/JCO.2007.12.9791 [DOI] [PubMed] [Google Scholar]
- 13.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015; 16: e173–80. doi: 10.1016/S1470-2045(14)71116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y-mei, Li J, Liu S, Cui J-fa, Zhan J-feng, Pang J, , Hao DP, Liu XJ, Gao CP, Tang GZ, et al. Mri-Based radiomics nomogram for differentiation of benign and malignant lesions of the parotid gland. Eur Radiol 2020; 266(Suppl 4. doi: 10.1007/s00330-020-07483-4 [DOI] [PubMed] [Google Scholar]
- 15.Nie P, Yang G, Wang Z, Yan L, Miao W, Hao D, et al. A CT-based radiomics nomogram for differentiation of renal angiomyolipoma without visible fat from homogeneous clear cell renal cell carcinoma. Eur Radiol 2020; 30: 1274–84. doi: 10.1007/s00330-019-06427-x [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Nie P, Wang Y, Xu W, Duan S, Chen H, et al. Radiomics nomogram for differentiating between benign and malignant soft‐tissue masses of the extremities. J Magn Reson Imaging 2020; 51: 155–63. doi: 10.1002/jmri.26818 [DOI] [PubMed] [Google Scholar]
- 17.Christe A, Waldherr C, Hallett R, Zbaeren P, Thoeny H. Mr imaging of parotid tumors: typical lesion characteristics in MR imaging improve discrimination between benign and malignant disease. AJNR Am J Neuroradiol 2011; 32: 1202–7. doi: 10.3174/ajnr.A2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RTH, Hermann G, et al. Ct-Based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiotherapy and Oncology 2015; 114: 345–50. doi: 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 2004; 22: 81–91. doi: 10.1016/j.mri.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 20.Tustison NJ, Avants BB, Cook PA, , Egan A, Yushkevich PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29: 1310–20. doi: 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depeursinge A, Foncubierta-Rodriguez A, Van De Ville D, Müller H. Three-Dimensional solid texture analysis in biomedical imaging: review and opportunities. Med Image Anal 2014; 18: 176–96. doi: 10.1016/j.media.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 22.Cook JA, Ranstam J. Overfitting. Br J Surg 2016; 103: 1814. doi: 10.1002/bjs.10244 [DOI] [PubMed] [Google Scholar]
- 23.Alhamzawi R, Ali HTM, . The Bayesian adaptive LASSO regression. Math Biosci 2018; 303: 75–82. doi: 10.1016/j.mbs.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Bian ZP, Hou J, Chau LP. Convolutional neural networks with dynamic regularization. IEEE Trans Neural Netw Learn Syst 2020;. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Lu G, Li J, Xu Y, Zhang Z, Zhang D. Multiscale conditional regularization for Convolutional neural networks. IEEE Trans Cybern 2020;: 1–15. doi: 10.1109/TCYB.2020.2979968 [DOI] [PubMed] [Google Scholar]
- 26.Motoori K, Yamamoto S, Ueda T, Nakano K, Muto T, Nagai Y. Inter- and intratumoral variability in magnetic resonance imaging of pleomorphic adenoma: an attempt to interpret the variable magnetic resonance findings. J Comput Assist Tomogr 2004; 28: 233–46. [DOI] [PubMed] [Google Scholar]
- 27.Wang C-W, Chu Y-H, Chiu D-Y, Shin N, Hsu H-H, Lee J-C, et al. Journal Club: the Warthin tumor score: a simple and reliable method to distinguish Warthin tumors from pleomorphic adenomas and carcinomas. American Journal of Roentgenology 2018; 210: 1330–7. doi: 10.2214/AJR.17.18492 [DOI] [PubMed] [Google Scholar]
- 28.Pinkston JA, Cole P. Cigarette Smoking and Warthin’s Tumor. Am J Epidemiol 1996; 144: 183–7. doi: 10.1093/oxfordjournals.aje.a008906 [DOI] [PubMed] [Google Scholar]
- 29.Maiorano E, Lo Muzio L, Favia G, Piattelli A. Warthin’s tumour: a study of 78 cases with emphasis on bilaterality, multifocality and association with other malignancies. Oral Oncol 2002; 38: 35–40. doi: 10.1016/S1368-8375(01)00019-7 [DOI] [PubMed] [Google Scholar]
- 30.Schmidt RL, Hall BJ, Wilson AR, Layfield LJ. A systematic review and meta-analysis of the diagnostic accuracy of fine-needle aspiration cytology for parotid gland lesions. Am J Clin Pathol 2011; 136: 45–59. doi: 10.1309/AJCPOIE0CZNAT6SQ [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y-F, Li H, Wang X-M, Cai Y-F. Sonoelastography for differential diagnosis between malignant and benign parotid lesions: a meta-analysis. Eur Radiol 2019; 29: 725–35. doi: 10.1007/s00330-018-5609-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang JH, Kim DW, Kim KS, Lee SY. Mucosa-Associated lymphoid tissue lymphoma of the accessory parotid gland presenting as a simple cheek mass. Medicine 2019; 98: e17042. doi: 10.1097/MD.0000000000017042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yabuuchi H, Matsuo Y, Kamitani T, Setoguchi T, Okafuji T, Soeda H, et al. Parotid gland tumors: can addition of diffusion-weighted MR imaging to dynamic contrast-enhanced MR imaging improve diagnostic accuracy in characterization? Radiology 2008; 249: 909–16. doi: 10.1148/radiol.2493072045 [DOI] [PubMed] [Google Scholar]
- 34.Yerli H, Agildere AM, Aydin E, Geyik E, Haberal N, Kaskati T, et al. Value of apparent diffusion coefficient calculation in the differential diagnosis of parotid gland tumors. Acta radiol 2007; 48: 980–7. doi: 10.1080/02841850701501717 [DOI] [PubMed] [Google Scholar]
- 35.Li L, Wang K, Ma X, Liu Z, Wang S, Du J, et al. Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur J Radiol 2019; 118: 81–7. doi: 10.1016/j.ejrad.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 36.Nie P, Yang G, Guo J, Chen J, Li X, Ji Q, et al. A CT-based radiomics nomogram for differentiation of focal nodular hyperplasia from hepatocellular carcinoma in the non-cirrhotic liver. Cancer Imaging 2020; 2020(1):20. doi: 10.1186/s40644-020-00297-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng B, Chen X, Chen Y, Li Z, Hao Y, Zhang C, et al. Differentiating minimally invasive and invasive adenocarcinomas in patients with solitary sub-solid pulmonary nodules with a radiomics nomogram. Clin Radiol 2019; 74: 570.e1–570.e11. doi: 10.1016/j.crad.2019.03.018 [DOI] [PubMed] [Google Scholar]
- 38.Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015; 162: W1–73. doi: 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.