Abstract
Introduction
Recent advances in the detection of genomic DNA from plasma samples allow us to follow tumor DNA shedding in plasma during systemic treatment. Osimertinib is the standard of care for patients with NSCLC with acquired EGFR T790M mutations. We assessed changes in serial plasma cell-free circulating tumor DNA (ctDNA) genomic alterations to predict osimertinib efficacy.
Methods
We prospectively collected plasma from patients having EGFR-mutated advanced NSCLC previously treated with EGFR tyrosine kinase inhibitor therapy and with acquired EGFR T790M mutation detected by standard methods. Plasma samples were collected before starting osimertinib treatment, 4 weeks after osimertinib treatment, and on progression. ctDNA was analyzed using the Guardant360 assay.
Results
A total of 15 eligible patients received osimertinib. Before starting treatment, EGFR-activating mutations were detected in the ctDNA of all patients, and EGFR T790M was detected in 93% of the cases. Osimertinib treatment was associated with an objective response rate of 53% and a median progression-free survival of 7.3 months. A total of 12 of the 15 patients had undetectable plasma T790M and decreased activating mutation allelic frequency (AF) at week 4. None of the 12 patients had disease progression within 16 weeks. For the remaining three patients, with detectable plasma T790M (n = 2) or increased activating mutation AF (n = 1) at week 4, two had progressive disease within 16 weeks (p = 0.03).
Conclusions
In patients with EGFR-mutated advanced NSCLC, persistent EGFR T790M or increasing activating mutation AF as detected in ctDNA 4 weeks after the start of osimertinib treatment may predict disease progression within 16 weeks.
Keywords: Non–small cell lung cancer, EGFR mutations, ctDNA, Osimertinib, Outcome prediction
Introduction
First- and second-generation EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib, erlotinib, and afatinib, are effective as first-line treatment of advanced NSCLC harboring EGFR-activating mutations (e.g., deletions in exon 19 and exon 21 L858R mutation).1, 2, 3, 4, 5, 6, 7 The EGFR T790M mutation develops in 55% of tumors after EGFR TKI therapy and is the most common mechanism of acquired resistance.8, 9, 10, 11 Osimertinib monotherapy currently is the recommended second-line treatment for EGFR T790M mutation–positive NSCLC.12, 13, 14 In contrast, resistance to osimertinib inevitably develops, with EGFR C797S being one example of a mutation associated with acquired resistance.15
In some patients with advanced NSCLC, it may be challenging to acquire tumor tissue to assess for EGFR T790M. Plasma cell-free circulating tumor DNA (ctDNA) analysis is an alternative method for detecting genomic alterations in tumor DNA. The ability to collect blood at multiple time points makes ctDNA analysis a convenient approach for following changes in tumor DNA during systemic therapy and at the time of disease progression. In recent years, ctDNA analysis (liquid biopsy) has been widely used to detect EGFR T790M and to identify mechanisms of resistance; however, the role of ctDNA analysis in early prediction of treatment efficacy is unknown.15, 16, 17
We used serial plasma ctDNA genomic alterations to predict osimertinib efficacy and searched for possible resistance mechanisms to osimertinib.
Materials and Methods
Patients
This study was approved by the Research Ethics Committee of the National Taiwan University Hospital (NTUH REC No. 201705042RIPC). All patients provided signed informed consent for this study. We prospectively collected plasma from patients with EGFR-mutated advanced NSCLC who harbored acquired EGFR T790M mutation detected by various standard hotspot tissue- or plasma-based methods (e.g., cobas EGFR mutation Test version 2 [Roche Molecular Systems, Pleasanton, CA]) after previous EGFR TKI therapy and planned to start osimertinib therapy. We collected the clinical data prospectively, including patient and tumor characteristics, previous anticancer treatments, methods to detect EGFR T790M–positive disease, and time of disease progression.
Plasma ctDNA Analysis
Plasma samples were collected before starting osimertinib treatment, 4 weeks after the initiation of osimertinib treatment, and on disease progression. ctDNA was extracted from the plasma samples, and, after bar coding and hybrid capture, subjected to digital next-generation sequencing (NGS) and analysis using Guardant360 (Guardant Health, Redwood City, CA). Detailed protocols of blood sample processing, ctDNA isolation, sequencing, data analysis, and detection limits have been described elsewhere.18
Statistical Analysis
Objective responses were evaluated on the basis of the Response Evaluation Criteria in Solid Tumors version 1.1 by the treating physicians. Progression-free survival (PFS) was defined as the time from the first dose of osimertinib to the date of radiological or clinical disease progression or death and was estimated using the Kaplan-Meier method. The data cutoff date was July 31, 2019. Differences in treatment outcomes between the compared groups were determined using Fisher’s exact test.
Results
A total of 15 patients were enrolled. EGFR-activating mutations consisted of EGFR exon 19 deletions (eight patients) and L858R mutation (seven patients). Erlotinib, afatinib, and gefitinib were administered in seven (47%), five (33%), and three (20%) patients, respectively. A total of 10 (66.7%) and five (33.3%) patients presented bone metastases and liver metastases before the commencement of osimertinib therapy, respectively. Other patient characteristics are illustrated in Table 1. All patients received osimertinib at a standard dose of 80 mg/day. A total of 12 patients concurrently participated in the single-arm PLASMA study (ClinicalTrials.gov Identifier: NCT02811354), which treated plasma EGFR T790M–positive patients detected by droplet digital polymerase chain reaction (PCR)–based technology (Sanomics, Hong Kong, People’s Republic of China) with osimertinib. Acquired T790M mutation was diagnosed by using plasma samples alone (cobas EGFR mutation test version 2 or droplet digital PCR) (n = 11), tissue or pleural effusion alone (n = 2), or a combination of both tissue and plasma samples (n = 2) (Fig. 1). Treatment decisions were based on the aforementioned T790M mutation test results. Guardant360 detected EGFR-activating mutations in all patients before starting osimertinib treatment; T790M was detected by Guardant360 in 93% (n = 14) of the patients. TP53 mutations, EGFR amplification, and MET amplification were detected in eight, eight, and one patient, respectively (Fig. 2).
Table 1.
Patient Characteristics
| Variable | N | % |
|---|---|---|
| Total | 15 | 100 |
| Sex | ||
| Male | 8 | 53 |
| Female | 7 | 47 |
| Age (y) | ||
| Median (range) | 62 (48–77) | |
| Smoking | ||
| Never-smoker | 12 | 80 |
| Histology | ||
| Adenocarcinoma | 15 | 100 |
| EGFR mutation | ||
| Deletion 19 | 8 | 53 |
| L858R | 7 | 47 |
| Metastasis | ||
| Brain | 5 | 33 |
| Bone | 10 | 67 |
| Liver | 5 | 33 |
| Pleural effusion | 5 | 33 |
| Prior systemic therapy | ||
| Platinuma | 3 | 20 |
| Pemetrexed | 3 | 20 |
| Gefitinib | 3 | 20 |
| Erlotinib | 7 | 47 |
| Afatinib | 5 | 33 |
Cisplatin or carboplatin.
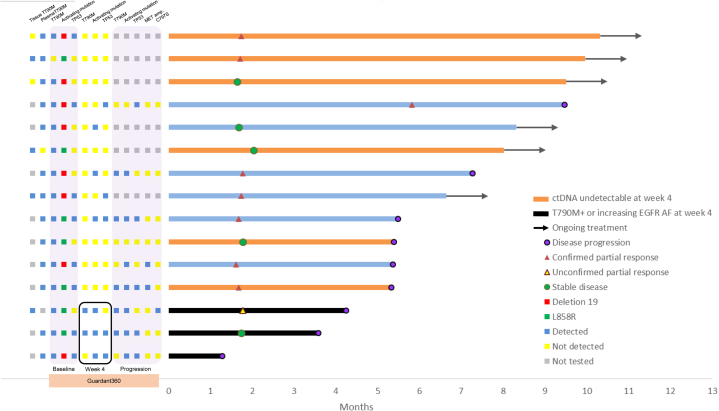
Figure 1.
PFS of patients receiving osimertinib for EGFR T790M–positive disease, with major Guardant360 results at baseline, week 4, and progression. AF, allelic frequency; ctDNA, circulating tumor DNA; PFS, progression-free survival.
Figure 2.
Heatmap summarizing the findings from tissue and plasma before starting osimertinib therapy and detailed Guardant360 results at three time points. AF, allelic frequency; ctDNA, circulating tumor DNA.
After osimertinib treatment, the objective response rate was 53%, and the median PFS was 7.3 months (Fig. 1). After 4 weeks of osimertinib, in six of the 15 patients, ctDNA was undetectable (Fig. 2). Among the 14 patients with EGFR T790M detected by Guardant360 at baseline, 12 had no detectable plasma EGFR T790M at week 4. Among these, eight had undetectable, three had decreased, and one (patient 15) had increased EGFR-activating mutations allelic frequency (AF). Two patients (patients 13 and 14) had persistent EGFR T790M at week 4 (Fig. 2).
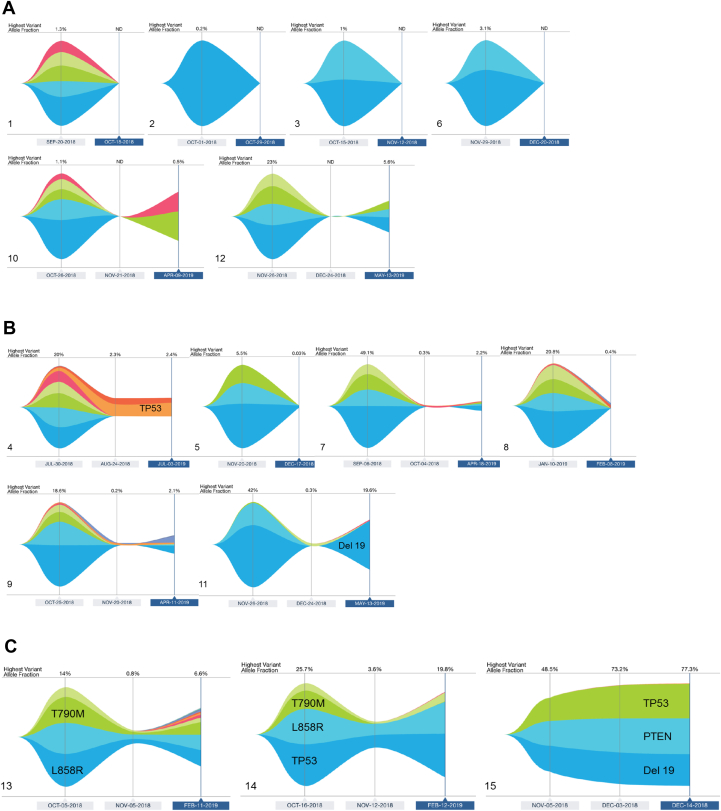
Among patients with persistent plasma EGFR T790M mutations at week 4, one presented disease progression at week 15 (patient 14) and one at week 18 (patient 13) (Figs. 1 and 2). Patient 15 had rapid progression on osimertinib treatment, with clinical progression at week 5. The initial EGFR T790M AF was low (0.2%) and was not detectable at week 4, but the EGFR-activating mutation AF increased at week 4 and furthermore on progression (48.5% at baseline, 66.5% at week 4, and 66.9% on progression). Alterations in TP53 and PTEN were also detected at baseline in this patient and increased at week 4 (Fig. 3C, panel 15). Patient 14 developed a new liver metastasis after stable disease as best response. This patient presented plasma TP53 and CDKN2A mutations detected at baseline, with AF of those mutations having decreased at week 4 (Fig. 3C, panel 14). Patient 13 presented an initial tumor response but later progressed with a resistant EGFR C797S mutation (Fig. 3C, panel 13).
Figure 3.
Patterns of Guardant360 tumor response maps. (A) Undetectable mutations of any kind at week 4. (B) No detectable EGFR T790M and no increase in EGFR-activating mutation AF at week 4. (C) Detectable EGFR T790M or increasing EGFR-activating mutation AF at week 4. AF, allelic frequency; APR, April; AUG, August; ctDNA, circulating tumor DNA; DEC, December; FEB, February; JAN, January; JUL, July; ND, not determined; NOV, November; OCT, October; SEP, September.
Among the eight patients harboring TP53 mutations at baseline, three (38%) with undetectable plasma TP53 mutation at week 4 achieved disease control (stable disease or partial response by Response Evaluation Criteria in Solid Tumors 1.1), whereas two of the three with detectable TP53 and EGFR-activating mutations at week 4 presented disease progression within 16 weeks (Figs. 1 and 2).
On the basis of the Guardant360 tumor response map, we classified the patterns into the following three categories: (1) undetectable mutations of any kind at week 4 (Fig. 3A); (2) no detectable EGFR T790M and no increase in EGFR-activating mutation AF at week 4, with or without mutations in other genes (Fig. 3B); and (3) detectable EGFR T790M or increasing EGFR-activating mutation AF at week 4 (Fig. 3C). Patients in category 3 had the worst osimertinib treatment outcome (Fig. 3), which, compared with the combined categories 1 and 2, was associated with early disease progression within 16 weeks (p = 0.03). Presence of liver metastases was not associated with early disease progression within 16 weeks (p = 0.07).
Among the nine patients with disease progression during the observation period, four presented documented resistance alterations, as follows: EGFR C797S (one), EGFR G724S (one), and MET amplification (two) (Fig. 2).
Discussion
In this study, we reported that, in patients with EGFR T790M mutation–positive NSCLC treated with osimertinib, undetectable EFGR T790M in ctDNA at 4 weeks after treatment initiation was associated with a favorable clinical outcome. In contrast, patients with detectable EGFR T790M or increasing AF for EGFR-activating mutations in ctDNA at the same time point had a less favorable prognosis.
The Guardant360 assay is a hybrid capture-based NGS technology, with high concordance with tissue-based genotyping in previously untreated patients with metastatic NSCLC.19 When used in combination with tissue-based NGS, this assay increases the detection rate of targetable mutations in patients with NSCLC.20 In patients previously treated with EGFR TKI therapy, this technology can detect resistance mutations15 and favorably affects clinical decision making at the time of disease progression in one-third of patients with NSCLC, including those with EGFR-mutated tumors.21 To the best of our knowledge, this is the first study to use this test during osimertinib therapy (at week 4) to identify possible genomic alterations or changes in AF to predict osimertinib treatment outcomes.
Plasma-based detection (liquid biopsy) of EGFR mutations is an alternative to tissue-based diagnosis.22 One example, the cobas assay, which uses PCR technology to evaluate mutations in EGFR, is approved by the U.S. Food and Drug Administration to identify appropriate patients for EGFR TKI treatment.16 A retrospective analysis of plasma samples from patients from the AURA3 study revealed that the plasma EGFR T790M–positive percentage agreement was higher using the Guardant360 NGS assay than with cobas plasma, using cobas tissue test results as a reference.23 Liquid biopsy can also be used in identifying resistance mechanisms to osimertinib.15 A recent report revealed that clearance of all EGFR mutations as assessed by cobas plasma analysis after second-line osimertinib treatment was a positive predictor of clinical outcome.24 An exploratory study of AURA3 study using droplet digital PCR technology also revealed that clearance of plasma EGFR mutations may predict response to osimertinib.25 In this study, we highlighted the role of plasma ctDNA analysis by using NGS technology in monitoring treatment efficacy. Most patients presented decreased and even undetectable EGFR ctDNA after 4 weeks of osimertinib treatment; however, the persistence of EGFR T790M mutation in ctDNA may predict a worse outcome. Nevertheless, in one patient in whom EGFR T790M was cleared from the ctDNA (patient 15), the original EGFR driver mutation remained and the AF increased at week 4, and the patient experienced rapid tumor progression. Hence, undetectable EGFR T790M alone may not be the optimal predictor of treatment outcome.
The presence of concurrent mutations in TP53 and EGFR before treatment was reported in 30% to 50% of patients with lung cancer, which may be a prognostic marker for diminished clinical benefit from first-generation EGFR TKIs.26, 27, 28 However, in our study, with 53% of cases having both EGFR and TP53 mutations, this did not seem to be a clinical disadvantage, consistent with previous findings for second-line osimertinib treatment.23,29 The increments and decrements of TP53 AF were in line with EGFR T790M changes at week 4 in all patients except for one (patient 4) who had undetectable T790M AF (20% to undetectable) and slightly increased TP53 AF (1.8%–2.3%) at week 4. This patient had a relatively longer PFS of 9.5 months on osimertinib treatment. Hence, the predictive role of TP53 remained elusive.
One patient (patient 15) in this study cohort, whose tumor presented a PTEN mutation (PTEN T319 frameshift) at baseline did not have a clinical response to osimertinib. The AFs of EGFR and PTEN mutations increased 4 weeks after osimertinib treatment and on disease progression. Concurrent mutations in EGFR and PTEN have been reported in 5% of patients with EGFR-mutated NSCLC at diagnosis.26 Alterations in PTEN, as detected in tissue samples, were reported as resistance mechanisms to erlotinib in EGFR mutation–positive NSCLC.30 It has also been reported as a mechanism of acquired and primary resistance to osimertinib therapy by analyzing osimertinib-resistant tumor tissues.31, 32, 33, 34 Primary resistance to osimertinib developed in 7.7% of patients with EGFR T790M–positive NSCLC. Mechanisms of primary resistance, such as small cell transformation, ERBB2 exon 16 skipping or amplification, MET amplification, BIM deletion polymorphism, EZH2 mutation, and KRAS G12D mutation, have been reported.33, 34, 35, 36, 37 To the best of our knowledge, this is the first report of inferring PTEN alteration as a potential mechanism of primary resistance by using plasma ctDNA analysis.
Regarding the plasma shedding status of tumor DNA, in one retrospective study, detection of EGFR T790M in plasma was more likely in patients with bone metastases or more than three metastatic sites by using various non-NGS techniques after progression on first- or second-generation EGFR TKIs.38 Furthermore, the retrospective analysis of plasma samples from patients from the AURA3 study revealed that plasma EGFR T790M detection was associated with the presence of extrathoracic disease and a larger tumor size.23 In the AURA3 study, osimertinib-treated patients with cobas plasma EGFR T790M–positive disease had shorter PFS as compared with patients with plasma EGFR T790M–negative disease (median, 8.3 versus 12.5 mo).23 In a real-world study, ctDNA was detectable by the Guardant360 assay in 65% of first- or second-generation EGFR TKI–pretreated patients with EGFR-mutated NSCLC who developed resistance.29 Among these shedders, 45% of them had EGFR T790M. In this study, most of the patients (66.7%) presented bone metastases. In this study, the two patients with disease limited to the lungs had the lowest plasma ctDNA AF. Of note, most of these selected patients had had positive plasma EGFR T790M detected by standard methods before participating this study; hence, these patients are not representative for the general patients with EGFR-mutated NSCLC.
This study has some limitations. First, the sample size was relatively small, and the results are more likely hypothesis-generating rather than definitive. Nevertheless, our findings support addressing these questions in a larger prospective study designed to identify the role of ctDNA monitoring during EGFR TKI treatment for advanced-stage NSCLC. Such a study could also address the role of earlier interventions for targetable resistance mutations. Second, standard testing for EGFR T790M in most patients was performed by a hotspot plasma-based test; only a few patients (n = 4) had tissue-based diagnosis of EGFR T790M–positive disease. This likely improved the sensitivity of Guardant360, as such cases were known to have DNA shedding before liquid-based NGS testing. As mentioned previously, shedders of ctDNA usually have extrathoracic disease and a larger tumor size.23 Patients with EGFR T790M–negative plasma still need a tumor rebiopsy to determine the presence and absence of EGFR T790M.22
In conclusion, in patients with EGFR-mutated advanced NSCLC, persistent EGFR T790M or increasing EGFR-activating mutation AF in ctDNA at week 4 after osimertinib treatment may predict early disease progression, defined in our study as within 16 weeks. On the basis of these findings, future studies could incorporate the evaluation of ctDNA at week 4 to assess the role of early treatment interventions for emerging resistance to osimertinib.
Acknowledgments
The authors thank all the patients and their families who have participated in this study. The authors acknowledge the efforts made by the study coordinators, Ms. Hsiang-Chun Lin and Ms. Ya-Ying Bai. Support for Guardant360 testing was provided by Guardant Health. The authors also thank Editage (editage.com) for English language editing. The authors attest that this study was not supported by external funding sources. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Disclosure: Dr. Yang has received personal fees from Boehringer Ingelheim, Eli Lilly, Bayer, Roche-Genentech, Chugai Pharmaceuticals, Astellas, Merck Sharp & Dohme, Merck Serono, Pfizer, Novartis, Celgene, Merrimack, Yuhan Pharmaceuticals, Bristol-Myers Squibb, Ono Pharmaceuticals, Daiichi Sankyo, Hansoh Pharmaceuticals, Takeda Pharmaceuticals, Blueprint Medicines, AstraZeneca, and G1 Therapeutics; has served on advisory boards for the aforementioned companies; and has received honorariums from Boehringer Ingelheim, Eli Lilly, Bayer, Roche-Genentech, Chugai Pharmaceuticals, Merck Sharp & Dohme, Pfizer, Novartis, Bristol-Myers Squibb, Ono Pharmaceuticals, and AstraZeneca. Dr. Liao has received honorariums/speaker fees from Boehringer Ingelheim, Roche, Chugai Pharmaceuticals, Merck Sharp & Dohme, Pfizer, Novartis, Ono Pharmaceuticals, and AstraZeneca. Dr. Lee has received honorariums for participation in compensated advisory boards of Novartis and Takeda Oncology. Dr. Liao has received honorariums from AstraZeneca, Roche, Boehringer Ingelheim, Eli Lilly, Pfizer, Merck Sharp & Dohme Oncology, Novartis, and Bristol-Myers Squibb and travel/accommodations and meeting expenses from AstraZeneca, Roche, Boehringer Ingelheim, Merck Sharp & Dohme, and Bristol-Myers Squibb. Dr. Ho has received grants from AstraZeneca; has received honorariums/speaker fees from Boehringer Ingelheim, Eli Lilly, Roche-Genentech, Chugai Pharmaceuticals, Merck Sharp & Dohme, Pfizer, Novartis, Bristol-Myers Squibb, and Ono Pharmaceuticals. Dr. Lin has served on advisory boards for Blueprint Medicines, Boehringer Ingelheim, Daiichi Sankyo, Novartis, and Takeda; has received honorariums from Bristol-Myers Squibb, Daiichi Sankyo, Novartis, and Roche; and has received travel grant from BeiGene and Eli Lilly. Dr. Shih has received speaking honorariums from AstraZeneca, Roche, Boehringer Ingelheim, Pfizer, Novartis, Bristol-Myers Squibb, Merck Sharp & Dohme, and Eli Lilly and has been paid for fulfilling consulting or advisory roles by AstraZeneca, Roche, Boehringer Ingelheim, Novartis, Merck Sharp & Dohme, AbbVie, and Chugai Pharmaceuticals. Dr. Soo has received grants and personal fees from AstraZeneca and Boehringer Ingelheim and personal fees from Amgen, Bristol-Myers Squibb, Eli Lilly, Merck, Novartis, Pfizer, Roche, Taiho Pharmaceuticals, Takeda, and Yuhan Pharmaceuticals. The remaining authors declare no conflict of interest.
References
- 1.Yang J.C., Hirsh V., Schuler M. Symptom control and quality of life in LUX-Lung 3: a phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3342–3350. doi: 10.1200/JCO.2012.46.1764. [DOI] [PubMed] [Google Scholar]
- 2.Sequist L.V., Yang J.C., Yamamoto N. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y.L., Cheng Y., Zhou X. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 4.Maemondo M., Inoue A., Kobayashi K. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T., Morita S., Yatabe Y. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R., Carcereny E., Gervais R. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C., Wu Y.L., Chen G. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 8.Yu H.A., Arcila M.E., Rekhtman N. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westover D., Zugazagoitia J., Cho B.C., Lovly C.M., Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018;29(suppl 1):i10–i19. doi: 10.1093/annonc/mdx703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.John T., Akamatsu H., Delmonte A. EGFR mutation analysis for prospective patient selection in AURA3 phase III trial of osimertinib versus platinum-pemetrexed in patients with EGFR T790M-positive advanced non-small-cell lung cancer. Lung Cancer. 2018;126:133–138. doi: 10.1016/j.lungcan.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Wu S.G., Liu Y.N., Tsai M.F. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget. 2016;7:12404–12413. doi: 10.18632/oncotarget.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J.C., Ahn M.J., Kim D.W. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017;35:1288–1296. doi: 10.1200/JCO.2016.70.3223. [DOI] [PubMed] [Google Scholar]
- 13.Goss G., Tsai C.M., Shepherd F.A. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016;17:1643–1652. doi: 10.1016/S1470-2045(16)30508-3. [DOI] [PubMed] [Google Scholar]
- 14.Mok T.S., Wu Y.L., Ahn M.J. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thress K.S., Paweletz C.P., Felip E. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins S., Yang J.C., Ramalingam S.S. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2017;12:1061–1070. doi: 10.1016/j.jtho.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Lin C.C., Shih J.Y., Yu C.J. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med. 2018;6:107–116. doi: 10.1016/S2213-2600(17)30480-0. [DOI] [PubMed] [Google Scholar]
- 18.Odegaard J.I., Vincent J.J., Mortimer S. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–3549. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 19.Leighl N.B., Page R.D., Raymond V.M. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non–small cell lung cancer. Clin Cancer Res. 2019;25:4691–4700. doi: 10.1158/1078-0432.CCR-19-0624. [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal C., Thompson J.C., Black T.A. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019;5:173–180. doi: 10.1001/jamaoncol.2018.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laufer-Geva S., Rozenblum A.B., Twito T. The clinical impact of comprehensive genomic testing of circulating cell-free DNA in advanced lung cancer. J Thorac Oncol. 2018;13:1705–1716. doi: 10.1016/j.jtho.2018.07.101. [DOI] [PubMed] [Google Scholar]
- 22.Oxnard G.R., Thress K.S., Alden R.S. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadimitrakopoulou V.A., Han J.Y., Ahn M.J. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non–small cell lung cancer. Cancer. 2020;126:373–380. doi: 10.1002/cncr.32503. [DOI] [PubMed] [Google Scholar]
- 24.Boysen Fynboe Ebert E., McCulloch T., Holmskov Hansen K., Linnet H., Sorensen B., Meldgaard P. Clearing of circulating tumour DNA predicts clinical response to osimertinib in EGFR mutated lung cancer patients. Lung Cancer. 2020;143:67–72. doi: 10.1016/j.lungcan.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd F.A., Papadimitrakopoulou V., Mok T. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib in the AURA3 trial. J Clin Oncol. 2018;36(suppl 15) 9027–9027. [Google Scholar]
- 26.VanderLaan P.A., Rangachari D., Mockus S.M. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: correlation with clinical outcomes. Lung Cancer. 2017;106:17–21. doi: 10.1016/j.lungcan.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canale M., Petracci E., Delmonte A. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 28.Labbé C., Cabanero M., Korpanty G.J. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC) Lung Cancer. 2017;111:23–29. doi: 10.1016/j.lungcan.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Zugazagoitia J., Gómez-Rueda A., Jantus-Lewintre E. Clinical utility of plasma-based digital next-generation sequencing in oncogene-driven non-small-cell lung cancer patients with tyrosine kinase inhibitor resistance. Lung Cancer. 2019;134:72–78. doi: 10.1016/j.lungcan.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 30.Sos M.L., Koker M., Weir B.A. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR [published correction appears in Cancer Res. 2015;75:1922] Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T.M., Song A., Kim D.W. Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol. 2015;10:1736–1744. doi: 10.1097/JTO.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 32.Bordi P., Del Re M., Minari R. From the beginning to resistance: study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC. Lung Cancer. 2019;131:78–85. doi: 10.1016/j.lungcan.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Hong M.H., Kim M.H., Kim S.Y. Molecular landscape of osimertinib resistance revealed by targeted panel sequencing and patient-derived cancer models in non-small cell lung cancer patients. Ann Oncol. 2018;29(suppl 8):viii516. [Google Scholar]
- 34.Xu C., Wang W., Zhu Y. Potential resistance mechanisms using next generation sequencing from Chinese EGFR T790M+ non-small cell lung cancer patients with primary resistance to osimertinib: a multicenter study. Ann Oncol. 2019;30(suppl 2):ii38–ii68. [Google Scholar]
- 35.Hsu C.C., Liao B.C., Liao W.Y. Exon 16-skipping HER2 as a novel mechanism of osimertinib resistance in EGFR L858R/T790M-positive non-small cell lung cancer. J Thorac Oncol. 2020;15:50–61. doi: 10.1016/j.jtho.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Minari R., Bordi P., Del Re M. Primary resistance to osimertinib due to SCLC transformation: issue of T790M determination on liquid re-biopsy. Lung Cancer. 2018;115:21–27. doi: 10.1016/j.lungcan.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Chang L.K., Chang Y.L., Shih J.Y. Primary resistance to osimertinib despite acquired T790M. Respirol Case Rep. 2020;8 doi: 10.1002/rcr2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minari R., Mazzaschi G., Bordi P. Detection of EGFR-activating and T790M mutations by liquid biopsy in patients with EGFR-mutated non-small-cell lung cancer who have progressed to first- and second-generation tyrosine kinase inhibitors: a multicenter real-life retrospective study. Clin Lung Cancer. 2020;21:e464–e473. doi: 10.1016/j.cllc.2020.02.021. [DOI] [PubMed] [Google Scholar]