Abstract
The glycoprotein B (gB) and D (gD) genes from five ruminant alphaherpesviruses, bovine herpesvirus 1 (BHV-1), bovine herpesvirus 5 (BHV-5), caprine herpesvirus 1 (CapHV-1), cervine herpesvirus 1, and rangiferine herpesvirus 1, were partially sequenced. The nucleotide sequence alignments revealed a highly conserved gB gene, with homologies ranging between 87.2 and 99.6%, and a more variable gD gene, with homologies ranging between 71.3 and 98.9%. The phylogenetic analysis of the gB and gD nucleotide and deduced amino acid sequences revealed that BHV-5 is the most closely related virus to the BHV-1 subtype 1 and BHV-1 subtype 2 cluster and that CapHV-1 is the most distantly related virus. The phylogenetic data showed a close relationship of all the studied viruses with suid herpesvirus 1. On the basis of sequence data for the gB gene, a nested PCR combined with restriction enzyme analysis (REA) of the PCR products was developed for the simultaneous detection and identification of the viruses that were studied. Nested primers from highly conserved sequence stretches were selected in order to amplify a region of 294 bp in all five viruses, and a subsequent REA of the PCR products allowed specific identification. A mimic molecule that served as an internal standard of the amplification efficiency was constructed. The practical diagnostic applicability of the assay was evaluated with clinical samples consisting of semen and organ specimens from experimentally infected animals.
Bovine herpesvirus 1 (BHV-1) is a major pathogen of cattle that is distributed worldwide and that causes various syndromes like infectious bovine rhinotracheitis (IBR), infectious pustular vulvovaginitis (IPV), and infectious pustular balanoposthitis (IPB). In general, BHV-1 subtype 1 (BHV-1.1) represents strains that cause IBR, while BHV-1 subtype 2 (BHV-1.2) includes viruses that cause IPV and IPB (10, 16, 38).
Methodologies for the routine detection of BHV-1 are well advanced; however, their efficiencies are compromised by cross-reactions with other closely related alphaherpesviruses of ruminants. Four related herpesviruses which cause serological cross-reactions with BHV-1 may eventually interfere with IBR eradication and control programs. These viruses are bovine herpesvirus 5 (BHV-5), caprine herpesvirus 1 (CapHV-1), cervine herpesvirus 1 (CerHV-1), and rangiferine herpesvirus 1 (RanHV-1). These alphaherpesviruses cause various disorders in ruminants. Outbreaks in calves of fatal meningoencephalitis caused by BHV-5 have been reported in several countries, like Australia, Hungary, the United States, Argentina, and Italy and sporadically in other countries around the world (2, 3, 12, 14). CapHV-1 is distributed worldwide (18) and causes generalized disease in young kids, in which it mainly affects the digestive tract (23), while in adult goats the infection remains unapparent or may cause abortion, vulvovaginitis, or balanoposthitis (19). CerHV-1 was first isolated in 1982 from an outbreak of ocular disease in a red deer farm in Scotland (17, 24). The virus is widespread in free-living and farmed red deer (24). RanHV-1 was isolated from reindeer in Finland (7, 8), and serological evidence of infection with a virus related to BHV-1 has been reported in caribou in Canada (9) and in reindeer in the United States (4). Although they differ considerably in their pathogenicities, all these viruses are closely related to each other and share many common epitopes (11, 20, 21, 25, 26, 30, 36).
Relatively poor information concerning the ability of the related alphaherpesviruses to cross the host species barrier and establish infection in heterologous animal species is available. However, there is experimental evidence that BHV-1 can produce a primary infection in goats (11, 28, 37), and CapHV-1 may infect cattle without any demonstrable latency (11). In addition, RanHV-1 may infect cattle and produce mild rhinitis (25), and CerHV-1 has been associated with abortive infections in calves (29). The existence of these alphaherpesvirus infections in various ruminant species other than bovine species is a major threat for eradication schemes because confusion can arise when a ruminant is falsely identified as BHV-1 positive, even though it is infected with a related but distinct alphaherpesvirus. It is thus imperative to have specific diagnostic tests which can clearly detect and differentiate the various alphaherpesviruses of ruminants in order to facilitate safe conditions for animal health and international trade. No published routine diagnostic method which could be used for the simultaneous differentiation of these five alphaherpesvirus infections is available. The lack of a specific diagnostic assay is partially due to the poor knowledge of the genetic backgrounds of viruses related to BHV-1. Today, few published sequences are available for BHV-5 and CapHV-1, and none are available for CerHV-1 and RanHV-1.
The aim of the work described here was to study the genetic relationship of BHV-1 and the serologically related bovine, caprine, cervine, and rangiferine herpesviruses in order to better understand their epidemiology and taxonomic status and to develop a diagnostic system which can be used for the simultaneous detection and differentiation of all five viruses.
MATERIALS AND METHODS
Virus strains and isolates.
The virus strains and isolates used in the present studies are outlined in Table 1. Viruses were grown by our standard routine protocols (5).
TABLE 1.
Viral strains and isolates used in PCR experiments
| Virus | Strain or isolatea | PCR result | REA band size (bp)b |
|---|---|---|---|
| Ruminant alphaherpes viruses | |||
| BHV-1 | Jura, Cooper, LA, K22, Shelef, HB03, HB107, HB111, HB112, HB120, HB132, HB144, HB151, HB152, HB154, HB156, HB164, HB179, HB191 | + | 195 |
| BHV-5 | N569, Na67, HB184 | + | 217 |
| CapHV-1 | E/CH | + | 117 |
| CerHV-1 | Banffshire 82 | + | 240 |
| RanHV-1 | Salla 82 | + | 141 |
| Other related herpesviruses: | |||
| PrV | Phylaxia, three clinical isolatesc | − | |
| BHV-2 | Clinical isolate | − | |
| EHV-1 | Clinical isolate | − | |
| EHV-4 | Clinical isolate | − | |
| FHV-1 | Clinical isolate | − |
HB, from the BHV-1 collection of A. Bartha, Budapest, Hungary.
Only visible bands on agarose gels above 100 bp.
Clinical isolate; from the virus collection of the National Veterinary Institute, Uppsala, Sweden.
Clinical samples.
Semen specimens were collected from five bulls experimentally infected with a field type strain of BHV-1 (34). After 44 days, all the animals were treated for 5 consecutive days with dexamethasone (0.1 mg/kg of body weight) to reactivate latent BHV-1. Semen specimens were collected over a period of 107 days and were sent to our laboratory as frozen extended semen. Detailed data about the experimental infection and virus isolation from these samples are described in a separate article (34).
Specimens of trigeminal ganglia from cattle with experimental BHV-1 (strain Jura) and BHV-5 (strain N569) infections, goats with experimental CapHV-1 (strain E/CH) infections, red deer with experimental CerHV-1 (strain Banffshire 82) infections, and reindeer with experimental RanHV-1 (strain Salla 82) infections were collected as frozen tissue and were provided by collaborating colleagues. Data about these experimental infections will be published in separate articles. The organs were homogenized, 1 g of tissue in 8 ml of phosphate-buffered saline (pH 7.4) without calcium and magnesium, and were stored at −70°C.
DNA extraction.
Total DNA was extracted from the viral suspensions and from the homogenized tissues, by the proteinase K-sodium dodecyl sulfate–phenol-chloroform method as described by Sambrook et al. (33). The following procedure was followed for extraction of DNA from the semen samples. A volume of 50 μl of frozen extended semen was diluted with 50 μl of the phosphate-buffered saline, and Nonidet P-40 was added to a final concentration of 1%. The sample was shortly vortexed and centrifuged for 5 min at 12,000 × g. The supernatant (seminal fluid) was incubated for 30 min at 37°C in the presense of 1 mg of proteinase K per ml and 0.1% sodium dodecyl sulfate. After incubation, 100 μl of distilled H2O was added and the DNA was extracted by the phenol-chloroform procedure (33). The DNA concentration and purity were measured with a spectrophotometer (GeneQuant; Pharmacia LKB Biochrom Ltd., Cambridge, United Kingdom), and amounts not exceeding 500 ng of total DNA were used in the PCR.
Sequencing, phylogeny, and primer design.
The DNAs extracted from viral suspensions of BHV-1 Jura and K22, BHV-5 N565, CapHV-1 E/CH, CerHV-1 Banffshire 82, and RanHV-1 Salla 82 were used in PCR experiments in order to amplify a region from the glycoprotein B (gB) and D (gD) genes in all these viruses. The gB region was amplified from BHV-1, BHV-5, and CerHV-1 with the primers CR39 (5′-CACGACCTGGGCGGGCAGCAC-3′; sense; 5′ end, position 55,939) and CR40 (5′-CTGCAACGCGAAGGTGTGGCTGTC-3′; antisense; 5′ end, position 56,641) and from CapHV-1 and RanHV-1 with the primers JH1 (5′-CACGGACCTGGTGGACAAGAAG-3′; sense; 5′ end, position 56,017) and OIBR22 (5′-TCTCGTCTCGCAGCATTTCGT-3′; antisense; 5′ end, position 56,564). The gD region was amplified from all viruses studied with the primers CR52 (5′-CCCGMYGCCGCGATACAACTAC-3′; sense; 5′ end, position 119,000) and CR54 (5′-CTTGTGTGCCTCCTGCGGGTA-3′; antisense; 5′ end, position 119,615). The positions of the primers are based on the complete BHV-1 genome (GenBank accession no. BHV1CGEN). Primers CR52 and CR54 are consensus primers selected from a region of homology between BHV-1 and BHV-5 gD sequences (BHV-1, Tikoo et al. [35]; BHV-5, Abdelmagid et al. [1]). The resulting PCR products were sequenced with an ABI PRISM device with dye terminators (Applied Biosystems Inc., Foster City, Calif.). The nucleotide and deduced amino acid sequences were aligned with the aid of the multiple program DNASTAR (DNASTAR Inc., Madison, Wis.) by using the clustal method (15). In the alignment the sequences of suid herpesvirus 1 (Aujeszky’s disease virus; alias pseudorabies virus [PrV]) were included (the gB sequence is from Robbins et al. [31], and the gD sequence is from Petrovskis et al. [27]). Phylogenetic trees were constructed by using the PHYLIP package of computer programs (13) and the neighbor-joining method (32), with PrV used as the outgroup. The parsimony method was also used. The trees were inferred from the molecular sequences from the gB and gD regions. The bootstrap technique was applied for statistical analysis of the phylogenetic trees by using 1,000 replicates. The programs DRAWGRAM and DRAWTREE from PHYLIP were used to produce graphic outputs of the phylogenetic trees.
In order to develop a diagnostic PCR assay that can be used to detect all the ruminant alphaherpesviruses, the gB sequence data obtained from all the viruses studied were used to select nested consensus primers. The primers were chosen with the help of the Oligo program (National Biosciences Inc., Plymouth, Minn.) to amplify a region of 294 bp from all five herpesviruses. The primers used for the first round of amplification were as follows: CR30 (5′-TCGAARGCCGAGTACCTGCG-3′; sense; 5′ end, position 56,051) and CR31, (5′-CCAGTCCCAGGCRACCGTCAC-3′; antisense; 5′ end, position 56,494). Those used for the second round were as follows: CR32 (5′-TGGTGGCCTTYGACCGCGAC-3′; sense; 5′ end, position 56,085) and CR33 (5′-GCTCCGGCGAGTAGCTGGTGTG-3′; antisense; 5′ end, position 56,378). Positions are based on the complete BHV-1 genome (GenBank accession no. BHV1CGEN).
Construction of an internal control.
An internal control of the amplification efficiency, also termed a “mimic” molecule, was constructed. Two oligonucleotides were used to amplify a region of 426 bp from the BHV-1 genome between nucleotides 1,507 and 1,932 (according to GenBank accession no. BHV1CGEN). This region was selected because it contained restriction sites for the two enzymes used for identification of the PCR products. In this way, the mimic molecule could also serve as an internal control of the digestion efficiency. The sense oligonucleotide had CR30 and CR32 as 5′ overhangs, while the antisense oligonucleotide had CR31 and CR33 as 5′ overhangs. Consequently, the PCR produced mimic molecules of 509 bp with target sequences for the diagnostic primers at both ends. The mimic molecule was cloned in pMOSBlue T-vector (Amersham, Little Chalfont, United Kingdom). Serial dilutions were tested in the PCR to study the interaction with the target during amplification, especially when a low target copy number was used. The dilution near the sensitivity limit which ensured a reproducible amplification was used for the assay.
In vitro amplification by PCR.
The first round of amplification was performed in a 50-μl reaction mixture containing 5 μl of the sample, each deoxynucleotide at a concentration of 0.2 mM, 15 pmol of each primer (CR30 and CR31), 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 2 mM MgCl2, 10% dimethyl sulfoxide (DMSO), and 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.). Two droplets of mineral oil (Sigma, St. Louis, Mo.) were added onto the reaction mixtures to prevent evaporation. An initial incubation at 94°C for 9 min was used to activate the polymerase enzyme. The thermal profile was as follows: 95°C for 1 min for denaturation, 60°C for 1 min for primer annealing, and 72°C for 1 min for primer extension. This cycling profile was repeated 35 times, with a final extension at 72°C for 7 min. Subsequently, 1-μl amounts of the PCR mixtures were transferred to new tubes containing freshly added reagents and the two internal primers CR32 and CR33. The conditions of the second round of amplification were identical to those of the first round, except that the MgCl2 concentration was 1.5 mM and the primer annealing temperature was 62°C. The PCR products were electrophoresed in 2% agarose (Sigma) and were visualized by ethidium bromide staining under UV light.
Specificity and sensitivity of the PCR assay.
To test the specificity of the PCR assay, the most closely related herpesviruses, as judged by phylogenetic studies of alphaherpesviruses based on the gB sequences (22) and listed in Table 1, were analyzed. For sensitivity studies, serial 10-fold dilutions of purified BHV-1 DNA, ranging from 100 pg to 0.5 fg, were used in the nested PCR assay. Dilutions of less than 75 fg were tested two times by PCR. The experiment was carried out in parallel with and without the mimic molecule. Prior to dilution, the DNA concentration was measured with a spectrophotometer (GeneQuant).
REA.
In order to specifically identify the viruses, a restriction enzyme analysis (REA) system was developed. Without further purification, the PCR products were digested with the restriction enzymes Fnu4HI and Sau96I (New England Biolabs Inc., Beverly, Mass.) in the same reaction tube by following the manufacturer’s recommendations. The resulting products were separated by electrophoresis in 3% agarose gels (NuSieve [FMC] and agarose [Kodak], 3:1) and were visualized as described above. The consistency of the identification system based on REA of the PCR products was tested with a large number of BHV-1 isolates (Table 1).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in GenBank (National Center Biotechnology Information, Bethesda, Md.) under accession nos. AF078724 to AF078735.
RESULTS
Phylogenetic analysis.
In this study, the gB and gD genes from BHV-1.1 Jura, BHV-1.2 K22, BHV-5 N565, CapHV-1 E/CH, CerHV-1 Banffshire 82, and RanHV-1 Salla 82 were partially sequenced. The lengths of the sequenced regions varied between 652 and 467 nucleotides for the gB gene and between 573 and 537 nucleotides for the gD gene. A consistent blunt-end alignment provided by 467 nucleotides in the gB gene and 537 nucleotides in the gD gene (Fig. 1) revealed high degrees of homology between all six herpesviruses (when each of the two subtypes of BHV-1 is considered to be a separate virus) in the gB gene, while the gD gene was more variable (Fig. 2). Sequence alignments in the gB region revealed a close relationship of all the studied viruses with PrV (Fig. 2). The phylogenetic trees inferred by the gB and gD nucleotide sequences by either the neighbor-joining or the parsimony methods revealed that BHV-5 is the virus most closely related to the BHV-1.1 and BHV-1.2 cluster and that CapHV-1 is the most distantly related. CerHV-1 appeared to be more closely related to BHV-1 and BHV-5 than to RanHV-1 and CapHV-1. The virus most closely related to RanHV-1 is CerHV-1 (Fig. 3). The trees were supported by high bootstrap numbers. The same topography and high bootstrap numbers were obtained when the trees were constructed on the basis of the amino acid sequence data (not shown).
FIG. 1.
Alignment of the deduced amino acid sequences of a region from the gB and gD proteins in six ruminant alphaherpesviruses. The sequence of PrV is also included in the alignment; the gB sequence is from Robbins et al. (31), and the gD sequence is from Petrovskis et al. (27). A thick line under the sequences indicates highly conserved regions (more than 4 consecutive amino acids).
FIG. 2.
Nucleotide and amino acid sequence similarities for the gB and gD regions of ruminant alphaherpesviruses and PrV.
FIG. 3.
Phylogenetic trees of ruminant alphaherpesvirus based on the nucleotide sequences of the gB and gD regions. Tree A was generated by the neighbor-joining method, and tree B was generated by the parsimony method. The lengths of the branches in the tree inferred by the neighbor-joining method reflect phylogenetic distances. The numbers on trees A and B indicate the percentage of times that the particular group was predicted after statistical analysis by bootstrapping. PrV was used as the outgroup.
PCR performance.
The specific 294-bp PCR product was consistently obtained as a clear electrophoretic band from all ruminant alphaherpesviruses studied (Fig. 4A). The other related herpesviruses, PrV, bovine herpesvirus 2, equine herpesvirus 1 and 4, and feline herpesvirus 1, were not amplified (Table 1). The use of the enhancer DMSO at a concentration of 10% was found to be essential, because any specific amplification could be observed without DMSO. Also, the initial denaturation step at 94°C for 9 min for activation of the polymerase enzyme (AmpliTaq Gold; Perkin-Elmer Cetus) was found to be beneficial, since poor amplification efficiencies were obtained when Amplitaq (Perkin-Elmer Cetus) was used without the first long predenaturation step.
FIG. 4.
(A) PCR amplification of ruminant alphaherpesvirus DNA with the mimic molecule and posterior identification by REA. Lane 1, positive PCR product from a ruminant alphaherpesvirus (BHV-1 as a model) before REA; lanes 2 to 6, positive PCR products after REA; lane 2, CapHV-1; lane 3, RanHV-1; lane 4, BHV-1; lane 5, BHV-5; lane 6, CerHV-1. The bands under 100 bp in length are minor residual digestion products. Lanes M, marker, 100-bp ladder. (B) PCR and REA of ruminant alphaherpesvirus DNA amplified directly from trigeminal ganglia. Lanes 2, 6 and 10, BHV-1; lanes 1 and 8, BHV-5; lanes 3 and 4, CapHV-1; lanes 5 and 11, CerHV-1; lanes 7, 9, and 12, RanHV-1. In each lane the upper band corresponds to the digested amplified mimic molecule and the lower band corresponds to the digested amplified viral DNA. Lanes M, marker, 100-bp ladder.
The internal standard of amplification was consistently amplified together with the specific target. The different size of the mimic molecule, 467 bp, in contrast to the size of the target, 294 bp, allowed for the easy differentiation of the products on agarose gels.
The nested PCR assay detected as few as 10 to 15 fg of purified BHV-1 DNA or approximately 60 to 90 genome copies (data not shown). Even six genome copies could be detected, but at this dilution the results were inconsistent. The mimic molecule did not affect the amplification efficiency at the lowest dilutions of the target, as observed in parallel experiments with and without the mimic molecule (data not shown).
Identification by REA.
The direct digestion of the PCR products with the restriction enzymes Fnu4HI and Sau96I in the same reaction tube allowed the easy discrimination of the amplified viral sequences on agarose gels (Table 1; Fig. 4A and B). The digestion of the PCR products generated distinct restriction patterns, depending on the origin of the amplified viral DNA: 117 bp for CapHV-1, 141 bp for RanHV-1, 195 bp for BHV-1, 217 bp for BHV-5, and 240 bp for CerHV-1 (Fig. 4A). The mimic molecule was also digested and produced a band of 237 bp, which is similar to the position of the band of 240 bp of CerHV-1. REA generated other minor products of less than 100 bp, but these were poorly visible and were not considered for the identification (Fig. 4A). The consistency of the diagnostic system based on REA of the PCR products has been tested with 19 strains and isolates of the BHV-1.1 and BHV-1.2 subtypes, and all of them had the same restriction patterns (Table 1). Distinction between the two subtypes of BHV-1 was not possible due to the high degree of homology (99.6%) in the sequenced region of the gB gene.
Clinical samples.
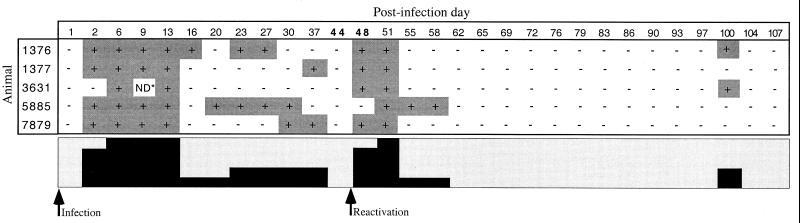
The practical diagnostic applicability of the nested PCR-REA assay was evaluated with clinical samples consisting of organ and semen specimens from experimentally infected animals. The results of the PCR-REA with BHV-1-infected semen showed that BHV-1 was periodically shed in the semen of infected bulls for up to 100 days postinfection (d.p.i.) for two of the five bulls (Fig. 5). At between 2 and 13 d.p.i., virus shedding was detected in most of the animals. A second peak in which BHV-1 was detected in most of the animals occurred between 48 and 51 d.p.i., after dexamethasone treatment for 5 consecutive days (between 44 and 48 d.p.i.). During the third month postinfection, BHV-1 was not detected in the semen from any of the bulls (Fig. 5). Comparison with virus isolation showed that the PCR assay could detect the virus in more samples and during longer periods of time. By virus isolation BHV-1 was detected in the semen from all the animals only during the first 9 d.p.i. and in the semen from an animal at 48 d.p.i. Detailed results of virus isolation will be published in a separate article (34).
FIG. 5.
PCR results for frozen extended semen samples from five bulls experimentally infected with BHV-1. Postinfection days in boldface type indicate the days of dexamethasone treatment. ND, not done. The lower box expresses the results as a diagram.
By using the PCR assay, viral DNA could be detected in the homogenized specimens of trigeminal ganglia from the experimentally infected animals. The REA performed directly with the PCR products showed which viral DNA was the origin of the amplification (Fig. 4B).
DISCUSSION
The aim of the present studies was to improve the control of IBR and diseases caused by other related alphaherpesviruses by (i) studying the genetic relationship of BHV-1, BHV-5, CapHV-1, CerHV-1, and RanHV-1 and (ii) developing a specific diagnostic method for the detection and identification of all of them.
Although the genome sequence of BHV-1 is available today, the related alphaherpesviruses have been poorly characterized so far, hampering the development of a specific diagnostic system for this group of related viruses. The present studies are the first to characterize genomic sequences of CerHV-1 and RanHV-1 and are among the first ones to provide such information for BHV-5 and CapHV-1. Comparison of the sequences revealed a highly conserved gB gene, especially in the central part of the sequences. The region sequenced in the gD gene was much more variable (Fig. 1 and 2). The phylogenetic analysis showed a consistent group, referred as the BHV-1 group, formed by BHV-1.1, BHV-1.2, BHV-5, and CerHV-1. This grouping was supported by high bootstrap numbers. RanHV-1 was located between this group and the more distantly related CapHV-1 but was still closer to the BHV-1 group than to CapHV-1. The present results are mostly in agreement with those reported by Lyaku et al. (20), whose results were based on serology, although BHV-5 was not included in that study. In their report they stated that the cervine, caprine, and rangiferine viruses are more closely related to BHV-1 than they are to each other; however, we found that RanHV-1 is more closely related to CerHV-1 than to BHV-1 and that CapHV-1 is slightly more closely related to CerHV-1 and RanHV-1 than to BHV-1. In addition, our genetic studies revealed a close relationship in the analyzed region of the gB gene from all ruminant alphaherpesviruses and PrV, in agreement with the previous work of McGeoch and Cook (22), in which PrV was found to be the closest relative of BHV-1.
Highly conserved stretches of sequences of the gB gene were used to select nested consensus primers for a diagnostic PCR. Only a few ambiguities had to be introduced into the primer sequences in order to fit the sequences of all viruses studied. In order to increase the amplification efficiency, the size of the amplified region was kept under 300 bp. Furthermore, the guanine-plus-cytosine content was selected on the basis of the average for the BHV-1 genome, considering that a high G+C content promotes secondary structures which might affect both primer annealing and extension. Accordingly, DMSO, which plays a role in the loosening up of complex secondary structures, has been found to be essential for the amplification reaction. To increase the reliability of the system, a mimic molecule was used as an internal standard to avoid false-negative results due to inhibitors or reaction failures. This is particularly important for the detection of BHV-1 in complex specimens like semen.
Discrimination between the different viruses could be achieved by a simple digestion with Fnu4HI and Sau96I in the same reaction tube. After cleavage, all the amplified products generated a single specific band above 100 bp. The mimic molecule was also constructed to serve as a control for the efficiency of digestion of the two restriction enzymes. The unique major product that originated after digestion of the mimic molecule migrated like the product from CerHV-1, resulting in a single band (Fig. 4). However, discrimination between the mimic molecule and CerHV-1 is easily achieved since the the PCR products are electrophoresed before REA. Distinction between the two subtypes of BHV-1 was not possible due to the high degree of sequence conservation in the gB sequences. In fact, distinction between the two subtypes only has epidemiological value and it is not absolutely necessary for the routine detection of BHV-1. Furthermore, the correlation of BHV-1.1 as the agent of IBR and BHV-1.2 as the agent of IPV and IPB is not exact, since the two subtypes may cause heterologous symptoms (38).
The results indicate that the gB gene is a valuable target for diagnostic PCR because the highly conserved sequences allow the amplification of geographically and chronologically distant isolates. However, since sequence variation among BHV-1 strains exists, as demonstrated by genomic restriction enzyme patterns (38), it is important to examine a large number of strains and isolates before considering the general applicability of a newly developed PCR assay. In total, 19 strains and isolates of BHV-1, including BHV-1 subtypes 1.1 and 1.2, were analyzed, and all of them were amplified and showed the same restriction patterns after REA. Among the rest of related ruminant alphaherpesviruses, only one strain (three strains in the case of BHV-5) could be analyzed because these are poorly characterized viruses and few strains are available. However, the conserved nature of the gB gene, especially the regions whose sequences were used for the construction of the primers that were selected, would help to minimize the possibility that certain isolates would be missed by this assay.
The practical diagnostic applicability of the PCR-REA system has been evaluated with clinical samples consisting of organ specimens from ruminants experimentally infected with BHV-1, BHV-5, CapHV-1, CerHV-1, and RanHV-1. The results indicated that the system can be used for the detection and identification of all five ruminant alphaherpesviruses directly from sites of herpesvirus replication as well as from sites of latency, i.e., trigeminal ganglia (Fig. 4B). The diagnostic applicability of the system has also been evaluated with semen specimens from cattle experimentally infected with BHV-1. Semen is known to contain inhibitory factors and is cytotoxic for cell cultures (6, 39). Accordingly, we have found that a thorough procedure for sample preparation is essential to achieve consistent results. For this reason, we extracted and purified the viral DNA prior to amplification and used an internal standard to detect false-negative results. The results with the semen samples illustrate the intermittent course of BHV-1 shedding in the semen. Comparison with virus isolation showed that PCR detected the virus in more samples and during longer periods of time. Furthermore, the PCR results were obtained in 2 working days, while virus isolation usually took up to 2 weeks to obtain a diagnosis.
In summary, in the present studies the phylogenetic relationship of five ruminant alphaherpesviruses, BHV-1, BHV-5, CapHV-1, CerHV-1, and RanHV-1, has been determined by comparing molecular sequences from the gB and gD genes. The novel data allowed the development of a rapid and effective diagnostic system based on PCR and REA for the detection and discrimination of all five of these related alphaherpesviruses.
ACKNOWLEDGMENTS
We thank M. E. Riquelme for excellent technical assistance, R. Harring (Holland Genetics, Arnhem, The Netherlands) for providing the semen samples from experimentally infected bulls, and A. Bartha (Budapest, Hungary) for providing the BHV-1 strains. Thanks are due to E. Thiry and G. Meyer (Liége, Belgium), M. Engels and A. Six (Zürich, Switzerland), H. Reid and I. Campbell (Edinburgh, United Kingdom), C. Ek-Komonen (Helsinki, Finland), and M. Banks (Addlestone, United Kingdom) for providing the samples from experimentally infected animals and M. Engels for critical reading of the manuscript.
This work was financially supported by the Commission of the European Communities, Agriculture and Fisheries (FAIR)-specific RTD programme, PL95-0316, “Improved methods for the diagnosis of ruminant alphaherpesvirus infections in relation to the control of infectious bovine rhinotracheitis (IBR).” The work performed with BHV-1-infected semen specimens was financially supported by the Swedish Farmers’ Foundation for Agricultural Research (Stiftelsen Lantbruksforskning, projects 9730015 and 9830027).
REFERENCES
- 1.Abdelmagid O Y, Minocha H C, Collins J K, Chowdhury S I. Fine mapping of bovine herpesvirus-1 (BHV-1) glycoprotein D (gD) neutralizing epitopes by type-specific monoclonal antibodies and sequence comparison with BHV-5 gD. Virology. 1995;206:242–253. doi: 10.1016/s0042-6822(95)80039-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartha A, Hajdu G, Aldasi P, Paczolay G. Occurrence of encephalitis caused by infectious bovine rhinotracheitis virus in calves in Hungary. Acta Vet Hung. 1969;19:145–151. [PubMed] [Google Scholar]
- 3.Carrillo B J, Pospischil A, Dahme E. Pathology of bovine viral necrotizing encephalitis in Argentina. Zentbl Veterinarmed B. 1983;30:161–168. doi: 10.1111/j.1439-0450.1983.tb01829.x. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich R A. Respiratory viruses. In: Dietrich R A, editor. Alaskan wildlife diseases. Fairbanks: University of Alaska; 1981. pp. 28–29. [Google Scholar]
- 5.Dinter Z. In: Diagnostic virology. Moreno-Lopez G, editor. Uppsala: Swedish University of Agricultural Sciences; 1989. pp. 95–110. [Google Scholar]
- 6.Dyer J R, Gilliam B L, Jr. Eron J J, Grosso L, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA with Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 7.Ek-Kommonen C, Veijalainen P, Rantala M, Neuvonen E. Neutralizing antibodies to bovine herpesvirus 1 in reindeer. Acta Vet Scand. 1982;23:565–569. doi: 10.1186/BF03546775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ek-Kommonen C, Pelkonen S, Nettleton P F. Isolation of a herpesvirus serologically related to bovine herpesvirus 1 from a reindeer (Rangifer tarandus) Acta Vet Scand. 1986;27:299–301. doi: 10.1186/BF03548174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Azhary M, Roy R S, Fréchette J L. Serological evidence of IBR and BVD infection in caribou (Rangifer tarandus) Vet Rec. 1979;105:336. doi: 10.1136/vr.105.14.336. [DOI] [PubMed] [Google Scholar]
- 10.Engels M, Steck F, Wyler R. Comparison of the genomes of bovine rhinotracheitis and infectious pustular vulvovaginitis virus strains by restriction endonuclease analysis. Arch Virol. 1981;67:169–174. doi: 10.1007/BF01318601. [DOI] [PubMed] [Google Scholar]
- 11.Engels M, Palatini M, Metzler A E, Probst U, Kihm U, Ackermann M. Interactions of bovine and caprine herpesviruses with the natural and foreign hosts. Vet Microbiol. 1992;33:69–78. doi: 10.1016/0378-1135(92)90036-s. [DOI] [PubMed] [Google Scholar]
- 12.Eugster A K, Angulo A B, Jones L P. Herpesvirus encephalitis in range calves. Proc Annu Meet Am Assoc Vet Lab Diagn. 1974;17:267–281. [Google Scholar]
- 13.Felsenstein J. PHYLIP inference package, version 3.5. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 14.French E L. A specific virus encephalitis in calves: isolation and characterization of the causal agent. Aust Vet J. 1962;38:216–221. [Google Scholar]
- 15.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 16.House J A. Bovine herpesvirus IBR-IPV. Strain differences. Cornell Vet. 1972;62:431–453. [PubMed] [Google Scholar]
- 17.Inglis D M, Bowie J M, Allan M J, Nettleton P F. Ocular disease in red deer calves associated with a herpesvirus infection. Vet Rec. 1983;113:182–183. doi: 10.1136/vr.113.8.182. [DOI] [PubMed] [Google Scholar]
- 18.Kao M, Leiskau T, Koptopoulos G, Papadopoulos O, Horner G W, Hyllseth B, Fadel M, Gedi A H, Straub O C, Ludwig H. Goat herpesvirus infections: a survey on specific antibodies in different countries. In: Pastoret P-P, Thiry E, Saliki, editors. Immunity to herpesvirus infections of domestic animals. Report EUR 9737 EN. Luxembourg: Commission of the European Communities; 1985. pp. 93–97. [Google Scholar]
- 19.Koptopoulos G. Goat herpesvirus 1 infection: a review. Vet Bull. 1992;62:77–84. [Google Scholar]
- 20.Lyaku J R S, Nettleton P F, Marsden H. A comparison of serological relationships among five ruminant alphaherpesviruses by ELISA. Arch Virol. 1992;124:333–341. doi: 10.1007/BF01309813. [DOI] [PubMed] [Google Scholar]
- 21.Martin W B, Castrucci G, Frigeri F, Ferrari M. A serological comparison of some animal herpesviruses. Comp Immunol Microbiol Infect Dis. 1990;13:74–84. doi: 10.1016/0147-9571(90)90519-y. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J, Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 23.Mettler F, Engels M, Wild P, Bivetti A. Herpesvirus-Infektion bei Zicklein in der Schweiz. Schweiz Arch Tierheilkd. 1979;121:655–662. [PubMed] [Google Scholar]
- 24.Nettleton P F, Sinclair J A, Herring J A, Inglis D M, Fletcher T J, Ross H M, Bonniwell M A. Prevalence of herpesvirus infection in British red deer and investigations of further disease outbreaks. Vet Rec. 1986;8:267–270. doi: 10.1136/vr.118.10.267. [DOI] [PubMed] [Google Scholar]
- 25.Nettleton P F, Thiry E, Reid H, Pastoret P-P. Herpesvirus infections in Cervidae. Rev Sci Tech Off Int Epizool. 1988;7:977–988. doi: 10.20506/rst.7.4.384. [DOI] [PubMed] [Google Scholar]
- 26.Nixon P, Edwards S, White H. Serological comparison of antigenically related herpesviruses in cattle, red deer and goats. Vet Res Commun. 1988;12:355–362. doi: 10.1007/BF00343256. [DOI] [PubMed] [Google Scholar]
- 27.Petrovskis E A, Timmins J G, Armentrout M A, Marchioli C C, Yancey R J, Jr, Post L E. DNA sequence of the gene for pseudorabies virus gp50, glycoprotein without N-linked glycosylation. J Virol. 1986;59:216–223. doi: 10.1128/jvi.59.2.216-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirak M, Thiry E, Brochier B, Pastoret P-P. Infection experimentale de la chévre par le virus de la rhinotracheite infectieuse bovine (bovine herpesvirus 1) et tentative de reactivation virale. Rec Med Vet. 1983;159:1103–1106. [Google Scholar]
- 29.Reid H W, Nettleton P F, Pow I, Sinclair J A. Experimental infection of red deer (Cervus elaphus) and cattle with a herpesvirus isolated from red deer. Vet Rec. 1986;118:156–158. doi: 10.1136/vr.119.6.156. [DOI] [PubMed] [Google Scholar]
- 30.Rimstad E, Krona R, Hyllseth B. Comparison of herpesviruses isolated from reindeer, goats, and cattle by restriction endonuclease analysis. Arch Virol. 1992;123:389–397. doi: 10.1007/BF01317272. [DOI] [PubMed] [Google Scholar]
- 31.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbour joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook S, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Smits, C. B., C. van Maanen, R. D. Glas, A. L. W. De Gee, T. Dijkstra, J. T. van Oirschot, and F. A. M. Rijsewijk. Comparison of three polymerase chain reaction methods for routine detection of bovine herpesvirus 1 DNA in fresh bull semen. Submitted for publication. [DOI] [PubMed]
- 35.Tikoo S K, Fitzpatrick D R, Babiuk L A, Zamb T J. Molecular cloning, sequencing, and expression of functional bovine herpesvirus 1 glycoprotein gIV in transfected bovine cells. J Virol. 1990;64:5132–5142. doi: 10.1128/jvi.64.10.5132-5142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanderplasschen A, Bublot M, Pastoret P-P, Thiry E. Restriction maps of the DNA of cervid herpesvirus 1 and cervid herpesvirus 2, two viruses related to bovine herpesvirus 1. Arch Virol. 1993;128:379–388. doi: 10.1007/BF01309448. [DOI] [PubMed] [Google Scholar]
- 37.Wafula J S, Mushi E Z, Wamwayi H. Reaction of goat to infection with infectious bovine rhinotracheitis virus. Res Vet Sci. 1985;39:84–86. [PubMed] [Google Scholar]
- 38.Wyler R, Engels M, Schwyzer M. Infectious bovine rhinotracheitis/vulvovaginitis (BHV-1) In: Wittmann G, editor. Herpesvirus diseases of cattle, horses and pigs. Boston, Mass: Kluwer Academic Publishers; 1989. pp. 1–72. [Google Scholar]
- 39.Xia J Q, Yason C V, Kibenge F S. Comparison of dot blot hybridization, polymerase chain reaction, and virus isolation for detection of bovine herpesvirus-1 (BHV-1) in artificially infected bovine semen. Can J Vet Res. 1995;59:102–109. [PMC free article] [PubMed] [Google Scholar]