Abstract
Introduction
Although treatment with osimertinib confers survival benefits in patients with lung cancer with the EGFR T790M mutation, the mechanism of acquired resistance to osimertinib remains poorly understood. We conducted a prospective observational study to identify the mechanism on the basis of repeated tissue biopsies.
Methods
Patients with EGFR-mutated advanced lung cancer with a T790M mutation detected on a tissue biopsy underwent a rebiopsy after developing acquired resistance to osimertinib. Nucleic acids extracted from the biopsy samples were subjected to targeted resequencing (Oncomine Comprehensive Assay), and circulating cell-free DNA (ccfDNA) was analyzed by CAncer Personalized Profiling by deep Sequencing (AVENIO ctDNA Surveillance Kit).
Results
Between November 2016 and March 2020, a total of 87 patients were screened. Among them, 44 developed acquired resistance. Of these, 19 samples from rebiopsies and 12 from preosimertinib biopsies were able to be analyzed by an Oncomine Comprehensive Assay. A ccfDNA analysis was performed in 16 patients. Regarding the mechanisms of acquired resistance, structural change in EGFR, namely, C797S, G796S, or L792V, was the most frequent alteration, being observed in 57.9% of the cases. MET gain was observed in 31.6% of the cases, and gains in cell cycle genes were observed in 26.3% of the cases. In addition, we identified GAS6 gain and an ATM mutation in a patient with small-cell transformation and a BRAF V600E mutation in a patient with oligoprogressive disease.
Conclusions
A repeated tissue biopsy and a ccfDNA analysis were useful in analyzing the mechanisms underlying acquired resistance. A long treatment history of EGFR TKIs may result in a high percentage of EGFR structural change.
Keywords: Osimertinib, Targeted resequencing, Acquired resistance, EGFR T790M
Introduction
Lung cancer is a leading cause of cancer death, with 1.76 million cases worldwide and 74,300 cases in Japan.1,2 Recent advances in precision medicine using genetic profiling of tumors have enabled patients with advanced lung cancer with driver mutations to survive for more than 1 year.3, 4, 5, 6, 7 EGFR tyrosine kinase inhibitors (TKIs) have been developed for patients with advanced lung cancer with EGFR-activating mutations, which are the most frequent mutations, being found in up to 55% of Japanese and 15% of non-Asian patients with lung adenocarcinoma.8
In 2005, the EGFR T790M mutation was identified as a mechanism of acquired resistance to first-generation EGFR TKIs,9 and osimertinib was subsequently developed to target the EGFR T790M mutation. Osimertinib targets both T790M and sensitive mutations, but it has minimal sensitivity to wild-type EGFR.10,11 Furthermore, osimertinib has substantial benefit not only for patients with advanced lung cancer with the EGFR T790M mutation12, 13, 14 but also for EGFR TKI-naive patients in clinical trials.15
Although a repeated tissue biopsy has been the most effective and reliable procedure for clarifying mechanisms underlying resistance and deciding on consequent proper treatment, performing biopsy on heavily treated tumors can be challenging, owing to the fibrous changes in the tumor microenvironment.16
In this study, we explored the mechanisms underlying acquired resistance to second-line osimertinib in patients with lung cancer with EGFR T790M mutations through repeated tissue biopsies. We also considered the benefits of a circulating cell-free DNA (ccfDNA) analysis in this setting.
Materials and Methods
Patients
Patients with advanced NSCLC with an EGFR mutation who developed EGFR T790M after treatment with first- or second-generation EGFR TKIs were included. Hematoxylin and eosin (HE)-stained slides, prepared from the same formalin-fixed, paraffin-embedded (FFPE) blocks from which the T790M mutation had been detected through routine clinical practice usually with multiplex polymerase chain reaction, were screened centrally by pathologists (KTag and KN) to determine whether or not the samples were adequate for next-generation sequencing (NGS) analyses. Cases were registered in the study when the tissue area on the HE slide exceeded 5 mm2 and the tumor cell counts exceeded 30% of the tissue. Cell blocks from fluid samples were allowed if they met the above-mentioned criteria. Patients were excluded if they were EGFR TKI-naive or had already been treated with osimertinib or if the patients’ EGFR T790M was proven only by a liquid biopsy. The planned sample size was 80 for this prospective observational biomarker study, with 20 patients assumed to receive a successful rebiopsy. The study (LOGIK1607) was conducted at 25 institusions in the Lung Oncology Group in Kyushu (LOGIK), and was organized by the Clinical Research Support Center Kyushu (CReS-Kyushu), Fukuoka, Japan.
Rebiopsy and Sample Collection
The registered patients were prospectively followed during osimertinib treatment, and a rebiopsy was planned once the patient developed progressive disease after a durable response (acquired resistance) to osimertinib. The definition of acquired resistance was reported previously.17 The rebiopsy was performed after receiving written informed consent and before the next systemic therapy was implemented for lung cancer. Biopsy on progressing lesions was done on a physician’s decision. The HE slides of the rebiopsy specimen were checked centrally again by the pathologists. The paired unstained slides (from one block for which EGFR T790M had been proven before treatment with osimertinib and from another block obtained by a rebiopsy, both mandatory), at least 10 slides per specimen, were then collected for an NGS analysis. Corresponding EGFR TKI-naive unstained slides were also collected in cases in which tissue samples obtained at the diagnosis (before any EGFR TKI treatment) were available (optional). Plasma samples from 10 mL of blood were also collected at the same time as the rebiopsy and subjected to a ccfDNA analysis.
NGS of FFPE Tissue Specimens and Plasma Samples
Nucleic acids were extracted from the FFPE tumor sections and plasma samples, and an NGS analysis for tissues with ion PGM using an Oncomine Comprehensive Assay (OCA), version (v) 1 or v3 (since September 2018) (Thermo Fisher Scientific, Waltham, MA), was performed at an institution certified with the Clinical Laboratory Improvement Amendments (SRL Inc., Tokyo, Japan). The OCA panel consisted of a DNA and RNA panel capable of detecting 143 (v1) or 161 (v3) genetic alterations, including single-nucleotide variations (SNVs), indels, copy number variations (CNVs), and fusions. The Torrent Suite software program (Thermo Fisher Scientific) was used for the data alignment, and binary alignment and matching files were analyzed using the ionReporter 5.12 software program (Thermo Fisher Scientific). Filters equipped in ionReporter 5.12, such as Oncomine extended 5.12, Confident Somatic CNVs—CNVs only 5.12, and Default Fusions View 5.12, were used to filter SNVs, CNVs, and fusions, respectively. Filtered SNVs were visually inspected with an integrated genomic viewer to exclude amplicon errors. The integrated genomic viewer was also used to examine whether or not the allelic location of EGFR T790M was cis or trans to the C797S, G796S, or L792V mutation. The ccfDNA analysis was performed with NextSeq (Illumina, San Diego, CA) using the AVENIO ctDNA Surveillance Kit (Roche Diagnostics, Basel, Switzerland) at Kindai University (KS and KN). The panel was developed to detect 197 genetic alterations, including SNVs and copy number gains, using the CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) technology and its algorithm to quantify small numbers of reads while suppressing amplicon errors.18,19 SNVs found at a rate exceeding 0.01% from the Exome Aggregation Consortium, 1000 Genomes Project, Single Nucleotide Polymorphism Database, or Human Genetic Variation Database were excluded to filter out possible germline mutations.
Statistical Analyses
Statistical significance between groups was determined using Fisher’s exact test and the Mann-Whitney U test. Wilcoxon’s signed rank test with continuity correction was used for pre- and postcomparisons within the groups. A Muller plot was generated from variant allele frequency data of nonsynonymous mutations detected at each time point using the MullerPlot, R package version 0.1.2, to reveal population and frequency dynamics of mutations in case number 9. p values less than 0.05 were considered to indicate statistical significance.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol (UMIN00002529020) was approved by a central institutional review board (16-E11, Clinical Research Network Fukuoka, Fukuoka, Japan) or local institutional review boards, depending on the regulation of each institution. Written informed consent was obtained from all patients. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Results
Cases Analyzed
Between November 2016 and March 2020, a total of 87 patients from 25 institutions were registered. Among the 72 eligible patients, 23 were still on osimertinib, and 44 had developed acquired resistance. The remaining five patients experienced disease progression before meeting the criteria of a durable response. Among the 44 patients who developed acquired resistance, 21 underwent a rebiopsy (Fig. 1). There were no reported complications from the rebiopsy. Furthermore, 20 paired tissue samples from rebiopsies and preosimertinib (baseline) biopsies were subjected to an OCA, and 19 samples (95%) from rebiopsies and 12 samples (60%) from baseline biopsies were able to be analyzed by the OCA. The DNA analysis was not successful in one case (case number 20), so that case was not included in the analysis result. A ccfDNA analysis was performed for all 16 patients who agreed to provide a plasma sample (Table 1). Optional analyses performed on the tissue biopsy sample obtained before any EGFR TKI treatment were successful in two of the six samples collected (33%, data not found). Nonsynonymous mutations with the Catalogue of Somatic Mutations in Cancer identifiers and CNVs of interest from each case are also summarized in Table 1. Filtered variant call format (VCF) data are available in Supplementary Tables 1 to 3.
Figure 1.
Flow diagram of the study. A total of 87 patients from 25 institutions were screened. A total of 19 patients with paired tumor tissue biopsy samples were analyzed. HE, hematoxylin and eosin; Mar., March; PD, progression disease.
Table 1.
Biopsy Data, Treatment History of EGFR TKIs, and Representative Genetic Alterations From Individual Cases
| Case | EGFR TKI(s) Used Before Osimertinib |
Osimertinib Duration (m) | Tissue Biopsy |
OCA Ver. | Representative Genetic Alterations at Rebiopsy (Tissue) |
ccf DNA (Plasma) | Representative Genetic Alterations at Rebiopsy (Plasma) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EGFR TKI(s) | Duration (m) | Baseline | Rebiopsy | EGFR Common | T790M | C797S, etc. | SNV | CN Gain | EGFR Common | T790M | C797S, etc. | SNV | CN Gain | ||||
| 1 | A | 15.4 | 11.6 | DR | DR | 1 | Del19 | + | C797Sa | D | Del19 | + | C797S | KEAP1 | EGFR, MET | ||
| 2 | G, E | 27.1 | 11.1 | DR | DR | 1 | Del19 | + | C797S | NF1, GATA3 | EGFR | D | Del19 | + | C797S | KRAS | EGFR, MET |
| 3 | A | 20.6 | 12.6 | DR | DR | 1 | Del19 | + | C797S | EGFR | D | Del19 | + | C797S | ERBB2 | EGFR, MET | |
| 4 | G | 46 | 28.6 | DR | DR | 1 | L858R | + | C797S | EGFR | D | L858R | + | C797S | EGFR | ||
| 5 | A | 40.7 | 29.1 | DR | DR | 3 | Del19 | + | C797S | MYCN | D | Del19 | |||||
| 6 | G, BE | 18.9 | 17.6 | DR | DR | 1 | L858R | + | G796S | D | L858R | ||||||
| 7 | G | 31.6 | 19.8 | DR | DR | 1 | L858R | + | C797S | CDK6 | NA | ||||||
| 8 | G | 24.2 | 13.4 | D | DR | 1 | L858R | EGFR, MET, CDK4 | D | L858R | EGFR, MET | ||||||
| 9 | G | 24.4 | 8.8 | D | DR | 3 | ND | BRAF, IDH2 | D | ND | + | ||||||
| 10 | G, E, A | 25.3 | 19.4 | DR | DR | 3 | Del19 | CDK4b | D | Del19 | |||||||
| 11 | G, BE | 24.7 | 18.5 | DR | DR | 1 | L858R | NA | |||||||||
| 12 | G, E, A, E, A | 25.2 | 12.6 | DR | DR | 1 | L858R | ATM | GAS6 | D | L858R | EGFR | |||||
| 13 | G, A | 18.6 | 19.8 | X | DR | 3 | Del19 | + | C797Sc | PTENd | D | Del19 | + | C797S | EGFR, MET | ||
| 14 | G | 7.6 | 22.9 | X | D | 1 | Del19 | + | C797S | CTNNB1, SMAD4d | D | Del19 | + | C797S | |||
| 15 | G, E | 19.4 | 24.3 | X | DR | 3 | L858R | + | L792V | EGFR, CDK4d | D | L858R | + | L792V | EGFR | ||
| 16 | G | 11.7 | 17 | R | DR | 3 | Del19 | + | C797S | CDKN2A, IDH2d | NA | ||||||
| 17 | S | 27.9 | 18.2 | X | DR | 3 | Del19 | + | D | Del19 | + | EGFR, MET | |||||
| 18 | E, INV | 26.1 | 23.7 | X | DR | 1 | Del19 | + | PIK3CAd | D | Del19 | + | ERBB2 | ||||
| 19 | E | 35.4 | 19.1 | X | DR | 1 | Del19 | PIK3CAd | CDK4, CCND1d | D | Del19 | ||||||
| 20 | G | 33.1 | 23.1 | R | R | 1 | NA | ||||||||||
A, afatinib; BE, bevacizumab + erlotinib; ccfDNA, circulating cell-free DNA; CN, copy number; D, DNA; Del19, exon 19 deletion; DR, DNA and RNA; E, erlotinib; G, gefitinib; INV, investigational agent; NA, not available; ND, not detected; OCA, Oncomine Comprehensive Assay; R: RNA; SNV, single nucleotide variant; Ver., version; X: no data.
Cis/trans to T790M.
Also detected in baseline data.
Trans to T790M.
Possible mechanism, baseline data not available.
The background characteristics of the 19 cases analyzed were as follows (Table 2): seven males and 12 females, median age 66 years old (range: 42–84 y old), and exon 19 deletion (Del19) in 12 patients and L858R in seven patients. The EGFR TKIs used before osimertinib were gefitinib in 13 patients, erlotinib in eight patients, and afatinib in seven patients. Eight patients received more than one EGFR TKI. The median duration on osimertinib was 18.5 months (ranged from 8.8 to 29.1 mo), and the cumulative duration on any EGFR TKI before osimertinib (median) was as long as 24.7 months (ranged from 7.6 to 46 mo).
Table 2.
Patients’ Characteristics
| Factors | N |
|---|---|
| Sex | |
| Male | 7 |
| Female | 12 |
| Age (y), median (range) | 66 (42–84) |
| Smoking history | |
| Positive | 6 |
| EGFR mutation before EGFR TKI | |
| Del19 | 12 |
| L858R | 7 |
| Median duration on osimertinib, mo (range) | 18.5 (8.8–29.1) |
| TKI(s) before osimertinib | |
| Gefitinib | 13 |
| Erlotinib | 8 |
| Afatinib | 7 |
| Received more than one TKI before osimertinib | 8 |
| Cumulative duration on TKI(s) before osimertinib | 24.7 (7.6–46) |
Del19, exon 19 deletion; TKI, tyrosine kinase inhibitor.
EGFR Alterations and Relationship With Clinical Background Factors
All 12 patients had original EGFR-sensitizing mutations and T790M mutations at the baseline (preosimertinib) biopsy. Of 19 patients, 18 maintained the original EGFR-sensitizing mutation at rebiopsy, and 13 still carried the T790M mutation, whereas the other five lost the T790M mutation. One patient who did not have a sensitizing mutation either in the rebiopsy tissue or in the ccfDNA had T790M only from the ccfDNA analysis. This patient was included in the T790M-maintained group (total 14 patients, 73.7%). Additional EGFR mutations, such as C797S, G796S, and L792V, were found in those patients who maintained T790M mutations. C797S was found in nine patients, and G796S or L792V was found in one patient each, resulting in 11 patients who developed additional EGFR mutations that caused structural changes in the EGFR protein (57.9%). The allelic locations of these mutations in relation with T790M were cis in nine patients, trans in one patient, and cis and trans in one patient. Additional EGFR mutations of C797S, G796S, and L792V were not found in the five patients who lost T790M (Table 1). Baseline biopsy samples were available in three of five patients who developed innate resistance. Among these three patients, the OCA analysis was successful for two samples at baseline, in which only EGFR-sensitizing mutations, not the T790M mutation, were detected (data not found).
Several studies have reported that the existence of EGFR T790M is more frequent in patients with EGFR exon 19 deletion (Del19) than in those with EGFR L858R and is associated with a longer treatment time with EGFR TKIs.21 It is also reported that the tumor mutational burden is related to the treatment efficacy of EGFR TKIs.22 Therefore, we compared the patient background characteristics between EGFR C797S, G796S, or L792V positivity and negativity. Other than the fact that EGFR C797S, G796S, or L792V only arose from patients in the T790M-maintained group, there were no significant differences regarding patients’ sex, age, duration of previous EGFR TKI treatment, duration of osimertinib treatment, or number of concurrent genetic alterations (nonsynonymous mutation and CNV) (Table 3 and Fig. 2A).
Table 3.
Relationship Between Patients’ Background and C797S Status at Re-bx
| Factors | C797S+ (Range) | C797S− (Range) | p Value | ||
|---|---|---|---|---|---|
| EGFR T790M | |||||
| Positive | 11 | 78.6% | 3 | 21.4% | |
| Negative | 0 | 0.0% | 5 | 100% | 0.01 |
| EGFR common mutation | |||||
| Del19 | 7 | 58.3% | 5 | 41.7% | |
| L858R | 4 | 57.1% | 3 | 42.9% | NS |
| CN gain | |||||
| EGFR | 6 | 66.7% | 3 | 33.3% | NS |
| MET | 4 | 66.7% | 2 | 33.3% | NS |
| Cell cycle | 2 | 40.0% | 3 | 60.0% | NS |
| Age | |||||
| Median | 66 | (42–82) | 68 | (60–84) | NS |
| Sex | |||||
| Male | 3 | 42.9% | 4 | 57.1% | |
| Female | 8 | 66.7% | 4 | 33.3% | NS |
| TKI durations (mo) | |||||
| Osimertinib | 19.8 | (11.1–29.1) | 18.35 | (8.8–23.7) | NS |
| Before osimertinib | 19.4 | (7.6–46) | 25.25 | (24.2–35.4) | NS |
| TKI used | |||||
| 1 | 7 | 63.6% | 4 | 36.4% | |
| 2< | 4 | 50.0% | 4 | 50.0% | NS |
| Number of other mutations | |||||
| Tissue at re-biopsy | 4 | (0–9) | 3.5 | (1–8) | NS |
| ccfDNA after re-biopsy | 4 | (2–9) | 5 | (2–17) | NS |
Note: Including G796S and L792V.
ccfDNA, circulating cell-free DNA; CN, copy number; Del19, exon 19 deletion; mut, mutation; NS, not significant; TKI, tyrosine kinase inhibitor.
Figure 2.
Co-occurring mutations were plotted. Nonsynonymous mutations in all 19 cases. Recurrent mutations observed more than once were plotted. (A) The combined result from OCA and CAPP-Seq is illustrated in a cumulative bar plot according to the presence or absence of an EGFR C797S mutation. (B) A comparison of nonsynonymous mutation count between the baseline and rebiopsy is plotted from the data of 12 patients for whom paired tissue biopsy samples were available. The OCA results are revealed in a cumulative bar plot according to the mutation appearance. The gray bar indicates mutations observed only from a biopsy sample obtained before osimertinib treatment, and the white bar indicates the mutation observed only in a sample obtained after acquiring resistance to osimertinib. (C) The black bar indicates that the mutation was observed in both instances. A Muller plot generated using data from tissue biopsies of case # 9 reveals that BRAF V600E arose as a new clone with an NF1, IDH2, and MTOR mutations at the rebiopsy, whereas both EGFR Del19 and T790M disappeared from the tumor. SF3B1 and ATM mutations were omitted from the plot because the VAFs of those mutations were unchanged throughout the clinical course. †Case #2, ¶case #9, ‡case #5. #, number; CAPP-Seq, CAncer Personalized Profiling by deep Sequencing; Del19, exon 19 deletion; OCA, Oncomine Comprehensive Assay; TKI, tyrosine kinase inhibitor; VAF, variant allele frequency.
Co-Occurring Mutations
Nonsynonymous mutations, aside from EGFR alterations, were found in all cases with acquired resistance to osimertinib. The TP53 mutation was the most frequently observed co-alteration and was found in 11 patients (57.9%), followed by mutations in NOTCH1 (eight patients, 42.1%), CTNNB1, RB1, and NF1 (four patients each, 21.1%) (Fig. 2A). The number of mutations was higher in baseline biopsy samples (ranged from 1 to 21, median = 4.5) than in rebiopsy samples obtained after acquiring resistance to osimertinib (ranged from 0 to 9, median = 4, p = 0.022; Fig. 2B). Many of those mutation had already been present in the baseline biopsy samples. Among them, mutations in PIK3CA and the ATM signaling pathway, both of which have been reported as negative predictors of EGFR TKI treatment efficacy,23 were observed independent of C797S, G796S, or L792V mutation, suggesting independent mechanisms of resistance to osimertinib treatment. Some of those mutations, such as NF1, ATM, BRAF, GATA3, NADP(+) (IDH)2, MTOR, and MYCN, were earned after the tumor acquired resistance to osimertinib and arose simultaneously in several individual patients (Fig. 2B).
A BRAF V600E mutation was found in one patient from a rebiopsy sample whose EGFR mutation had only been found by ccfDNA at the rebiopsy. Although the patient had EGFR Del19 at the baseline, it was not detected from either the rebiopsy sample or the ccfDNA; only EGFR T790M was detected in ccfDNA. This patient is still on osimertinib treatment for 10 months after having received surgical resection for a solitary progressive lesion, in which a BRAF V600E mutation had been detected (case number 9; Table 1 and Fig. 2C and D).
Copy Number Variations
The copy number gain of MET, which was the first reported mechanism of resistance to EGFR TKI,24 was found in six cases after resistance to osimertinib had been acquired. Furthermore, all patients with the MET gain exhibited EGFR copy number gain, and four also had a C797S mutation. ERBB2 gain was observed in one case who had lost EGFR T790M (Table 1).
Copy number gain of genes related to the cell cycle, CDK4, CDK6, and CCND1, was detected in five cases with acquired resistance to osimertinib. Among them, one case already had CDK4 gain before osimertinib treatment, resulting in four cases possibly having copy number gain in cell cycle genes that might be related to a mechanism underlying acquired resistance to osimertinib (Table 1).
A copy number gain of GAS6, a ligand of AXL, was detected in the patient with small-cell transformation (Table 1). Although this patient had no evidence of genetic alterations in TP53 or RB1, ATM Q2615R was acquired after the patient developed resistance to osimertinib.
Discussion
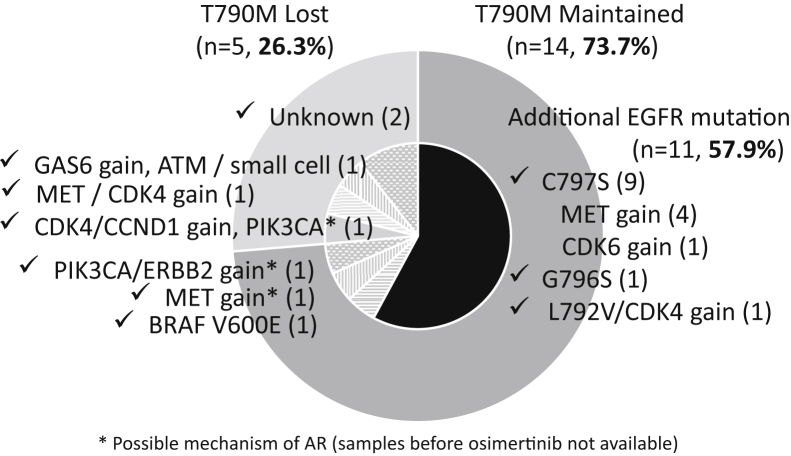
The overview of acquired resistance mechanisms to osimertinib in this study is presented as a pie chart (Fig. 3). Structural change in EGFR, namely, C797S, G796S, or L792V, was the most frequent alteration, observed in 57.9% of the cases. MET gain was the second-most frequent alteration, observed in 31.6% of the cases. Gains in cell cycle genes, CDK4, CDK6, and CCND1, were observed in 26.3% of the cases. ERBB2 gain and the PIK3CA mutation were also potential mechanisms of acquired resistance. In addition, we identified GAS6 gain and an ATM mutation in a patient with small-cell transformation and BRAF V600E mutation in a patient with oligoprogressive disease.
Figure 3.
The mechanisms of AR to osimertinib. The overview of AR mechanisms to osimertinib is presented as a pie chart. Structural change in EGFR was the most frequent alteration, observed in 57.9% of the cases. MET gain was the second-most frequent alteration, observed in 31.6% of the cases. Gains in cell cycle genes, CDK4, CDK6, and CCND1, were observed in 26.3% of the cases. ERBB2 gain and the PIK3CA mutation were also potential mechanisms of AR. In addition, we identified GAS6 gain and an ATM mutation in a patient with small-cell transformation and BRAF V600E mutation in a patient with oligoprogressive disease. ∗Possible mechanism of AR (samples before osimertinib not available). AR, acquired resistance.
The main mechanisms of acquired resistance to first- and second-generation EGFR TKIs are as follows: (1) an EGFR-dependent mechanism, represented by the EGFR T790M mutation, which inhibits first- and second-generation EGFR TKIs to compete adenosine triphosphate binding to EGFR-sensitive mutation9; (2) bypass track alteration, in which cancer cells use alternate pathways to bypass the EGFR pathway, for example, MET or HER2 amplification24,25 and PTEN deleted from chromosome 10 down-regulation26,27; and (3) morphologic transformation, in which cancer cells have morphologic changes, such as small-cell transformation or epithelial-mesenchymal transformation.28
Recent studies have identified mechanisms of acquired resistance to osimertinib. When osimertinib is administered to EGFR T790M-positive patients, the additional EGFR mutation C797S is the most frequently found, being detected in 20% to 30% of patients.29, 30, 31 In contrast, when osimertinib is used in the first-line setting, the C797S mutation is less common, and various other mechanisms, including the KRAS mutation or fusion driver alterations, are identified.32, 33, 34 In this study, there were more cases with EGFR-dependent resistance than were found in previous reports. Because the EGFR T790M mutation is thought to be a result of a durable response to first- and second-generation EGFR TKIs,35 a C797S mutation may also be a result of a durable response to osimertinib.29 Indeed, the patients included in this study had been receiving osimertinib for a relatively long time. Furthermore, all C797S mutation, including G796S, and L792V mutations were detected along with the T790M mutation, and in many cases, they occurred cis to T790M. This suggests that the C797S mutation arises more often from T790M mutations than wild types, when tumors with EGFR T790M are still dependent on the EGFR pathway.36,37
Coexisting genetic alterations are also important when considering the resistance mechanisms. In a list of recurrent genetic alterations from all 19 patients, NOTCH1 and CTNNB1 mutations were relatively frequently found, being noted in 8 and 4 patients, respectively (Fig. 2A). Among the 12 patients with paired biopsy data available, six already had NOTCH1 mutation, and four already had CTNNB1 mutation before osimertinib treatment (three had overlapping mutations, Fig. 2C). Recently, genetic alterations related to NOTCH and WNT, β-catenin signaling were reported to play an important role in EGFR TKI resistance. EGFR-mutant proteins, especially T790M mutant, bind to phosphorylated β-catenin and are reportedly stabilized by EGFR TKI treatment.38,39 STAT3, located downstream of NOTCH signaling, is also phosphorylated by EGFR TKI treatment,40 which induces a stem cell-like phenotype or epithelial-to-mesenchymal transition phenotype. These findings indicate that pre-existing alterations in NOTCH and CTNNB1 might be involved in resistance mechanisms to EGFR TKI treatment. In fact, both patients whose resistance mechanisms were unknown (T790M lost; Fig. 3) exhibited NOTCH or CTNNB1 mutations in their tissue before osimertinib treatment. Inactivation of NF1 is also implicated in both intrinsic and acquired resistance to EGFR TKI by activating the downstream signaling of RAS-MEK-ERK.41
Similar to previous studies, MET gain, ERBB2 gain, and PIK3CA mutation30,42,43 were also observed in this study. Copy number gain in CDK4 and CDK6, which has also been reported to be related to osimertinib resistance, was found in one-fourth of the cases.44
Small-cell transformation has also been reported as a mechanism of acquired resistance to osimertinib, often accompanied by RB1 and TP53 alterations.30,45 We failed to detect either RB1 or TP53 alteration in the present study, but we did find an ATM mutation and GAS6 copy number gain in the patient with small-cell transformation. Patients with small-cell transformation respond to chemotherapy directed at SCLC but not to the extent found in patients with actual advanced small-cell lung cancer. Therefore, adding molecular-targeted therapy (e.g., ATM inhibitor and AXL inhibitor) to chemotherapy may be an option that should be tested in a clinical setting.
Overall, NGS using OCA panel from re-biopsied sample was feasible and useful to analyze resistance mechanisms, although some samples were too old to analyze because they had been obtained by a biopsy several years ago. Furthermore, a ccfDNA analysis using CAPP-Seq covered almost all meaningful genomic alterations, such as SNV and CNV gain. The CAPP-Seq technique is one of the most promising techniques for detecting genomic alterations from a small fraction of genomic DNA.18 This technique is able to suppress sequencing errors while maximizing mutation detection, which has made it possible to detect somatic mutations in less than 0.02% of DNA.19 Because the spatial heterogeneity of the tumor is one of the main mechanisms underlying resistance to EGFR TKI,46,47 a ccfDNA analysis outperforms a tissue biopsy with respect to detecting somatic mutations from various lesions of a patient’s body. Our finding that the number of nonsynonymous mutations was smaller in the rebiopsy samples than in the baseline samples also supports the idea of clonal selection by osimertinib therapy.
A discrepancy was noted in the present study between the tissue and liquid biopsy findings in one patient with an EGFR T790M and G796S mutations. In that patient, neither the EGFR T790M mutation nor the G796S mutation was detected from the ccfDNA analysis. Nevertheless, because the allele frequency of EGFR L858R in the ccfDNA of that patient was as low as 0.46%, this result may have merely been a false negative caused by the quantity of ccfDNA derived from osimertinib-resistant cells being too low to evaluate properly.
EGFR-sensitizing mutations Del19 and L858R were found in all but one patient by both a tissue biopsy and ccfDNA analysis. The exceptional case had oligoprogression disease that was being treated by surgery and was still on osimertinib treatment. EGFR mutations in ccfDNA reportedly fall below the limit of detection soon after the initiation of EGFR TKIs.48 Considering the tumor burden the exceptional case had, it is not surprising that an EGFR T790M mutation was barely detected in ccfDNA (allele frequency: 0.10%). Because this patient had a BRAF V600E mutation, which has been reported to be a rare mechanism of osimertinib resistance,49 a therapeutic strategy targeting BRAF (e.g., dabrafenib and trametinib) may be a treatment option for this patient. Considering the technical difficulty and invasiveness of a rebiopsy, ccfDNA may come to replace a tissue biopsy when evaluating the mechanisms underlying resistance and deciding on further therapy.
Several ongoing clinical trials are attempting to inhibit acquired resistance to osimertinib. For example, the EGFR C797S mutation can be overcome by mutant-selective allosteric inhibitors,50,51 and more interestingly, by first-generation TKIs. Combination treatment with osimertinib and gefitinib in EGFR TKI-naive patients is ongoing (NCT03122717). Because of the heterogeneity in the mechanisms of resistance to osimertinib, a biomarker-directed phase 2 study is ongoing (NCT03944772). This study includes an umbrella trial for known resistant mechanisms allocating savolitinib for MET amplification and necitumumab for EGFR amplification. More recently, the fourth-generation EGFR TKI BLU-945 was developed to target triple-mutant EGFR harboring either the activating L858R or Del19 mutations combined with the acquired T790M and C797S mutations; it was reported to have antitumor activity in vivo.52 The publication of the results of these studies is awaited.
The major limitation of this study is the small number of cases analyzed (n = 19), which is caused by the difficulty of performing a tissue biopsy for heavily treated patients with lung cancer. To resolve this issue, a ccfDNA analysis may be useful. Furthermore, because a tissue biopsy requires additional efforts to be made before advancing to the next treatment approach, it is difficult to perform a tissue biopsy in patients with rapid disease progression. This may have resulted in patients who undertook a biopsy include a series of patient with slow progression of disease. Indeed, many patients had a long treatment history of EGFR TKIs and had received osimertinib for a long time.
In conclusion, we investigated the mechanisms underlying acquired resistance to osimertinib in patients with EGFR T790M mutations from paired tissue biopsies. EGFR structural change was the most common cause of acquired resistance to osimertinib in patients with lung cancer with an EGFR T790M mutation. ccfDNA analyses may replace tissue biopsies for detecting mechanisms of resistance.
Acknowledgments
This work was supported by AstraZeneca. The authors thank all collaborators in the Lung Oncology Group in Kyushu (LOGIK) listed subsequently and Mr. Brian Quinn for his critical comments on the manuscript: Hiroyuki Yamaguchi, Nagasaki University Hospital, Nagasaki; Sho Saeki, Kumamoto University Hospital, Kumamoto; Akira Nagashima, Kitakyushu Municipal Medical Center, Kitakyushu; Naoya Yokomakura, Kagoshima University Hospital, Kagoshima; Koichiro Watanabe, Tsurumi Hospital, Beppu; Masayuki Ishida, Chikamori Hospital, Kochi; Yukihiro Sugimoto, Fukuoka Seishukai Hospital, Kasuya; Tomofumi Yohena, Okinawa Hospital, Ginowan; Shinji Ise, Omuta National Hospital, Omuta; Megumi Inaba, Kumamoto Chuo Hospital, Kumamoto; Masanobu Ishigaki, Urasoe Sogo Hospital, Urasoe; Haruhiro Saito, Kanagawa Cancer Center, Yokohama; and Takatoshi Fujishita, Matsuyama Red Cross Hospital, Matsuyama. The authors also thank Ms. Oouraji at CRes-Kyushu for her assistance. The authors have completed the material design analysis reporting checklist.
Footnotes
Cite this article as: Osoegawa A, Yamaguchi M, Nakamura T, et al. High incidence of c797s mutation in patients with long treatment history of EGFr tyrosine kinase inhibitors including osimertinib. jto clin res rep 2021;2:100191.
Disclosure: All authors have completed the uniform disclosure form of the International Committee of Medical Journal Editors. Drs. Osoegawa, Tanaka, and Kashiwabara report receiving honorarium related to this study from AstraZeneca. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2021.100191.
Data Sharing Statement
The variant call format data are available as supplementary files.
Supplementary Data
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ministry of Health, Labour and Welfare Overview of vital statistics (fixed number) in 2018. https://www.mhlw.go.jp/toukei/saikin/hw/jinkou/kakutei18/index.html Accessed December 25, 2020.
- 3.Maemondo M., Inoue A., Kobayashi K. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T., Morita S., Yatabe Y. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Park K., Tan E.H., O’Byrne K. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17:577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R., Carcereny E., Gervais R. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C., Wu Y.L., Chen G. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 8.Kohno T., Nakaoku T., Tsuta K. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4:156–164. doi: 10.3978/j.issn.2218-6751.2014.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi S., Boggon T.J., Dayaram T. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 10.Cross D.A., Ashton S.E., Ghiorghiu S. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay M.R., Anderton M., Ashton S. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem. 2014;57:8249–8267. doi: 10.1021/jm500973a. [DOI] [PubMed] [Google Scholar]
- 12.Goss G., Tsai C.M., Shepherd F.A. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016;17:1643–1652. doi: 10.1016/S1470-2045(16)30508-3. [DOI] [PubMed] [Google Scholar]
- 13.Jänne P.A., Yang J.C., Kim D.W. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 14.Mok T.S., Wu Y.-L., Ahn M.-J. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soria J.C., Ohe Y., Vansteenkiste J. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 16.Nosaki K., Satouchi M., Kurata T. Re-biopsy status among non-small cell lung cancer patients in Japan: a retrospective study. Lung Cancer. 2016;101:1–8. doi: 10.1016/j.lungcan.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Jackman D., Pao W., Riely G.J. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman A.M., Bratman S.V., To J. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman A.M., Lovejoy A.F., Klass D.M. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UMN UMIN-CTR clinical trial. https://upload.umin.ac.jp/cgi-open-bin/ctr_e/ctr_view.cgi?recptno=R000029086 Accessed December 25, 2020.
- 21.Lin Y.T., Chen J.S., Liao W.Y. Clinical outcomes and secondary epidermal growth factor receptor (EGFR) T790M mutation among first-line gefitinib, erlotinib and afatinib-treated non-small cell lung cancer patients with activating EGFR mutations. Int J Cancer. 2019;144:2887–2896. doi: 10.1002/ijc.32025. [DOI] [PubMed] [Google Scholar]
- 22.Offin M., Rizvi H., Tenet M. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res. 2019;25:1063–1069. doi: 10.1158/1078-0432.CCR-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Y., Bao H., Le X. Distinct co-acquired alterations and genomic evolution during TKI treatment in non-small-cell lung cancer patients with or without acquired T790M mutation. Oncogene. 2020;39:1846–1859. doi: 10.1038/s41388-019-1104-z. [DOI] [PubMed] [Google Scholar]
- 24.Engelman J.A., Zejnullahu K., Mitsudomi T. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 25.Yano S., Wang W., Li Q. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 26.Uramoto H., Shimokawa H., Hanagiri T., Kuwano M., Ono M. Expression of selected gene for acquired drug resistance to EGFR-TKI in lung adenocarcinoma. Lung Cancer. 2011;73:361–365. doi: 10.1016/j.lungcan.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Osoegawa A., Gills J.J., Kawabata S., Dennis P.A. Rapamycin sensitizes cancer cells to growth inhibition by the PARP inhibitor olaparib. Oncotarget. 2017;8:87044–87053. doi: 10.18632/oncotarget.19667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niederst M.J., Sequist L.V., Poirier J.T. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oxnard G.R., Hu Y., Mileham K.F. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piotrowska Z., Isozaki H., Lennerz J.K. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin C.C., Shih J.Y., Yu C.J. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med. 2018;6:107–116. doi: 10.1016/S2213-2600(17)30480-0. [DOI] [PubMed] [Google Scholar]
- 32.Ramalingam S.S., Yang J.C., Lee C.K. Osimertinib as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer. J Clin Oncol. 2018;36:841–849. doi: 10.1200/JCO.2017.74.7576. [DOI] [PubMed] [Google Scholar]
- 33.Ou S.I., Horn L., Cruz M. Emergence of FGFR3-TACC3 fusions as a potential by-pass resistance mechanism to EGFR tyrosine kinase inhibitors in EGFR mutated NSCLC patients. Lung Cancer. 2017;111:61–64. doi: 10.1016/j.lungcan.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrock A.B., Zhu V.W., Hsieh W.S. Receptor tyrosine kinase fusions and BRAF kinase fusions are rare but actionable resistance mechanisms to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2018;13:1312–1323. doi: 10.1016/j.jtho.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 35.Oxnard G.R., Arcila M.E., Sima C.S. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Zhang X.C., Yang J.J. EGFR L792H and G796R: two novel mutations mediating resistance to the third-generation EGFR tyrosine kinase inhibitor osimertinib. J Thorac Oncol. 2018;13:1415–1421. doi: 10.1016/j.jtho.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 37.Thress K.S., Paweletz C.P., Felip E. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama S., Sng N., Carretero J. β-Catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 2014;74:5891–5902. doi: 10.1158/0008-5472.CAN-14-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Togashi Y., Hayashi H., Terashima M. Inhibition of β-catenin enhances the anticancer effect of irreversible EGFR-TKI in EGFR-mutated non-small-cell lung cancer with a T790M mutation. J Thorac Oncol. 2015;10:93–101. doi: 10.1097/JTO.0000000000000353. [DOI] [PubMed] [Google Scholar]
- 40.Bousquet Mur E., Bernardo S., Papon L. Notch inhibition overcomes resistance to tyrosine kinase inhibitors in EGFR-driven lung adenocarcinoma. J Clin Invest. 2020;130:612–624. doi: 10.1172/JCI126896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bruin E.C., Cowell C., Warne P.H. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Planchard D., Loriot Y., André F. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol. 2015;26:2073–2078. doi: 10.1093/annonc/mdv319. [DOI] [PubMed] [Google Scholar]
- 43.Ortiz-Cuaran S., Scheffler M., Plenker D. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016;22:4837–4847. doi: 10.1158/1078-0432.CCR-15-1915. [DOI] [PubMed] [Google Scholar]
- 44.Blakely C.M., Watkins T.B.K., Wu W. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim T.M., Song A., Kim D.W. Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol. 2015;10:1736–1744. doi: 10.1097/JTO.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 46.Suda K., Murakami I., Sakai K. Heterogeneity in resistance mechanisms causes shorter duration of epidermal growth factor receptor kinase inhibitor treatment in lung cancer. Lung Cancer. 2016;91:36–40. doi: 10.1016/j.lungcan.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Hashimoto T., Osoegawa A., Takumi Y. Intratumoral heterogeneity of copy number variation in lung cancer harboring L858R via immunohistochemical heterogeneous staining. Lung Cancer. 2018;124:241–247. doi: 10.1016/j.lungcan.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Iwama E., Sakai K., Hidaka N. Longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer. 2020;126:219–227. doi: 10.1002/cncr.32481. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y., Gan J., Guo K., Deng Y., Fang W. Acquired BRAF V600E mutation mediated resistance to osimertinib and responded to osimertinib, dabrafenib, and trametinib combination therapy. J Thorac Oncol. 2019;14:e236–e237. doi: 10.1016/j.jtho.2019.05.040. [DOI] [PubMed] [Google Scholar]
- 50.Jia Y., Yun C.H., Park E. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchibori K., Inase N., Araki M. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun. 2017;8:14768. doi: 10.1038/ncomms14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schalm S., Dineen T., Lim S. 1296P BLU-945, a highly potent and selective 4th generation EGFR TKI for the treatment of EGFR T790M/C797S resistant NSCLC. Ann Oncol. 2020;31(suppl 4):S839. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.